Abstract
Eya proteins are essential co-activators of the Six family of homeobox transcription factors and also contain a unique protein tyrosine phosphatase activity, belonging to the haloacid dehalogenase family of phosphatases. The phosphatase activity of Eya is important for a subset of Six1-mediated transcription, making this a unique type of transcriptional control. It is also responsible for directing cells to the repair instead of apoptosis pathway upon DNA damage. Furthermore, the phosphatase activity of Eya is critical for transformation, migration, invasion, and metastasis of breast cancer cells. Thus, inhibitors of the Eya phosphatase activity may be anti-tumorigenic and anti-metastatic, as well as sensitize cancer cells to DNA damage inducing therapies. In this paper, we identified a previously unknown chemical series using high throughput screening that inhibits the Eya2 phosphatase activity with IC50s ranging from 1.8 to 79 μM. Compound activity was confirmed using an alternative malachite green assay and H2AX, a known Eya substrate. Importantly, these Eya2 phosphatase inhibitors show specificity and do not significantly inhibit several other cellular phosphatases. Our studies identify the first selective Eya2 phosphatase inhibitors that can potentially be developed into chemical probes for functional studies of Eya phosphatase or into anti-cancer drugs in the future.
Keywords: Phosphatase, Eyes Absent 2, Eya2, Eya2 inhibitor, Six1
INTRODUCTION
The Eya proteins are mammalian homologues of the Drosophila Eyes Absent genes and were first identified as essential co-activators of members of the Six family of transcription factors, including Six1. The Six1 homeoprotein is essential for the development of many organs, including the muscle, kidney, olfactory epithelium, and inner ear1. It is typically down-regulated after organ development is complete, and its expression level is low or absent in most adult tissues. However, Six1 is over-expressed in numerous cancers, such as breast, ovarian, cervical, and hepatocellular carcinomas, as well as rhabdomyosarcomas, Wilms’ tumors, and leukemias1,2. Six1 expression has been linked to transformation, tumor growth, and metastasis in multiple tumor types, including breast cancer1,3-5. Experimentally lowering Six1 levels significantly decreases cancer cell proliferation1 and metastasis1,5 in different cancer models.
Given that Six1 does not have an intrinsic activation or repression domain, it requires co-activators such as the Eya family of proteins to mediate its transcriptional activity, both in normal development1,6 and in various disease processes1,6,7. Eya proteins have been linked to many types of cancer in which Six1 is over-expressed1,8,9. Examination of the Wang and Van de Vijver public breast cancer microarray datasets10,11 demonstrated that over-expression of Six1 and Eya together significantly predict shortened time to relapse and metastasis and shortened survival, whereas each gene individually does not9. Furthermore, Eya2 knockdown in Six1 over-expressing MCF7 cells inhibits the ability of Six1 to induce TGF-β signaling, epithelial-mesenchymal transition, and tumor initiating cell characteristics, properties that are associated with Six1-induced tumorigenesis and metastasis9. These data provide strong support that Six1 and Eya2 cooperate to induce tumorigenic and metastatic properties.
The Eya proteins have a C-terminal Eya Domain (ED)12 that contains signature motifs of the haloacid dehalogenase (HAD) hydrolases, a diverse collection of enzymes including phosphatases1,12,13. Eya proteins and other HAD family of phosphatases use an Asp as their active site residue instead of the more commonly used Cys in cellular phosphatases14. A few other HAD phosphatases (for example, Scp1 and Chronophin) target proteins, however, most HAD phosphatases do not have protein phosphatase activity12. All other known HAD protein phosphatases are Ser/Thr phosphatases (such as Scp1), while the Eya domain of Eya targets phosphorylated Tyr15. Recent evidence demonstrates that mouse Eya proteins can utilize their intrinsic phosphatase activity to switch the Six1 transcriptional complex from a repressor to an activator complex for some Six1-induced genes1, although the mechanism of this switch remains unclear. In Drosophila, while the phosphatase activity of Eya is not globally required for the ability of Six1 to induce transcription, it is required to induce transcription of a subset of genes16. The Eya proteins therefore represent the first transcription factor with intrinsic phosphatase activity that modulates transcriptional complexes1,12,13. Importantly, using two different breast cancer cell lines and both over-expression and knockdown systems, and examining multiple different Eyas, Hegde and colleagues demonstrated that Eya proteins, and in particular their Tyr phosphatase activity, are critical for transformation, migration, invasion, and metastasis of breast cancer cells8 although the specific mechanism by which Eya's phosphatase activity induces the metastatic phenotype is still unclear.
In addition to its role in Six-mediated transcription, Eya proteins have recently been shown to play a role in DNA repair. In response to DNA damage, Eya proteins dephosphorylate a phospho-Tyr on histone variant H2AX, and this dephosphorylation is critical for directing cells to the repair instead of apoptotic pathway15,17. Thus, knockdown of Eya proteins leads to a significant increase in apoptotic cells in response to hypoxia or ionizing radiation15,17. Currently, about half of all people with cancer are treated with radiation therapy, either alone or with other cancer treatment, to kill cancer cells and reduce tumor burden. Selectively sensitizing tumor tissue by engaging the apoptotic program of a cell is of great interest to the field of radiation oncology18. It is foreseeable that inhibitors of Eya's phosphatase activity may greatly increase the efficiency of radiation therapy, or of any DNA damaging related therapy (many cancer therapies use a combination of both), in cancers that are known to express Eya, including breast cancers, Wilms’ tumor, and ovarian carcinomas1,9.
