Abstract
The intracellular localization and target of the napyradiomycin congeners CNQ525.510B and A80815C were explored using an immunoaffinity fluorescence (IAF) approach. Semi-synthetic methods were used to prepare probes from napyradiomycin CNQ525.510B and derivative A80815C. The results of confocal microscopy indicated that probes from both natural products localized predominantly within the endoplasmic reticulum (ER) of HCT-116 human colon carcinoma cells. Parallel immunoaffinity precipitation efforts using a monoclonal antibody designed against the IAF tag, resulted in the isolation of an Hsp90 family member. This protein was identified as human Grp94 (hGrp94), by its specific mass spectral signature. This observation was validated by Western blot analyses and by the result of an in vitro Grp94 binding assay. The fact that the napyradiomycins CNQ525.510B and A80815C bind to hGrp94, and their associated probes localize within the ER, suggest the use of these materials as molecular probes for monitoring ER-based chaperone function.
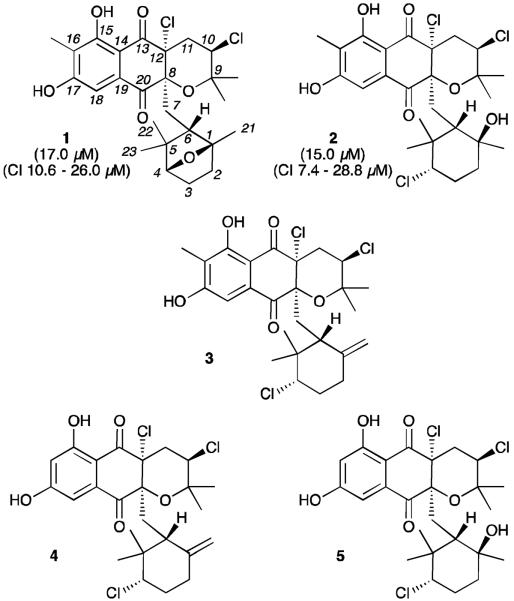
Marine actinomycetes provide robust access to unusual secondary metabolites.1 In the course of microbial studies of ocean sediments, we isolated an actinomycete, strain CNQ-525, from sediment sample collected in 152 m of water off the coast of La Jolla, CA.2a From cultures of this strain, we isolated four new compounds, including napyradiomycin CNQ525.510B (1) and the previously described compounds A80915C (2) and A80915A (3).2b These meroterpenoids belong to a larger class of chlorodihydroquinones (Fig. 1), including napyradiomycin B1 (4) and B4 (5).2–9 As part of an on-going effort to explore the modes of antitumor activity of marine microbial metabolites, we recently examined the cancer cell cytotoxicities and cell cycle properties of several napyradiomycin class meroterpenoids.2b In this report, we provide evidence that the antiproliferative effects of the napyradiomycins are derived by binding to the intracellular target protein Grp94.
Figure 1.
Structures of napyradiomycin CNQ525.510B (1), A80915C (2), A80915A (3), napyradiomycin B1 (4), and napyradiomycin B4 (5). IC50 cytotoxicity values against HCT-116 colon carcinoma and values at 95% confidence (CI) are provided for compounds 1 and 2 in parentheses.
In the past, the napyradiomycin family of chlorodihydroquinones has served as a starting point for both drug discovery3–13 and biosynthetic studies.14–16 In recent biosynthetic efforts, studies in the Moore laboratory unveiled the napyradiomycin biosynthetic cluster15 and mined the resulting enzymes to identify a new vanadium-dependent choroperoxidase.16 Subsequently, efforts from the Snyder laboratory provided an excellent illustration as to how biosynthetic knowledge facilitated the total synthesis of (–)-napyradiomycin A1.17
Earlier antimicrobial screening efforts indicated that several members of this family, including napyradiomycin B1 (4) and B4 (5),3 A80915A (3),5 and their biosynthetic precursors,9 displayed activity against Gram-positive bacteria. More recently, in depth kinetic analyses indicated that metabolite A80915A(3) displays a potent and rapid bactericidal activity against potent methicillin-resistant Staphylococcus aureus (MRSA) strains.18
Studies have also been conducted to examine selected bioactivities in mammalian cells. In 1991, a team at Lilly Research Laboratories, reported that A80915A (3) inhibited gastric (H+-K+) ATPases using a series of enzymatic assays.19 A few years later, screening efforts at Fujisawa Pharmaceutical Co. Ltd. demonstrated that napyradiomycins A and B1 (4) acted as non-steroidal estrogen receptor antagonists.10 And, more recently napyradiomycin A1 was shown to inhibit mitochondrial complexes I and II.20 To date, however, the activity of members of this family of natural products have not been evaluated in terms of their specificity in a proteomic cell target context.
