Abstract
The mTOR protein is a master regulator of cell growth and proliferation, and inhibitors of its kinase activity have the potential to become new class of anti-cancer drugs. Starting from quinoline 1, which was identified in a biochemical mTOR assay, we developed a tricyclic benzonaphthyridinone inhibitor Torin1(26), which inhibited phosphorylation of mTORC1 and mTORC2 substrates in cells at concentrations of 2 nM and 10 nM, respectively. Moreover, Torin1 exhibits 1000-fold selectivity for mTOR over PI3K (EC50 = 1800 nM) and exhibits 100-fold binding selectivity relative to 450 other protein kinases. Torin1 was efficacious at a dose of 20 mg/kg in a U87MG xenograft model, and demonstrated good pharmacodynamic inhibition of downstream effectors of mTOR in tumor and peripheral tissues. These results demonstrate that Torin1 is a useful probe of mTOR-dependent phenomena and that benzonaphthridinones represent a promising scaffold for the further development of mTOR-specific inhibitors with the potential for clinical utility.
Introduction
The mammalian target of Rapamycin (mTOR) is a key node in the PI3K/Akt/mTOR signaling pathway that is often deregulated in human cancer.1 In conjunction with PI3K and Akt/PKB, mTOR integrates signals derived from extracellular cues, such as growth factors, energy, stress and nutrients, and regulates growth-related cellular processes, including mRNA translation, ribosome biogenesis, autophagy, and metabolism. The mTOR serine/threonine protein kinase shares a structurally similar catalytic domain with other PIKK family members, including PI3Ks, DNA-PK, ATM, ATR and SMG-1.2 mTOR exists in at least two distinct protein complexes in cells: mTOR Complex 1 (mTORC1) and mTOR Complex 2 (mTORC2). mTORC1 regulates mRNA translation through the phosphorylation of S6K1 and 4EBP1 while mTORC2 regulates cell survival through the phosphorylation of Akt/PKB and other AGC-family kinases, such as SGK. Hyperactivation of the mTOR signaling pathway is often implicated in cellular growth deregulation in cancer, and therefore, there has been a substantial effort to develop mTOR inhibitors for potential clinical application.3,4
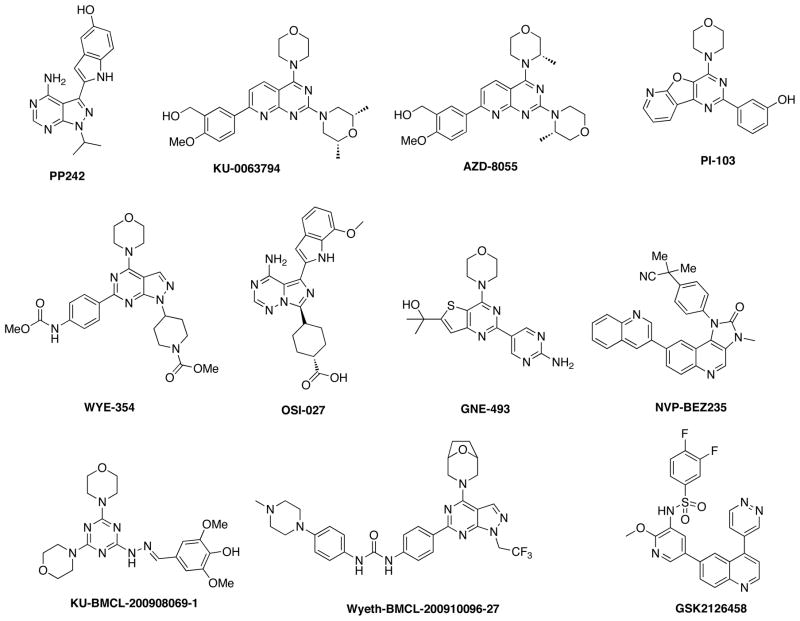
The development of ATP-competitive mTOR inhibitors was not viewed as a priority until recently because of rapamycin and its analogs (known as rapalogs), which were widely considered highly potent and selective allosteric inhibitors of mTORC1. In some cell types, rapamycin can also inhibit mTORC2.5 Unfortunately, clinical success of these compounds has been limited to a small number of relatively rare cancers, including mantle cell lymphoma, renal cell carcinoma, and endometrial cancer.6 Several explanations for this limited efficacy have been proposed: 1) rapamycin may only be effective in tumors where it is also capable of inhibiting mTORC2; 2) the induction by rapamycin of a feedback signal that leads to the hyperactivation of PI3K signaling might undermine the anti-proliferative effect of mTORC1 inhibition; or 3) in many cell types rapamycin fails to significantly inhibit mTORC1 kinase activity towards substrates, such as 4E-BP1, that are major regulators of proliferation. 7,8 It is currently unknown whether any or all of these explanations account for the apparently limited clinical efficacy of rapamycin and its derivatives. Nonetheless, these potential deficiencies in rapamycin-based therapies have spurred the rapid development of ATP-competitive mTOR inhibitors that target both mTOR complexes. Recently, a number of selective ATP-competitive mTOR inhibitors have been reported including Torin17, PP242,8 Ku63794,9 WYE-354,10 OSI-027,11 AZD-8055,12 KU-BMCL-200908069-1,13 Wyeth-BMCL-200910096-2714 as well as a number of compounds that are capable of inhibiting both PI3K and mTOR and other PIKK-family kinases such as PI-103,15 NVP-BEZ235,16 and GNE-49317, GSK212645818 (Figure 1). Here we describe our screening and medicinal chemistry efforts that resulted in the identification of benzonapthridinones exemplified by 26 as highly potent and selective mTOR inhibitors.
Figure 1.
Chemical structures of reported ATP-competitive mTOR inhibitors
Results and discussion
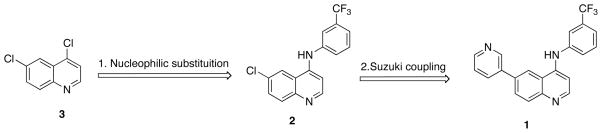
In order to identify compounds capable of inhibiting mTOR kinase activity, we established a medium-throughput biochemical assay utilizing purified mTORC1 from mammalian cells.19 We screened a compound library consisting of both known kinase inhibitors as well as heterocycles that would be anticipated to have the propensity to bind to the ATP binding pocket of kinases.20 This screening effort resulted in the identification of several classes of compounds, however, based on the criteria of the scaffold selectivity, we focused our attention on quinoline hit compound 1. Although compound 1 exhibited only moderate biochemical inhibitory activity (mTORC1 IC50 = 5 μM) and was unable to inhibit phosphorylation of the cellular mTORC1 substrate S6K at a concentration of 10 μM, it did exhibit selectivity for PIKK-family kinases when screened in binding assays against a panel of approximately 400 kinases at a concentration of 10 μM (KinomeScan™). In addition, the quinoline and isosteric quinazoline scaffolds are ‘privileged’ ATP-site binders and several highly selective kinase inhibitors such as HKI-27221 and lapatinib22 are built from these heterocyclic cores.
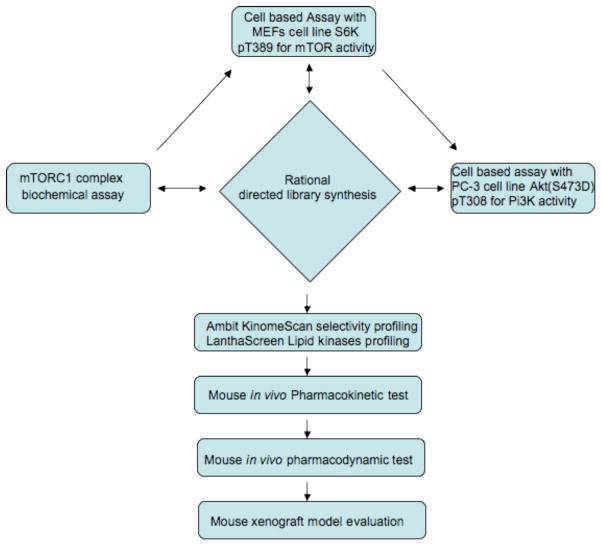
In order to optimize the cellular potency and selectivity of ‘hit’ compound 1, we established a streamlined flowchart of biochemical and cellular assays to explore the structure-activity relationship (Figure 2). Newly synthesized compounds were initially screened for their ability to inhibit mTORC1 activity by monitoring the inhibition of phosphorylation of T389 of S6K1 by immunoblot analyses in mouse embryonic fibroblasts (MEFs). The cellular assay was primarily used to prioritize compounds and, in parallel, compounds were also tested using an in vitro mTORC1 kinase assay. Compounds that exhibited promising cellular mTORC1 activity were assessed for selectivity versus inhibition of cellular PI3K activity by monitoring inhibition of Akt T308 in PC-3 cells that stably express the phosphomimetic S473D mutant of Akt7. The S473D mutant of Akt was used to achieve a more pure PI3K-dependent phosphorylation read-out by eliminating the effect of mTORC2 phosphorylation at that site, which normally facilitates phosphorylation at T308 by PDK1.23 The most potent and selective candidates to emerge from this reiterative process between the chemical modification and biological evaluation were subject to broad kinase selectivity profiling using the Ambit KinomeScan™ and LifeTechnologies LanthaScreen™ platforms. Compounds that passed our criteria for triage were further profiled for their in vitro and in vivo drug metabolism and pharmacokinetic properties (DMPK) and finally tested in a U87MG tumor xenograft model.