Although it has traditionally been difficult to identify specific phosphatase inhibitors, the fact that Eya proteins belong to the HAD family of protein phosphatases that use an Asp instead of the more commonly used Cys as their active site residue, provides a unique opportunity to potentially identify specific Eya phosphatase inhibitors. We report here the identification and characterization of a previously unknown chemical series that specifically inhibit the Eya2 phosphatase. These novel compounds can be used by the research community as chemical probes to further study the function of Eya's phosphatase activity and its role in Six1-mediated breast tumorigenesis and metastasis. There is also the exciting possibility of developing this series of compounds into potential anti-cancer drugs in the future.
MATERIALS AND METHODS
Protein Expression and Purification
Human Eya2 ED (residue 253-538) that contains the Eya phosphatase activity was sub-cloned into the pGEX-6P1 vector(GE Healthcare) using the BamHI and XhoI sites and confirmed by DNA sequencing. Plasmids containing these constructs were transformed into E. coli strain XA90. Cells were grown until OD600 reached 0.8-1.0 and protein expression was induced at 20°C with 0.2 mM IPTG for 20 hours. Cell pellets were lysed by sonication in buffer L (50 mM Tris, pH 7.5, 250 mM NaCl, 5% glycerol, 1 mM DTT) containing protease inhibitors pepstatin A, leupeptin, and PMSF. Lysates were cleared via centrifugation (2 × 45 minutes at 18,000 × g). The supernatant containing GST-Eya2 ED proteins was loaded via gravity on glutathione-Sepharose 4B resin (GE Healthcare) and thoroughly washed with buffer L. ED protein was cleaved from the glutathione resin with PreScission protease at 4°C for 16 hours, eluted, and concentrated. ED protein was further purified on a Superdex 200 size exclusion column (GE Healthcare) using buffer L. Purified protein was aliquoted and stored at −80°C.
OMFP-based Eya Phosphatase assay
The activity of ED was measured in 50 μL reactions in black, 96-well, half-volume microtiter plates (Greiner Bio-one) with OMFP (3-O-methylfluorescein phosphate, Sigma-Aldrich) as the substrate. Upon dephosphorylation, OMFP is converted to a fluorescent product OMF. Enzyme and substrate concentrations were optimized to have a linear response during the assay, consume less than 15% of substrate after one hour, and have substrate concentration below the Km. The final assay condition is 50 mM MES, pH 6.5, 50 mM NaCl, 5 μM MgCl2, 0.05% BSA, 1 mM DTT, and the reaction contained 50 nM Eya2 ED and 100 μM OMFP. Reactions were started by the addition of OMFP and were continued for 1 hour at room temperature and terminated by the addition of 75 mM EDTA. ED is stable and retains its activity after at least an hour at room temperature (data not shown); therefore room temperature was chosen as the reaction temperature for convenience. Fluorescence intensity was measured at 485/515 nm excitation/emission on a Fluoromax-3 plate reader (Horiba Jobin Yvon).
Miniaturized Eya phosphatase assay for HTS
The OMFP-based Eya2 ED phosphatase assay were miniaturized and optimized in 1536-well black assay plates (Greiner Bio-one). 1.5 μL/well of 200 nM Eya2 ED was incubated with or without 23 nL compound or DMSO control for 10 minutes followed by an addition of 1.5 μL/well of 50 μM OMFP and incubated for 30 minutes. The resulting fluorescence intensity was measured on a Viewlux plate reader (PerkinElmer) with an excitation wavelength of 485 nm and emission of 525 nm. Since there were no Eya2 inhibitors available during the assay development, we used EGTA as a positive control to access the assay quality.
Compound library and instruments for liquid handling
The LOPAC library (Library of Pharmacologically Active Compounds, Sigma-Aldrich) consisting of 1,280 compounds was used for the assay validation. The collection of 331,609 compounds for the primary screen was provided by the NIH's Molecular Library Initiative (http://pubchem.ncbi.nlm.nih.gov/). All compounds were dissolved in DMSO as 10 mM stock solutions. All compounds were serially diluted at 1:5 ratio in DMSO in 384-well plates for 4 concentrations using a CyBi®-Well dispensing station with a 384-well head (Cybio) and then reformatted into 1536-well plates at 7 μL/well. An automated dispensing station (BioRAPTR FRD, Beckman Coulter) was used to dispense reagents into 1536-well plates at volumes of 1-3 μL/well. Compounds were transferred to 1536-well assay plates in 23 nL/well using an automated pin-tool station (Kalypsys®). The purity of all compounds in the library is greater than 98% as analyzed by HPLC. Further analyses by 1H NMR spectroscopy and mass spectrometry confirmed their structural identity. In addition, the bioactivity of 10 mM stock samples recapitulates in the primary assay after several months of storage. One of the key compounds (MLS000544460) has been incubated with pH 4, 7 and 10 buffer and is stable over extended periods of time.
HTS Data analysis
The primary screening data was analyzed as previously described19. IC50 values were calculated from the fluorescence signal intensity using the Prism software (Graphpad Software, Inc.). The Z’ factor index of assay quality control20 was defined as 1-(3*SSR/R), where SSR is the summation of the standard deviation of positive inhibition controls and negative inhibition controls and R is the mean of the positive controls minus the mean of negative controls. All values were expressed as mean +/− SD.
pH2AX-based Eya phosphatase assay
pH2AX phosphatase assays (50 μL) were carried out in transparent, 96-well, half-area microplates (Greiner Bio-One). The assay was performed at pH 6.0 in 50 mM MES, 50 mM NaCl, 5 μM MgCl2, 0.05% BSA, 1 mM DTT and contained 3.9 μM ED and 50 μM pH2AX peptide (KATQASQEpY, Abgent). Because of the lower sensitivity of the malachite green assay compared to the OMFP-based phosphatase assay, a much higher enzyme concentration (3.9 μM) is used to achieve sufficient assay signal/background. Reactions were allowed to proceed for 40 minutes at 37°C and terminated by the addition of 100 μL malachite green solution (Millipore). The free phosphate released from dephosphorylation forms a green complex with malachite green and molybdate that can be monitored using absorbance at 620 nm. After a 20-minute incubation at room temperature, the absorbance was measured using a Spectramax PLUS 384 plate reader (Molecular Devices).
