Abstract
Autophagy is an intracellular homeostatic mechanism important for the degradation of waste components from the cytoplasm in acidic lysosomal compartments. Originally, surplus parts of the cytoplasm that acted as targets for autophagy were thought to comprise cellular organelles and proteins, but this has now extended to include a range of pathogens with particular emphasis on intracellular bacteria. The finding that autophagy can sequester intracellular bacteria and mediate their destruction has opened the door to a wider role for autophagy as an effector arm of the immune system. In innate immunity, autophagy works downstream of pattern recognition receptors where it facilitates a number of effector responses, including cytokine production and phagocytosis. Autophagy is also able to intersect pathways of innate and adaptive immunity through its potential to deliver antigens for antigen presentation. Autophagy provides a substantial source of antigens for loading onto MHC class II molecules and it may be important in dendritic cells for cross-priming to CD8+ T cells. In lymphocytes, autophagy is essential for cell survival and homeostasis, particularly in T cells. In the thymus, autophagy can modulate the selection of certain CD4+ T-cell clones while in the bone marrow autophagy is needed for B-cell development at specific stages. However, large holes exist in our knowledge as to how autophagy regulates, and is regulated by, the immune system and it is important to now apply what we have gleaned from in vitro studies to how autophagy operates in vivo in the setting of natural infection.
Keywords: adaptive immunity, antigen presentation, autophagy, innate immunity, intracellular bacteria
Introduction
Eukaryotic cells have evolved two effective mechanisms for the breakdown of intracellular constituents: proteasomes and autophagy. Although multiple autophagic pathways exist in mammalian cells (microautophagy and chaperone-mediated autophagy) each specializing in their mode of degradation, only in the case of macroautophagy (herein referred to as ‘autophagy’) has a well-defined role in immunity been described. Unlike proteasomes – which degrade ubiquitin-tagged, small, short-lived proteins – autophagy can remove large protein aggregates and entire organelles through their delivery to the lysosome.1 Controlled by a number of autophagy-related (Atg) genes, autophagy engulfs its target cargo into a double-membraned vesicle, termed an autophagosome, which subsequently fuses with the lysosome to form a single-membraned autolysosome. Here, the luminal contents are degraded and recycled back to the cytoplasm (Fig. 1).
Figure 1.
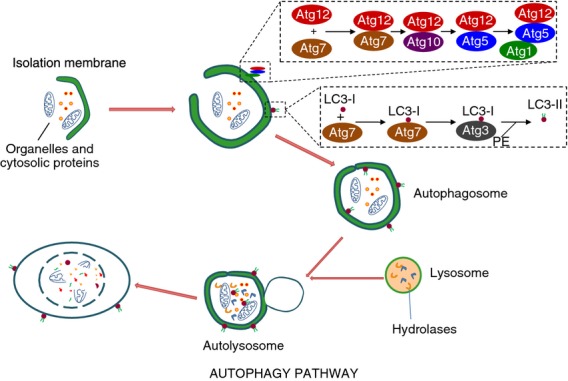
Schematic diagram of the autophagy pathway. During autophagy, cytoplasmic constituents are enclosed in an isolation membrane that is elongated mainly through the action of two ubiquitin-like conjugation systems into a double-membraned autophagosome. Autophagosomes fuse with lysosomes to form autolysosomes where breakdown of the vesicle contents takes place along with the autophagosome inner membrane.
During autophagy initiation, activation of UNC-51-like kinase 1/2 (called Atg1 in yeast) in cooperation with the type III phosphatidylinositol 3-kinase hVPS34, Beclin 1 and Atg14 leads to the formation of an isolation membrane at the site of the endoplasmic reticulum.2,3 Phosphatidylinositol 3-phosphate (PI3P) produced by hVPS34 recruits the PI3P-binding effectors WIPI-1 and WIPI-2 that serve to further promote the formation of the isolation membrane.1 This nascent autophagosome is subsequently elongated following the action of microtubule-associated light chain 3 (LC3), a product of two protein conjugation systems and the prototypic marker of autophagosomes. In one, the covalent conjugate Atg5–Atg12 complexes to Atg16L1 to form an E3-like enzyme that aids the conjugation of LC3 to phosphotidylethanolamine (PE) to make LC3-PE (also referred to as LC3-II). The Atg5–Atg12–Atg16L1 complex also facilitates the localization of the second conjugation system to the isolation membrane. Here, LC3 is first activated by Atg7 and then coupled to PE by Atg3. LC3-II can then insert into the isolation membrane and facilitate its elongation into a closed, double-membraned autophagosome. In the final step, autophagosomes fuse with late endosomal or lysosomal organelles to form the terminal digestive compartment, the autolysosome.1,4
Functionally, autophagy was first described as a cellular response in rats to numerous stressors such as potassium deficiency and starvation.5 However, there is now good reason to believe that autophagy forms the most ancient of immune defences. In the early evolution of eukaryotic cells its action was probably pivotal in protecting the cytosol from invading foreign organisms. An evolutionary hangover of this primitive immune function can perhaps be observed in the effective removal of mitochondria by autophagy. Mitochondria are organelles that have evolved from a Rickettsia-like α-protobacterium that at one time were foreign to the cytoplasm of eukaryotic cells.1 With the onset of complex life and intricate vertebrate immune systems, autophagy has evolved a multifaceted role becoming a bona fide regulator and effector of the immune system.
