Abstract
Substrate recognition by the Neurospora Varkud satellite ribozyme depends on the formation of a magnesium-dependent kissing-loop interaction between the stem-loop I (SLI) substrate and stem-loop V (SLV) of the catalytic domain. From mutagenesis studies, it has been established that this I/V kissing-loop interaction involves three Watson–Crick base pairs and is associated with a structural rearrangement of the SLI substrate that facilitates catalysis. Here, we report the NMR structural characterization of this I/V kissing-loop using isolated stem-loops. NMR studies were performed on different SLI/SLV complexes containing a common SLV and shiftable, preshifted, or double-stranded SLI variants. These studies confirm the presence of three Watson–Crick base pairs at the kissing-loop junction and provide evidence for the structural rearrangement of shiftable SLI variants upon SLV binding. NMR structure determination of an SLI/SLV complex demonstrates that both the SLI and SLV loops adopt U-turn structures, which facilitates intermolecular Watson–Crick base pairing. Several other interactions at the I/V interface, including base triples and base stacking, help create a continuously stacked structure. These NMR studies provide a structural basis to understand the stability of the I/V kissing-loop interaction and lead us to propose a kinetic model for substrate activation in the VS ribozyme.
The U-turn fold is widespread in RNA structures. It was first identified in the anticodon hairpin loop of yeast phenylalanine tRNA1 and later observed in several hairpin loops, internal loops, and helical junctions of a variety of functional RNAs.2−11 The UNR sequence (U = uracil, N = any base, and R = purine) typical of this fold facilitates the formation of a sharp backbone turn at the UN step and promotes stacking of the bases after the turn in a favorable position for Watson–Crick base pairing. As a result, U-turn structures are often key contributors to RNA folding and tertiary interactions. In addition, U-turn structures are known to initiate rapid RNA/RNA pairing, as found for codon–anticodon recognition12,13 and antisense RNA targeting in prokaryotes,14 and such pairing is known to trigger conformational changes in macromolecular complexes involving RNA.14,15 However, despite the importance of U-turn structures in RNA function, few structural studies have thoroughly examined their contribution to RNA/RNA interactions and associated conformational changes.
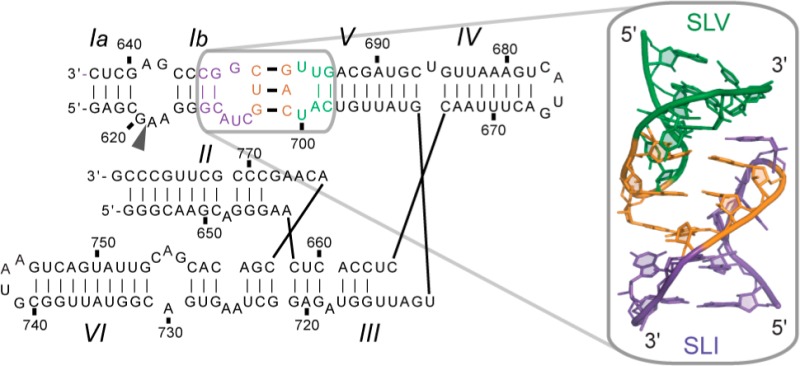
The Neurospora Varkud satellite (VS) ribozyme provides a simple model system for investigating the role of U-turn structures in an RNA/RNA interaction. The VS ribozyme is derived from the VS RNA, a circular single-stranded RNA satellite found in certain strains of Neurospora that depends on the cleavage and ligation activities of the VS ribozyme for its replication.16−18 In vitro, the minimal region of the VS RNA that maintains catalytic activity contains six stem-loop domains (SLI-SLVI), with stem-loop I (SLI) corresponding to the substrate and stem-loops II to VI (SLII-SLVI) forming the catalytic domain (Figure 1A).19,20 Of particular interest, a unique kissing-loop interaction between SLI and SLV (Figure 1A) plays a central role in substrate recognition and is very important for in vitro cleavage and ligation activities.21−26 Mutagenesis and chemical probing data indicate that this I/V kissing-loop forms in the presence of magnesium (Mg2+) ions and involves three Watson–Crick (W–C) base pairs.21,23 In addition, formation of the I/V kissing-loop is accompanied by a structural rearrangement in SLI, best described as a helix shift (Figure 1B), that activates the substrate for catalysis.27−29 Interestingly, the I/V kissing-loop and associated helix shift can be reproduced biochemically using isolated stem-loops (SLI and SLV).29
Figure 1.
Primary and secondary structures of RNAs used in this study. (A) The catalytic domain of the Neurospora VS ribozyme containing helical domains II–VI and an SLI substrate (SLIps) containing stems Ia and Ib. The cleavage site is indicated by an arrowhead. The I/V kissing-loop interaction involves Watson–Crick base pairs (thick black lines) between residues G630, U631, and C632 of SLI (shaded in purple) and residues C699, A698, and G697 of SLV (shaded in green). (B) Formation of the I/V kissing-loop is accompanied by a structural rearrangement of the SLI substrate from an unshifted (free SLIsb) to a shifted (bound SLIsb) conformation. (C) The SLV and SLI RNAs used for investigation of SLI/SLV complexes by NMR spectroscopy. In (B) and (C), the cleavage site of the SLI substrates (SLIsb, SLIps, SLIpsΔIa, SLIsbΔIa, and SLIsbΔIaGU) is between residues −1 and +1.
High-resolution NMR studies of stem-loop domains derived from the VS ribozyme have provided initial structural insight into the I/V kissing-loop and the associated helix shift. NMR structures of the SLI cleavage site internal loop were determined that correspond to both the unshifted and shifted conformations30−32 and help explain the structural changes that likely occur at the cleavage site as a result of the helix shift. NMR structures of an isolated SLV domain indicate that the SLV loop forms a U-turn structure,33,34 as predicted from the loop sequence and mutational studies.20 Furthermore, this U-turn structure becomes more compact in the presence of Mg2+ ions.33,34 Although a U-turn structure was also predicted in SLI,20 NMR studies of an isolated SLI domain indicate that the SLI loop is mostly disordered in both the absence and the presence of Mg2+ ions.30 On the basis of these NMR structures and biochemical data, three-dimensional models of the I/V kissing-loop interaction were derived.25,35 In addition, a model of the SLI/SLV interaction was incorporated in a low-resolution solution structure of the full VS ribozyme obtained by small-angle X-ray scattering (SAXS).36 However, these models of the SLI/SLV complex lack the high level of precision that can only be achieved through high-resolution structural studies.
In this manuscript, we perform NMR structural studies of SLI/SLV complexes to gain additional structural insights into substrate recognition and activation by the VS ribozyme. We obtain structural evidence for formation of three W–C base pairs at the kissing-loop junction and for the helix shift in SLI as a result of SLV binding. In addition, our NMR data provide novel insights into the structure of the I/V kissing-loop that help explain the stability of this interaction and allow us to propose a kinetic model of helix shifting in SLI.
Experimental Procedures
Preparation of RNA for NMR Studies
All RNAs used in this study (SLV, SLIps, SLIpsΔIa, SLIsb, SLIsbΔIa, SLIsbΔIaGU, SLIds1, and SLIds2; Figure 1B,C) were synthesized in vitro using T7 RNA polymerase (prepared in house), single-stranded DNA templates (IDT), and NTPs (13C/15N- or 15N-NTPs prepared in house or unlabeled NTPs from Sigma-Aldrich), and they were purified as previously described.25,33 The purified RNAs were first exchanged in NMR buffer (10 mM Tris-d11 at pH 7.0, 50 mM NaCl, and 0.05 mM NaN3 in 90% H2O/10% D2O) by use of Amicon Ultra-4 3K ultrafiltration devices and then refolded by heating to 95 °C for two min and then cooling in ice water. Prior to complex formation, the RNA samples were transferred to NMR buffer supplemented with either 5 mM or 20 mM MgCl2 99.995% (Sigma-Aldrich) using the ultrafiltration device. In the case of SLIsbΔIaGU, 20 mM MgCl2 was added directly to the sample without the ultrafiltration device to ensure that it maintained its hairpin conformation. Using these procedures, the SLI and SLV RNAs formed hairpin structures, because only this conformation and not the duplex conformation was detected by native gels (Figure S1 of Supporting Information).
Formation of SLI/SLV complexes was generally achieved by titrating the isotopically labeled stem-loop with the unlabeled stem-loop in ratios of 1:0, 1:0.25, 1:0.5, 1:0.75, and 1:1, and the resulting complex was concentrated to a final volume of either 350 or 500 μL. For imino NMR studies of complex formation, the following complexes were prepared at concentrations of 0.8–1.4 mM: SLIsbΔIa/15N-SLV, 15N-SLIsbΔIa/SLV, 15N-SLIsbΔIa/15N-SLV, 15N-SLIsbΔIaGU/SLV, SLIps/15N-SLV, 15N-SLIpsΔIa/SLV, 15N-SL1ds1/SLV, and 15N-SLIds2/SLV. For NMR structure determination, the following complexes were prepared at concentrations of 1.0–2.2 mM in NMR buffer supplemented with 5 mM MgCl2: SLIds2/15N-SLV, SLIds2/13C15N-SLV, 15N-SLIds2/SLV, 13C15N-SLIds2/SLV, 13C15N-SLIds1/SLV, 15N-SLIsbΔIa/15N-SLV, SLIsbΔIa/15N-SLV, and SLIsbΔIa/13C15N-SLV. Moreover, two complexes were similarly prepared, in which 13C/15N-labeling of SLIds2 was restricted to either guanines or to guanines and cytosines: 13C15N G-SLIds2/SLV and 13C15N CG-SLIds2/SLV. For NMR studies in D2O, the samples were lyophilized and resuspended in 99.996% D2O a minimum of four times.