To evaluate the effect of a compound on ED's phosphatase activity, various concentrations of the compound were incubated with ED for 10 minutes prior to the addition of pH2AX that starts the reaction. Dose response curves were generated and IC50 values calculated using Prism.
Phosphatase assays of PTP1B, PPM1A, and Scp1
The phosphatase assay of PTP1B was conducted in 30 mM Tris, pH 7.5, 75 mM NaCl, 1.25 mM MgCl2, 1 mM EDTA, 0.033% BSA, 1 mM DTT using 20 nM PTP1B (R&D Systems) and 100 μM OMFP as the substrate. To evaluate the effect of a compound on PTP1B's phosphatase activity, various concentrations of the compound were incubated with PTP1B for 10 minutes prior to the addition of OMFP to start the reaction. Reactions were in 50 μL volumes and were set up in black, 96-well, half area microplates using an automated liquid handling system (Janus) and bulk liquid dispenser (Biotek). Reactions proceeded for 60 minutes at room temperature followed by the addition of Na3VO4 to terminate the reaction. Fluorescence intensity was measured using an EnVision plate reader (Perkin Elmer).
The phosphatase assay of PPM1A (ProSpec) was conducted in 50 mM Tris, pH 7.5, 0.1 mM EDTA, 0.5 mM DTT, 1.25 mM MgCl2 using 10.7 nM PPM1A and 100 μM OMFP. The effect of compounds on PPM1A's phosphatase activity was evaluated similarly as for PTP1B, except that EDTA was used to terminate the reaction.
The phosphatase assay of Scp1 (a gift from Dr. Jessie Zhang, UT-Austin) was conducted in 50 mM MES, pH 5.5, 25 mM NaCl, 1.25 mM MgCl2 using 120 nM Scp1 and 100 μM OMFP. The effect of compounds on Scp1's phosphatase activity was evaluated similarly as for PTP1B, except that EDTA was used to terminate the reaction.
Eya2 kinetic experiments
Due to the solubility limitations and relatively high Km values of the OMFP substrate, we developed a second fluorogenic Eya2 phosphatase assay using a different substrate, fluorescein diphosphate (FDP, Anaspec), to measure compounds’ effect on enzyme kinetics. All kinetic experiments were performed in 384 black medium binding plates (Greiner Bio-one). Using the FDP substrate we were able to achieve an FDP stock solution of 100 mM in Eya2 ED buffer, therefore allowing us to test Eya2 kinetic experiments at the appropriate substrate versus compound concentrations to calculate an accurate km value and to generate a Lineweaver-Burke plot for Eya2 ED. Briefly, 2.5 μl of compound (MLS000544460) at 100, 40, 20, 10, 5 or 0 μM was incubated with 2.5 μl of 2 μM Eya2 ED for 10 minutes. Subsequently, 5 μl of FDP substrate at 20, 12, 7.2, 4.3, 2.6, 1.6, 1, or 0 mM was added and plates were incubated for an additional 30 minutes before measured on the Viewlux plate reader with excitation wavelength of 485 nm and emission wavelength of 525 nm. Kinetic analysis was performed using GraphPad Prism 4 (GraphPad Software, Inc.).
RESULTS
Known phosphatase inhibitors do not significantly inhibit Eya2's phosphatase activity
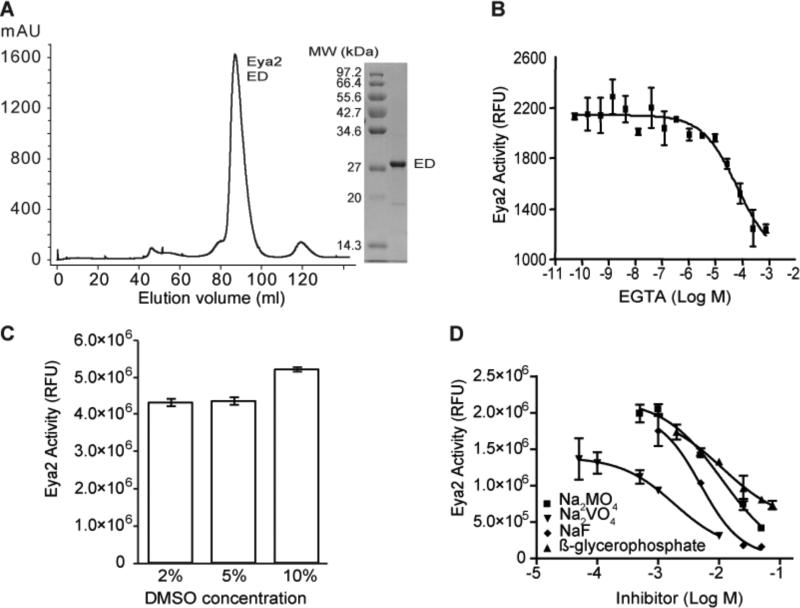
We chose the catalytic domain of human Eya2 (ED), which contains the Tyr phosphatase activity, for our studies. This choice in part is because that the full-length human Eya2 is unstable and cannot be expressed and purified from E. coli. In addition, Eya2 has a N-terminal Ser/Thr phosphatase domain with an unrelated and largely uncharacterized function21. Carrying out the HTS using just the ED avoids complication from this additional phosphatase domain at the N-terminus of Eya2. We expressed the human Eya2 ED in E. coli as a GST-fusion protein. The protein was purified on glutathione resin, cleaved off the GST tag, and further purified using a Superdex-200 column. Eya2 ED elutes as a monomer on the Superdex-200 column at ~30 kDa (Fig. 1A). We can obtain greater than 6 mg purified ED per liter of bacterial culture at a purity of > 99% (Fig 1A).