Autophagy in innate immunity
Autophagy and pattern recognition receptors
Pattern recognition receptors (PRRs) comprise a group of innate receptors responsible for detection of invading pathogens through the recognition of motifs specific to the foreign microbe. These motifs are collectively termed pathogen-associated molecular patterns (PAMPs) and stimulate a myriad of effector responses when bound to their cognate PRR.6 Toll-like receptors (TLRs) are a group of PRRs strongly connected to the autophagy pathway. TLR4 is able to induce autophagy in murine macrophages following stimulation with lipopolysaccharide (LPS).7 Similarly, LPS stimulation of TLR4 was shown to increase the clearance of Mycobacterium tuberculosis by autophagy.8 Induction of autophagy by TLR9 following stimulation with CpG-rich DNA has also been reported.9,10 Autophagy not only acts downstream of TLR signalling, but can also play a role in facilitating recognition of PAMPS by TLRs. Lee et al. showed that autophagy was able to deliver viral ligands to TLR7 in plasmacytoid dendritic cells (pDCs) following vesicular stomatitis virus and Sendai virus infection resulting in type I interferon production.11
The NOD-like receptors (NLRs) are a group of innate receptors responsible for intracellular bacteria sensing and represent a second class of PRRs with links to autophagy. Activation of the NLRs nucleotide-binding oligomerization domain 1 (NOD1) and NOD2 in mice leads to recruitment of Atg16L1 at the plasma membrane to the site entry of invading Shigella flexneri and Listeria monocytogenes, resulting in their sequestration in autophagosomes and subsequent destruction.12 In humans, the treatment of DCs with the NOD2 ligand muramyldipeptide (MDP) has been shown to induce autophagy and this effect was lost in MDP-treated DCs from patients with Crohn's disease with a polymorphism in NOD2.13 The inability of DCs to induce NOD2-mediated autophagy led to increased bacterial burden with Salmonella enterica serovar Typhimurium. Similarly, autophagy is induced in murine peritoneal macrophages after NOD2 stimulation following infection with adherent-invasive Escherichia coli. As in human DCs, absence of NOD2 signalling resulted in increased bacterial burden, attributed to poor autophagy induction.14 However, a recent study by Benjamin et al. contradicts the importance of NOD2 for autophagy induction in response to S. Typhimurium. Intestinal epithelial cell autophagy was shown to be essential for intestinal defence against invasive bacteria such as S. Typhimurium and Enterococcus faecalis and this was dependent on MyD88 signalling, not NOD2.15 In contrast to NOD1 and NOD2, the NLR NLRP4 can inhibit autophagy through its ability to bind and inhibit the action of Beclin 1. In the presence of group A streptococcus, NLRP4 is recruited to bacteria-containing phagosomes where it transiently dissociates with Beclin 1, perhaps providing a further NLR-mediated mechanism for autophagy induction in the presence of bacterial infection.16
Autophagy can also act downstream of virus-sensing pathways mediated by retinoic acid inducible gene-like receptors (RLRs). Tormo et al. showed that treatment with the double-stranded RNA mimic polyinosine-polycytidylic acid, acting through the RLR MDA-5, is able to induce autophagy in melanoma cells resulting in autophagy-dependent cell death.17 The relationship of RLRs with autophagy is notable as multiple reports document the negative regulation of RLRs by autophagy. For example, Atg5-deficient murine macrophages have increased RLR signalling mediated by enhanced reactive oxygen species production owing to an increase in mitochondrial volume.18 In the same study, loss of Atg5 in mouse embryonic fibroblasts (MEFs) was associated with improved resistance to vesicular stomatitis virus infection. Hence, autophagy exhibits a complex regulatory role in the context of infection-sensing pathways via PRRs, acting in some cases to mediate the effective sensing and removal of intracellular bacteria, but can function in a negative regulatory capacity in the setting of viral recognition through RLRs.