Native Gel Electrophoresis Studies of RNA Hairpins
The RNA samples were prepared by heating and snap-cooling 5 μg of each RNA in 20 μL of buffer containing 10 mM Tris, pH 7.0, 50 mM NaCl, and 20 mM MgCl2. Native polyacrylamide gel electrophoresis was carried out on 7.5% polyacrylamide gels (37.5:1 acrylamide:bisacylamide) and run in TB buffer (20 mM Tris borate at pH 8). The gels were prerun for 30 min at 200 V and 4 °C, loaded with RNA samples (SLI and SLV variants), and then run for 2 h at 250 V and 4 °C. The gels were stained with Stains All (Sigma-Aldrich).
NMR Spectroscopy
NMR data were acquired on Varian UnityINOVA 500 and 600 MHz NMR spectrometers equipped with a pulse-field gradient unit and actively shielded z-gradient probes, either a 1H{13C/15N} triple-resonance probe (standard or cold probe) or a 1H{15N–31P} indirect detection probe. For imino NMR studies of complex formation, NMR data were acquired at either 25 °C (for complexes of SLIps, SLIsbΔIa, SLIpsΔIa, SLIds1, and SLIds2 with SLV) or 5 °C (for the SLIsbΔIaGU/SLV complex). The following NMR spectra were collected: 1D 15N-decoupled 1H watergate37 and its 15N-filtered and 15N-edited versions, imino-optimized 2D 1H–15N HSQC,38 2D HNN-COSY,39 and 2D 15N(t1/t2)-decoupled 1H–1H flip-back watergate NOESY (2D NOESY)37,40 with mixing times of 150 ms or 250 ms. For NMR structure determination, NMR data were collected at 25 °C on SLIsbΔIa/SLV and SLIds1/SLV, SLIds2/SLV complexes. For assignment of exchangeable protons and their attached nitrogens, the following experiments were collected in H2O: imino and amino-optimized 2D 1H–15N HSQC ;38 G-specific 2D H(NC)-TOCSY-(C)H41 (for SLIsbΔIa/15N-SLV only); 2D HNN-COSY;39 2D 1H–15N CPMG-NOESY42 (for SLIsbΔIa/SLV and SLIds2/SLV only); 2D NOESY;37,40 3D 15N-edited NOESY-HSQC43 and 3D 15N/13C-edited NOESY-HSQC44 with a mixing time of 150 ms (for SLIds2/SLV only). For assignment of nonexchangeable protons and their attached carbons, the following experiments were collected in D2O: 2D 1H–13C constant-time-HSQC (CT-HSQC);45,46 3D CT-HCCH–COSY;47 3D HCCH-TOCSY47 (for SLIsbΔIa/SLV and SLIds2/SLV only) and 3D 13C-edited HMQC-NOESY48 collected with mixing times of 70 or 140 ms. In addition, 2D long-range 1H–15N HMQC49 spectra optimized for transfers via J = 21 Hz were collected in D2O for the assignment of purines N7 and N9. NMR data were processed using the NMRPipe/NMRDraw package50 and analyzed with the CCPNMR suite.51
Structural Restraints
Structural restraints were collected for structure determination of the SLIds2/SLV complex. Two-dimensional HNN-COSY spectra were collected to detect 2JNN couplings across hydrogen bonds in W–C base pairs. Distance restraints were extracted from 2D NOESY,37,40 2D 1H–15N CPMG-NOESY,42 3D 15N-edited NOESY-HSQC,43 and 3D 15N/13C-edited NOESY-HSQC44 spectra recorded in H2O and 3D 13C-edited HMQC-NOESY48 spectra collected in D2O. The NOE-derived distance restraints were separated in four classes: very strong (1.8–3.3 Å), strong (1.8–4.5 Å), medium (1.8–5.5 Å), and weak (2.8–7.5 Å). Based on NMR evidence for specific base pairing obtained from NOESY and HNN-COSY spectra, canonical distance restraints were employed to define the hydrogen bonds and planarity of standard W–C and G–U wobble base pairs. In addition, cross-strand interphosphate distance constraints were introduced for each of these base pairs (≥18 Å) to help define A-form geometry in the SLIds2 and SLV stems. Based on NMR data, all glycosidic angles adopt an anti conformation, which was defined using H1′-H6/H8 distance restraints of 3.2–3.6 Å for pyrimidines and 3.4–3.8 Å for purines. Based on NMR evidence, backbone torsion angles of residues in helical regions (1I-6I, 16I-22I in SLIds2 and 1V-7V, 15V-21V in SLV) were restrained to A-form values (±15°), whereas a selected few were also restrained to A-form values (±30°) in the kissing-loop region (α and β of 13I and 12V, γ and δ of 12I, 13I, 11V, 12V, and ε and ζ of 12I and 11V). Based on the N7 chemical shift values of G11I and A11V, which are characteristic of the U-turn motif, two specific distance restraints of 1.8–3.0 Å were defined (G11I N7 to U9I 2′–OH and A11V N7 to U9V 2′–OH). Finally, based on the C7I NH2 chemical shifts, which are characteristics of G–C base pairs, an ambiguous distance restraint (1.8–2.2 Å) was defined between C7 I NH2 and the O6 of either G14I or G15I.
Residual Dipolar Coupling Measurements
NMR studies were performed at 25 °C with an 15N-SLIds2/15N-SLV sample (0.2–0.3 mM RNA in NMR buffer supplemented with 5 mM MgCl2) titrated with a concentrated solution of Pf1 filamentous phage to yield Pf1 phage concentrations of 0, 14.6, 17.2, 18.9, and 20.5 mg/mL.52 For each of these samples, values of the 2H splitting of D2O were measured from 1D 2H NMR spectra, and imino 1H–15N splittings were measured from 2D 1H–15N HSQC spectra38 collected with active JNH coupling in t1. The 1H–15N splittings were measured from fitted peaks using NMRWish50 in the NMRPipe/NMRDraw package. Residual dipolar coupling (RDC) values were calculated for H1–N1 (G) and H3–N3 (U) signals detected at the different Pf1 phage concentrations. For each H–N signal, the RDC values were plotted with respect to Pf1 phage concentration in order to obtain a linear fit from which a precise RDC value was determined at 17.2 mg phage/mL.