Interested in further exploring this activity, we applied a streamlined immunoaffinity fluorescent approach shown in our laboratories to provide a rapid evaluation of cellular and molecular targeting in tumor cell lines.21–23 We began by applying conventional cytostatic and cytotoxicity assays. As indicated in Fig. 1, we found IC50 values in HCT-116 cells from 15–17 μM. Application of fluorescence-activated cell sorting (FACS) analysis using Yo-Pro staining24 indicated that napyradiomycin CNQ525.510B (1) and A80915C (2) induced apoptosis in a dose dependent manner (Fig. 2a), thereby providing a reliable phenotypic marker. Confirmation of the apoptotic effect was achieved through Western blot analysis. As shown in Fig. 2a, caspase 3 activation25 was observed in lysates from cells treated with 2 (Fig. 2b) or 1 (Fig. 2c).
Figure 2.
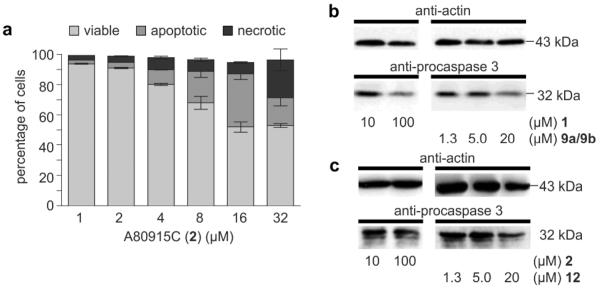
Apoptotic activity. (a) Apoptosis induction measured by flow cytometry after incubating HCT-116 cells with A80915C (2) for 24 h. (b–c) Degradation of pro-caspase 3 in response to incubation with A80915C (2) napyradiomycin CNQ525.510B (1), probes 9a/9b and probe 12. Actin levels were evaluated using an anti-actin antibody as a standard with 20 μg loaded for each lane.
In previous studies, we had shown that structural variations among the napyradiomycin class congeners resulted in significant reproducible cytostatic activity within the HCT-116 cell line.2 These structure activity relationship (SAR) data suggested that the cytotoxicity of members of this family was attenuated by structural features within a specific biomolecular environment such as a protein binding pocket.
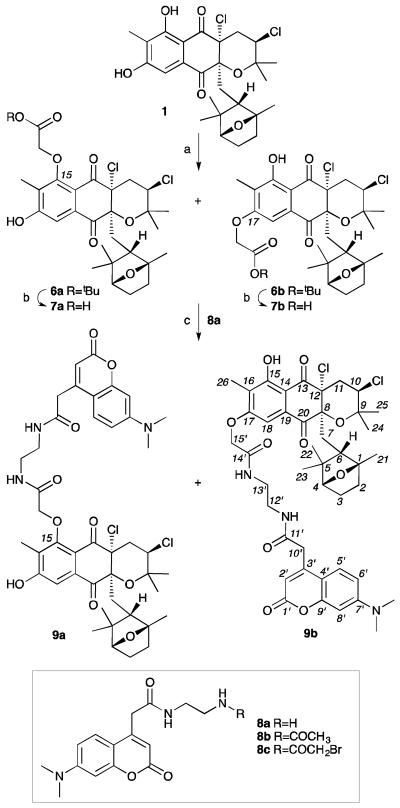
Using available labelling methods,26 we turned our attention to preparation of an IAF tag by esterification of the hydroxyl group at C15 or C17. We found that preparation of a probe from 1 using the corresponding α-bromoamide IAF tag (8c) was complicated by a low yield (~26%) of a regiochemical mixture of the C15-substituted 9a and C17-substituted 9b probes. Unfortunately, these regioisomers could not be separated via multiple attempts using pTLC, flash chromatography or RP C-18 HPLC.