Figure 2.
Screening flowchart for development of mTOR inhibitors
In order to begin the SAR exploration of compound 1, we generated a number of analogs by varying the side chains at 4 and 6-positions of the quinoline. A few representative compounds that emerged from this effort are illustrated by compounds 4, 5, and 6 (Table 1). Unfortunately these substitutions generally resulted in very minor changes in biochemical IC50 values and none of the compounds inhibited mTOR or PI3K activity in cellular assays at submicromolar concentrations (Table 1).
Table 1.
Enzymatic and cellular activities profile of acyclic quinoline seriesa
| Entry | Structure | mTOR IC50(nM) mTORC1 | mTOR IC50(nM) Cellular | PI3K IC50(nM) Cellular |
|---|---|---|---|---|
| 4 |
|
1700 | >300 | >300 |
| 5 |
|
966 | >300 | >300 |
| 6 |
|
3810 | >300 | >300 |
the IC50 determinations are the mean of two of independent measurement with standard error of <20% For cellular assay, 300 nM was the highest concentration tested
A substructure search of compound 1 identified a broad class of tricyclic quinoline- derived dual PI3K/mTOR inhibitors exemplified by NVP-BEZ235 as containing a similar substructure24,25 (Figure 1). Tricylic core structure have been exploited in the development of numerous other kinase inhibitors targeting EGFR,26a PDGFR,26b Aurora,26c CK2,26d IKK26e. NVP-BEZ235 was predicted to bind to PI3K using the quinoline nitrogen to form a key hydrogen bond to the kinase ‘hinge-region’.25 The cyclic urea moiety was predicted to be important for constraining the pendant phenyl ring to the presumed bio-active conformation. We decided to introduce a similar constraint by introducing a 6-membered lactam that would result in a tricyclic scaffold which would facilitate additional diversification points as well as introduce the potentially important carbonyl oxygen for the potency.
The introduction of this additional ring constraint resulted in benzonaththridinone compound 7, which exhibited a dramatic 1000-fold improved mTOR cellular potency compared to the screening hit compound 1. Compound 7 possessed a biochemical IC50 of 5.4 nM against mTORC1 enzyme and an EC50 of 3 nM for inhibition of S6K phosphorylation in cells without any detectable inhibitory activity on PI3K up to a concentration of 300 nM.
In order to rationalize the potency of 7 in a structural context, we docked compound 7 into the ATP-binding site of an mTOR homology model built using the related Pi3Kγ crystal structure (PDB code: 3DBS) (Figure 3). Similar to the published homology model of NVP-BEZ235 with Pi3Kγ,27 a hydrogen bond was predicted to form between the quinolione nitrogen of compound 7 and Val2240 within the kinase hinge region.16 A comparison to the proposed binding mode of morpholine-derived mTOR inhibitors such as PI-103, suggests that the morpholine oxygen makes a similar hydrogen bond. The quinoline moiety of compound 7 was predicted to reside within the inner hydrophobic pocket formed by Glu2190, Leu2192, Asp2195, Tyr2225, Asp2357, Phe2358, Gly2359, and Asp2360. The phenyl piperazine side chain was oriented below a loop defined by residues from Ile2183 to Gln2187, which occupy a similar region of the ATP-binding site as the ‘P-loop’ of protein kinases. Using this homology model as a guide, we embarked on a further elaboration of the benzonaphthridione scaffold at the quinoline (R1) and lactam phenyl (R2) positions. Previous efforts to optimize PI3K inhibitors have suggested that modification at either of these positions could modulate potency while the R1 position may provide a stronger contribution to specificity versus other PIKK-family kinases.27 In addition, to improve the extremely poor water-solubility of this compound class, we continued our SAR exploration with R2 fixed as a phenyl piperazine, which introduces a water solubility enhancing basic secondary nitrogen versus the acyl piperazine present in our library compounds (Figure 4). The target compounds were synthesized using a four-step sequence starting from dichloroquinoline scaffold S1 (Scheme 2). Introduction of substituted anilines in the first step was followed by construction of the 6-membered lactam using a Horner-Emmon’s Wadsworth reaction. The R2 side chain was then introduced via palladium-mediated coupling with the desired boronic acid. The biological activities of the compounds that resulted from this effort are illustrated in Table 3 and 4.
Figure 3.
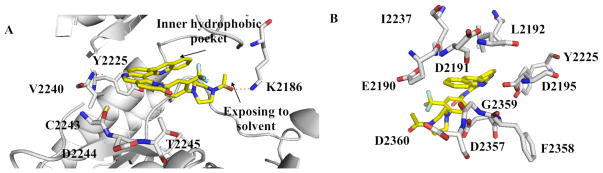
Model demonstration of compound 7 binding with mTOR
Figure 4.
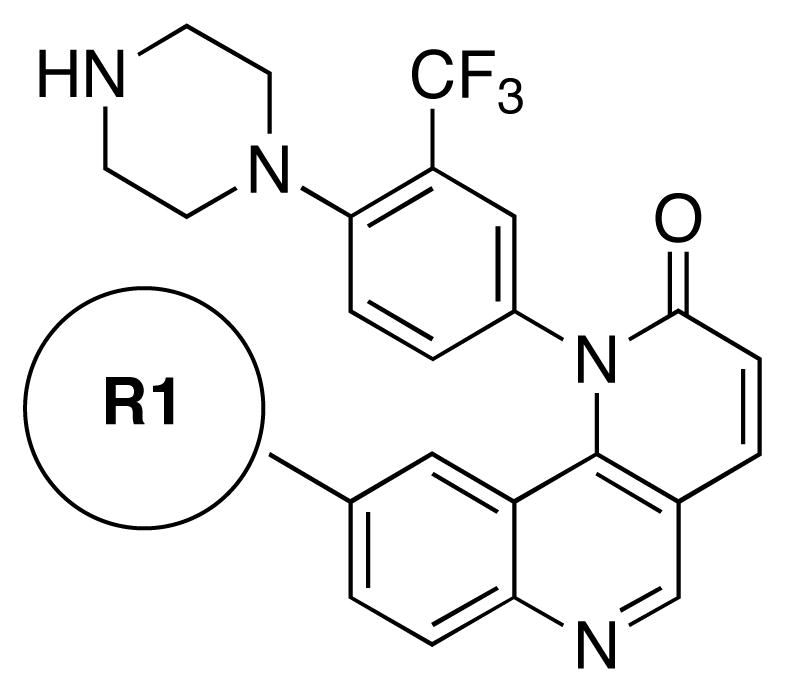
Representative structure of variations of R1
Scheme 2.