FIGURE 1.
Purification and characterization of the phosphatase activity of Eya2 ED. (A) Superdex-200 elution profile of Eya2 ED and SDS PAGE of purified Eya2 ED. (B) EGTA effectively inhibits Eya2 phosphatase activity with an IC50 of 65 μM and can be used as a positive control for inhibition. (C) DMSO (up to 10%) has no effect on the phosphatase activity of Eya2 ED. (D) Common phosphatase inhibitors do not significantly inhibit Eya2 ED. Compounds showing inhibition display high IC50 values: 11.3 mM for Na2MnO4, 8.2 mM for β-glycerophosphate, 6.6 mM for NaF, and 1.8 mM for Na3VO4. Okadaic acid, L-phenylalanine, cyclosporine A, 1,10-phenanthroline, and phenylarsine oxide do not show inhibition at concentrations tested.
A phosphatase assay was then developed using purified human Eya2 ED and the small molecule OMFP (3-O-methylfluorescein phosphate) as a substrate. OMFP is dephosphorylated to yield a fluorescent product (3-O-methylfluorescein) that can be detected at 485/515 nm excitation/emission wavelengths. Like other HAD family members, EDs are Mg2+-dependent phosphatases12,13 and EDTA and EGTA can both inhibit the phosphatase activity of ED. Fig 1B demonstrates the inhibition of the ED phosphatase activity by EGTA with an IC50 of 65 μM. Therefore, we used either EGTA or EDTA as positive controls for HTS assay development and screening. Furthermore, DMSO tolerance tests demonstrated that the phosphatase activity of Eya2 ED is not affected by up to 10% DMSO concentration (Fig 1C).
We reasoned that existing phosphatase inhibitors would likely not inhibit the phosphatase activity of ED, because the ED of Eya is a unique HAD phosphatase22 and there are no known specific HAD phosphatase inhibitors. Thus, we tested nine common phosphatase inhibitors against the Eya2 ED including okadaic acid (inhibitor of Ser/Thr phosphatase PP2A), L-phenylalanine (intestinal alkaline phosphatase inhibitor), cyclosporine A (calcineurin inhibitor through binding to cyclophilin), (1, 10)-phenanthroline, phenylarsine oxide (protein tyrosine phosphatase inhibitor), NaF (phospho-serine or threonine inhibitor), Na3VO4 (protein phosphotyrosyl phosphatase inhibitor), Na2MnO4 (protein phosphotyrosyl phosphatase inhibitor), and β-glycerophosphate. Five inhibitors (okadaic acid, L-phenylalanine, cyclosporine A, 1,10-phenanthroline, and phenylarsine oxide) were inactive when tested against ED's phosphatase activity at concentrations that completely inhibit their cognitive phosphatases (data not shown). Four other inhibitors only inhibited ED's phosphatase activity at very high concentrations, with IC50s of 11.3 mM for Na2MnO4, 8.2 mM for beta-glycerophosphate, 6.6 mM for NaF, and 1.8 mM for Na3VO4 (Fig. 1D). These data suggest that known phosphatase inhibitors do not significantly inhibit Eya's phosphatase activity. This, in combination with the functional significance of Eya's phosphatase activity in tumorigenesis, metastasis, and DNA repair, prompted us to carry out a large scale HTS to identify potent and specific inhibitors of the Eya phosphatase activity.
The OMFP-based phosphatase assay is suitable for HTS
The OMFP-based phosphatase assay was optimized and miniaturized in a 1536-well format for adaptation to HTS. The Eya2 enzyme concentration was first titrated in the assay using OMFP as a substrate. The enzyme activity linearly increased with the increase of enzyme concentration (data not shown). To minimize the consumption of enzyme in later large scale primary screening, 100 nM Eya2 concentration was selected that produced sufficient assay signal. The signal-to-basal ratio at this enzyme concentration was ~8 fold, which reproduced in multiple experiments. In the substrate concentration titration experiment, we found that the solubility of the OMFP substrate in the assay buffer was limited and thus the plateau was not reached due to insufficiently high substrate concentrations (data not shown). The OMFP substrate dose response curve was still in the linear portion at the highest available substrate concentration of 1.25 mM. Based on these results, 25 μM OMFP concentration was selected for further experiments that yielded sufficient assay signal window.
The OMFP-based Eya2 phosphatase assay was miniaturized into a 3 μl/well assay in the 1536-well plate format for HTS. A DMSO plate was first used to assess the assay performance. The signal-to-basal ratio was 7.75 and the Z’ factor20 was 0.7 (Fig. 2A), indicating a robust assay that is suitable for HTS. We then carried out a pilot screen using the LOPAC library of 1,280 compounds with pharmacologically known activities (Sigma-Aldrich) as well as a ~2800-compound approved drug set (NIH Chemical Genomics Center (NCGC) collection). Each compound was screened in a seven-concentration titration ranging from 0.12 to 76.6 μM as previously described23. The average signal-to-basal ratio was 7 fold and the Z factor was 0.7 (Fig. 2B), similar to what was observed in the DMSO plate test. After eliminating obvious metal chelators, three compounds were identified as primary hits with a hit selection criteria of IC50 < 35 μM and maximal inhibition > 80 %, (Fig. 2C, D). The hit rate from this test screen was 0.55 %, which is an acceptable rate for HTS. Furthermore, these pilot compounds displayed identical potencies between low throughput and high throughput assay formats (data not shown).
FIGURE 2.