We then examined a 3-step protocol (Scheme 1). Using this approach, we were able install the tert-butyl 2-bromoacetate in good yield (72%), but again, we obtained an ~1:1 inseparable mixture of the C15-isomer 6a and C17-isomer 6b. While we were able to effectively convert this material into the corresponding mixture of probes 9a/9b in an overall yield of 50%, we were never able to obtain one isomer in a pure form (see copies of NMR spectral data provided in the Supporting Information).
Scheme 1.
Synthesis of napyradiomycin CNQ525.510B IAF probes 9a/9b. Reagents and conditions: (a) t-butyl-bromoacetate, K2CO3, DMF, rt, 72%; (b) TFA, CH2Cl2; (c) tag 8a,26 HATU, EtNiPr2, DMF, rt, 70%. Overall yield of 50%.
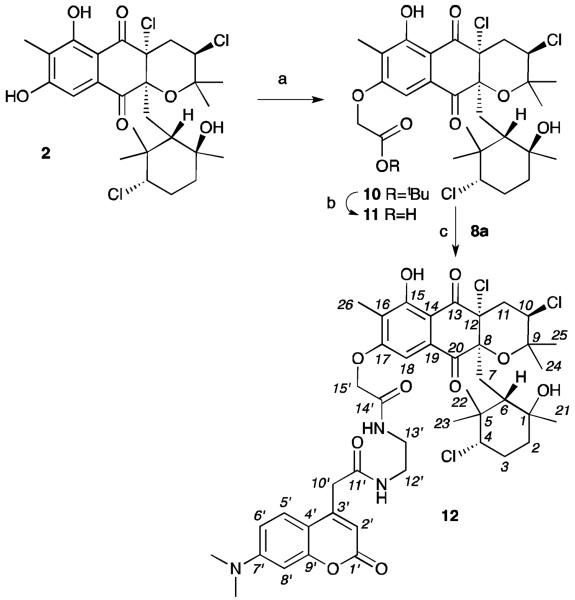
Additionally, we prepared a second probe 12 from A80915C (2). Using comparable methods, we were able to obtain a single mono-C-17 labelled adduct 10 in 65% yield from 2 (Scheme 2). The position of the tag was confirmed by the loss of the C-17 phenolic proton at 9.56 ppm, while the proton at C-15 was observed at 12.11 ppm and 12.12 ppm in 10 and 12, respectively (see Supporting Information).2,9 Ester 10 was then deprotected, and the resulting acid 11 coupled to IAF tag 8a to afford probe 12 in 34% overall yield, >98% purity, as a single isomer.
Scheme 2.
Synthesis of A80915C IAF probe 12. Reagents and conditions: (a) t-butyl-bromoacetate, K2CO3, DMF, rt, 65%; (b) TFA, CH2Cl2; (c) tag 8a,26 HATU, EtNiPr2, DMF, rt, 53%. Overall yield of 34%.
In order to ensure that probes maintained the activity of the parent natural products, probes 9a/9b and 12 were assessed for their ability to induce apoptosis.2 We elected to use this assay to validate the biological activity of probes 9a/9b and 12, as unlike an IC50 values, this screen provided a direct comparison with the phenotype of their parent natural products 1 or 2. As shown in Fig. 2, cells treated with napyradiomycin CNQ525.510B (1, Fig. 2c) and A80915C (2, Fig. 2b) showed a significant loss in the levels of procaspase 3.25 Comparable dose-dependant loss was also observed with probes 9a/9b (Fig. 2c) and 12 (Fig. 2b). Although the potency of the probes 9a/9b and 12 were less than their corresponding natural products 1 and 2, the core biological action was maintained. Moreover, the apoptotic activity was not seen when the cells were incubated with the IAF tag alone as in control 8b (data not shown).
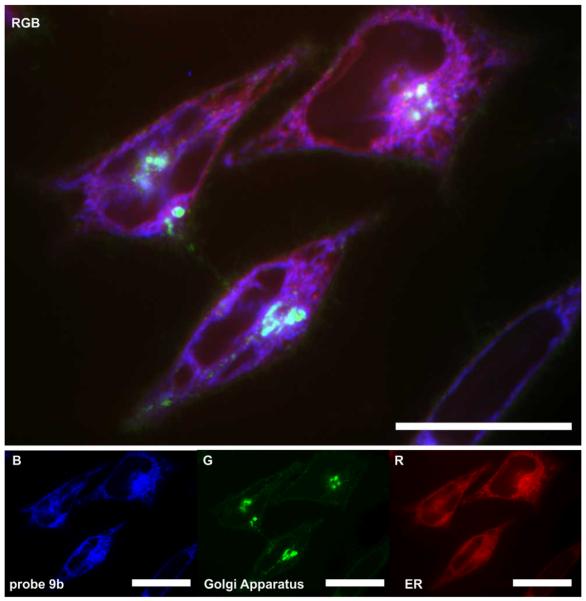
We then turned to confocal microscopy to visualize the subcellular location of probes 9a/9b and 12. HeLa cells were chosen for much of the visualization work due to the larger cytoplasmic area. Using time course microscopy, we found that uptake of the probes became apparent after 8 h and was optimally observed after 24 h. As depicted in Fig. 3, probe 12 appeared to localize in perinuclear space with the appearance of secondary structure.