Autophagy and bacteria handling
While autophagy was classically defined as an indiscriminate bulk-degradation pathway, specialized forms of autophagy have also evolved to promote the selective targeting of various cellular components such as mitochondria (mitophagy), peroxisomes (pexophagy) and intracellular bacteria (xenophagy) that have escaped the endocytic pathway to persist in the cytosol. The uptake of cytosolic bacteria by autophagy is mediated by a central adaptor protein called p62 (also known as SQSTM1), which recognizes polyubquitin tags on bacteria and links them to LC3-positive phagophores (growing autophagosomes) through its LC3-interaction region.1,19 In this way, the intracellular pathogen L. monocytogenes was found to be targeted to autophagosomes. When present in the cytosol, polyubiquitin-tagged L. monocytogenes is recognized by p62, which in turn binds LC3, coating the bacteria in polyubiquitin-p62-LC3, which permits the engulfment of L. monocytogenes by autophagosomes.19 The importance of autophagy in the removal of this specialized cytosolic pathogen is highlighted by the action of three Listeria-derived factors: listeriolysin, phospholipase C and actin polymerization protein A that all inhibit autophagy, facilitating prolonged survival of L. monocytogenes in the cytosol.19,20
Xenophagy can also mediate the removal of vacuolar bacteria. During bacille Calmette–Guérin (BCG) infection of the RAW cell line, autophagy facilitates the fusion of BCG-containing vacuoles with lysosomes to facilitate mycobacterial killing.20 Mycobacterium tuberculosis,21,22 Salmonella23 and Helicobacter pylori-containing phagosomes24 have all been shown to fuse or to be directly engulfed by autophagosomes offering an alternative method for the destruction of intracellular bacteria by autophagy. Recent reports have furthered this work by demonstrating a crucial role for autophagy in mycobacterial handling in vivo.25,26 When infected with M. tuberculosis, mice with a myeloid lineage-specific deletion of Atg5 exhibited enhanced mycobacterial burden compared with wild-type controls, alongside increased tissue necrosis and lung pathology.25 Watson et al. have identified the basis for this in vivo requirement: during M. tuberculosis infection of macrophages the bacterial ESX-1 secretion system initiates permeabilization of the phagosomal membrane that exposes bacterial DNA to components of the cytosolic DNA pathway, including STING. STING is responsible for the ubiquitin-tagging of bacteria that ultimately directs the bacteria to the autophagy machinery via the adaptors p62 and NDP52.26 Although most studies of bacterial-specific autophagy involve the capture of intracellular bacteria, autophagy can similarly eliminate extracellular bacteria. Yuan et al. demonstrated that Pseudomanas aeruginosa is able to induce autophagy in the alveolar macrophages cell line MH-S. Knockdown of beclin 1 with small interfering RNA or treatment with the autophagy inhibitor 3-MA resulted in a marked increase in bacterial load whereas treatment with the autophagy inducer rapamycin improved bacterial clearance.27
Recent studies have defined a function for autophagy in the formation of neutrophil extracellular traps (NETs), a process by which neutrophils capture and destroy invading bacteria. NETs are comprised of extracellular chromatin structures released by neutrophils during a form of specialized cell death distinct from apoptosis and necrosis called NETosis. PMA-induced NETosis was found to be dependent on autophagy and treatment of neutrophils with the autophagy inhibitor wortmannin was able to exert an inhibitory effect on NET formation.28 The importance of autophagy in NET formation was verified in subsequent studies that also showed that pharmacological manipulation of autophagy could induce or inhibit NET formation and mediate the delivery of factors important for NET function.29,30 Table 1 offers several further examples as to how autophagy interacts with invading bacteria, including the relationship with some viruses and parasites.
Table 1.
Select examples of pathogen interaction with the autophagy pathway
| Pathogen | Autophagic interaction | Reference |
|---|---|---|
| Bacteria | ||
| Mycobacterium tuberculosis | Bacteria-containing phagosomes fuse with autophagosomes | 21,22 |
| Salmonella typhimurium | Damaged phagosomes are targeted by autophagy | 23 |
| Helicobacter pylori | Autophagy targets bacteria-containing autophagosomes | 24 |
| Pseudomonas aeruginosa | Infection induces autophagy, which is required for controlling bacterial load | 27 |
| Escherichia coli | Adherent-invasive strain recruits autophagy machinery to the site of phagocytosis | 14 |
| Bacillus anthracis | Autophagy degrades anthrax lethal toxin | 58,59 |
| Listeria monocytogenes | Listeriolysin, phospholipase C, and actin polymerization protein A inhibit autophagy. Autophagy adaptors target cytosolic bacteria to autophagy | 19,20,60 |
| Shigella flexneri | P62 and NDP52 target bacteria to autophagy | 60 |
| Vibrio cholerae | Cholera toxin inhibits autophagy | 59 |
| Viruses | ||
| Sindbis virus | Autophagy degrades viral capsid subsequent to viral-induced autophagy | 61 |
| Vesicular stomatitis virus | Autophagy delivers viral ligands to TLR7 during infection of plasmacytoid dendritic cells | 11 |
| Human immunodeficiency virus | HIV can induce autophagy-dependent cell death in bystander T cells via gp41. The virus can also inhibit autophagy in dendritic cells by activating mammalian target of rapamycin | 62,63 |
| Herpes simplex virus 1 | HSV-1 protein ICP34.5 inhibits autophagy through interaction with Beclin 1 | 64 |
| Human cytomegalovirus | The hCMV protein TRS1 inhibits autophagy through its interaction with Beclin1 | 65 |
| Measles virus | Viral infection induces autophagy, which is important for controlling infection | 66 |
| Protozoa | ||
| Toxoplasma gondii | CD40-dependent activation of macrophages induces autophagy resulting in T. gondii killing | 67 |
Autophagy and cytokines