HTS assay optimization for the Eya2 phosphatase assay using OMFP as a substrate. (A) Scattered plot of a DMSO plate test of the Eya2 phosphatase assay in a 1536-well plate. The wells in column 2 contain 1 mM EGTA that was used as a positive control. All other wells contain DMSO as the negative (no inhibition) control. (B) Z’ and signal/background (S/B) values of a pilot screen using the LOPAC library. (C) The pilot screen revealed three compounds that inhibit Eya2 phosphatase activity. (D) Chemical structures of the three compounds in C. (E) Two pilot screen compounds inhibit Eya2 phosphate activity using the phospho-peptide substrate, pH2AX.
A large scale primary screen identified a class of structurally related initial hits
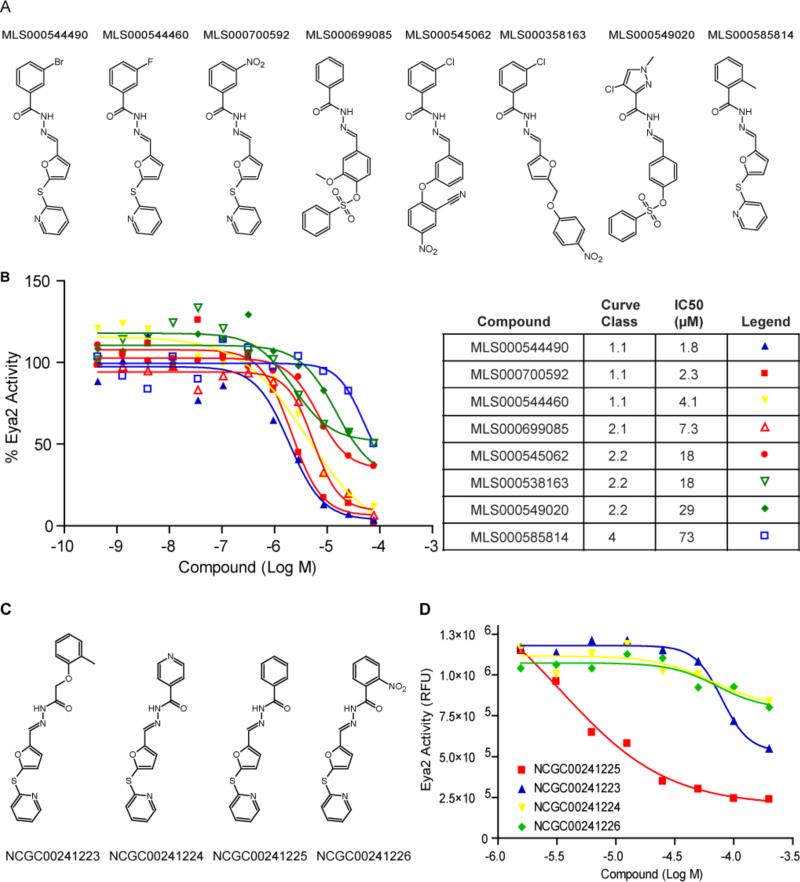
We next carried out a large-scale screen of 334,695 compounds from the MLPCN (Molecular Libraries Probe Centers Network) at four different concentrations ranging from 0.61 to 76.7 μM in a quantitative HTS format23. Dose responses were assigned to different curve classes to evaluate the results of qHTS23. Curve classes 1.1 and 2.1 both demonstrate high compound efficacy with either complete (class 1.1) or partial (class 2.1) dose response curves. Compounds in these two curve classes have the highest chances of reproducing and are typically considered the most promising compounds from a primary screen. A few compounds increased the phosphatase activity in the primary screen and may warrant further investigation as possible activators of the Eya2 phosphatase. In this paper, we decided to focus on compounds that inhibit the Eya2 phosphatase activity, with the hope that these may be promising leads for anti-cancer drugs. The overall hit rate of the primary screen was low and only 3 inhibitory compounds belonged to class 1.1 and 7 belonged to class 2.1 (Table 1). However, all three class 1.1 compounds, one class 2.1 compound, and four compounds from other curve classes, clearly belong to the same structural class characterized by an N-arylidenebenzohydrazide core (Fig. 3A). We decided to focus on this class of compounds for further characterization.
Table 1.
Results of large scale HTS. Curve classes are defined in Inglese et al.23. Compounds in class 1.1 and 2.1 have high efficacy with either complete (1.1) or partial (2.1) dose response curves and are typically considered the most promising compounds from the screen.
| Activity | Distribution | Curve Classification | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.1 | 1.2 | 1.3 | 1.4 | 2.1 | 2.2 | 2.3 | 2.4 | 3 | 4 | 5 | ||
| Activation | Cmpd No | 10 | 0 | 0 | 0 | 119 | 74 | 0 | 36 | 115 | ||
| % library | 0.003% | 0.00% | 0.00% | 0.00% | 0.04% | 0.02% | 0.00% | 0.01% | 0.03% | 329717 | 52 | |
| Inhibition | Cmpd No | 3 | 0 | 0 | 0 | 7 | 497 | 0 | 735 | 3330 | 98.51% | 0.02% |
| % library | 0.001% | 0.00% | 0.00% | 0.00% | 0.002% | 0.15% | 0.00% | 0.22% | 0.99% | |||
FIGURE 3.
A new series of Eya2 ED phosphatase inhibitors were identified in the HTS that inhibit Eya2's phosphatase activity. (A) Structures of compounds that demonstrate inhibitory activity in the large scale HTS. (B) Activity of this series was confirmed in 12-point dose response curves. IC50 values of each compound are listed. (C) Structures of four additional commercially available compounds that have the same N-arylidenebenzohydrazide core. (D) The four compounds tested display varying levels of inhibition towards Eya2 ED.