Figure 3.
Confocal fluorescent image depicting the localization of probe 12 in the endoplasmic reticulum (ER) of HeLa cells. HeLa cells were incubated with 20 nM probe 12 for 24 h at which point the cells were imaged live. Probe 12 was blue (B) fluorescent. Cells were counterstained for their ER (red, R) and Golgi apparatus (green, G) using ER-tracker Red27 and BODIPY-FL C5-ceramide28, respectively. The three-colour (RGB) image depicts the overlay of each stain. Bars denote 10 μm.
Next, we applied orthogonal co-stains to validate the subcellular localization (Fig. 3). The ER-tracker red stain (BODIPY TR glibenclamide)27 showed a strong correlation to the localization of the compound suggesting that the target of the molecule is predominantly in the ER, as best illustrated with probe 12 (Fig. 4). BODIPY-FL C5-ceramide,28 a green stain for the Golgi apparatus, also faintly stained the plasma membrane of the cells, and provided further support for the ER localization.
Figure 4.
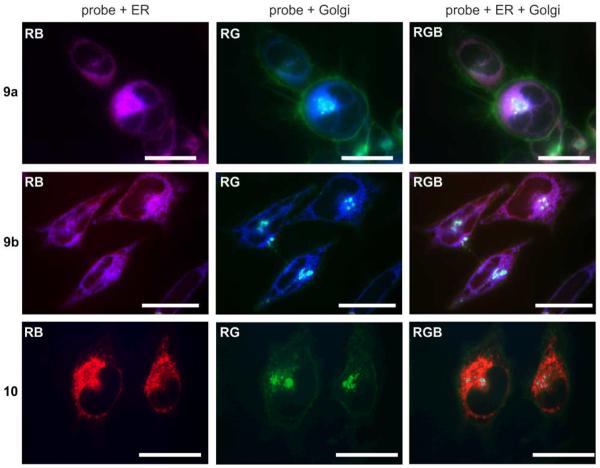
Subcellular localization of probe mixture 9a/9b and pure probe 12 in HCT-116 cells. Live cell images depicting HCT-116 cells that were incubated with media containing 20 nM probes 9a/9b, 20 nM probe 12 or 10 μM tag control 8b for 24 h. Two and three-color imaging depict overlay of the probes and ER stains (RB), probes and Golgi stains (GB) or all three channels (RGB). Bars denote 10 μm. Expanded and single channel images provided herein and in Figure S1 in the Supporting Information clearly demonstrate lack of Golgi localization.
Further confirmation of the subcellular localization was established by examining probes 9a/9b, which showed a similar staining pattern to the probe 12. As shown in Fig. 4, there was a clear overlap of the red (ER stain) and blue (probe) channels (RB, Fig. 3) for both the probe mixture 9a/9a and pure probe 12, but not for the Golgi apparatus (RG, Fig. 3). An IAF control 8b showed no specific staining and therefore supported the thesis that the localization of 9a/9b and 12 was due to the functionality of the cytotoxic natural product.