As previously discussed, autophagy is invoked during infection following the detection of PAMPs by PRRs. However, autophagy is also under the control of cytokines and immunologically relevant cell surface receptors. The T helper type 1 (Th1) cytokine interferon-γ is a potent inducer of autophagy while the Th2 cytokines interleukin-4 (IL-4) and IL-13 have been shown to have inhibitory effects. This observation suggests a role for autophagy as an effector arm of Th1-mediated immunity and may partly explain why Th1 cytokines afford protection against intracellular bacteria.1 Autophagy also has a role in the biogenesis and secretion of various pro-inflammatory cytokines. Atg16L1-deficient macrophages exhibit enhanced IL-1β and IL-18 secretion following stimulation with LPS.4 Human peripheral blood mononuclear cells with a Crohn's disease-associated Atg16L1 variant display a similar phenotype, with increased IL-1β secretion in response to MDP stimulation.31 Zhou et al. demonstrated that a block in autophagy results in the accumulation of reactive oxygen species and the release of DNA from damaged mitochondria. This was shown to drive activation of the NLRP3 inflammasome and ultimately in excessive caspase-1 activation resulting in IL-1β and IL-18 production.32 Interestingly, IL-1β can itself induce autophagy in macrophages, suggesting that IL-1β might regulate its own production through autophagy.33 Production of multiple other cytokines is enhanced in the absence of autophagy, increases in IL-1α, IL-12, IL-17 and CXCL1 were all observed in Atg5 fl/fl LysM-Cre+ mice compared with wild-type controls in response to M. tuberculosis infection.25 Tumour necrosis factor-α and IL-6 secretion are also regulated by autophagy; knockdown of Atg16L1 and the autophagy-related gene IRGM by small interfering RNA resulted in significantly increased production in human THP-1 macrophages following infection with adherent-invasive E. coli.14 Inhibition of autophagy with 3-MA or knockdown of beclin 1 and Atg7 promotes IL-1α, IL-1β and IL-23 secretion by macrophages and DCs and this in turn augments innate secretion of IL-17, IFN-γ and IL-22 by γδ T cells.34
Autophagy in adaptive immunity
Autophagy and antigen presentation
MHC class I presentation
In the presence of infection, T cells recognize foreign antigen presented in the context of MHC molecules. Antigen presentation refers to the pathways involved in the effective delivery of antigens to MHC molecules, often requiring a complex interplay between intracellular factors and compartments. CD8+ T cells recognize peptides presented in the context of MHC class I molecules expressed on the surface of all nucleated cells. MHC class I molecules present antigen derived from various intracellular sources such as viral proteins, endogenous tumour antigens and cytoplasmic and nuclear self-antigens.35 During MHC Class I presentation antigens are processed by the proteasome into peptides before being transported into the endoplasmic reticulum by the transporter associated with antigen-processing, where they are loaded onto MHC Class I molecules before cell surface expression via the golgi apparatus. Limited evidence exists that autophagy plays a role in the conventional MHC class I pathway. Schmid et al. could not demonstrate improved MHC class I presentation of a viral epitope when it was conjugated to LC3, a mechanism that was able to enhance the presentation of MHC class II antigens.36 Multiple other studies have also failed to report a requirement for autophagy in MHC class I presentation.37–39 However, inhibition of autophagy was found to decrease MHC class I surface expression in B16 murine melanoma cells and subsequent tumour cell cytolysis by CD8+ T cells.40 Autophagy has been implicated in an alternative pathway of MHC class I presentation that exists in DCs and macrophages, termed cross-presentation. Cross-presentation provides the means to present extracellular-sourced antigens on MHC class I molecules that would normally be routed through the MHC class II pathway to induce a CD4+ T-cell response. Via this mechanism CD8+ T cells may respond to exogenous antigens and phagocytosed material.35 Li et al. demonstrated how the tumour-specific CD8+ T-cell response could be enhanced using dendritic cells that had phagocytosed tumour cells treated with rapamycin or starvation to induce autophagy.41 Immunization of mice with purified tumour-derived autophagosomes could also induce the tumour-specific T-cell response.
MHC class II presentation
CD4+ T cells recognize antigen in the context of MHC class II molecules expressed on professional antigen-presenting cells and epithelial cells.36 Unlike endogenous MHC class I peptides, which are generated by proteasomes in the cytosol, exogenous antigens are processed in endolysosomal compartments where they are degraded by lysosomal proteases and loaded onto MHC class II molecules that have trafficked from the endoplasmic reticulum to MHC class II compartments (MIICs). Because of the prominent role of the lysosomal system in MHC class II presentation many studies have linked autophagy to this form of antigen presentation. Traditionally, MHC class II antigens were believed to be sourced from the extracellular space following phagocytosis by antigen-presenting cells. However, autophagy may comprise a significant source of MHC class II antigens derived from intracellular sources through the delivery of material to the lysosome. Schmid et al. showed in primary monocyte-derived DCs that autophagosomes frequently fused with MIICs. More than 50% of MIICs were observed to receive input from autophagosomes.36 In a separate study, analysis of the human B lymphoblastoid cell line showed that some MHC class II epitopes were derived from intracellular sources and starvation-induced autophagy could enhance the presentation of intracellular antigens on MHC class II molecules.42 Use of starvation and rapamycin in macrophages and DCs can also increase MHC class II expression of a mycobacterial antigen following phagocytosis of BCG.43 The secreted antigen, Ag85B, was found colocalized with LC3+ autophagosomes suggesting that autophagy may capture antigens following their escape from the phagosome and deliver them to the lysosome to prime CD4+ T cells. Indeed, the immunization of mice with rapamycin-treated DCs infected with mycobacteria could enhance the priming of mycobacteria-specific CD4+ T cells.43 Conjugation of the influenza matrix protein 1 to LC3 could also enhance the priming of antigen-specific CD4+ T cells36; whereas mice with a DC-specific deletion of Atg5 have impaired CD4+ T-cell priming in response to herpes simplex virus infection.37 The intersection of autophagy with antigen-processing pathways may be even closer than originally presumed. A recent study reported the formation of autophagosome-like structures emanating from MIICs in DCs. These structures contain the molecular machinery involved in antigen-processing as well as the autophagosome markers LC3 and Atg16L144. These observations offer the interesting prospect of antigen-presenting cells carrying out a specialized form of autophagy specific to the setting of antigen presentation, but add further complexity to the role that autophagy plays in the delivery of antigens to MHC molecules. What is clear is that autophagy provides a prominent source of antigens that can be routed into antigen-processing pathways through the fusion of autophagosomes with antigen-loading compartments (Fig. 2). As such, autophagy can be thought of as a key regulator of T-cell priming by influencing the repertoire of epitopes presented and thereby the nature and intensity of the ensuing T-cell response.