A 12-point dose response study was then carried out using re-sourced material and validated all eight compounds as inhibitors of ED's phosphatase activity in the OMFP-based phosphatase assay (Fig. 3B). We also carried out the inhibition assay under high Mg2+ (30 μM-1.25 mM) to exclude metal chelators, in the absence of DTT to remove non-specific covalent modifiers, and by adding compounds immediately before plate reading to rule out quenchers that interfere with readout of the assay (data not shown). All eight compounds passed these false positive tests and were thus considered highly promising primary screen hits.
Following our initial analysis, we identified four other commercially available compounds that have the same core structure as this class of initial hits (Fig. 3C). We tested these compounds using the OMFP-based phosphatase assay (Fig. 3D). Compound NCGC00241225 is the most active with an IC50 of 3.5 μM. Compound NCGC00241223 has an IC50 of 79.1 μM while compounds NCGC00241224 and NCGC00241226 are essentially inactive.
The best compounds from this series are active in a pH2AX-based secondary phosphatase assay
To further confirm that this class of compounds are authentic inhibitors of the Eya2 phosphatase, we developed a malachite green-based secondary phosphatase assay using phosphorylated H2AX peptide (pH2AX), a known physiological substrate of the Eya phosphatase15,17. In this assay, inorganic phosphate released from the pH2AX peptide upon dephosphorylation by ED forms a colored complex with malachite green and molybdate, of which the absorbance can be measured at 620 nm. We demonstrate that two of the class 1.1 compounds (another class 1.1 compound was not tested due to limited availability of the compound) and another structurally highly related compound (NCGC00241225) are active in the pH2AX assay with IC50 in the range of 16.8 to 45.2 μM (Fig. 4). Several other compounds with higher IC50s in the OMFP assay (Fig. 3B, D) do not significantly inhibit ED's phosphatase activity in the pH2AX assay (Fig. 4), maintaining the rank order of IC50s observed in the primary OMFP assay (Fig. 3).
FIGURE 4.
A secondary, malachite green based phosphatase assay using a phospho-H2AX peptide as the substrate confirmed the inhibition of Eya2's phosphatase activity by the best compounds of this class.
In addition, we tested the three hits from the pilot screen and two of the compounds (NCGC00181091 and NCGC00093729) are active in the pH2AX-based phosphatase assay with IC50s of 32.0 and 18.3 μM, respectively (Fig. 2E). These compounds provide additional chemical scaffold for future optimization.
The N-arylidenebenzohydrazide compounds do not inhibit other cellular phosphatases
A counter screen was then carried out on the most potent compounds to evaluate their specificity against a number of other cellular phosphatases. The other phosphatases examined include PTP1B, a prototypic protein tyrosine phosphatase that does not require Mg2+ for catalytic activity; PPM1A, a Ser/Thr phosphatase that, like Eya2, requires Mg2+ for catalytic function24; and Scp1, another HAD family protein phosphatase25,26. Scp1 is an Mg2+-dependent phosphatase that, like Eya2, uses a catalytic aspartic acid as the nucleophile in the dephosphorylation reaction. However, Scp1 dephosphorylates phospho-Serine, unlike Eya2, the only known HAD member that dephosphorylates phospho-Tyrosine. We tested the effect of compounds MLS000700592, MLS000544460, and NCGC00241225 on the activity of PTP1B, PPM1A, and Scp1 using an OMFP-based phosphatase assay. The assay conditions for each enzyme were derived from previous reports24-26. This inhibitor series does not significantly inhibit the above phosphatases tested (Fig. 5A-C), demonstrating specificity against the Eya2 phosphatase.
FIGURE 5.
Identified lead compounds specifically inhibit the Eya2 Tyr phosphatase activity, and do not significantly inhibit other cellular phosphatases, including the Mg2+-dependent phosphatase PPM1A (A), protein tyrosine phosphatase 1B (PTP1B) (B), and HAD family member Scp1 (C) in an OMFP-based phosphatase assay.
Enzyme kinetic analyses suggest that the N-arylidenebenzohydrazide compounds are mixed mode inhibitors
We further carried out kinetic competition experiments using the more soluble FDP as a substrate (Fig. 6). The classical Lineweaver-Burke analysis of the inhibitor suggests a mixed mode behavior, between competitive and non-competitive inhibition, indicating that the enzyme is able to partially accommodate the inhibitor and the substrate at the same time.
FIGURE 6.
Enzymatic kinetic experiments indicate that the compounds are mixed-mode inhibitors.
DISCUSSION
Inhibitors of the Eya phosphatase activity can potentially inhibit breast tumorigenesis/metastasis and/or serve as sensitizers for therapeutics that act by inducing DNA damage. Although many phosphatases have been described as attractive targets for drug discovery27,28, the difficulty in obtaining small molecules that inhibit phosphatases in a selective manner is well known and is mostly due to the high degree of similarity between the catalytic domains of the enzymes. The Eya proteins, as novel HAD family phosphatases that target phospho-Tyr, offer us a unique opportunity to potentially identify specific Eya phosphatase inhibitors.
To that end, we developed a fluorescent HTS phosphatase assay using the Eya2 ED and small molecule OMFP as a substrate. It is standard practice to use artificial, small molecule substrates for HTS phosphatase assays since these assays are sensitive, robust, and inexpensive compared to assays using phosphorylated peptide or protein substrates29. The catalytic domain of Eya2 (ED) was chosen over full length protein due to its ease of purification as full length Eya2 is not stable. In addition, using only the ED also avoids the complication of the additional Ser/Thr phosphatase domain at the N-terminus of Eya221. We carried out a qHTS of over 330,000 compounds that allowed for the identification of a series of small molecule inhibitors of Eya2 phosphatases. Analogs within this chemotype ranged from no activity to low micromolar IC50s, indicating inherent SAR within the series. For example, the benzohydrazide substitution was tolerated in the meta position but not the ortho position. The furanyl-2-thio-2-pyridine substituent of the N-arylidene functional group provided best activity although other benzylidene substituents seemed to be tolerated.