We then applied the immunoaffinity properties of the IAF tag (Fig. 5). A 200 μL aliquot HCT-116 cell lysate, prepared with 1 mg/mL of total protein, was incubated with 0.5 μM to 50.0 μM of 9a/9b or 12 at 4 °C. After 4 h, 25 μL of an Affigel-10 resin conjugated with ~4 mg/mL of an anti-IAF antibody, XRI-TF-35, was added. After mild shaking at 4 °C for 12 h, the solution was removed and the resin was washed twice with 1 mL of ice-cold wash buffer (phosphate buffered saline (PBS) pH 7.2, 5 mM EDTA, 1% NP-40, and 0.1% SDS). The bound materials were eluted by incubation by addition of 35 μL of 100 μM 7-dimethylamino-4-coumarin acid in PBS at rt for 30 min followed by centrifugation at 15,000 RPM for 5 min. The resulting solution was removed, diluted with 4x SDS-PAGE gel loading buffer (200 mM Tris-Cl pH 6.8, 400 mM DTT, 8% SDS, 0.4% bromophenol blue and 40% glycerol) and evaluated on a 4–14% gradient Bis-Tris SDS PAGE gel (Invitrogen).
Figure 5.
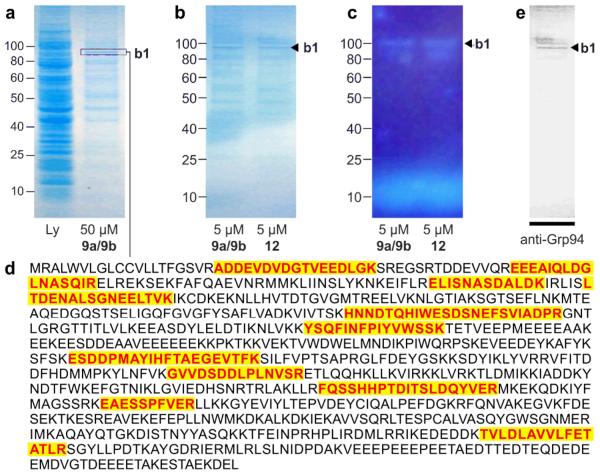
Immunoprecipitation (IP) of hGrp94. (a) A Coomassie G-250 blue silver stained SDS PAGE gel depicting cell lysate from HCT-116 cells that were cultured for 24 h in media containing 50 μM probe 12. Lanes denote lysate (Ly) and the immunoprecipitated product (50 μM 9a/9b). Band b1 denotes the major protein. (b) A Coomassie G-250 blue silver stained SDS PAGE gel depicting the immunoprecipitated product obtained when using 5 μM probes 9a/9b or 5 μM probe 12. (c) A fluorescent image of the gel in (b) depicting band b1. The band at the bottom of the gel corresponds to 9a/9b or 12. (d) Trypsin-digestion followed LC-MS/MS analysis identified 11 peptides associated with hGrp94. Observed peptides are coloured and shaded. (e) While from the same protein family, the observed peptides would not be derived from cytosolic Hsp90 alpha member 1. (f) Western blot identifying the major band from the immunoprecitation with probe 12 as hGrp94. This blot depicts a Western blot using an anti-hGrp94 mAb.
As depicted in Fig. 5a, a major band b1 was observed at ~95 kDa along with several minor bands observed. Further analysis with a ten-fold reduction in probe concentration returned comparable results with significant enhancement of the intensity of the b1 band at ~95 kDa (Fig. 5b) as compared to other protein bands. Further analysis by imaging the gel before fixation and staining (Fig. 5c) indicate that band b1 was blue fluorescent, suggesting covalent modification by an IAF tag. This was the only band within the gel apparent (Fig. 5c).
Convinced that the 95 kDa band was significant, we turned to mass spectral methods to identify this protein. A sample of a band stained by Coomassie blue silver was excised and submitted to trypsin-digestion followed by LC-MS/MS analysis. Eleven peptides were found that matched hGrp94 (Fig. 5d), a protein whose molecular weight was consistent with that of band b1.
We then validated this observation by confirming the presence of hGrp94 within the immunoprecipitated product. As shown in Fig. 5e, Western blot analysis with a rabbit polyclonal antibody against Grp94 (G4545, Sigma Aldrich) confirmed that band b1 was hGrp94, indicating that both 9a/9b and 12 targeted hGrp94.