Structure Calculation
Three-dimensional structures were calculated with restrained molecular dynamics and simulated annealing in X-PLOR-NIH version 2.19 using a two-stage protocol, as previously described.53 Briefly, an initial set of 50 structures was calculated at stage one from structures with randomized backbone angles. At stage two, the structures were refined with RDC restraints using a single alignment tensor. The axial and rhombic components of the alignment tensor were explored with MODULE 1.0 and a grid search procedure to obtain reliable values of Da and R. The grid search was repeated using RDC values from the SLIds2 stem (residues 1–6, 16–21) and the SLV stem (residues 2–7, 15–20), independently. All grid search calculations converged on similar values of Da and R [values of Da(NH) = 17.7 Hz and R = 0.25 were used]. A final set of 500 structures was calculated, from which the 20 lowest-energy structures that satisfied the experimental restraints (no distance violation > 0.2 Å, no torsion angle violation > 5° and no RDC violation > 0.1 Hz) were selected for analysis. These 20 lowest-energy structures were used to calculate an average structure that was minimized against NOE and dihedral restraints. All structures were visualized with PyMOL Molecular Graphics System, Version 1.3 Schrödinger, LLC and analyzed with PyMOL and Curves+. Interhelical parameters were calculated as previously described.35
Results
NMR Evidence for Formation of the I/V Kissing-Loop Interaction
It was previously established that the SLV RNA used in this study (Figure 1C) adopts a stable hairpin that is compatible with SLI/SLV complex formation.25 Thus, we next focused on identifying suitable SLI RNAs for our investigations of SLI/SLV complexes. Several SLI variants (Figure 1C) were characterized by imino NMR spectroscopy and native gel electrophoresis (Figure S1 of Supporting Information) to identify those that form a stable hairpin to be tested for complex formation. Initially, we used shiftable SLI substrates with a stable stem Ia (e.g., SLIsb in Figure 1B) as found in the wild-type VS ribozyme, but such SLI variants do not form a stable SLI/SLV complex (not shown), in agreement with biochemical studies showing that stem Ia hinders formation of the I/V kissing-loop interaction.27,54−56
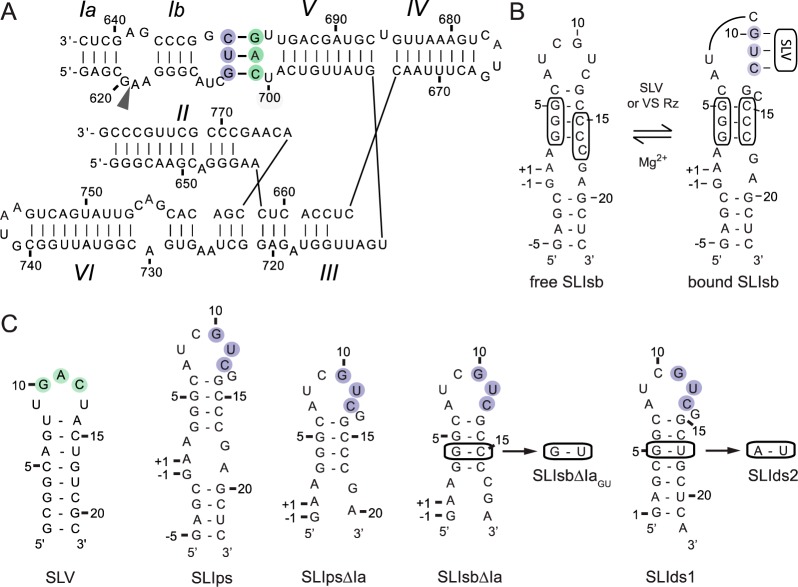
Next, we investigated complex formation with a shiftable SLI substrate that does not contain stem Ia (SLIsbΔIa; Figure 2A). The SLIsbΔIa variant also contains a G627A mutation (A7I in SLIsbΔIa) compared to the natural sequence of the VS ribozyme (Figure 2A). This variant was selected because a similar SLI substrate with the natural G at this position did not form a stable homogeneous complex with SLV for NMR studies (data not shown). Importantly, the G627A mutation is compatible with both cleavage and ligation activities of the VS ribozyme,24,25,35 and it slightly increases the affinity of the SLI substrate for the VS ribozyme.24 Formation of the SLI/SLV complex with SLIsbΔIa was monitored by 1D imino proton NMR (Figure 2B). In addition, imino proton assignments of SLIsbΔIa and SLV were obtained from 2D NOESY spectra of both their free (not shown) and bound forms (Figure 2C). Interestingly, upon addition of SLIsbΔIa to SLV, three new imino proton signals are observed (Figure 2B), including one for SLV (G H1 at 12.11 ppm) and two for SLIsbΔIa (G H1 at 12.92 ppm and U H3 at 14.14 ppm; Figure 2C), which indicates that complex formation stabilizes three W–C base pairs, one A–U, and two G–C base pairs. Interestingly, two NOE signals involving these new imino protons are observed (Figure 2C), which support stacking of the A–U base pair between the two G–C base pairs (Figure 2A). Further evidence for intermolecular W–C base pairing was obtained from 2D HNN-COSY39 spectra collected on three SLIsbΔIa/SLV complexes prepared with differential 15N labeling (Figure S2 of the Supporting Information). These spectra confirm W–C base pairing in the SLIsbΔIa stem (15N-SLIsbΔIa/SLV complex), the SLV stem (SLIsbΔIa/15N-SLV complex), and at the kissing-loop junction (15N-SLIsbΔIa/15N-SLV complex). Taken together, the imino NMR data support a secondary structure model for the SLIsbΔIa/SLV complex (Figure 2A) that includes the three proposed W–C base pairs (G11I-C12V, U12I-A11V, and C13I-G10V) at the kissing-loop junction.
Figure 2.
NMR evidence for formation of the SLIsbΔIa/SLV complex. (A) Proposed secondary structure of SLIsbΔIa, a shiftable SLI variant, and SLV in their free (left) and bound (right) forms. The boxed nucleotides in SLIsbΔIa are those involved in the helix shift associated with SLV binding. (B) Imino region of 1D 1H NMR spectra recorded for the titration of 15N-labeled SLV with unlabeled SLIsbΔIa. 1D NMR spectra were collected under the same conditions and plotted on the same vertical scale but were adjusted to take into account sample dilution during the titration. Imino proton assignment is provided for both SLIsbΔIa and SLV in their free forms. The imino proton signals marked with an asterisk in the 1:1 SLIsbΔIa/SLV complex represent a small excess of free SLV. (C) Imino region of the 2D NOESY spectrum of the SLIsbΔIa/15N-SLV complex. On top, 1D 15N-filtered (purple; SLIsbΔIa) and 15N-edited (green; SLV) spectra of the complex annotated with imino proton assignments. Annotations in (B) and (C) are used to identify imino signals within SLIsbΔIa (purple), within SLV (green), and at the kissing-loop junction (orange), and these imino signals provide evidence for the base pairs shaded with the corresponding colors in (A).
NMR Evidence for the Helix Shift in SLI upon Interaction with SLV
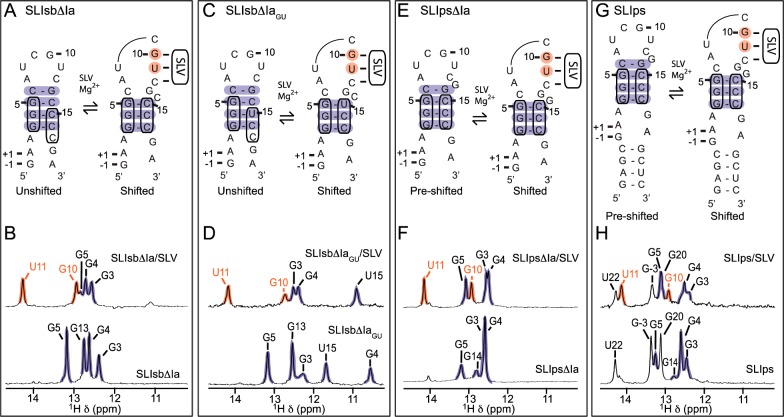
It was previously demonstrated biochemically that SLI undergoes a structural rearrangement from an unshifted to a shifted conformation upon interaction with SLV, and this helix shift activates the substrate for catalysis.28,29 In SLIsbΔIa, this helix shift should involve three guanines (G3I, G4I, and G5I) that, upon SLV binding, change their base pairing partners in stem Ib, thereby excluding C14I from the helix (Figure 2A). To provide NMR evidence for this structural change, the 1D imino 1H NMR spectrum of SLIsbΔIa was compared to the 1D 15N-filtered imino 1H spectrum of the SLIsbΔIa/15N-SLV complex, which allows for selective detection of the SLI signals from the complex (Figure 3AB). Four imino signals are observed for free SLI, in agreement with the four proposed G–C base pairs in the stem involving G3I, G4I, G5I, and G13I. In contrast, five imino signals are observed for the SLV-bound SLI, two originating from U11I and G10I forming W–C base pairs at the kissing-loop junction and three from G3I, G4I, and G5I forming G–C base pairs in the SLI stem (Figure 3A,B). The imino signal of G13I, detected for free SLIsbΔIa, is absent in the spectrum of the SLIsbΔIa/SLV complex (Figure 3B), suggesting that SLV binding destabilizes the SLI loop closing base pair (C4I–G13I in Figure 3A). Importantly, there are significant chemical shift changes for the imino signals of G3I and G5I in the stem, which reflect important changes in their chemical environment upon complex formation (Figure 3B and Table S1 of the Supporting Information). Taken together, changes in 1D 1H imino spectra of free and SLV-bound SLI are compatible with the helix shift in SLI upon complex formation, although it does not provide evidence for the proposed C4I-G13I base pair closing the loop.28,29
Figure 3.
NMR evidence of structural rearrangements in SLI variants. Upper panels: Proposed secondary structures of the free and SLV-bound forms of shiftable SLI [(A) SLIsbΔIa and (C) SLIsbΔIaGU] and preshifted SLI [(E) SLIpsΔIa and (G) SLIps] variants. Lower panels: Imino region of 1D 1H NMR spectra of (B) SLIsbΔIa, (D) SLIsbΔIaGU, (F) SLIpsΔIa, and (H) SLIps (bottom spectra), along with the 1D 1H 15N-filtered or 15N-edited NMR spectra of the (B) SLIsbΔIa/15N-SLV, (D) 15N-SLIsbΔIaGU/SLV, (F) 15N-SLIpsΔIa/SLV, and (H) SLIps/15N-SLV complexes for detection of SLI imino proton signals only (top spectra). The shaded imino proton signals of residues from the kissing-loop interaction (orange) and the adjacent SLI stem (purple) provide evidence for the base pairs shaded with the corresponding colors in the proposed secondary structures shown in (A), (C), (E), and (G).
To provide unambiguous evidence for base-pair shifting in the SLI stem, complex formation was investigated using a variant of SLIsbΔIa carrying a C15I to U15I change that transforms the G4I–C15I base pair of the free substrate to a G–U base pair (SLIsbΔIaGU; Figure 3C). The imino NMR data of the SLIsbΔIaGU/SLV complex confirm formation of the three W–C base pairs at the kissing-loop junction (Figure S3 of the Supporting Information). Comparison of the imino 1H spectra of SLIsbΔIaGU in its free and SLV-bound forms is compatible with the helix shift in SLI (Figure 3CD). In particular, the imino protons of G13I and G5I observed in free SLIsbΔIaGU are not observed for the SLV-bound SLIsbΔIaGU, in agreement with destabilization of the loop-closing base pairs upon complex formation. Nevertheless, U15I presents a large chemical shift change (from 11.69 to 10.91 ppm; Figure 3D and Table S1 of the Supporting Information) consistent with formation of a G–U base pair closing the loop.57 More importantly, NOE signals confirm that G4I forms a G–U base pair in the free SLIsbΔIaGU but forms a G–C base pair in the SLIsbΔIaGU/SLV complex (Figure S3 of the Supporting Information), providing definite NMR evidence for helix shifting in SLI as a result of complex formation.