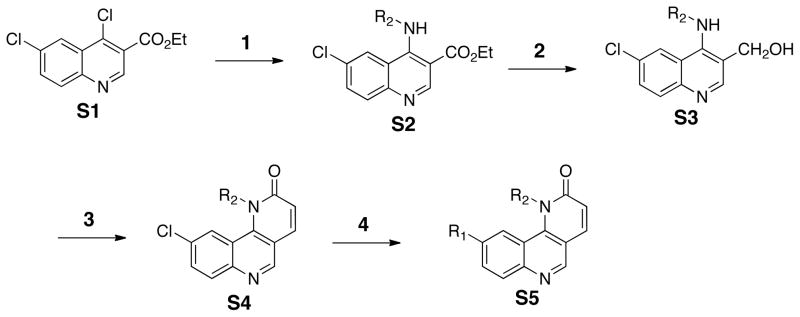
Reagents and conditions: 1. aniline (R2NH2), 1,4-dioxane, 100 °C, 4h; 2. LAH, THF, 0°C-rt, 4h; 3. a) MnO2, CH2Cl2, rt, 6h, b) triethyl phosphonoacetate, K2CO3, EtOH, 100 °C, 12h; 4. R1B(OH)2, PdCl2(Ph3P)2, t-Bu-Xphos, Na2CO3, 1,4-dioxane, 100 °C, 12h
Table 3.
Enzymatic and cellular activities profile of compounds varying R1
| Entry | R1 | mTOR IC50(nM) mTORC1 | mTOR IC50(nM) Cellular |
|---|---|---|---|
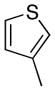
| 8 |
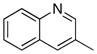
|
13.5 | 40 |
| 9 |
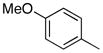
|
78.9 | 100 |
| 10 |
|
93.8 | >300 |
| 11 |
|
3440 | >300 |
| 12 |
|
33.2 | >300 |
| 13 |
|
662 | >300 |
| 14 |
|
213 | >300 |
| 15 |
|
66.5 | 150 |
| 16 |
|
82.6 | 40 |
| 17 |
|
45.9 | >300 |
| 18 |
|
214 | >300 |
IC50 determinations are the mean of two of independent measurement with standard error of <20%, None of the tested compounds exhibited IC50 for inhibition of cellular PI3K activity of less than 300 nM.
Table 4.
Enzymatic and cellular activities profile of compounds varying R2
| Entry | R2 | mTOR IC50(nM) mTORC1 | mTOR IC50(nM) Cellular |
|---|---|---|---|
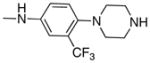
| 8 |
|
13.5 | 40 |
| 19 |
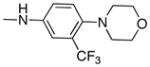
|
25.8 | 35 |
| 20 |
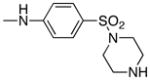
|
239 | >300 |
| 21 |
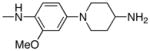
|
95 | 200 |
| 22 |
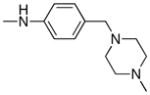
|
66 | 250 |
| 23 |
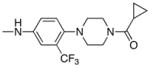
|
6.84 | 1 |
| 24 |
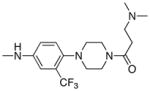
|
3.6 | 50 |
| 25 |
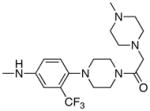
|
0.57 | 5 |
| 26 |
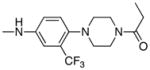
|
0.29 | 2 |
| 27 |
|
389 | >300 |
| 28 |
|
1.26 | 25 |
| 29 |
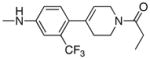
|
21.4 | 30 |
| 30 |
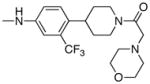
|
4.25 | 5 |
IC50 determinations are the mean of two of independent measurement with standard error of <20%, none of the tested compounds exhibited an IC50 for inhibition of cellular PI3K activity of less than 300 nM.
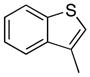
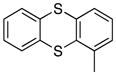
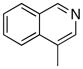
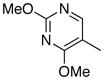
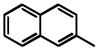
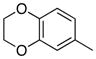
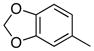
Removal of the acyl group of compound 7 to yield compound 8 resulted in a 2–10 fold reduction in potency in both biochemical and cellular assays. Based on the homology model this could be rationalized by the potential loss of a hydrogen bond between the amide carbonyl oxygen and epsilon amine of Lys2186. Removal of the quinoline nitrogen of compound 8 to yield compound 14 resulted in more than a 20-fold loss in cellular mTOR potency, which may be attributable to the loss of a hydrogen bond between the quinoline N and the phenol of Tyr2225. Introduction of the tricyclic thianthrene side chain (11) significantly reduced the cellular mTOR potency providing a potential upper limit on the size of the functionality that can be accommodated within the inner hydrophobic pocket. Most of the bicyclic side chains such as benzothiophene (10), isoquinoline (12), naphthalene (14), benzodioxane (15) and benzodioxole (16) maintained reasonable enzymatic activities but only those with appropriately placed hydrogen-bond accepting potential (15, 16) displayed good cellular activity. Monocyclic R1 groups with substitution at the para-position (9) possessed activity in the same range as the bicyclic ring systems. The dramatic loss of activity observed between compound 13 and 9 may be attributed to the lack of tolerance for alpha-substitution, which is consistent with the observations made for the GNE-493 series.17 Cumulatively, these data suggest that a combination of hydrophobic and hydrogen bonding interactions are critical for achieving potent cellular inhibition of mTOR and that the quinoline moiety was the preferred core for this pharmacophore. None of the compounds in this series displayed cellular activity against PI3K up to a concentration of 300 nM. A number of discordances were observed between the biochemical and cellular IC50 measurements that may be attributed to differential cellular permeability and/or the inability of the biochemical assay to accurately reflect the functionally relevant intracellular mTOR. Based on this observation, the cell based IC50 was used as a guide for the chemical optimization process.
Following the initial optimization steps, we introduced a variety of side chains to the R2 position while fixing the R1 side chain as a quinoline (Figure 5 and Table 4). Introducing sulfonyl (20) or methyl spacer (22) before the piperazine functionality resulted in a loss in mTOR potency. Introducing an ortho-methoxy substitution (21 verus 8) resulted in a 6-fold loss in cellular mTOR inhibition. Replacement of an R2 phenyl moiety with small aliphatic side chains (represented by 27) resulted in inactive compounds. Most compounds that retained the meta-CF3 substituted aniline and also those derivatized at the para-position with piperazinyl, phenyl, and piperidinyl groups retained similar or superior potency relative to compound 7. Introducing a carbonyl moiety in the form of an acetyl group to the piperazine nitrogen (7 versus 8) resulted in a 13-fold improvement in cellular potency, potentially due to the introduction of a hydrogen bond with Lys2187. (Figure 3) To avoid an 1,4-dianiline moiety that could potentially represent a metabolic liability in compound 7, several piperazine replacements were explored such as reversed piperidine (30), unsaturated piperidine (29) or phenyl (28) linkage, however, they all resulted in weaker biochemical and cellular mTOR inhibition. Further exploration of the piperazine acyl moiety, which the modeling predicted to be directed towards the solvent area, resulted in the identification of the propyl amide (26, named Torin1) as conferring the greatest potency and selectivity. Compound 26 is a picomolar inhibitor of mTORC1 enzymatic activity and single digit nanomolar inhibitor of cellular mTOR activity. Although most compounds in this series have over 100-fold selectivity relative to inhibition of cellular PI3K activity, a more detailed study of 26 revealed that it has more than 800-fold selectivity between mTOR and PI3K 7.
Figure 5.
Representative structures of variations of R2
We next investigated the SAR of the lactam ring using Torin1 as a staring point. The resultant SAR indicated that the rigid 6-membered lactam was preferred, as urea, carbamate, and pyrimidinedione analogs lost more than 500-fold activity against the mTORC1 complex (Table 5). Introduction of a methyl group to either the alpha(31) or beta position(32) of the lactam ring significantly diminished potency in both the biochemical and cellular assays, suggesting these positions make a close contact to the ATP-binding site.
Table 5.
Enzymatic and cellular activities profile of fixed conformation compounds.a
| Entry | Structure | mTOR IC50(nM) mTORC1 | mTOR IC50(nM) Cellular |
|---|---|---|---|
| 26 |
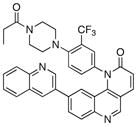
|
0.29 | 2 |
| 31 |
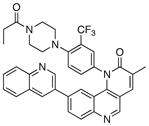
|
26.6 | 100 |
| 32 |
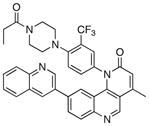
|
51.9 | >300 |
| 33 |
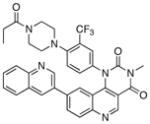
|
230 | 300 |
| 34 |
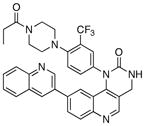
|
396 | >300 |
| 35 |
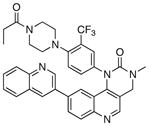
|
287 | 100 |
| 36 |
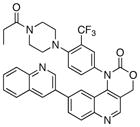
|
336 | >300 |
IC50 determinations are the mean of two of independent measurements with standard error of <20%, Torin1 (26) has a cellular IC50 of 1800 nM for PI3K. None of other compounds exhibited a cellular IC50 for PI3K of less than 300 nM.