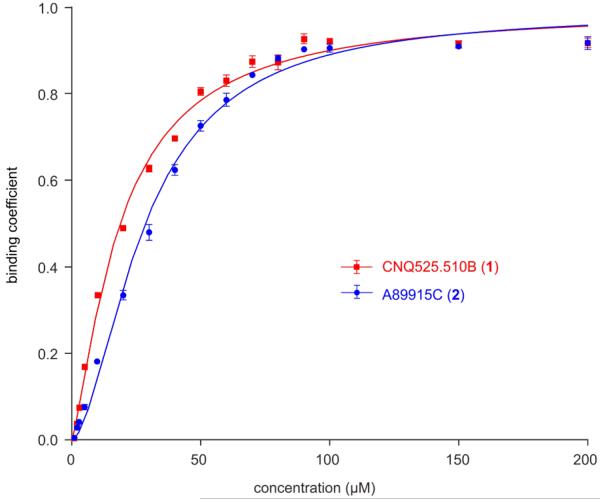
Given this evidence, we then applied conventional binding methods to confirm the binding between the napyradiomycins 1 and 2 and recombinantly expressed N-terminal His10-tagged hGrp94 (see Supporting Information). We developed a method using a combination of LC-MS analysis and equilibrium microdialysis29 to monitor the binding to hGrp94. Using this method, we found that 1 and 2 both bound to hGrp94 with a binding constant Kd of 18.7±1.5 μM and 28.2±1.7 μM respectively (Fig. 6).
Figure 6.
Equlibrium Microdialysis. The binding of napyradiomycins CNQ525.510B (1) or A80915C (2) with recombinant hGrp94 was determined by equilibrium microdialysis. LC-MS analysis using UV-Vis detection at 380 nm and peaks were validated by MS analysis. The LC-MS method was standardized with known concentrations prior to use. Three repetitions were conducted at each concentration and the deviation between these experiments depicted by error bars. Experiments were conducted with 20 μM hGrp94.
Conclusions
This study identified the targeting of napyradiomycins CNQ525.510B (1) and A80941C (2) to hGrp94, a heat shock protein found within the ER of mammalian cells. In this study, we demonstrated the preparation of biologically active IAF probes 9a/9b and 12 from 1 and 2, respectively. Using time course confocal microscopy, we determined that both probes 9a/9b and 12 localized within the ER, similar to that of hGrp94. We then applied the affinity features of our IAF probes to identify hGrp94, as a predominant protein binding to 9a/9b and 12. This observation was then validated by Western blot analyses, and subsequent binding studies using recombinant hGrp94 confirming that compounds 1 and 2 indeed bound to hGrp94. However, hGrp94 was not the sole protein identified during immunopreciptation studies. This lack of selective isolation of hGrp94 can be explained by a number of reasons including: off-targets, lack of probe selectivity, or the co-immunoprepcitation of hGrp94 binding proteins. The fact that 1 and 2 bind to hGrp94, and that their corresponding probes localize subcellularly within the ER, as does hGrp94, provides strong support for the fact that 1 and 2 target hGrp94 in live cells.
This discovery provides an excellent addition to the set of natural products including radicicol29 and geldanamycin30 known to bind to Hsp90 proteins. Most recently, efforts in the Gewirth and Blagg laboratories31–35 have led to a detailed structural understanding of the interaction between radicicol, geldanamycin, as well as a synthetically-derived hybrid molecule and the N-terminal ATP binding site of canine Grp94 (cGrp94).34 These studies have recently suggested the potential for discovery of paraform–selective inhibitors.35 Efforts are now underway to evaluate the mode of binding of napyradiomycins CNQ525.510B (1, Fig. 2c) and A80915C (2, Fig. 2b) with the goal of determining if these molecules share the same the ATP binding site as occupied by radicicol and geldanamycin.
Supplementary Material
Acknowledgements
This work was supported by the US National Institutes of Health under grant R37 CA044848 (from the National Cancer Institute). Samples of the XRI-TF35 mAb and IAF tags were provided by the Xenobe Research Institute. We thank Drs. Nicole Coufal, Majid Ghassemian, Yongxuan Su and Anthony Mrse for assistance with confocal microscopy, protein mass spectroscopy, small molecule mass spectroscopy, and NMR spectroscopy services, respectively.