Complex formation was also investigated with two preshifted SLI substrates, one with a stem Ia (SLIps) and one without a stem Ia (SLIpsΔIa; Figure 3E–H). The imino NMR data of the SLIps/SLV and the SLIpsΔIa/SLV complexes confirm formation of the three W–C base pairs at the kissing-loop junction (Figures S4 and S5 of the Supporting Information). For the stem residues of SLIpsΔIa and SLIps, there are only minor changes in chemical shifts as a result of SLV binding (Figure 3E–H and Table S1 of the Supporting Information), indicating that the structure of stem Ib is not largely affected by complex formation. However, we note the disappearance of G14I upon complex formation in both cases (Figure 3E–H), which is in agreement with destabilization of the SLI loop-closing base pair. In summary, imino NMR data obtained with shiftable versus preshifted substrates provide structural validation for helix shifting upon complex formation with the shiftable SLI substrates and the lack of such phenomenon with preshifted SLI substrates.
Ideal Complex for NMR Structure Determination
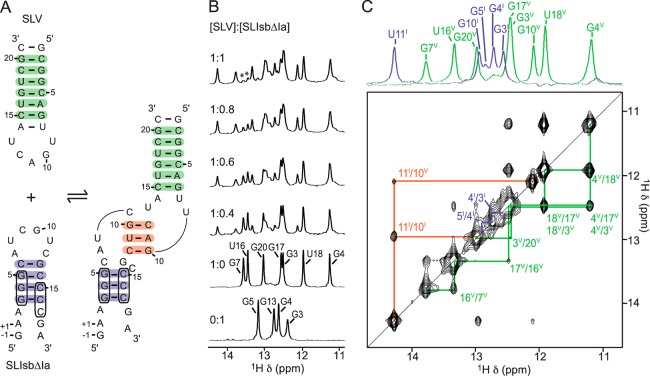
An ideal SLI/SLV complex for NMR structure determination should present a high affinity, a minimal size, and stable stems associated with a large number of detectable imino protons. In this regard, we determined that the SLI variants considered thus far are suboptimal for NMR structure determination aimed at providing a high-resolution understanding of the I/V kissing-loop interaction. Thus, two additional small SLI variants were prepared, SLIds1 (Figure S6 of the Supporting Information) and SLIds2 (Figure 4), that each contains a unique stem Ib that is extended compared to the wild-type SLI. Although these SLI variants are not substrates of the VS ribozyme, their extended stem Ib makes them particularly useful for NMR structure determination. Particularly, they should facilitate NMR resonance assignments and structure refinement using residual dipolar couplings (RDC). As observed with preshifted substrates, imino NMR studies indicate that there is little structural change in stem Ib of SLIds1 and SLIds2 upon complex formation with SLV (Figure 4 and Figure S6 and Table S1 of the Supporting Information). Of note, a weak G imino proton signal, assigned to G15I, is observed in the 1D imino 1H NMR spectra of free SLIds2, and this signal disappears completely upon complex formation (Figure 4B), in agreement with destabilization of the SLI loop-closing base pair. Furthermore, the SLIds1/SLV and SLIds2/SLV complexes both give high-quality imino NMR data, including the imino signals and NOE interconnectivities that are characteristic of the I/V kissing-loop junction (Figure 4C and Figure S6C of the Supporting Information). We pursued NMR structure determination with the SLIds2/SLV complex, which provides superior imino proton dispersion for both SLI and SLV within a complex of reasonable size (43 nt) and of affinity similar to that of the two complexes formed with preshifted SLI substrates [Kd = 0.4–0.7 μM (Bouchard, P. and Legault, P., Département de Biochimie et Médecine Moléculair, Université de Montréal, 2013, unpublished data)].
Figure 4.
NMR evidence for formation of the SLIds2/SLV complex. (A) Proposed secondary structure of SLIds2, a double-stranded SLI variant, and SLV in their free (left) and bound (right) forms. (B) Imino region of 1D 1H NMR spectra recorded for the titration of 15N-labeled SLIds2 with unlabeled SLV. 1D NMR spectra were collected under the same conditions and plotted as described in Figure 2. Imino proton assignment is provided for both SLIds2 and SLV in their free forms. (C) Imino region of the 2D NOESY spectrum of the 15N-SLIds2/SLV complex. The signals marked with an asterisk are decoupling artifacts. On top, 1D 15N-filtered (green; SLV) and 15N-edited (purple; SLIds2) spectra of the complex annotated with imino proton assignment. Annotations in (B) and (C) are used to identify imino signals within SLIds2 (purple), SLV (green), and at the kissing-loop junction (orange), which provide evidence for the base pairs shaded with the corresponding colors in (A).
NMR Structure Determination of the SLIds2/SLV Complex
Multidimensional heteronuclear NMR methods were employed to determine the structure of the SLIds2/SLV complex using several combinations of unlabeled and uniformly labeled (15N or 13C/15N) SLIds2 and SLV RNAs. In addition, complexes incorporating 13C and 15N labeling of specific residue types within SLIds2 and complexes containing different SLI variants (SLIsbΔIa/SLV and SLIds1/SLV, Figure 1) were used to facilitate and confirm spectral analysis. The sequential resonance assignment mainly relied on 2D and 3D NOESY-type spectra of these complexes (see Experimental Procedures), allowing almost complete assignment of the observable resonances (1H, 15N, and 13C) from the bases and the C1′-H1′ and C2′-H2′ resonances from the ribose moieties (Tables S3 and S4 of the Supporting Information).
Three-dimensional structures were calculated using distance, dihedral angle, and RDC restraints (Figure 5 and Table 1). The overall structure is well-defined by the NMR data, with a heavy-atom RMSD of 2.02 ± 0.77 Å for the 20 lowest-energy structures (Figure 5 and Table 1). The stems of both SLI and SLV form well-defined A-form helices (RMSDs of 0.5–0.6 Å). The SLV loop is also well-defined (RMSD of 1.01 ± 0.20 Å), except for the extruded U13V, whereas the SLI loop is not as well-defined, mainly due to a limited number of restraints for residues A8I, U9I, and G15I (RMSD of 1.78 ± 0.69 Å). In terms of interhelical parameters (Figure S7 of the Supporting Information), the majority of structures have an average interhelical angle (θI–V) ∼130° [θI–V = 131° ± 3° with an axis displacement (dI–V) = 15 ± 3 Å], although values ∼160° are also observed in 6 of the 20 structures [θI–V = 161° ± 9° with dI–V = 19 ± 7 Å].
Figure 5.

Stereoview of the NMR structures of the SLIds2/SLV kissing-loop complex. Superposition of the 20 lowest-energy structures on the minimized average structure (not shown). Only heavy atoms of SLIds2 and SLV residues 2–20 were used for the superposition and are shown. Residues are shown in the prevailing coloring scheme (SLIds2, purple; SLV, green; W–C base pairs at the kissing-loop junction, orange).
Table 1. Structural Statistics.
| SLIds2/SLV | |
|---|---|
| number of NOE-derived distance restraints | 507 |
| intranucleotide | 181 |
| internucleotide | 291 |
| intermolecular | 32 |
| ambiguous | 3 |
| hydrogen-bond restraints | 89 |
| base-pair planarity restraints | 32 |
| dihedral angle restraints | 173 |
| residual dipolar coupling (1DNH) restraints | 16 |
| P–P distance restraints | 19 |
| total number of restraints | 836 |
| RMSD from experimental restraints | |
| NOE (Å; none > 0.2 Å) | 0.069 ± 0.008 |
| dihedral (°; none > 5°) | 0.008 ± 0.001 |
| residual dipolar coupling (Hz; none >0.1 Hz) | 0.0018 ± 0.0005 |
| RMSD from idealized geometry | |
| bonds (Å) | 0.00441 ± 0.00005 |
| angles (°) | 0.995 ± 0.001 |
| impropers (°) | 0.385 ± 0.001 |
| heavy-atom RMSD to the minimized average structure | |
| overall (SLIds2: 2–20, SLV: 2–20) | 2.02 ± 0.77 |
| overall SLI (SLIds2: 2–20) | 1.82 ± 0.77 |
| overall SLV (SLV: 2–20) | 1.15 ± 0.27 |
| stem I (SLIds2: 2–6, 16–20) | 0.59 ± 0.10 |
| stem V (SLV: 2–7, 15–20) | 0.49 ± 0.07 |
| loop I (SLIds2: 6–16) | 1.78 ± 0.69 |
| loop V (SLV: 7–15) | 1.01 ± 0.20 |
| kissing-loop stem (SLIds2: 11–13, SLV: 10–12) | 0.73 ± 0.19 |
| kissing-loop junction (SLIds2: 6–16, SLV: 7–15) | 1.70 ± 0.51 |
Standard U-Turns at the Kissing-Loop Junction
As previously proposed, both the SLI and SLV loops within the SLI/SLV complex adopt a U-turn structure (Figure 6).21 The consensus UNR sequence of the U-turn is represented in SLIds2 by residues U9I, C10I, and G11I and in SLV by residues U9V, G10V, and A11V. These loop structures are characterized by a sharp turn in the ribose-phosphate backbone after the first U (U9I or U9V), which is supported by several NOEs for both SLIds2 and SLV, namely NOEs between U9 H1′/H2′ and R11 H8 and between U9 H1′ and U/C12 H6/H5. As found in other U-turn structures, there are several base-stacking interactions after the sharp turn, which are supported by sequential NOEs characteristic of A-form geometry. These stacking interactions involve C10I, G11I, U12I, C13I, and G14I in SLIds2 and G10V, A11V, C12V, and A14V in SLV (Figure 6).