Kinase selectivity
Biochemical characterization of 26 at varying ATP-concentrations demonstrated that it is an ATP competitive inhibitor of mTOR. The selectivity of 26 was assessed using the KinomeScan™ methodology across a panel of 442 human kinases at a concentration of 10 μM. Only three kinases displayed tight binding to 26 (Ambit scores of less than 2) and dissociation constants were determined for mTOR, PI3Kα and PI3Kα (I800L) (Table 6). This analysis revealed that 26 is extremely selective for PIKK family kinases relative to all other serine/threonine and tyrosine kinases. To further investigate these observed binding events, we performed enzymatic assays across most of the known PIKK family kinases using the LanthaScreen™ technology. This analysis revealed that 26 is very selective relative to other PIKK family kinases with the exception of DNA-PK (Table 7). The activity against DNA-PK was further characterized using a radiometric kinase assay where the compound exhibited an IC50 of approximately 1 μM.7 The origin of the 150-fold discrepancy between the LanthaScreen IC50 for DNA-PK versus the radiometric assay is currently unknown.
Table 6.
Kinome Scan™ profile of 26a
| Kinase | % control | Kd(nM) | Kinase | % control | Kd(nM) |
|---|---|---|---|---|---|
| mTOR | 0 | 0.42 | PIK3CA(E545K) | 0.6 | N.D |
| PIK3CA(C420R) | 0.7 | N.D | PIK3CA(I800L) | 0 | 14 |
| PIK3CA | 1.3 | 23 | MRCKA | 0.65 | 10000 |
Compound 26 was profiled at a concentration of 10 μM against a diverse panel of 442 kinases by Ambit Biosciences
Table 7.
LanthaScreen™ profile of 26 in invitrogen lipid kinase panela
| Kinase | IC50(nM) | Kinase | IC50(nM) |
|---|---|---|---|
| PI4K α | >10000 | P110 α/p85α | 250 |
| PI4K β | 6680 | P110δ/P85α | 564 |
| PI3K-C2 α | 176 | P110γ | 171 |
| PI3K-C2β | 549 | DNA-PK | 6.34 |
| hVPS34 | 533 | mTOR | 4.32 |
Kinase targets were tested with biochemical enzymatic kinase assays using the SelectScreen® Kinase Profiling Service (Life Technologies Corporation, Madison, WI) to determine IC50 values as shown in Table 7. The compounds were assayed at 10 concentrations (three-fold serial dilutions starting from 1 μM) at an ATP concentration equal to the ATP Km,app for the assay following the detailed procedures described in the SelectScreen® Customer Protocol and Assay Conditions documents located at www.invitrogen.com/kinaseprofiling.
Pharmacokinetic data
The pharmacokinetic properties of 26 were next evaluated both in vitro and in vivo. In both human and mouse liver microsome stability studies, 26 was rapidly consumed with a half-life of 4 minutes (Table 8). The significant difference in human liver microsome metabolism observed in the presence and absence of NADPH, indicates a NADPH dependent metabolism mechanism. The in vivo pharmacokinetic parameters of 26 in mice were determined following the intravenous administration of 1 mg/kg, oral administration of 10 mg/kg or intraperitoneal administration of 10 mg/Kg of 26 (Table 9). As expected from the in vitro studies, 26 exhibited a short T1/2 of 0.5 hours that may partially due to the rapid first-pass metabolism. 26 also exhibited low exposure and limited bioavailability following the oral administration, possibly suggestive of low absorption or high first pass metabolism. The better exposure and much longer half life achieved following intraperitineal administration further confirmed the instability of 26 to the liver metabolism.
Table 8.
Human and mouse liver microsome stability data of 26a
| Species | MR (nmol/min/mg) | T1/2 (min) | %R at 60 min | %R at 60min (-NADPH) |
|---|---|---|---|---|
| Human | 1.30 | 4 | 1 | 69 |
| Mouse | 1.53 | 4 | 0.2 | 1 |
human liver microsome (7 males+2 females, cat: 452165) and male mouse liver microsomes(cat. 452220) from BD Gentest.
Table 9.
Mouse PK data of 26a
| Route | Cmax (ng/mL) | Tmax (h) | AUC (h*ng/mL) | T1/2 (h) | MRT (h) | CL (mL/min/Kg) | Vss (L/Kg) | F (%) |
|---|---|---|---|---|---|---|---|---|
| IV | 2757 | ND | 720 | 0.5 | 0.43 | 23.0 | 0.59 | ND |
| PO | 223 | 0.25 | 396 | 0.79 | 1.51 | ND | ND | 5.49 |
| IP | 5121 | 0.08 | 5718 | 4.52 | ND | ND | ND | ND |
C57BL/6 male mouse was dosed at 1 mg/Kg for the intravenous (IV), 10 mg/Kg for the oral (PO) and 10 mg for the intraperitineal (IP) studies. I. formulation used 10% N-Methyl pyrrolidone (NMP) and 50% polyethylene glycol (PEG-200) in water. P.O. formulation used 0.5% w/v NaCMC with 0.1% w/v Tween-80. IP administration formulation was 10% v/v dimethylacetimate (DMA); 20% v/v polyethylene glycol (PEG-200); 20% v/v PG in water
Pharmacodynamic data
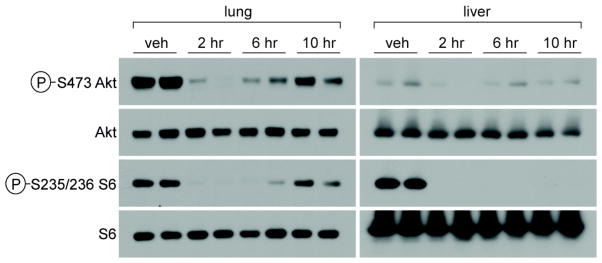
In order to assess the pharmacodynamic effects of 26, mice were treated with a single intraperitoneal dose of 20 mg/kg 26 and the phosphorylation of S6 at S235/236 and of Akt1 at S473 were measured by Western-blot in lysates derived from liver and lung. 26 suppressed Akt1 phosphorylation at S473, the phosphorylation site of mTORC2, for approximately 2–3 hours, after which the phosphorylation gradually returns to baseline levels. Inhibition of S6 at S235/236, an indirect substrate of mTORC1, was more markedly suppressed with complete inhibition of phosphorylation observed up to 6 hours in the lung and to greater than 10 hours in the liver (Figure 6). These pharamodynamic studies indicate that despite the poor pharmacological properties of 26, suppression of both mTORC1 and mTORC2 dependent outputs can be achieved in mice.
Figure 6.
Pharmacodynamic inhibition of Akt S473 and S6 S235/236 phosphorylation in lung and liver of mice treated with a single intraperotineal dose of 20 mg/Kg 26.a
Anti-tumor in vivo efficacy
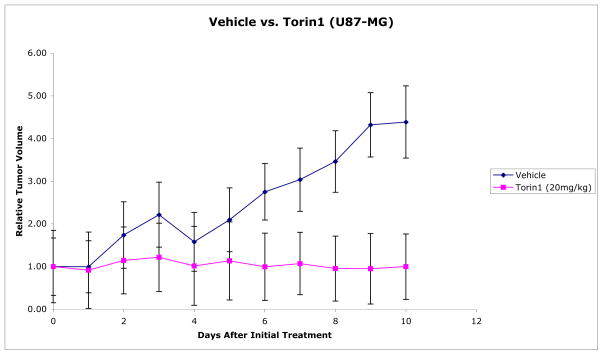
The results from the pharmacodynamic studies encouraged us to investigate whether 26 would inhibit growth in a U87MG xenograft model. U87MG is a PTENnull glioblastoma cell line that exhibits strong activation of the PI3K-mTOR-Akt pathway and has been commonly used for xenograft experiments to evaluate the efficacy of PI3K and mTOR kinase inhibitors.11,28 Due to the relatively poor pharmacokinetic properties, we decided to perform once daily IP dosing of 20 mg/kg of 26. Continuous dosing for 10 days resulted in greater than 99% inhibition of tumor growth as assessed by caliper measurements. (Figure 7) Upon cessation of drug treatment, the tumors continued to grow suggesting that 26 treatment is primarily cytostatic and that a significant number of tumor cells remain viable during treatment (data not shown).