Footnotes
† Electronic Supplementary Information (ESI) available: [procedures for the probe synthesis, spectroscopic and NMR data, as well as biological methods and protocols]. See DOI: 10.1039/b000000x/
Notes and references
- 1.Fenical W, Jensen PR. Nat. Chem. Biol. 2006;2:666. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]
- 2.a) Soria-Mercado IE, Prieto-Davó A, Jensen PR, Fenical W. J. Nat. Prod. 2005;68:904. doi: 10.1021/np058011z. [DOI] [PubMed] [Google Scholar]; b) Farnaes L, Coufal NG, Kauffman CA, Rheingold AL, DiPasquale AG, Jensen PR, Fenical W. J. Nat. Prod. 2013 doi: 10.1021/np400466j. in revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shiomi K, Iinuma H, Hamada M, Manabe M, Matsuki C, Takeuchi T, Umezawa H. J. Antibiot. (Tokyo) 1986;39:487. doi: 10.7164/antibiotics.39.487. [DOI] [PubMed] [Google Scholar]
- 4.Shiomi K, Nakamura H, Iinuma H, Naganawa N, Isshiki K, Takeuchi T, Umezawa H, Iitaka Y. J. Antibiot. (Tokyo) 1986;39:494. doi: 10.7164/antibiotics.39.494. [DOI] [PubMed] [Google Scholar]
- 5.Shomura T, Gomi S, Ito M, Yoshida J, Tanak E, Amano S, Watabe H-O, Ohuchi S, Itoh J, Sezaki M, Takebe H, Uotani K. J. Antibiot. (Tokyo) 1987;40:732. doi: 10.7164/antibiotics.40.732. [DOI] [PubMed] [Google Scholar]
- 6.Gomi S, Ohuchi S, Sasaki T, Itoh J, Sezaki M. J. Antibiot. (Tokyo) 1987;40:741. doi: 10.7164/antibiotics.40.740. [DOI] [PubMed] [Google Scholar]
- 7.Shiomi K, Nakamura H, Iinuma H, Naganawa N, Takeuchi T, Umezawa H, Iitaka Y. J. Antibiot. (Tokyo) 1987;40:1213. doi: 10.7164/antibiotics.40.1213. [DOI] [PubMed] [Google Scholar]
- 8.Shiomi K, Iinuma H, Naganawa N, Isshiki K, Takeuchi T, Umezawa H, Iitaka Y. J. Antibiot. (Tokyo) 1987;40:1740. doi: 10.7164/antibiotics.40.1740. [DOI] [PubMed] [Google Scholar]
- 9.Fukuda DS, Mynderse JS, Baker PJ, Berry DM, Brock LD, Yao RC, Mertz FP, Nakatsukasa WM, Mabe J, Ott J, Counter FT, Ensminger PW, Allen NE, Alborn WE, Hobbs JN. J. Antibiot. (Tokyo) 1990;43:623. doi: 10.7164/antibiotics.43.623. [DOI] [PubMed] [Google Scholar]
- 10.Hori Y, Abe Y, Shigematsu N, Goto T, Okuhara M, Kohsaka M. J. Antibiot. (Tokyo) 1993;46:1890. doi: 10.7164/antibiotics.46.1890. [DOI] [PubMed] [Google Scholar]
- 11.Umezawa K, Masuoka S, Oshe T, Naganawa H, Kondo S, Ikeda Y, Kinoshita N, Hamada M, Sawa T, Takeuchi T. J. Antibiot. (Tokyo) 1995;48:604. doi: 10.7164/antibiotics.48.604. [DOI] [PubMed] [Google Scholar]
- 12.Motohashi K, Sue M, Furihata K, Ito S, Seto H. J. Nat. Prod. 2008;71:595. doi: 10.1021/np070575a. [DOI] [PubMed] [Google Scholar]
- 13.Izumikawa M, Nagai A, Hashimoto J, Takagi M, Shin-Ya K. J. Antibiot. (Tokyo) 2010;63:729. doi: 10.1038/ja.2010.120. [DOI] [PubMed] [Google Scholar]
- 14.Shiomi K, Iinuma H, Naganawa H, Isshiki K, Takeuchi T, Umezawa H. J. Antibiot. (Tokyo) 1987;40:1740. doi: 10.7164/antibiotics.40.1740. [DOI] [PubMed] [Google Scholar]
- 15.Winter JM, Moffitt MC, Zazopoulos E, McAlpine JB, Dorrestein PC, Moore BS. J. Biol. Chem. 2007;282:16362. doi: 10.1074/jbc.M611046200. [DOI] [PubMed] [Google Scholar]
- 16.Bernhardt P, Okino T, Winter JM, Miyanaga A, Moore BS. J. Am. Chem. Soc. 2011;133:4268. doi: 10.1021/ja201088k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Snyder SA, Tang TZ, Gupta R. J. Am. Chem. Soc. 2009;131:5744. doi: 10.1021/ja9014716. [DOI] [PubMed] [Google Scholar]