Figure 6.
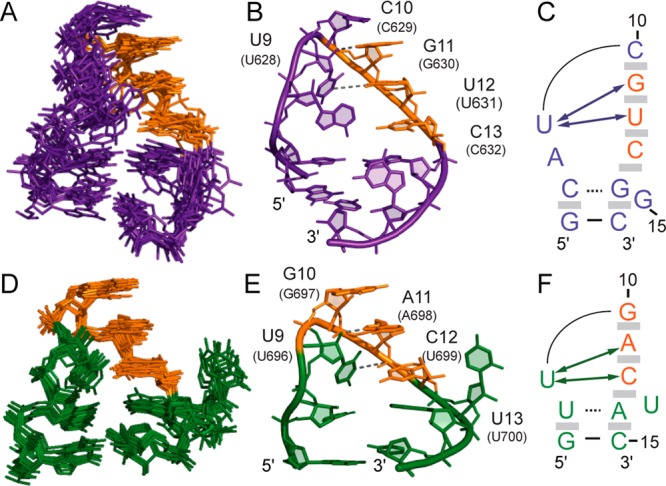
U-turn structures in the SLIds2 and SLV loops of the SLIds2/SLV complex. Superposition of (A) the SLIds2 and (D) SLV loops of the 10 lowest-energy structures on the minimized average structure (not shown). Only heavy atoms of SLIds2 residues 6–16 or SLV residues 7–15 were used for the superposition and are shown. Minimized average structure of the (B) SLIds2 and (E) SLV loops showing base stacking after the sharp turn in the backbone, which is also represented by shaded gray boxes in (C) and (F). Dashed lines represent hydrogen bonds characteristic of a U-turn motif between U9I 2′–OH and G11I N7 and between U9I H3 and G11I 3′-phosphate in SLIds2 as well as between U9V 2′–OH and A11V N7 and between U9V H3 and A11V 3′-phosphate in SLV. Schematic representation of the (C) SLIds2 and (F) SLV loops in the complex. The arrows indicate observed NOEs that are characteristic of the U-turn motif, including those between U9I H1′/H2′ and G11I H8 and between U9I H1′ and U12I H6/H5 in SLIds2 and the equivalent NOEs in SLV.
U-turn structures are generally stabilized by two hydrogen bonds involving the required U residue: U 2′–OH to R N7 and U H3 to R 3′-phosphate.34 For both the SLIds2 and SLV loops, a downfield-shifted signal was observed for R N7 (G11I at 228.5 ppm and A11V at 221.5 ppm, Table S2 of the Supporting Information) that has been previously associated with hydrogen bonding to U 2′–OH in the U-turn.25,58 On this basis, a distance restraint was defined between the U 2′–OH and R N7 (1.8–3.0 Å) that contribute to bring these residues within hydrogen-bonding distances in the ensemble of structures. The hydrogen bond between U H3 and R 3′-phosphate could not be similarly defined; nevertheless, the distances of U N3 to R 3′-P in the ensemble of structures are between 3.88 and 6.19 Å for SLIds2 and 4.02–5.65 Å for SLV, which is compatible with hydrogen bonding as observed in other U-turn structures.2,5,8,15,34,59
Both the SLIds2 and SLV loops present an extruded nucleotide, G15I in SLIds2 and U13V in SLV (Figure 6). The imino protons of G15I and U13V are not detected in the 1D 1H spectra of the complex and very few NOEs are observed with these bases as expected for an extruded conformation. For SLIds2, the conformation of G15I is not well-defined, but this nucleotide is most often extruded from the loop fold and potentially forms a hydrogen bond between G15I N7 and G14I 2′–OH as well as perpendicular π stacking between G15I and G14I, in which either the NH2 or the 2′–OH group of G14I can form a hydrogen bond with the base of G15I.60−62 In contrast, the extruded U13V of SLV is better defined with its W–C edge oriented toward the solvent in all 20 lowest-energy structures. Most of the U13V signals in the 2D 1H–13C CT-HSQC are observed at higher intensities and significantly different 1H and 13C chemical shifts compared to signals of other residues, which is indicative of fast dynamics for U13V.
Previous NMR studies of the free SLI and SLV hairpins derived from the VS ribozymes have provided a wealth of structural information for the SLI and SLV loops in their free forms.30,33,34 Although the free SLI loop is mostly disordered,30 the free SLV loop adopts a loose U-turn structure in the absence of Mg2+ ions (SLVfree)33 that becomes more compact in the presence of Mg2+ ions (SLVMg).34 The superpositions of these SLV loops with the SLV loop from the SLIds2/SLV complex demonstrate that in the complex, the structure of the SLV loop more closely matches the compact structure of the SLVMg loop (Figure S8 of the Supporting Information). However, the base of U9V is tilted in the complex such that it precludes its stacking with the 5′-phosphate of A11V, as found in SLVMg and several U-turn structures.34 In summary, upon formation of the I/V kissing-loop complex, loop I undergoes a significant structural change from a disordered state to a compact fold stabilized by a U-turn structure, whereas loop V undergoes only minor structural changes within its compact U-turn fold.
Kissing-Loop Junction Stabilized by Base-Pairing and Stacking Interactions
U-turn structures typically expose the W–C edges of bases after the turn, making them available for base pairing. In the SLIds2/SLV complex, these exposed bases from SLI and SLV interact with each other to create the kissing-loop junction (Figure 7A). The G11I, U12I, and C13I bases of SLIds2 and C12V, A11V, and G10V bases of SLV, respectively, form the three proposed W–C base pairs that stack on each other to adopt a short A-form-like helix that is well-defined by several characteristic NOEs (Figures 3 and 7 and Table S5 of the Supporting Information).
Figure 7.
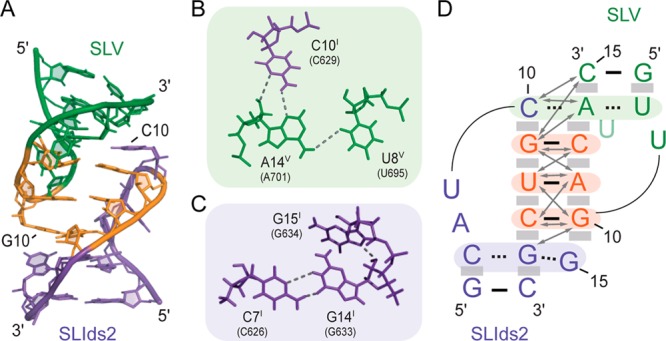
Structural characteristics of the I/V kissing-loop junction. (A) Minimized average structure showing the kissing-loop interaction. (B)(C) Base triples in the minimized average structure. Dashed lines represent the following hydrogen bonds defined on the basis of short distances in the ensemble of structures (given in parentheses): A14V H61 and U8V O2 (2.85–4.88 Å), A14V N3 and C10I H41 (2.57–3.43 Å), A14V O2′ and C10I N3 (3.06–4.56 Å), C7I H41 and G14I O6 (1.70–2.74 Å), C7I N3 and G14I H1 (2.85–4.52 Å in 19 structures). (D) Schematic representation of the kissing-loop interaction between SLIds2 and SLV. The arrows indicate intermolecular NOEs observed between residues at the kissing-loop junction (Table S2 of the Supporting Information). Residues shaded in orange form Watson–Crick base pairs at the kissing-loop junction, those shaded in green and purple are involved in base triples, and shaded gray boxes illustrate base stacking. For clarity, U13V was drawn in a lighter green color.
The kissing-loop junction is stabilized by additional hydrogen-bonding interactions, which contribute to formation of two base triples, one at each end of the kissing-loop helix (Figure 7). The C10I–A14V–U8V base triple is observed in all of the 20 lowest-energy structures and involves a two-hydrogen-bond base pair between the W–C edge of C10I and the sugar edge of A14V and a one-hydrogen-bond pair between the W–C edges of U8V and A14V (Figure 7B). The C7I–G14I–G15I base triple is defined by three hydrogen bonds observed simultaneously in the average structure (Figure 7C) and in 11 of the 20 lowest-energy structures: C7I NH2 to G14I O6, C7I N3 to G14I H1 and G14I 2′–OH to G15I N7.
Stacking interactions also play an important role in stabilizing the kissing-loop interactions, since both C10I and A14V stack on the G11I–C12V base pair at one end of the kissing-loop helix, while at the other end both G14I and G15I can potentially stack on the C13I–G10V base pair (Figure 7D). Furthermore, the noncanonical A14V–U8V base pair is stacked on the W–C G7V–C15V base pair of stem V, and the noncanonical G14I–C7I base pair is stacked on the W–C G6I–C16I base pair of stem I. Thus, stacking interactions at both ends of the kissing-loop helix propagate in both directions toward the stems of SLV and SLI to create a continuously stacked structure throughout the complex.