Figure 7.
In vivo efficacy of 26 in U87MG xenografts.a
aMice model was established with 6 week old immunodeficient mice by subcutaneous injection of U87MG cells. 26 as a solution /suspension in vehicle (20% N-mentyl-2-pyrrolidone, 40% PEG400 and 40% water) or vehicle was delivered by intraperitoneal injection once daily after the tumor reached 1 cm3 in size and continued for 10 days.
Conclusion
Starting from a quinoline screening ‘hit’ compound 1 discovered using a medium-throughput biochemical mTORC1 assay, a focused medicinal chemistry effort facilitated by molecular modeling and other known PIKK-family inhibitors resulted in the identification of a novel benzonapthridinone scaffold exemplified by 26. Compound 26 inhibits phosphorylation of both mTORC1 and mTORC2 at single-digit nanomolar concentrations while exhibiting 800-fold selectivity relative to cellular PI3K inhibitory activity. Compound 26 exhibited a short in vivo half-life and low oral bioavailability but displayed pharmacodynamic inhibition of both mTORC1 and mTORC2 outputs in lung and liver. Compound 26 dosed once a day at 20 mg/kg for 10 days demonstrated efficacy in a U87MG xenograft mouse model. Compound 26 is a valuable tool for investigating mTOR mediated signal pathways and provides a valuable starting-point for identification of compounds with improved ‘drug-like’ properties.
Experimental Procedures
Chemistry
All solvents and reagents were used as obtained. 1HNMR spectra were recorded with a Varian Inova 600 NMR spectrometer and referenced to dimethyl sulfoxide. Chemical shifts are expressed in ppm. In the NMR tabulation, s indicates singlet; d, doublet; t, triplet; q, quartet; m, multiplet; and br, broad peak. Mass spectra were measured with Waters Micromass ZQ using an ESI source coupled to a Waters 2525 HPLC system operating in reverse mode with an Waters Sunfire™ C18 5μm 4.6×50 mm column. Purification of compounds was performed with either a Teledyne ISCO combiflash Rf system or a Waters Micromass ZQ preparative system. The purity of all compounds was ≥95%. The purity was analyzed on an above mentioned Waters LC-MS Symmetry (C18 column, 4.6 × 50 mm, 5μM) using a gradient of 5–95% acetonitrile in water containing 0.05% trifluoacetic acid (TFA) over 8 min (10 min run time) at a flow rate of 2 mL/min.
General procedure for the preparation of compounds 4–6
To a solution of compound 3 (4,6-dichloroquinoline, 1 eq.) in 1,4-dioxane at room temperature in a sealed tube was added aniline (1 eq). The resultant solution was heated to 100–120 °C for 4–12 h. After cooling to room temperature, a solution of NaOH (1 N) was added to neutralize the solution followed by dilution with water and extraction with ethyl acetate. After drying with Na2SO4, the solvents were removed and the residue was purified by ISCO to furnish compound 2.
To a solution of compound 2 (1 eq.) in 1,4-dioxane at room temperature was added PdCl2(Ph3P)2 (0.1 eq.), t-Bu-Xphos (0.1 eq.), Na2CO3 (3 eq., 1 N) and quinoline-3-boronic acid. After degassing, the resultant mixture was heated to 100 °C for 6 h before cooling down to room temperature and filtration through celite. Upon removal of the solvents, the residue was purified by column chromatography to furnish compounds 4–6.
1-(4-(4-([3,6′-biquinolin]-4′-ylamino)-2-(trifluoromethyl)phenyl)piperazin-1-yl)ethanone (4)
20% overall yield from 2.1H NMR (600 MHz, DMSO- d6) δ 10.12 (s, 1H), 9.49 (d, J = 2.4 Hz, 1H), 9.05 (d, J = 1.8 Hz, 1H), 8.84 (d, J = 2.4 Hz, 1H), 8.67 (s, 1H), 8.42 (dd, J = 8.4, 2.4 Hz, 1H), 8.26 (dd, J = 8.4, 2.4 Hz, 1H), 8.19 (d, J = 2.4 Hz, 1H), 8.10 – 8.12 (m, 2H), 7.96 (d, J = 8.4 Hz, 1H), 7.82 (ddd, J = 7.2, 6.6, 1.8 Hz, 1H), 7.69 (ddd, J = 8.4, 7.8, 1.2 Hz, 1H), 7.63 (d, J = 8.4 Hz, 1H), 3.54 – 3.56 (m, 4H), 2.87 (t, J = 4.8 Hz, 2H), 2.81 (t, J = 4.8 Hz, 2H), 2.04 (s, 3H). MS (ESI): m/z (M+H)+ 542.87.
General procedure for compounds 7–31
To a solution of compound S1 (ethyl 4,6-dichloroquinoline-3-carboxylate, 1 eq.) in 1,4-dioxane at room temperature was added aniline (1 eq.) at room temperature. The reaction mixture was heated to 100 °C for 4 h, followed by cooling to room temperature and the addition of NaOH (1 N) to neutralize the solution. The resultant solution was diluted with water and extracted with ethyl acetate. After removal of the solvents, the residue was purified by flash chromatography to afford compound S2.
To a solution of compound S2 (1 eq.) in THF at 0 °C was added LAH (3 eq.). After 15 min, the solution was warmed to room temperature and stirred for 1-4 h before carefully quenching with methanol and water. Subsequent filtration through celite furnished crude S3, which was used in the next step without further purification.
To a solution of compound S3 in CH2Cl2 (1 eq.) at room temperature was added MnO2 (10 eq. mass). After 4 h, the reaction mixture was filtered through celite. The filtrate was concentrated in a sealed tube and dissolved in dry EtOH after which, K2CO3 (3 eq.) and triethyl phosphonoacetate (triethyl 2-phosphonopropionate for compound 32) were added. The resultant mixture was heated to 100 °C for 12 h before cooling to room temperature. Upon removal of the solvents under vacuum, the residue was diluted with water following by extraction with ethyl acetate. Purification of the residue by column chromatography provided compound S4.
To a solution of compound S4 in 1,4-dioxane at room temperature was added subsequently PdCl2(Ph3P)2 (0.1 eq.), t-Bu-Xphos (0.1 eq.), Na2CO3(3 eq., 1 N) and boronic acids or boronate pinacol esters. After degassing, the resultant mixture was heated to 100 °C for 6 h before cooling to room temperature and filtration through celite. Upon removal of the solvents, the residue was subjected to column purification to furnish the desired compounds (7–31).
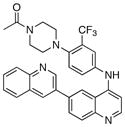
1-(4-(4-acetylpiperazin-1-yl)-3-(trifluoromethyl)phenyl)-9-(quinolin-3-yl)benzo[h][1,6]naphthyridin-2(1H)-one (7)
15% overall yield from S1. 1H NMR (600 MHz, DMSO-d6) δ 9.20 (s, 1H), 8.58 (d, J = 1.8 Hz, 1H), 8.34 (d, J = 9.6 Hz, 1H), 8.27 (s, 1H), 8.20 (d, J = 8.4 Hz, 1H), 8.16 (d, J = 7.2 Hz, 1H), 8.04 (d, J = 8.4 Hz, 1H), 7.97 – 7.98 (m, 2H), 7.79 – 7.82 (m, 2H), 7.67 – 7.71 (m, 2H), 7.11 (s, 1H), 6.95 (d, J = 9.0 Hz, 1H), 3.28 (m, 4H), 2.65 – 2.70 (m, 2H), 2.59 (m, 2H), 1.98 (s, 3H). MS (ESI): m/z (M+H)+ 594.81.