Figure 2.
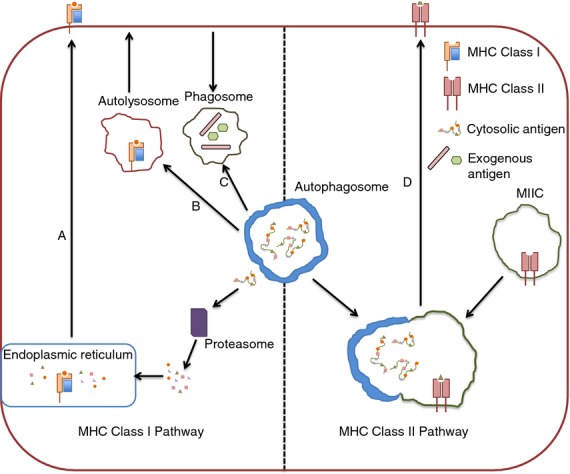
Autophagy and antigen-presenting pathways. The role of autophagy in MHC class I antigen presentation is controversial, but is postulated to occur through a number of potential mechanisms. Antigen may escape from autophagosomes into the cytosol where it can then be processed via the conventional MHC class I pathway that involves degradation by the proteasome before peptide loading in the endoplasmic reticulum (a). MHC class I molecules may also be loaded in autolysosomes themselves before trafficking to the cell surface (b). For cross-presentation, autophagosomes may intersect with phagosomes bearing phagocytosed exogenous antigen that can then be routed into the MHC class I pathway to prime CD8+ T cells (c). During MHC class II presentation, autophagosomes regularly fuse with MHC Class II loading compartments, thereby acting as a system for the delivery of cytosolic antigens to MHC class II molecules (d).
Autophagy in T cells
Although relatively small cells with limited cytoplasm, T cells have been extensively shown to perform autophagy and express autophagy genes.45–47 Autophagy is carried out constitutively to low levels in murine and human CD4+ and CD8+ T cells and can be induced following T-cell receptor stimulation45,47 and HIV infection48 in vitro. High levels of autophagy have been observed in cortical thymic epithelia cells of the murine thymus, suggesting that autophagy may have a role in the development and selection of T cells.49,50 Thymi with an Atg5 deletion in the stroma exhibit altered selection of certain CD4+ T-cell specificities alongside multi-organ inflammation, indicating a function for autophagy in T-cell selection and central tolerance.50 In contrast, no change in the selection of CD8+ T cells was observed. A number of genetic model systems have been employed to investigate a specific role for autophagy in T cells in vivo. In, Atg5−/− fetal liver chimeras, Atg7flox/flox Lck-Cre and Vps34flox/flox CD4-Cre mice, autophagy-deficient T cells develop normally within the thymus47,51–53. The effect of losing autophagy in the peripheral T-cell compartment is much more pronounced, however. There is a significant reduction in T-cell numbers in the spleen and lymph nodes of mice with a T-cell-specific deletion in Atg5−/−, Atg7−/−, Atg3−/− and Vps34−/−, with increased levels of apoptosis observed.46,47,51–53 Atg5−/−, Atg7−/− and Vps34−/− T cells also fail to proliferate effectively following activation.47,53 These defects appear to stem from an inability of autophagy-deficient T cells to regulate organelle quality control. An increase in mitochondrial load has been observed in autophagy-deficient T cells and this has been associated with enhanced levels of reactive oxygen species and cell death.46,51 Atg7−/− T cells also display defective calcium influx following T-cell receptor stimulation due to dysregulated endoplasmic reticulum homeostasis.46 Autophagy has recently been found to regulate energy metabolism in T cells. Blocking autophagy with lysosomal inhibitors was shown to inhibit the increase in ATP production normally observed following T-cell activation. When an exogenous energy source was provided to autophagy-deficient T cells in the form of methyl pyruvate, some of the defects in T-cell function could be reversed.54 Interestingly, a role for autophagy has recently been described in inducible natural killer T (iNKT) development. Mice with a T-cell-specific deletion of Vps34 exhibited a block in the early stages of iNKT development (stage 0) in the thymus.53 The same study also reported an essential role for autophagy in Foxp3+ regulatory T cell homeostasis and function, suggesting that the influence of autophagy in T-cell biology extends to multiple subsets and settings. Although much remains to be explained as to why the loss of autophagy impacts on T cells so severely, the evidence to date suggests a critical role for autophagy in T-cell homeostasis and function.