Discussion
Structural Characteristics of the Kissing-Loop Interaction
NMR studies of different SLI/SLV complexes performed in the presence of Mg2+ ions provide direct evidence that the SLI/SLV interaction of the VS ribozyme involves three consecutive W–C base pairs at the kissing-loop junction. In addition, the NMR structure of an optimal SLI/SLV complex formed with SLIds2 and SLV clearly demonstrates that the kissing-loop junction involves two U-turn structures, one in each loop, that enable formation of intermolecular W–C base pairs. In agreement with these NMR data, site-directed mutagenesis studies previously revealed that residues in the SLI and SLV loops must both fit the UNR consensus of U-turn motifs and accommodate W–C base pairing at the I/V kissing-loop junction.20,21,27 Chemical modifications experiments are also in agreement with the observed U-turn structures in both SLI and SLV, in particular the two hydrogen bonds commonly found in U-turn structures: U 2′–OH to R N7 and U H3 to R 3′-phosphate.22,27,63−66
In addition to the three W–C base pairs, other interactions contribute to the stability of the I/V kissing-loop interaction. The NMR structure of the SLIds2/SLV complex contains two previously uncharacterized base triples that provide additional hydrogen bonding and stacking interactions at the kissing-loop junction. The C10I–A14V–U8V base triple (C629–A701–U695 in the VS ribozyme) is particularly well-defined by the NMR data and helps rationalize the preference for a cytidine at the N position of the SLI U-turn.21 Although several studies have highlighted the importance of C62920,27−29 in the SLI/SLV interaction, its role had not been clearly established. As shown from the NMR structure and previous models of the SLI/SLV complex,25,35 the U-turn fold in SLI brings C629 to stack with G630 in proximity of A701.25 A cytidine is likely optimal at this position because its small size prevents steric hindrance and its unique W–C edge allows hydrogen bonding with the sugar edge of A701 in SLV. Both deletion of C629 and 2′-deoxy modification at A701 significantly reduce the VS ribozyme self-cleavage and ligation activities,27,63,65 in agreement with the importance of these residues at the I/V kissing-loop interface. In contrast, the C7I–G14I–G15I base triple (C626–G633–C634 in the VS ribozyme) is less well-defined by the NMR data, and the identity of the residues involved is not strictly required for cleavage and ligation activities.20,21,28
Given the Mg2+-dependence for formation of the SLI/SLV complex,21,23 it is likely that Mg2+ ions specifically associate with the kissing-loop junction. Previous phosphorothioate interference and manganese rescue experiments indicate that the 5′-phosphates of U631 (SLI) and C699 (SLV) each coordinate a metal ion.22 In addition, four Mg2+-ion binding sites were previously identified within the free SLV loop,34 and these may also be present in the SLI/SLV complex. In particular, the Mg2+ ion that associates with the 5′-phosphate of C699 in the free SLV is likely present in the complex. By analogy, a Mg2+ ion is likely associated with the 5′-phosphate of U631, which occupies a similar position in SLI as the 5′-phosphate of C699 in SLV. Additionally, Mg2+ ion(s) may help bridge phosphates from SLI and SLV that come in proximity at the kissing-loop junction, namely those of the two Us from the UNR motifs of SLI and SLV. Future studies are needed to precisely locate Mg2+-ion binding sites at the I/V kissing-loop junction.
RNA loops containing U-turn structures often include an extruded nucleotide,4,7,9,11 but the role of this residue is not always well understood. In the SLIds2/SLV complex, both the SLI and SLV loops contain an extruded nucleotide. For SLV, the extruded U13V (U700 in the VS ribozyme) can be substituted by any other standard base, but its phosphate backbone is important for cleavage activity and SLI binding.25 U700 may play a dual role in the kissing-loop by providing a flexible linker to SLV as well as a Mg2+-binding site, as previously proposed from kinetic studies of U700 variants.25 For SLIds2, the extruded G15I (C634 in the VS ribozyme) can be substituted by any other standard base,20,28 and its location is not precisely defined by the NMR data. Thus, C634 may be intrinsically dynamic in the VS ribozyme and thereby provide the necessary loop flexibility to optimize interactions at the kissing-loop interface. As discussed below, C634 likely plays a pivotal role in transducing part of the energy provided by formation of the kissing-loop interaction into the energy required for substrate activation.
Although the SLIds2/SLV complex forms a continuously stacked structure that appears essentially linear, the stems of SLI and SLV are not coaxially stacked within the complex. Rather, the kissing-loop junction causes a helical displacement of 10–24 Å between stem I and stem V associated with an interhelical angle of 127–175°. The arrays of helical displacements and interhelical angles observed in the 20 lowest-energy structures may reflect to some extent the relative range of motion of these stems. First of all, several residues in SLI (C7I, A8I, U9I, C13I, G14I, and G15I) and SLV (U8V and U9V) give 1H and 13C NMR signals of low relative intensity (data not shown), suggesting local dynamics at the kissing-loop junction. Moreover, it appears from the ensemble of structures that the observed range of interhelical angles may largely result from less-well-defined local structure for residues C7I–U9I and C13I–G15I in SLI. Interestingly, we previously kinetically characterized SLI/ribozyme systems with helix-length variations in SLI and SLV and built SLI/SLV models compatible with the kinetic data that display helical displacements of 18–21 Å and interhelical angles of 165–171°.35 The helical parameters of these models fall within the range detected in the ensemble of NMR structures,35 suggesting that within the spectrum of interhelical parameters observed for the SLIds2/SLV complex, some are compatible with an active VS ribozyme architecture. Similarly, the interhelical parameters previously reported for the SAXS model of the full VS ribozyme are more or less compatible with that of the SLIds2/SLV complex (helical displacement of ∼25 Å and interhelical angle of ∼154°).35,36
Conformational Changes in SLI and SLV upon Formation of the Kissing-Loop Interaction
NMR studies of different SLI/SLV complexes also provide direct structural evidence that the SLI/SLV interaction involves a helix shift in stem Ib, as previously shown from biochemical experiments.28,29 As expected, this helix shift is observed only with shiftable SLI variants but neither with preshifted variants nor with double-stranded variants. NMR studies of SLI/SLV complexes involving these SLI variants together with available NMR structures of the free SLI30 and SLV33,34 allow us to examine structural changes that occur in SLI and SLV as a result of complex formation. Given that the formation of the SLI/SLV complex and the associated helix shift in SLI observed in the VS ribozyme can be reproduced with isolated hairpins,29 it is likely that the structural changes observed with isolated stem-loops hold in the intact ribozyme. The SLV loop adopts a compact U-turn structure in its free form, and only a minor structural change is observed in the SLI/SLV complex.35 In contrast, SLI loop is mostly disordered in its free form,30 and this disordered state may help facilitate recognition by the compact SLV loop and trigger a structural transition in SLI upon interaction with SLV. Upon SLV binding, the SLI loop undergoes a disorder-to-order conformational transition that impacts on the loop closing base pairs of stem Ib. In particular, the NMR data indicate that the stem Ib of shiftable substrates contains four stable W–C base pairs in their free form but only three stable W–C base pairs in their SLV-bound forms. A fourth base pair closing the loop was predicted to form in the SLV-bound form according to biochemical studies,28,29 but the imino NMR data indicate that such a base pair is not stably formed in the complex. For SLIsbΔIa, for example, the C6I–G13I closing base pair (C626–G633 in the VS ribozyme) is destabilized upon complex formation such that the G13I imino proton is no longer observable in the SLIsbΔIa/SLV complex.
Interestingly, destabilization of the SLI loop closing base pair is also observed upon formation of SLI/SLV complexes with preshifted substrates and the double-stranded substrate SL1ds2. In these cases, the imino proton of the closing base pair (G14I for SLIpsΔIa and SLIps and G15I for SLIds2) is observed in the free SLI but not in the SLI/SLV complex. These NMR data obtained with shiftable and preshifted SLI substrates indicate that destabilization of the loop-closing G–C base pair is not a consequence of helix shifting in stem Ib but instead results from formation of the kissing-loop interaction.
Detailed NMR studies of SLIds2 demonstrate that the C7I–G15I loop-closing base pair is present in the free SLI but that G15I is preferentially extruded from the loop fold in the complex, with C7I forming an unstable and distorted W–C base pair with G14I (C626–G633 base pair in the VS ribozyme). Chemical modification data indicate that C626 remains mostly inaccessible to DMS modification upon binding of SLI to the VS ribozyme,28,29 whereas mutational analysis indicate that disruption of the C626–G633 base pair is compatible with self-cleavage and ligation activities of the VS ribozyme.20,21,28 Although these previous results appear contradictory, they both agree with the structure of the SLIds2/SLV complex, where the C7I–G14I base pair corresponding to C626–G633 is destabilized, but could still remain inaccessible to chemical modifications. Interestingly, the extrusion of G15I is similar to the one occurring upon formation of the I/V kissing-loop complex with shiftable SLI substrates, where C634 (equivalent to G15I) becomes extruded and accessible to DMS modification.28,29 Thus, the observation of a similar conformational change for residue G15I in SLIds2, which is not a shiftable SLI variant, strongly supports the concept that formation of the kissing-loop interaction is generally responsible for structural rearrangements involving G633 and G634 within stem Ib.