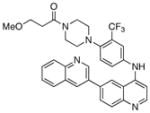
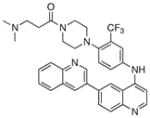
1-(4-(4-propionylpiperazin-1-yl)-3-(trifluoromethyl)phenyl)-9-(quinolin-3-yl)benzo[h][1,6]naphthyridin-2(1H)-one (26)
7 % overall yield from S1. 1H NMR (600 MHz, DMSO-d6) δ 9.19 (s, 1H), 8.58 (d, J = 2.4 Hz, 1H), 8.34 (d, J = 9.6 Hz, 1H), 8.26 (d, J = 1.8 Hz, 1H), 8.19 (d, J = 9.0 Hz, 1H), 8.14 (dd, J = 9.0, 1.8 Hz, 1H), 8.03 (d, J = 9.0 Hz, 1H), 7.96 – 7.98 (m, 2H), 7.77 – 7.82 (m, 2H), 7.66 – 7.70 (m, 2H), 7.09 (d, J = 1.8 Hz, 1H), 6.94 (d, J = 9.6 Hz, 1H), 3.40 –3.42 (m, 4H), 2.60 – 2.65 (m, 4H), 2.28 (q, J = 7.8 Hz, 2H), 0.98 (t, J = 7.8 Hz, 3H). MS (ESI): m/z (M+H)+ 608.23.
General procedure for the preparation of compounds 32–36
Compound S2 was subjected to Pd mediated coupling with quinoline 3-boronic acid under the conditions mentioned above to furnish compound S2a (ethyl 4-(3-(trifluoromethyl)-4-(4-propionylpiperazin-1-yl)phenylamino)-6-(quinolin-3-yl)quinoline-3-carboxylate). Compound S2a was dissolved in a mixture of MeOH/H2O (1:1) and treated with NaOH (1 N, 5 eq.) at room temperature. The reaction mixture was then stirred overnight. The pH of the solution was adjusted to 4, followed by extraction with ethyl acetate to afford the crude carboxylic acid S2b. Acid S2b (1 eq.) was dissolved in THF at room temperature and treated with triphosgene (0.4 eq.). After 2 h, methylamine (2 eq.) was added and the resulting mixture was heated at 40 °C for 1 h before quenching with saturated Na2CO3. Extraction with ethyl acetate followed by column chromatography afforded compound 33.
To a solution of compound S2b in THF was added methyl methoxyamine (2 eq.), Et3N (3 eq.) and HATU (2 eq.). The resultant solution was stirred at room temperature for 4 h before extraction with ethyl acetate. Column purification afforded Weinreb amide S2c, which was then dissolved in THF and treated with methyl magnesium bromide (3 eq.) at room temperature. The resulting solution was stirred for 4 h before quenching with sat. Na2CO3. Column chromatography afforded methyl ketone S2d, which was further elaborated to compound 33 in the same fashion as the synthesis of compound 8.
To a solution of compound S3 (1 eq.) in THF at room temperature was added triphosgene (0.4 eq.). The resultant solution was stirred at room temperature for 2 h before it was quenched with sat. Na2CO3. Column chromatography afforded compound 36.
To a solution of compound S3 in CH2Cl2 (1 eq.) at room temperature was added MnO2 (10 eq. mass). The mixture was stirred for 4 h before being filtered through celite. The filtrate was concentrated, dissolved in THF and treated with methyl amine (2 eq. for compound 35) or 2,4-dimethoxylbenzylamine (2 eq. for compound 34), NaBH(OAc)3 (5 eq.) and AcOH (1 drop) at room temperature. The reaction mixture was stirred for 4 h before it was quenched with sat. Na2CO3. After compound purification, the corresponding product was subjected to the same procedure as in the preparation of compound 33 to afford compounds 34 and 35.
Characterization of in vitro biochemical activity with the mTORC1 complex
Human mTORC1 complex was obtained as previously described (7,29). In vitro mTORC1 activity was assayed using the Lanthascreen time-resolved FRET assay (Invitrogen). Briefly, mTORC1 (0.1 μg each) was incubated with serially diluted inhibitors (3-fold, 11 points) for 30 min in 5 μL kinase buffer (25 mM HEPES, pH 7.4, 10 mM MgCl2, 4 mM MnCl2, 50 mM KCl) in a 384 well low volume plate (Corning). The kinase reaction was initiated by the addition of an equal volume of kinase buffer containing 0.8 μM GFP-labeled 4E-BP1 and 100 μM ATP at room temperature, and after 1 hour, the reaction was stopped with 10 μL of solution containing 20 mM EDTA and 4 nM Tb-labeled anti-phospho 4E-BP1 (T46) antibody. After incubation for 30 min, the FRET signal between Tb and GFP within the immune complex was read using an Envision plate reader (PerkinElmer). Each data point was duplicated and IC50 values were calculated using Prism4 software (GraphPad).
mTOR and PI3K Cellular Assays
Cellular IC50 values for mTOR were determined using p53−/− MEFs. Cells were treated with vehicle or increasing concentrations of compound for 1 h and then lysed. Phosphorylation of S6K1 Thr-389 was monitored by immunoblotting using a phospho-specific antibody. Meanwhile, cellular IC50 values for PI3Ka were determined based on phosphorylation of Akt Thr-308 in p53−/−/mLST8−/− MEFs or human PC3 cells expressing the S473D mutant of Akt1 as previously described.7
Ambit in vitro KinomeScan™ kinase selectivity profile
26 was profiled at a concentration of 10 μM against a diverse panel of 442 kinases by Ambit Biosciences. Scores for primary screen hits are reported as a percent of the DMSO control (% control). For kinases where no score is shown, no measurable binding was detected. The lower the score, the lower the Kd is likely to be, such that scores of zero represent strong hits. Scores are related to the probability of a hit, but are not strictly an affinity measurement. At a screening concentration of 10 μM, a score of less than 10% implies that the false positive probability is less than 20% and the Kd is most likely less than 1 μM. A score between 1–10% implies that the false positive probability is less than 10%, although it is difficult to assign a quantitative affinity from a single-point primary screen. A score of less than 1% implies that the false positive probability is less than 5% and the Kd is most likely less than 1 μM.
In vivo pharmacokinetic studies
The study was performed in Sai Advantium Pharma Limited company (India) with male Swiss Albino mice following single intravenous bolus and oral administration. A group of eighteen male mice were divided into two groups (Group 1: IV, and Group 2: PO), with each group comprised of nine mice. Animals in Group 1 were dosed intravenously via the tail vein at 1 mg/kg of 26 solution (7.5% v/v N-methyl pyrrolidone and 40% v/v polyethylene glycol-400 in normal saline). Group 2 animals were dosed orally at 10 mg/kg with a suspension formulation (0.5% w/v Na CMC with 0.1% v/v Tween-80 in water) of 26. Blood samples were collected at 0, 0.08 (for IV only), 0.25, 0.5, 1, 2, 4, 6, (For PO only) 8, 12 and 24 hours for the IV and PO groups. The blood samples were collected from sets of three mice at each time point in labeled microcentrifuge tubes containing K2EDTA as an anticoagulant. Plasma samples were separated by centrifugation and stored below −70 °C until bioanalysis. All samples were processed for analysis by precipitation using acetonitrile and analyzed with a partially validated LC/MS/MS method (LLOQ - 1.138 ng/mL). Pharmacokinetic parameters were calculated using the non-compartmental analysis tool of WinNonlin® Enterprise software (version 5.2).
Anti-tumor (U87MG model) efficacy studies
Immunodeficient mice (NCR nude, nu/nu; Taconic Laboratories) were maintained in a pathogen-free facility and were given autoclaved food and water ad libitum. U87-MG glioblastoma cells were xenografted into six-week old immunodeficient mice. Briefly, 2 × 106 U87MG cells were resuspended in 100 μL of media that had been pre-mixed with matrigel, and was injected subcutaneously in the upper flank region of mice that had been anaesthetized with isoflurane. Tumors were allowed to grow to 1 cm3 in size, and the animals were randomized into two treatment groups: vehicle and 26. For 26 injections, 26 powder was first dissolved at 25 mg/ml in 100% N-methyl-2-pyrrolidone and subsequently diluted 1:4 with sterile 50% PEG400 to a final concentration of 5 mg/ml 26. Vehicle or 26 was delivered by IP injection at the indicated dosage once daily. Tumors were measured with calipers in two dimensions every other day. Tumor volumes were estimated with the formula: volume = (2a × b)/2, where a = short and b = long tumor axes, respectively, in millimeters. All animal studies were performed according to the official guidelines from the MIT Committee on Animal Care and the American Association of Laboratory Animal Care.