Autophagy in B cells
With use of Atg5−/− Rag1−/− chimeric mice and mice with a B-cell-specific deletion of Atg5 it was shown that autophagy-deficient B-cell progenitors fail to transition between pro- and pre-B-cell stages in the bone marrow, suggesting a requirement for autophagy in B-cell development.55 Like T cells, B cells are also severely affected by the absence of autophagy in the periphery. For example, deletion of Atg7 in the haematopoietic system results in reduced numbers of peripheral B cells,56 while mice lacking Atg5 in the B-cell lineage showed a particular requirement for autophagy in B-1a B-cell homeostasis55,57. Pengo et al. have recently elucidated the role of autophagy in B cells responding to antigen in an elegant study. In response to LPS stimulation ex vivo, Atg5-deficient B cells could effectively differentiate into plasma cells (PCs), exhibiting normal proliferation, cell enlargement and CD138 expression (a marker of PCs in mice). However, Atg5−/− PCs displayed increased intracellular immunoglobulin content and IgM production in vitro in response to LPS. The absence of autophagy was shown to increase endoplasmic reticulum stress, leading to an increase in Blimp-1 expression, which subsequently drives IgH expression and ultimately increased immunoglobulin production. However, when Atg5flox/flox CD19-Cre mice were immunized with a pneumococcal vaccine the antibody response in vivo was significantly decreased. The enhanced antibody production observed in vitro was negated by a requirement for autophagy in PC viability in vivo57.
Conclusion
We now know that autophagy and immunity are inexplicably linked and many aspects of the immune system exploit autophagy to regulate and mediate an array of effector functions. From the evidence available this observation is most pronounced in the innate immune system. Here, autophagy is implicated in pathogen sensing, phagocytosis, the removal of intracellular pathogens, and cytokine production. However, the capacity for autophagy to intercede in antigen processing and presentation pathways places it at the fulcrum of innate and adaptive immunity. More is known of how autophagy interjects into the MHC class II pathway, but a role for it in MHC class I presentation is still unclear and requires further work. In the adaptive immune system, autophagy is essential for the homeostasis of lymphocytes. T cells show an exquisite sensitivity to the loss of autophagy when compared to other immune cells, some of which fair relatively well without it. What are the reasons for these cell-specific differences? An answer may lie in the differing ability of cells to cope with increased reactive oxygen species levels observed in autophagy-deficient cells; or a less stringent requirement for autophagy-dependent organelle homeostasis in larger immune cells. In the case of B cells, we have finally begun to delineate the role that autophagy plays in lymphocytes in response to infection. For T cells this is still largely lacking. Much work has been done to investigate the role of autophagy in T cells in the steady state, but we must now define what part, if any, it has to play in antigen-specific T cells in vivo. In what way would T cells require classical degradative autophagy to respond to antigen? Perhaps T cells use novel roles for autophagy proteins that remain undiscovered. Although autophagy detection in primary cells ex vivo is challenging, use of mouse models with lineage-specific deletions in autophagy genes will help to answer some of these pressing questions. The ability to modulate the innate immune system so effectively through manipulation of autophagy suggests that targeting autophagy in the setting of the adaptive immunity may yield benefits in key areas such as vaccination and ageing.
Acknowledgments
AKS is funded by the NIHR BRC Oxford. DJP is funded by The Wellcome Trust, The Sidney Perry Foundation, The Allan and Nesta Ferguson Charitable Trust and St Catherine's College, Oxford.
Disclosures
The authors declare that they have no competing financial interests.
References
- 1.Deretic V. Autophagy in infection. Curr Opin Immunol. 2010;22:252–62. doi: 10.1016/j.ceb.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS lett. 2010;584:1287–95. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–7. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 4.Kuballa P, Nolte WM, Castoreno AB, Xavier RJ. Autophagy and the immune system. Annu Rev Immunol. 2012;30:611–46. doi: 10.1146/annurev-immunol-020711-074948. [DOI] [PubMed] [Google Scholar]
- 5.Hruban Z, Spargo B, Swift H, Wissler RW, Kleinfeld RG. Focal cytoplasmic degradation. Am J Pathol. 1963;42:657–83. [PMC free article] [PubMed] [Google Scholar]
- 6.Takeuchi O, Akira S. Pattern recognition receptor and inflammation. Cell. 2010;140:805–20. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 7.Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal. 2010;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Y, Jagannath C, Liu XD, Sharafkbaneh A, Kolodzieska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–44. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanjuan MA, Dillon CP, Tait SW, et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 2007;450:1253–7. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 10.Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J. 2008;27:1110–21. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 12.Travassos LH, Carneiro LA, Ramjeet M, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2009;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 13.Cooney R, Baker J, Brain O, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–7. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 14.Lapaquette P, Bringer M, Darfeuille-Michaud A. Defects in autophagy favour adherent-invasive Escherichia coli persistence within macrophages leading to increased pro-inflammatory response. Cell Microbiol. 2012;14:791–807. doi: 10.1111/j.1462-5822.2012.01768.x. [DOI] [PubMed] [Google Scholar]