- 18.Haste NM, Farnaes L, Perera VR, Fenical W, Nizet V, Hensler ME. Mar. Drugs. 2011;9:680. doi: 10.3390/md9040680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danzig AH, Minor PL, Garrigus JL, Fukuda DS, Mynderse JS. Biochem. Pharmacol. 1991;42:2019. doi: 10.1016/0006-2952(91)90603-3. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto K, Tashiro E, Motohashi K, Seto H, Imoto M. J. Antibiot (Tokyo) 2012;65:211. doi: 10.1038/ja.2011.138. [DOI] [PubMed] [Google Scholar]
- 21.Hughes CC, MacMillan JB, Gaudêncio SP, Jensen PR, Fenical W. Angew. Chem., Int. Ed. 2008;48:725. doi: 10.1002/anie.200804890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hughes CC, Yang YL, Liu WT, Dorrestein PC, La Clair JJ, Fenical W. J. Am. Chem. Soc. 2008;131:12094. doi: 10.1021/ja903149u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu WL, Guizzunti G, Foley TL, Burkart MD, La Clair JJ. J. Nat. Prod. 2010;73:1659. doi: 10.1021/np100371k. [DOI] [PubMed] [Google Scholar]
- 24.Idziorek T, Estaquier J, De Bels F, Ameisen JC. J. Immunol. Methods. 1995;185:249. doi: 10.1016/0022-1759(95)00172-7. [DOI] [PubMed] [Google Scholar]
- 25.Snigdha S, Smith ED, Prieto GA, Cotman CW. Neurosci. Bull. 2012;28:14. doi: 10.1007/s12264-012-1057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alexander MD, Burkart MD, Leonard MS, Portonovo P, Liang B, Ding X, Joullié MM, Gulledge BM, Aggen JB, Chamberlin AR, Sandler J, Fenical W, Cui J, Gharpure SJ, Polosukhin A, Zhang HR, Evans PA, Richardson AD, Harper MK, Ireland CM, Vong BG, Brady TP, Theodorakis EA, La Clair JJ. Chembiochem. 2006;7:409. doi: 10.1002/cbic.200500466. [DOI] [PubMed] [Google Scholar]
- 27.Watkins S, Geng X, Li L, Papworth G, Robbins PD, Drain P. Traffic. 2002;3:461. doi: 10.1034/j.1600-0854.2002.30703.x. [DOI] [PubMed] [Google Scholar]
- 28.Chazotte B. CSH Protoc. 2008 doi: 10.1101/pdb.prot4930. prot4931. [DOI] [PubMed] [Google Scholar]
- 29.Tsai TH, Chromatogr J, B Analyt. Technol. Biomed. Life Sci. 2003;797:161. doi: 10.1016/j.jchromb.2003.08.036. [DOI] [PubMed] [Google Scholar]
- 30.Winssinger N, Fontaine JG, Barluenga S. Curr. Top. Med. Chem. 2009;9:1419. doi: 10.2174/156802609789895665. [DOI] [PubMed] [Google Scholar]
- 31.Gorska M, Popowska U, Sielicka-Dudzin A, Kuban-Jankowska A, Sawczuk W, Knap N, Cicero G, Wozniak F. Front. Biosci. 2012;17:2269. doi: 10.2741/4050. [DOI] [PubMed] [Google Scholar]
- 32.Soldano KL, Jivan A, Nicchitta CV, Gewirth DT. J. Biol. Chem. 2003;278:48330. doi: 10.1074/jbc.M308661200. [DOI] [PubMed] [Google Scholar]
- 33.Immormino RM, Dollins DE, Shaffer PL, Soldano KL, Walker MA, Gewirth DT. J. Biol. Chem. 2004;279:46162. doi: 10.1074/jbc.M405253200. [DOI] [PubMed] [Google Scholar]
- 34.Immormino RM, Metzger LE LE, 4th, Reardon PN, Dollins DE, Blagg BS, Gewirth DT. J. Mol. Biol. 2009;388:1033. doi: 10.1016/j.jmb.2009.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duerfeldt AS, Peterson LB, Maynard JC, Ng CL, Eletto D, Ostrovsky O, Shinogle HE, Moore DS, Argon Y, Nicchitta CV, Blagg BS. J. Am. Chem. Soc. 2012;134:9796. doi: 10.1021/ja303477g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.