In vivo pharmacodynamic studies
For pharmacodynamic experiments, 26 powder was first dissolved at 25 mg/ml in 100% N-methyl-2-pyrrolidone and then diluted 1:4 with sterile 50% PEG400 prior to injection. Six-week old male C57BL/6 mice were fasted overnight prior to drug treatment. The mice were treated with vehicle (for 10 hr) or 26 (20 mg/kg for 2, 6 or 10 hr) by IP injection, and then re-fed 1 h prior to sacrifice (CO2 asphyxiation). Tissues were collected and frozen on dry ice. The frozen tissue was thawed on ice and lysed by sonication in tissue lysis buffer (50 mM HEPES, pH 7.4, 40 mM NaCL, 2 mM EDTA, 1.5 mM sodium orthovanadate, 50 mM sodium fluoride, 10 mM sodium pyrophosphate, 10 mM sodium β-glycerophosphate, 0.1% SDS, 1.0% sodium deoxycholate and 1.0% Triton, supplemented with protease inhibitor cocktail tablets [Roche]). The concentration of clear lysate was measured using the Bradford assay and samples were subsequently normalized by protein content and analyzed by SDS-PAGE and immunoblotting.
Molecular modeling
An mTOR homology model was built with Modeller(9v6) based on a published PI3Kγ crystal structure complexed with GDC-0941 (PDB: 3DBS). 26 was docked into the model with Pair fitting followed by energy minimization with Discovery studio II.
Supplementary Material
Scheme 1.
Reagents and conditions: 1. appropriate anline, 1,4-dioxane, 100 °C, 6h; 2. appropriate boronic acid, PdCl-2(Ph3P)2, t-Bu-Xphos, Na2CO3, Dioxane, 100 °C, 4h-12h.
Table 2.
Enzymatic and cellular activities profile of compound 7
| Entry | Structure | mTOR IC50(nM) mTORC1 | mTOR IC50(nM) Cellular | PI3K IC50(nM) Cellular |
|---|---|---|---|---|
| 7 |
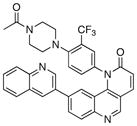
|
5.4 | 3 | >300 |
Acknowledgments
We thank Life Technologies Corporation, SelectScreen® Kinase Profiling Service for performing enzymatic biochemical kinase profiling. We also want to thank Ambit Bioscience for performing KinomeScan™ profiling and Dr. David Waller (Dana Farber Cancer Institute) for the useful proof reading.
Abbreviations
- mTOR
mammalian Target of Rapamycin
- Akt
v-akt murine thymoma viral oncogene homolog 1
- ATP
adenosine triphosphate
- PIKKs
PI3K related kinases
- DNA-PK
DNA activated protein kinase
- ATM
Ataxia Telangiectasia mutated kinase
- ATR
Ataxia Telangiectasia and Rad-3-related kinase
- SMG-1
Serine/threonine protine kinase -1
- 4EBP1
Eukaryotic translation initiation factor 4E-binding protein 1
- PK
Pharmacokinetics
- PD
Pharmacodynamics
- ADME
Absorption, Distribution, Metabolism, Excretion
- MEFs
Mouse Embryo fibroblasts
Footnotes
Six week old male C57BL/6 mice were fasted overnight prior to treatment with 26 as a suspension in 20% N-mentyl-2-pyrrolidone/40% PEG400/40% water or with vehicle by IP injection. Tissues were collected and analyzed at 2, 6 and 10 hours after dosing.
Supporting Information Available: Spectral data of 5-6, 8-25, 27-37. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 2.Abraham RT. PI 3-kinase related kinases: “big” players in stress-induced signaling pathways. DNA Repair. 2004;3:883–887. doi: 10.1016/j.dnarep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 3.Shor B, Gibbons JJ, Abraham RT, Yu K. Targeting mTOR globally in Cancer: thinking beyond rapamycin. Cell Cycle. 2009;23:3831–3837. doi: 10.4161/cc.8.23.10070. [DOI] [PubMed] [Google Scholar]
- 4.Liu Q, Thoreen C, Wang J, Sabatini D, Gray NS. mTOR mediated anit-cancer drug discovery. Drug Discovery today: Therapeutic Strategies. 2010 doi: 10.1016/j.ddstr.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sarbassov D, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 6.Lane HA, Breuleux M. Optimal targeting of the mTORC1 kinase in human cancer. Curr Opin Cell Biol. 2009;21:219–229. doi: 10.1016/j.ceb.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 7.Thoreen CC, Kang SA, Chang J, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive Mammalian Target of Rapamycin Inhibitor Reveals Rapamycin-resistant Functions of mTORC1. J Bio Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldman ME, Apsel B, Uotila A, Loewith R, Kinght ZA, Ruggero D, Shokat KM. Active-Site Inhibitors of mTOR Target Rapamycin-Resistant Outputs of mTORC1 and mTORC2. PLoS Biology. 2009;7:371–383. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-martinez JM, Moran J, Clarke RG, Gray A, Cosulich SC, Chresta CM, Alessi DR. Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR) Biochem J. 2009;421:29–42. doi: 10.1042/BJ20090489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu K, Toral-Barza L, Shi C, Zhang W, Lucas J, Shor B, Kim J, Verheijen J, Curran K, Malwitz DJ, Cole DC, Ellingboe J, Ayral-kaloustian S, Mansour TS, Gibbons JJ, Abraham RT, Nowak P, Zask A. Biochemical, Cellular, and In vivo Activity of Novel ATP-Competitive and Selective Inhibitors of the Mammalian Target of Rapamycin. Cancer Res. 2009;69:6232–6240. doi: 10.1158/0008-5472.CAN-09-0299. [DOI] [PubMed] [Google Scholar]
- 11.Barr S, Russo S, Buck E, Epstein D, Miglarese M. Co-targeting mTOR and IGF-1R/IR results in synergistic activity against a broad array of tumor cell lines, independent of KRAS mutation status. AACR 101st Annual meeting; Washington DC. April, 2010; p. 1632. [Google Scholar]
- 12.Chresta CM, Davies BR, Hickson I, Harding T, Cosulich lS, Critchlow SE, Vincent JP, Ellson R, Jones D, Sini P, james D, Howard Z, Dudley P, Hughes G, Smith L, Maguire S, Hummersone M, Malagu K, Menear K, Jenkins R, Jacobsen M, Smith GCM, Guichard S, Pass M. AZD8055 Is a Potent, Selective, and Orally Bioavailable ATP-Competitive Mammalian Target of Rapamycin Kinase Inhibitor with In vitro and In vivo Antitumor Activity. Cancer res. 2010;70:288–298. doi: 10.1158/0008-5472.CAN-09-1751. [DOI] [PubMed] [Google Scholar]
- 13.Menear KA, Gomez S, Malagu K, Balley C, Blackburn K, Cockcroft X, Ewen S, Fundo A, Gall AL, Hermann G, Sebastian L, Sunose M, Persnot T, Torode E, Hickson I, Martin NM, Smith GCM, Pike KG. Identification and optimization of novel and selective small molecular weight kinase inhibitors of mTOR. Bioorg Med Chem Lett. 2009;20:5898–5901. doi: 10.1016/j.bmcl.2009.08.069. [DOI] [PubMed] [Google Scholar]
- 14.Richard DJ, Verheijen JC, Curran K, Kaplan J, Troal-Barza L, Hollander I, Lucas J, Yu K, Zask A. Incorporation of water-solubilizing groups in pyrazolopyrimidine mTOR inhibitors: Discovery of highly potent and selective analogs with improved human microsomal stability. Bioorg Med Chem Lett. 2009;24:6830–6835. doi: 10.1016/j.bmcl.2009.10.096. [DOI] [PubMed] [Google Scholar]