- 15.Benjamin JL, Sumpter R, Jr, Levine B, Hooper LV. Intestinal epithelial autophagy is essential for host defence against invasive bacteria. Cell Host Microbe. 2013;13:723–34. doi: 10.1016/j.chom.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Journai N, Kobiyama K, Shiina M, Ogata K, Ishii KJ, Takeshita F. NLRP4 negatively regulates autophagic processes through an association with beclin1. J Immunol. 2011;18:1646–55. doi: 10.4049/jimmunol.1001654. [DOI] [PubMed] [Google Scholar]
- 17.Tormo D, Checinska A, Alonso-Curbela D, et al. Targeted activation of innate immunity for therapeutic induction of autophagy and apoptosis in melanoma cells. Cancer Cell. 2009;16:103–14. doi: 10.1016/j.ccr.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signalling. Proc Natl Acad Sci USA. 2009;106:2770–5. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshikawa Y, Ogawa M, Hain T, et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol. 2009;11:1233–40. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- 20.Birmingham L, Canadien V, Gouin E, Troy EB, Yoshimori T, Cossart P, Higgins DE, Brumell JH. Listeria monocytogenes evades killing by autophagy during colonization of host cell. Autophagy. 2007;3:442–51. doi: 10.4161/auto.4450. [DOI] [PubMed] [Google Scholar]
- 21.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–41. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 22.Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–66. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- 23.Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J Biol Chem. 2006;281:11374–83. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- 24.Wang YH, Wu JJ, Lei HY. The autophagic induction in Helicobacter pylori-infected macrophage. Exp Biol Med. 2009;234:171–80. doi: 10.3181/0808-RM-252. [DOI] [PubMed] [Google Scholar]
- 25.Castillo EF, Dekonenko A, Arko-Mensah J, et al. Autophagy protects against active tuberculosis by suppressing bacterial burden and inflammation. Proc Natl Acad Sci USA. 2012;109:E3168–76. doi: 10.1073/pnas.1210500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150:803–15. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan K, Huang C, Fox C, et al. Autophagy plays an essential role in the clearance of Pseudomonas aeruginosa by alveolar macrophages. J Cell Sci. 2012;125:507–15. doi: 10.1242/jcs.094573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Remijsen Q, Vanden Berghe T, Wirawan E, et al. Neutrophil extracellular trap cell death requires both autophagy and superoxide generation. Cell Res. 2011;21:290–304. doi: 10.1038/cr.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Itakura A, McCarty OJT. Pivotal role for the mTOR pathway in the formation of neutrophil extracellular traps (NETs) via regulation of autophagy. Am J Physiol Cell Physiol. 2013;305:C348–C354. doi: 10.1152/ajpcell.00108.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kambas K, Mitroulis I, Apostolidou E, et al. Autophagy mediates the delivery of thrombogenic tissue factor to neutrophil extracellular traps in human sepsis. PLoS ONE. 2012;7:e45427. doi: 10.1371/journal.pone.0045427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plantinga TS, Crisan TO, Oosting M, et al. Crohn's disease-associated ATG16L1 polymorphism modulates pro-inflammatory cytokine responses selectively upon activation of NOD2. Gut. 2011;60:1229–35. doi: 10.1136/gut.2010.228908. [DOI] [PubMed] [Google Scholar]
- 32.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–5. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 33.Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J Biol Chem. 2008;283:33175–82. doi: 10.1074/jbc.M804478200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peral de Castro C, Jones SA, Cheallaigh CN, et al. Autophagy regulates IL-23 secretion and innate T cell responses through effects on IL-1 secretion. J Immunol. 2012;189:4144–52. doi: 10.4049/jimmunol.1201946. [DOI] [PubMed] [Google Scholar]
- 35.Crotzer VL, Bum JS. Autophagy and adaptive immunity. Immunology. 2010;131:9–17. doi: 10.1111/j.1365-2567.2010.03321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmid D, Pypaert M, Munz C. Antigen-loading compartments for major histocompatibility complex class II molecules continually receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee HK, Mattei LM, Steinberg BE, et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 2010;32:227–39. doi: 10.1016/j.immuni.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nimmerjahn F, Milosevic S, Behrends U, Jaffee EM, Pardoll DM, Bornkamm GW, Mautner J. Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy. Eur J Immunol. 2003;33:1250–9. doi: 10.1002/eji.200323730. [DOI] [PubMed] [Google Scholar]
- 39.Leung CS, Haigh TA, Mackay LK, Rickinson AB, Taylor GS. Nuclear location of an endogenously expressed antigen, EBNA1, restricts access to macroautophagy and the range of CD4 epitope display. Proc Natl Acad Sci USA. 2010;107:2165–70. doi: 10.1073/pnas.0909448107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li B, Lei Z, Lichty BD, Li D, Zhang GM, Feng ZH, Wan Y, Huang B. Autophagy facilitates major histocompatibility complex class I expression induced by IFN-γ in B16 melanoma cells. Cancer Immunol Immunother. 2009;59:313–21. doi: 10.1007/s00262-009-0752-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Wang LX, Yang G, Hao F, Urba WJ, Hu HM. Efficient cross-presentation depends on autophagy in tumor cells. Cancer Res. 2008;68:6889–95. doi: 10.1158/0008-5472.CAN-08-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dengjel J, Schoor O, Fischer R, et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc Natl Acad Sci USA. 2005;102:7922–7. doi: 10.1073/pnas.0501190102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jagannath C, Lindsey DR, Dhandayuthapani S, Xu Y, Hunter RL, Jr, Eissa NT. Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat Med. 2009;15:267–76. doi: 10.1038/nm.1928. [DOI] [PubMed] [Google Scholar]