Kinetic Model for Substrate Recognition and Activation by the VS Ribozyme
Overall, the NMR data presented here are in agreement with previous biochemical studies, further supporting the value of using stem-loop domains to improve our understanding of substrate activation and recognition within the context of the full ribozyme. In-depth analysis of these NMR data reveals interesting conformational changes that are consistent with a kinetic model in which formation of an initial contact between the loops of SLI and SLV in the natural VS ribozyme destabilizes the SLI loop closing base pairs and triggers the helix shift in SLI. In this model, residues G697, A698, and C699 of the prefolded U-turn of SLV initially target residues C632, U631, and G630 of the unshifted SLI and thereby induce formation of the U-turn fold in the SLI loop. Upon formation of the U-turn fold in SLI, other hydrogen-bonding and stacking interactions form at the I/V kissing-loop junction to further stabilize the complex and thereby induce the rearrangement of the C626–G633 base pair and the extrusion of C634 within stem Ib. These structural changes likely lead to helix shifting in stem Ib and structural rearrangement in the cleavage site internal loop. Although this model will need further experimental validation, it is compatible with the ability of U-turn structures to initiate rapid RNA/RNA pairing and trigger structural changes in RNA complexes.14,15
Acknowledgments
We thank Dominique Chaussé for sample preparation, Nicolas Girard for structural analysis, and Ryan Richter for computer support. We also thank Alexandre Desjardins, Eric Bonneau, and James G. Omichinski for critical reading of the manuscript.
Glossary
ABBREVIATIONS
- 2D
two-dimensional
- 3D
three-dimensional
- bp
base pair
- HSQC
heteronuclear single quantum coherence
- NOE
nuclear Overhauser effect
- NOESY
NOE spectroscopy
- PDB
protein data bank
- SLI
stem-loop I
- SLV
stem-loop V
- RMSD
root mean square deviation
- SAXS
small-angle X-ray scattering
- W–C
Watson–Crick
Supporting Information Available
Supporting Figures: native gel electrophoresis of several RNAs investigated in this study (S1); NMR evidence for formation of the SLIsbΔIa/SLV complex (S2); of the SLIsbΔIaGU/SLV complex (S3), of the SLIps/SLV complex (S4), of the SLIpsΔIa/SLV complex (S5), and of the SLIds1/SLV complex (S6); interhelical angle distribution for the 20 lowest-energy structures of the SLIds2/SLV complex (S7); superposition of the loop structures of SLV free and bound to SLIds2 (S8). Supporting Tables: chemical shifts of detectable G/U imino protons for SLI variants either in their free form or bound to SLV RNA (S1); chemical shift of detectable G/U imino protons for SLV either in its free form or bound to SLI variants (S2); chemical shift assignments of exchangeable 1H and 15N for the SLIds2/SLV complex (S3); chemical shift assignments of nonexchangeable 1H and 13C for the SLIds2/SLV complex (S4); summary of intermolecular NOEs at the kissing-loop junction of the SLIds2/SLV complex (S5). This material is available free of charge via the Internet at http://pubs.acs.org.
Accession Codes
The NMR chemical shifts, structural restraints and structural coordinates of the SLIds2/SLV complex have been deposited through the RCSB Protein Data Bank and given BMRB entry code 19662 and PDB entry code 2MI0.
This work was supported by the Canadian Institutes for Health Research (CIHR) to P.L. [MOP-86502]. P.B. was supported by a Frederick Banting and Charles Best Canada Graduate Scholarship Ph.D. scholarship from CIHR and graduate scholarships from the Université de Montréal. P.L. holds a Canada Research Chair in Structural Biology and Engineering of RNA. Funding for open access charge: CIHR.
The authors declare no competing financial interest.
Supplementary Material
References
- Quigley G. J.; Rich A. (1976) Structural domains of transfer RNA molecules. Science 194, 796–806. [DOI] [PubMed] [Google Scholar]
- Pley H. W.; Flaherty K. M.; McKay D. B. (1994) Three-dimensional structure of a hammerhead ribozyme. Nature 372, 68–74. [DOI] [PubMed] [Google Scholar]
- Jucker F. M.; Pardi A. (1995) GNRA tetraloops make a U-turn. RNA 1, 219–222. [PMC free article] [PubMed] [Google Scholar]
- Huang S.; Wang Y.-X.; Draper D. E. (1996) Structure of a hexanucleotide RNA hairpin loop conserved in ribosomal RNAs. J. Mol. Biol. 258, 308–321. [DOI] [PubMed] [Google Scholar]
- Stallings S. C.; Moore P. B. (1997) The structure of an essential splicing element: stem loop IIa from yeast U2 snRNA. Structure 5, 1173–1185. [DOI] [PubMed] [Google Scholar]
- Gutell R. R.; Cannone J. J.; Konings D.; Gautheret D. (2000) Predicting U-turns in ribosomal RNA with comparative sequence analysis. J. Mol. Biol. 300, 791–803. [DOI] [PubMed] [Google Scholar]
- Huppler A.; Niksstad L. J.; Allman A. M.; Brow D. A.; Butcher S. E. (2002) Metal binding and base ionization in the U6 RNA intramolecular stem-loop structure. Nat. Struct. Biol. 9, 431–435. [DOI] [PubMed] [Google Scholar]
- Puglisi E. V.; Puglisi J. D. (1998) HIV-1 A-rich RNA loop mimics the tRNA anticodon structure. Nat. Struct. Biol. 5, 1033–1036. [DOI] [PubMed] [Google Scholar]
- Klosterman P. S.; Hendrix D. K.; Tamura M.; Holbrook S. R.; Brenner S. E. (2004) Three-dimensional motifs from the SCOR, structural classification of RNA database: extruded strands, base triples, tetraloops and U-turns. Nucleic Acids Res. 32, 2342–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sefcikova J.; Krasovska M. V.; Sponer J.; Walter N. G. (2007) The genomic HDV ribozyme utilizes a previously unnoticed U-turn motif to accomplish fast site-specific catalysis. Nucleic Acids Res. 35, 1933–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S.; Schroeder S. J. (2010) Nuclear magnetic resonance structure of the prohead RNA E-loop hairpin. Biochemistry 49, 5989–5997. [DOI] [PubMed] [Google Scholar]
- Dix D. B.; Wittenberg W. L.; Uhlenbeck O. C.; Thompson R. C. (1986) Effect of replacing uridine 33 in yeast tRNAPhe on the reaction with ribosomes. J. Biol. Chem. 261, 10112–10118. [PubMed] [Google Scholar]
- Ashraf S. S.; Ansari G.; Guenther R.; Sochacka E.; Malkiewicz A.; Agris P. F. (1999) The uridine in ″U-turn″: contributions to tRNA-ribosomal binding. RNA 5, 503–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franch T.; Petersen M.; Wagner E. G. H.; Jacobsen J. P.; Gerdes K. (1999) Antisense RNA regulation in prokaryotes: Rapid RNA/RNA interaction facilitated by a general U-turn loop structure. J. Mol. Biol. 294, 1115–1125. [DOI] [PubMed] [Google Scholar]
- Ogle J. M.; Brodersen D. E.; Clemons W. M. Jr.; Tarry M. J.; Carter A. P.; Ramakrishnan V. (2001) Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292, 897–902. [DOI] [PubMed] [Google Scholar]
- Collins R. A. (2002) The Neurospora Varkud satellite ribozyme. Biochem. Soc. Trans. 30, 1122–1126. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. (2004) The Varkud satellite ribozyme. RNA 10, 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T. J.; Lilley D. M. (2011) Do the hairpin and VS ribozymes share a common catalytic mechanism based on general acid-base catalysis? A critical assessment of available experimental data. RNA 17, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H. C. T.; Collins R. A. (1995) Efficient trans-cleavage of a stem-loop RNA substrate by a ribozyme derived from Neurospora VS RNA. EMBO J. 14, 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie T. L.; Olive J. E.; Collins R. A. (1995) A secondary-structure model for the self-cleaving region of Neurospora VS RNA. Proc. Natl. Acad. Sci. U. S. A. 92, 4686–4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi T.; Beattie T. L.; Olive J. E.; Collins R. A. (1996) A long-range pseudoknot is required for activity of the Neurospora VS ribozyme. EMBO J. 15, 2820–2825. [PMC free article] [PubMed] [Google Scholar]
- Sood V. D.; Beattie T. L.; Collins R. A. (1998) Identification of phosphate groups involved in metal binding and tertiary interactions in the core of the Neurospora VS ribozyme. J. Mol. Biol. 282, 741–750. [DOI] [PubMed] [Google Scholar]
- Hiley S. L.; Collins R. A. (2001) Rapid formation of a solvent-inaccessible core in the Neurospora Varkud satellite ribozyme. EMBO J. 20, 5461–5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamel R.; Collins R. A. (2002) Rearrangement of substrate secondary structure facilitates binding to the Neurospora VS ribozyme. J. Mol. Biol. 324, 903–915. [DOI] [PubMed] [Google Scholar]