- 15.Park S, Chapuis N, Bardet V, Tamburini J, Gallay N, Willems L, Knight ZA, Shokat KM, Azar N, Viguie F, IFrah N, Dreyfus F, Mayerus P, Lacombe C, Bouscary D. PI-103, a dual inhibitor of Class IA phosphatidylinositide 3-kinase and mTOR, has antileukemic activity in AML. Leukemia. 2008;22:1698–1706. doi: 10.1038/leu.2008.144. [DOI] [PubMed] [Google Scholar]
- 16.Serra V, Markman B, Scaltriti M, Eichhorn PJA, Valero V, Guzman M, Botero ML, Llonch E, Atzori F, Cosimo SD, Maira M, Garcia-Echeverria C, Parra JL, Arribas J, Baselga J. NVP-BEZ235, a Dual PI3K/mTOR Inhibitor, Prevents PI3K Signaling and Inhibits the Growth of Cancer Cells with Activating PI3K Mutations. Cancer Res. 2008;68:8022–8030. doi: 10.1158/0008-5472.CAN-08-1385. [DOI] [PubMed] [Google Scholar]
- 17.Sutherlin DP, Sampath D, Berry M, Castanedo G, Chang Z, Chuckowree I, Doston J, Folkes A, Freidman L, Cgolsmith R, Heffron T, Lee L, Lesnick J, Lewis C, Mathieu S, Nonomiya J, Olivero A, pang J, Prior WW, Salphati L, Sideris S, Tian, Tsui V, Wan N, Wang S, Wiesmann C, Wong S, Zhu B. Discovery of (Thienopyrimidin-2-yl)aminopyrimidines as Potent, Selective, and Orally Available Pan-PI3-Kinase and Dual Pan-PI3-Kinase/mTOR Inhibitors for the Treatment of Cancer. J Med Chem. 2010;53:1086–1097. doi: 10.1021/jm901284w. [DOI] [PubMed] [Google Scholar]
- 18.Knight SD, Adams ND, Burgess JL, Chaudhari AM, Darcy MG, Donatelli CA, Luengo JI, Newlander KA, Parrish CA, Ridgers L, Sarpong MA, Schmidt SJ, Van Aller GS, Carson JD, Diamond MA, Elkins PA, Gardiner CM, Garver E, Gilbert SA, Gontarek RR, Jackson JR, Kershner KL, Luo L, Raha K, Sherk CS, Sung C, Sutton D, Tummino PJ, wegrzyn RJ, Auger KR, Dhanak D. Discovery of GSK2126458, a Highly Potent Inhibitor of PI3K and the Mammalian Target of Rapamycin. ACS Med Chem Lett. 2010;1:39–43. doi: 10.1021/ml900028r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanack Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Peled LB, Sabatini DM. The Rag GTPases Bind Raptor and Mediate Amino Acid Signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding S, Gray NS, Wu X, Ding Q, Schultz PG. A combinatorial scaffold approach toward kinase-directed heterocycle libraries. J Am Chem Soc. 2002;124:1594–1596. doi: 10.1021/ja0170302. [DOI] [PubMed] [Google Scholar]
- 21.Rabindran SK, Discafani CM, Rosfjord EC, Baxter M, Floyd MB, Golas J, Hallett WA, Johnson BD, Nilakantan R, Overbeek E, Reich MF, Shen R, Shi X, Tsou HR, Wang YF, Wissner A. Antitumor activity of HKI-272, an orally active, irreversible inhibitor of the HER-2 tyrosine kinase. Cancer Res. 2004;11:3958–3965. doi: 10.1158/0008-5472.CAN-03-2868. [DOI] [PubMed] [Google Scholar]
- 22.Xia W, Mullin RJ, Keith BR, Liu LH, Ma H, Rusanak DW, Owens G, Alligood KJ, Spector NL. Anti-tumor activity of GW572016: a dual tyrosine kinase inhibitor blocks EGF activation of EGFR/erbB2 and downstream Erk1/2 and AKT pathways. Oncogene. 2002;41:6255–6263. doi: 10.1038/sj.onc.1205794. [DOI] [PubMed] [Google Scholar]
- 23.Aoki M, Batista O, Bellacosa A, Tsichlis P, Vogt PK. The Akt kinase: Molecular determinants of oncogenicity. Proc Natl Acad Sci USA. 1998;95:14950–14955. doi: 10.1073/pnas.95.25.14950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stowasser F, Baenziger M, Garad SD. Preparation of salts and crystalline forms of 2-methyl-2-[4-(3-methyl-2-oxo-8-quinolin-3-yl-2,3-dihydroimidazo[4,5-c]quinolin-1-yl)-phenyl]propionitrile and its use as a drug. WO2008064093 PCT Int Appl. 2008
- 25.Stauffer F, Maira SM, Furet P, Garcia-Echeverria C. Imidazo[4,5-c]quinolines as inhibitors of the PI3K/PKB-pathway. Bioorg Med Chem Let. 2008;18:1027–1030. doi: 10.1016/j.bmcl.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 26.(a) Rewcastle GW, Palmer BD, Bridges AJ, Showalter HDH, Sun L, Nelson J, McMichael A, Kraker AJ, Fry DW, Denny WA. Tyrosine Kinase Inhibitors. 9. Synthesis and Evaluation of Fused Tricyclic Quinazoline Analogues as ATP Site Inhibitors of the Tyrosine Kinase Activity of the Epidermal Growth Factor Receptor. J Med Chem. 1996;39:918–928. doi: 10.1021/jm950692f. [DOI] [PubMed] [Google Scholar]; (b) Gazit A, Yee K, Uecker A, Bohmer FD, Sjoblom T, Ostman A, Waltenberger J, Golomb G, Banai S, Heinrich MC, Levitzki A. Tricyclic Quinoxalines as Potent Kinase Inhibitors of PDGFR Kinase, Flt3 and Kit. Bioorg Med Chem. 2003;11:2007–2018. doi: 10.1016/s0968-0896(03)00048-8. [DOI] [PubMed] [Google Scholar]; (c) Courma MS, Wu J, Leou J, Tan U, Cahng c, Chang T, Lin W, Hsu JT, Chao Y, Wu S, Hsieh H. Aurora kinase A inhibitors: Identification, SAR exploration and molecular modeling of 6,7-dihydro-4H-pyrazolo- [1,5-a]pyrrolo[3,4-d]pyrimidine-5,8-dione scaffold. Bioorg Med Chem Lett. 2008;18:1623–1627. doi: 10.1016/j.bmcl.2008.01.068. [DOI] [PubMed] [Google Scholar]; (d) Moro S, Varano F, Cozza G, Pagano MA, Zagotto G, Chilin A, Guiotto A, Catarzi D, Calotta V, Pinna LA, Meggio F. Pyrazoloquinazoline Tricyclic System as Novel Scaffold to Design New Kinase CK2 inhibitors. Lett Drug Des Discovery. 2006;3:281–284. [Google Scholar]; (e) kempson J, Spergel SH, Guo J, Quesnelle C, Gill P, Belanger D, Dyckman AJ, Li T, Watterson SH, Langevine CM, Das J, Moquin RV, Furch JA, Marinier A, Dodier M, Martel A, Nirschl D, Kirk KV, Bruke JR, Pattoli MA, Gilloly K, McIntyre KW, Chen L, Yang Z, marathe PH, Wang-ierson D, Dodd HH, McKinnon M, Barrish JC, Pitts WJ. Novel Tricyclic Inhibitors of IKB Kinase. J Med Chem. 2009;52:1994–2005. doi: 10.1021/jm8015816. [DOI] [PubMed] [Google Scholar]
- 27.Maira S, Stauffer F, Brueggen J, Furet P, Schnell C, Fritsch C, Brachmann S, Chene P, Pover AD, Schoemaker K, Fabbro D, Gabriel D, Simonen M, Murphy L, Finan P, Sellers W, Garcia-Echeverria CG. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol Cancer Ther. 2008;7:1851–1863. doi: 10.1158/1535-7163.MCT-08-0017. [DOI] [PubMed] [Google Scholar]
- 28.Knight ZA, Gonzalez B, Feldman M, Zunder ER, Goldenberg DD, Willimas O, Loewith R, Stokoe D, Balla A, Toth B, Balla T, Weiss W, Willimas RL, Shokat KM. A Pharmacological Map of the PI3-K Family Defines a Role for p110α in Insulin Signaling. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yip CK, Murata K, Walz T, Sabatini DM, Kang SA. Structure of the Human mTOR Complex I and Its Implications for Rapamycin Inhibition. Mol Cell. 2010;5:768–774. doi: 10.1016/j.molcel.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.