- 44.Kondylis V, van Nispen tot Pannerden HE, van Dijk S, et al. Endosome mediated autophagy: an unconventional MIIC-driven autophagic pathway operational in dendritic cells. Autophagy. 2013;9:861–80. doi: 10.4161/auto.24111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerland LM. Autolysosomes accumulate during in vitro CD8+ T-lymphocyte aging and may participate in induced death sensitization of senescent cells. Exp Gerontol. 2004;39:789–800. doi: 10.1016/j.exger.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 46.Jia W, He Y. Temporal regulation of intracellular organelle homeostasis in T lymphocytes by autophagy. J Immunol. 2011;186:5313–22. doi: 10.4049/jimmunol.1002404. [DOI] [PubMed] [Google Scholar]
- 47.Pua H, Dzhagalov I, Chuck M, Mizushima N, He YW. A critical role for the autophagy gene Atg5 in T cell survival and proliferation. J Exp Med. 2007;204:25–31. doi: 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X, Gao Y, Tan J, et al. HIV-1 and HIV-2 infections induce autophagy in Jurkat and CD4+ T cells. Cell Signal. 2012;24:1414–19. doi: 10.1016/j.cellsig.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 49.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–11. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T cell repertoire and is essential for tolerance. Nature. 2008;455:393–400. doi: 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- 51.Pua H, Guo J, Komatsu M, He Y. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J Immunol. 2009;182:4046–55. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- 52.Willinger T, Flavell RA. Canonical autophagy dependent on the class III phosphoinositide-3 kinase Vps34 is required for naive T cell homeostasis. Proc Natl Acad Sci USA. 2012;109:8670–5. doi: 10.1073/pnas.1205305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parekh VV, Wu L, Boyd KL, et al. Impaired autophagy, defective T cell homeostasis, and a wasting syndrome in mice with a T cell-specific deletion of Vps34. J Immunol. 2013;190:5086–101. doi: 10.4049/jimmunol.1202071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hubbard VM, Valdor R, Patel B, Singh R, Cuervo AM, Macian F. Macroautophagy regulates energy metabolsim during effector T cell activation. J Immunol. 2010;185:7349–57. doi: 10.4049/jimmunol.1000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miller BC, Zhao Z, Stephenson LM, et al. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy. 2008;4:309–14. doi: 10.4161/auto.5474. [DOI] [PubMed] [Google Scholar]
- 56.Mortensen M, Ferguson DJ, Edelmann M, Kessler B, Morten KJ, Komatsu, Simon AK. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anaemia in vivo. Proc Natl Acad Sci USA. 2010;107:832–7. doi: 10.1073/pnas.0913170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pengo N, Scolari M, Oliva L, et al. Plasma cells require autophagy for sustainable immunoglobulin production. Nat Immunol. 2013;14:298–305. doi: 10.1038/ni.2524. [DOI] [PubMed] [Google Scholar]
- 58.Tan Y, Kusuma CM, St. John LJ, Vu HA, Alibek K, Wu A. Induction of autophagy by anthrax lethal toxin. Biochem Biophys Res Commun. 2009;379:293–7. doi: 10.1016/j.bbrc.2008.12.048. [DOI] [PubMed] [Google Scholar]
- 59.Shanazari S, Namolovan A, Mogridge L, Kim PK, Brumell JH. Baterial toxins can inhibit host cell autophagy through cAMP generation. Autophagy. 2011;7:957–65. doi: 10.4161/auto.7.9.16435. [DOI] [PubMed] [Google Scholar]
- 60.Mostowy S, Sancho-Shimizu V, Hamon M, Simeone R, Brosch R, Johansen T, Cossart P. p62 and NDP52 target intracytosolic Shigella and Listeria to different autophagy pathways. J Biol Chem. 2011;286:26987–95. doi: 10.1074/jbc.M111.223610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Orvedahl A, MacPherson S, Sumpter R, Talloczy Z, Zou Z, Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. 2010;7:115–27. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blanchet FP, Moris A, Nikolic DS, et al. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity. 2010;32:654–69. doi: 10.1016/j.immuni.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou D, Spector SA. Human immunodeficiency virus-1 infection inhibits autophagy. AIDS. 2008;22:695–9. doi: 10.1097/QAD.0b013e3282f4a836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Orvehdahl A, Alexander D, Talloczy Z, et al. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 65.Chaumorcel M, Lussignol M, Mouna L, Cavignac Y. The human cytomegalovirus protein TRS1 inhibits autophagy via its interaction with beclin 1. J Virol. 2012;86:2571–84. doi: 10.1128/JVI.05746-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Joubert PE, Meiffren G, Gregoire G, et al. Autophagy inhibition by the pathogen receptor CD46. Cell Host Microbe. 2000;6:354–66. doi: 10.1016/j.chom.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 67.Portillo JA, Okenka G, Reed E, et al. The CD40-autophagy pathway is needed for host protection despite IFN-γ-dependent immunity and CD40 induces autophagy via control of P21 levels. PLoS ONE. 2010;5:e14472. doi: 10.1371/journal.pone.0014472. [DOI] [PMC free article] [PubMed] [Google Scholar]