Furthermore, we carried out an orthogonal malachite green-based phosphatase assay monitoring phosphate release from the pH2AX peptide substrate, which confirmed the qHTS results. There is a reduction of IC50 values in the pH2AX assay compared to the OMFP assay, which may be a consequence of the different substrate used in the assays. Our specificity assays showed that these inhibitors do not significantly inhibit several other cellular phosphatases, including a prototypic protein tyrosine phosphatase PTP1B, an Mg2+-dependent Ser/Thr phosphatase PPM1A, and a Ser/Thr HAD family protein phosphatase Scp1. Kinetic studies reveal that the nature of this inhibition may be mixed mode. We plan to further evaluate whether these compounds inhibit the transformation, migration, and invasion properties of breast cancer cells and whether these compounds inhibit DNA repair mediated by H2AX under DNA damaging conditions. We also plan to explore its binding mechanism and carry out structure activity relationship studies of this series and further optimize these compounds to improve potency and specificity.
It is worth noting that Park et al. recently carried out a virtual screening using the crystal structure of the Eya2 ED and identified 7 compounds with IC50 ranging from 4.4 to 66.2 μM in a fluorescent phosphatase assay using small molecule 6,8-difluoro-4-methylumberlliferyl phosphate (DIFMUP) as a substrate30. These compounds were proposed to bind to the active site of Eya and chelate the active site Mg2+, although the specificity of these compounds against Eya has not been assessed. In general, efforts from our laboratories and others devoted to identifying potent and specific inhibitors of the Eya phosphatase may generate chemical probes to study the function of Eya phosphatase and one day lead to a new approach in the treatment of cancer.
ACKNOWLEDGEMENTS
We would like to thank Sam Michael and Paul Shinn for assistances on robotic screen and compound management, Jolene Ramsey and Daniel Zheng for technical assistance, Dr. Anton Simeonov for critical reading of the manuscript and suggestions, the Biophysics Core Facility at University of Colorado School of Medicine for use of the fluorometer, Dr. Robert Hodges (University of Colorado School of Medicine) for the use of the Spectramax PLUS 384 plate reader, the University of Colorado Skaggs School of Pharmacy and Pharmaceutical Sciences High Throughput and High Content Screening Core for use of liquid handling robots and EnVision plate reader, Dr. Jessie Zhang (UT-Austin) for the gift of Scp1 protein used in this study, and Debra Madden for giving us perspective on the relevant issues to breast cancer patients and for participating in this project. This research was supported by the Molecular Libraries Initiative of the NIH Roadmap for Medical Research, National Institutes of Health, through a grant from the NIH (R03DA030559) to R.Z. and H.L.F. Additional support for this work comes from a Department of Defense Synergistic Idea Award (BC084105), an AACR/BCRF (American Association for Cancer Research and Breast Cancer Research Foundation) grant, a State of Colorado Proof of Concept grant, and a Colorado Bioscience Discovery and Evaluation Grant to R.Z. and H.L.F. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Christensen KL, Patrick AN, McCoy EL, Ford HL. The six family of homeobox genes in development and cancer. Adv Cancer Res. 2008;101:93–126. doi: 10.1016/S0065-230X(08)00405-3. [DOI] [PubMed] [Google Scholar]
- 2.Behbakht K, Qamar L, Aldridge CS, Coletta RD, Davidson SA, Thorburn A, Ford HL. Six1 overexpression in ovarian carcinoma causes resistance to TRAIL-mediated apoptosis and is associated with poor survival. Cancer Res. 2007;67:3036–3042. doi: 10.1158/0008-5472.CAN-06-3755. [DOI] [PubMed] [Google Scholar]
- 3.McCoy EL, Iwanaga R, Jedlicka P, Abbey NS, Chodosh LA, Heichman KA, Welm AL, Ford HL. Six1 expands the mouse mammary epithelial stem/progenitor cell pool and induces mammary tumors that undergo epithelial-mesenchymal transition. J Clin Invest. 2009;119:2663–2677. doi: 10.1172/JCI37691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Micalizzi DS, Christensen KL, Jedlicka P, Coletta RD, Baron AE, Harrell JC, Horwitz KB, Billheimer D, Heichman KA, Welm AL, Schiemann WP, Ford HL. The Six1 homeoprotein induces human mammary carcinoma cells to undergo epithelial-mesenchymal transition and metastasis in mice through increasing TGF-beta signaling. J Clin Invest. 2009;119:2678–2690. doi: 10.1172/JCI37815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng KTP, Lee TKW, Cheng Q, Wo JYH, Sun CKW, Guo D-Y, Lim ZX, Lo C-M, Poon RTP, Fan S-T, Man K. Suppression of tumorigenesis and metastasis of hepatocellular carcinoma by shRNA interference targeting on homeoprotein Six1. International Journal of Cancer. 2010;127:859–872. doi: 10.1002/ijc.25105. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Knosp BM, Maconochie M, Friedman RA, Smith RJ. A comparative study of Eya1 and Eya4 protein function and its implication in branchio-oto-renal syndrome and DFNA10. J Assoc Res Otolaryngol. 