- Bouchard P.; Lacroix-Labonté J.; Desjardins G.; Lampron P.; Lisi V.; Lemieux S.; Major F.; Legault P. (2008) Role of SLV in SLI substrate recognition by the Neurospora VS ribozyme. RNA 14, 736–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAbreu D. M.; Olive J. E.; Collins R. A. (2011) Additional roles of a peripheral loop-loop interaction in the Neurospora VS ribozyme. Nucleic Acids Res. 39, 6223–6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie T. L.; Collins R. A. (1997) Identification of functional domains in the self-cleaving Neurospora VS ribozyme using damage selection. J. Mol. Biol. 267, 830–840. [DOI] [PubMed] [Google Scholar]
- Andersen A.; Collins R. A. (2000) Rearrangement of a stable RNA secondary structure during VS ribozyme catalysis. Mol. Cell 5, 469–478. [DOI] [PubMed] [Google Scholar]
- Andersen A. A.; Collins R. A. (2001) Intramolecular secondary structure rearrangement by the kissing interaction of the Neurospora VS ribozyme. Proc. Natl. Acad. Sci. U. S. A. 98, 7730–7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flinders J.; Dieckmann T. (2001) A pH controlled conformational switch in the cleavage site of the VS ribozyme substrate RNA. J. Mol. Biol. 308, 665–679. [DOI] [PubMed] [Google Scholar]
- Michiels P. J. A.; Schouten C. H. J.; Hilbers C. W.; Heus H. A. (2000) Structure of the ribozyme substrate hairpin of Neurospora VS RNA: A close look at the cleavage site. RNA 6, 1821–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann B.; Mitchell G. T.; Gendron P.; Major F.; Andersen A. A.; Collins R. A.; Legault P. (2003) NMR structure of the active conformation of the Varkud satellite ribozyme cleavage site. Proc. Natl. Acad. Sci. U. S. A. 100, 7003–7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell D. O.; Legault P. (2005) NMR structure of the Varkud satellite ribozyme stem-loop V RNA and magnesium-ion binding from chemical-shift mapping. Biochemistry 44, 4157–4170. [DOI] [PubMed] [Google Scholar]
- Campbell D. O.; Bouchard P.; Desjardins G.; Legault P. (2006) NMR structure of Varkud satellite ribozyme stem-loop V in the presence of magnesium ions and localization of metal-binding sites. Biochemistry 45, 10591–10605. [DOI] [PubMed] [Google Scholar]
- Lacroix-Labonte J.; Girard N.; Lemieux S.; Legault P. (2012) Helix-length compensation studies reveal the adaptability of the VS ribozyme architecture. Nucleic Acids Res. 40, 2284–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipfert J.; Ouellet J.; Norman D. G.; Doniach S.; Lilley D. M. (2008) The complete VS ribozyme in solution studied by small-angle X-ray scattering. Structure 16, 1357–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotto M.; Saudek V.; Skleñár V. (1992) Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 2, 661–665. [DOI] [PubMed] [Google Scholar]
- Kay L. E.; Keifer P.; Saarinen T. (1992) Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J. Am. Chem. Soc. 114, 10663–10665. [Google Scholar]
- Dingley A. J.; Grzesiek S. (1998) Direct observation of hydrogen bonds in nucleic acid base pairs by internucleotide 2JNN couplings. J. Am. Chem. Soc. 120, 8293–8297. [Google Scholar]
- Grzesiek S.; Bax A. (1993) The importance of not saturating water in protein NMR. Application to sensitivity enhancement of NOE measurements. J. Am. Chem. Soc. 115, 12593–12594. [Google Scholar]
- Simorre J. P.; Zimmermann G. R.; Mueller L.; Pardi A. (1996) Correlation of the guanosine exchangeable and non-exchangeable base protons in 13C/15N-labeled RNA with an HNC-TOCSY-CH experiment. J. Biomol. NMR 7, 153–156. [DOI] [PubMed] [Google Scholar]
- Mueller L.; Legault P.; Pardi A. (1995) Improved RNA structure determination by detection of NOE contacts to exchange-broadened amino groups. J. Am. Chem. Soc. 117, 11043–11048. [Google Scholar]
- Zhang O.; Kay L. E.; Olivier J. P.; Forman-Kay J. D. (1994) Backbone 1H and 15N resonance assignments of the N-terminal SH3 domain of drk in folded and unfolded states using enhanced-sensitivity pulsed field gradient NMR techniques. J. Biomol. NMR 4, 845–858. [DOI] [PubMed] [Google Scholar]
- Pascal S. M.; Muhandiram D. R.; Yamazaki T.; Forman-Kay J. D.; Kay L. E. (1994) Simultaneous acquisition of 15N and 13C-edited NOE spectra of proteins dissolved in H2O. J. Magn. Reson., Series B 103, 197–201. [Google Scholar]
- Vuister G. W.; Bax A. (1992) Resolution enhancement and spectral editing of uniformly 13C-enriched proteins by homonuclear broadband 13C decoupling. J. Magn. Reson. 98, 428–435. [Google Scholar]
- Santoro J.; King G. C. (1992) A constant-time 2D overbodenhausen experiment for inverse correlation of isotopically enriched species. J. Magn. Reson. 97, 202–207. [Google Scholar]
- Pardi A.; Nikonowicz E. P. (1992) Simple procedure for resonance assignment of the sugar protons in 13C-labeled RNAs. J. Am. Chem. Soc. 114, 9202–9203. [Google Scholar]
- Ikura M.; Kay L. E.; Tschudin R.; Bax A. (1990) Three-dimensional NOESY-HMQC spectroscopy of a 13C-labeled protein. J. Magn. Reson. 86, 204–209. [Google Scholar]
- Kay L. E.; Bax A. (1990) New methods for the measurement of NH---CαH coupling constants in 15N-labeled proteins. J. Magn. Reson. 86, 110–126. [Google Scholar]
- Delaglio F.; Grzesiek S.; Vuister G. W.; Zhu G.; Pfeifer J.; Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Vranken W. F.; Boucher W.; Stevens T. J.; Fogh R. H.; Pajon A.; Llinas M.; Ulrich E. L.; Markley J. L.; Ionides J.; Laue E. D. (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59, 687–696. [DOI] [PubMed] [Google Scholar]
- Hansen M. R.; Mueller L.; Pardi A. (1998) Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat. Struct. Biol. 5, 1064–1074. [DOI] [PubMed] [Google Scholar]
- Desjardins G.; Bonneau E.; Girard N.; Boisbouvier J.; Legault P. (2011) NMR structure of the A730 loop of the Neurospora VS ribozyme: insights into the formation of the active site. Nucleic Acids Res. 39, 4427–4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi T.; Collins R. A. (1998) Smaller, faster ribozymes reveal the catalytic core of Neurospora VS RNA. J. Mol. Biol. 277, 215–224. [DOI] [PubMed] [Google Scholar]
- McLeod A. C.; Lilley D. M. (2004) Efficient, pH-dependent RNA ligation by the VS ribozyme in trans. Biochemistry 43, 1118–1125. [DOI] [PubMed] [Google Scholar]
- Poon A. H.; Olive J. E.; McLaren M.; Collins R. A. (2006) Identification of separate structural features that affect rate and cation concentration dependence of self-cleavage by the Neurospora VS ribozyme. Biochemistry 45, 13394–13400. [DOI] [PubMed] [Google Scholar]
- Theimer C. A.; Jady B. E.; Chim N.; Richard P.; Breece K. E.; Kiss T.; Feigon J. (2007) Structural and functional characterization of human telomerase RNA processing and cajal body localization signals. Mol. Cell 27, 869–881. [DOI] [PubMed] [Google Scholar]
- Cabello-Villegas J.; Tworowska I.; Nikonowicz E. P. (2004) Metal ion stabilization of the U-turn of the A37 N6-dimethylallyl-modified anticodon stem-loop of Escherichia coli tRNAPhe. Biochemistry 43, 55–66. [DOI] [PubMed] [Google Scholar]
- Cabello-Villegas J.; Winkler M. E.; Nikonowicz E. P. (2002) Solution conformations of unmodified and A37N6-dimethylallyl modified anticodon stem-loops of Escherichia coli tRNAPhe. J. Mol. Biol. 319, 1015–1034. [DOI] [PubMed] [Google Scholar]
- Tewari A. K.; Dubey R. (2008) Emerging trends in molecular recognition: utility of weak aromatic interactions. Bioorgan. Med. Chem. 16, 126–143. [DOI] [PubMed] [Google Scholar]
- Steiner T.; Koellner G. (2001) Hydrogen bonds with pi-acceptors in proteins: frequencies and role in stabilizing local 3D structures. J. Mol. Biol. 305, 535–557. [DOI] [PubMed] [Google Scholar]
- Meyer E. A.; Castellano R. K.; Diederich F. (2003) Interactions with aromatic rings in chemical and biological recognition. Angew. Chem., Int. Ed. Engl. 42, 1210–1250. [DOI] [PubMed] [Google Scholar]
- Sood V. D.; Yekta S.; Collins R. A. (2002) The contribution of 2′-hydroxyls to the cleavage activity of the Neurospora VS ribozyme. Nucleic Acids Res. 30, 1132–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzokov S. B.; Murray I. A.; Grasby J. A. (2002) The role of magnesium ions and 2′-hydroxyl groups in the VS ribozyme-substrate interaction. J. Mol. Biol. 324, 215–226. [DOI] [PubMed] [Google Scholar]
- Jones F. D.; Strobel S. A. (2003) Ionization of a critical adenosine residue in the Neurospora Varkud satellite ribozyme active site. Biochemistry 42, 4265–4276. [DOI] [PubMed] [Google Scholar]
- Kovacheva Y. S.; Tzokov S. B.; Murray I. A.; Grasby J. A. (2004) The role of phosphate groups in the VS ribozyme-substrate interaction. Nucleic Acids Res. 32, 6240–6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.