2004;5:295–304. doi: 10.1007/s10162-004-4044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruf RG, Xu PX, Silvius D, Otto EA, Beekmann F, Muerb UT, Kumar S, Neuhaus TJ, Kemper MJ, Raymond RM, Jr., Brophy PD, Berkman J, Gattas M, Hyland V, Ruf EM, Schwartz C, Chang EH, Smith RJ, Stratakis CA, Weil D, Petit C, Hildebrandt F. SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proc Natl Acad Sci U S A. 2004;101:8090–8095. doi: 10.1073/pnas.0308475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pandey RN, Rani R, Yeo EJ, Spencer M, Hu S, Lang RA, Hegde RS. The Eyes Absent phosphatase-transactivator proteins promote proliferation, transformation, migration, and invasion of tumor cells. Oncogene. 2010;29:3715–3722. doi: 10.1038/onc.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farabaugh SM, Micalizzi DS, Jedlicka P, Zhao R, Ford HL. Eya2 is required to mediate the pro-metastatic functions of Six1 via the induction of TGF-beta signaling, epithelial-mesenchymal transition, and cancer stem cell properties. Oncogene. 2011 doi: 10.1038/onc.2011.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, Jatkoe T, Berns EM, Atkins D, Foekens JA. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 11.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 12.Rayapureddi JP, Kattamuri C, Steinmetz BD, Frankfort BJ, Ostrin EJ, Mardon G, Hegde RS. Eyes absent represents a class of protein tyrosine phosphatases. Nature. 2003;426:295–298. doi: 10.1038/nature02093. [DOI] [PubMed] [Google Scholar]
- 13.Tootle TL, Silver SJ, Davies EL, Newman V, Latek RR, Mills IA, Selengut JD, Parlikar BE, Rebay I. The transcription factor Eyes absent is a protein tyrosine phosphatase. Nature. 2003;426:299–302. doi: 10.1038/nature02097. [DOI] [PubMed] [Google Scholar]
- 14.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 15.Krishnan N, Jeong DG, Jung SK, Ryu SE, Xiao A, Allis CD, Kim SJ, Tonks NK. Dephosphorylation of the C-terminal tyrosyl residue of the DNA damage-related histone H2A.X is mediated by the protein phosphatase eyes absent. J Biol Chem. 2009;284:16066–16070. doi: 10.1074/jbc.C900032200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jemc J, Rebay I. Identification of transcriptional targets of the dual-function transcription factor/phosphatase eyes absent. Dev Biol. 2007;310:416–429. doi: 10.1016/j.ydbio.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cook PJ, Ju BG, Telese F, Wang X, Glass CK, Rosenfeld MG. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature. 2009;458:591–596. doi: 10.1038/nature07849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernier J, Hall EJ, Giaccia A. Radiation oncology: a century of achievements. Nat Rev Cancer. 2004;4:737–747. doi: 10.1038/nrc1451. [DOI] [PubMed] [Google Scholar]
- 19.Southall NT, Jadhav A, Huang R, Nguyen T, Wang Y. Enabling the Large Scale Analysis of Quantitative High Throughput Screening Data. In: Seethala R, Zhang L, editors. Handbook of drug screening. Informa Healthcare; New York: 2009. pp. 442–462. [Google Scholar]
- 20.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 21.Okabe Y, Sano T, Nagata S. Regulation of the innate immune response by threonine-phosphatase of Eyes absent. Nature. 2009;460:520–524. doi: 10.1038/nature08138. [DOI] [PubMed] [Google Scholar]
- 22.Jung SK, Jeong DG, Chung SJ, Kim JH, Park BC, Tonks NK, Ryu SE, Kim SJ. Crystal structure of ED-Eya2: insight into dual roles as a protein tyrosine phosphatase and a transcription factor. FASEB J. 2010;24:560–569. doi: 10.1096/fj.09-143891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, Zheng W, Austin CP. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci U S A. 2006;103:11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marley AE, Sullivan JE, Carling D, Abbott WM, Smith GJ, Taylor IW, Carey F, Beri RK. Biochemical characterization and deletion analysis of recombinant human protein phosphatase 2C alpha. The Biochemical journal. 1996;320( Pt 3):801–806. doi: 10.1042/bj3200801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosh A, Shuman S, Lima CD. The structure of Fcp1, an essential RNA polymerase II CTD phosphatase. Mol Cell. 2008;32:478–490. doi: 10.1016/j.molcel.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang M, Liu J, Kim Y, Dixon JE, Pfaff SL, Gill GN, Noel JP, Zhang Y. Structural and functional analysis of the phosphoryl transfer reaction mediated by the human small C-terminal domain phosphatase, Scp1. Protein science : a publication of the Protein Society. 2010;19:974–986. doi: 10.1002/pro.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jailkhani N, Chaudhri VK, Rao KV. Regulatory cascades of protein phosphatases: implications for cancer treatment. Anti-cancer agents in medicinal chemistry. 2011;11:64–77. doi: 10.2174/187152011794941253. [DOI] [PubMed] [Google Scholar]
- 28.Pereira SR, Vasconcelos VM, Antunes A. The phosphoprotein phosphatase family of Ser/Thr phosphatases as principal targets of naturally occurring toxins. Critical reviews in toxicology. 2011;41:83–110. doi: 10.3109/10408444.2010.515564. [DOI] [PubMed] [Google Scholar]
- 29.Tierno MB, Johnston PA, Foster C, Skoko JJ, Shinde SN, Shun TY, Lazo JS. Development and optimization of high-throughput in vitro protein phosphatase screening assays. Nat Protoc. 2007;2:1134–1144. doi: 10.1038/nprot.2007.155. [DOI] [PubMed] [Google Scholar]
- 30.Park H, Jung SK, Yu KR, Kim JH, Kim YS, Ko JH, Park BC, SJ. K. Structure-Based Virtual Screening Approach to the Discovery of Novel Inhibitors of Eyes Absent 2 Phosphatase with Various Metal Chelating Moieties. Chem Biol Drug Des. 2011;78:642–650. doi: 10.1111/j.1747-0285.2011.01192.x. [DOI] [PubMed] [Google Scholar]