Abstract
Prenatal detection of chromosome abnormalities has been offered for more than 40 years, first by amniocentesis in the early 1970s and additionally by chorionic villus sampling (CVS) in the early 1980s. Given the well-recognized association between increasing maternal age and trisomy,1–3 the primary utilization of prenatal testing has been by older mothers. This has drastically reduced the incidence of aneuploid children born to older mothers.4 Although younger women have relatively low risks of conceiving a child with aneuploidy, the majority of pregnant women are in their late teens, 20s, and early 30s. As such, most viable aneuploid babies are born to these younger mothers.5 Invasive prenatal diagnosis (CVS and amniocentesis) is not a feasible option for all low-risk mothers, as these procedures carry a small but finite risk and would ultimately cause more miscarriages than they would detect aneuploidy. For this reason, a number of noninvasive tests have been developed—including first-trimester risk assessment at 11 to 14 weeks, maternal serum analyte (quad) screening at 15 to 20 weeks, and sonographic fetal structural survey at 18 to 22 weeks—all of which are designed to give a woman an adjusted (more accurate) estimate of having an aneuploid fetus using as baseline her a priori age-related risk. Ultrasound and maternal serum analysis are considered screening procedures and both require follow up by CVS or amniocentesis in screen-positive cases for a definitive diagnosis of a chromosome abnormality in the fetus. The ability to isolate fetal cells and fetal DNA from maternal blood during pregnancy has opened up exciting opportunities for improved noninvasive prenatal testing (NIPT). Direct analysis of fetal cells from maternal circulation has been challenging given the scarcity of fetal cells in maternal blood (1:10,000–1:1,000,000) and the focus has shifted to the analysis of cell-free fetal DNA, which is found at a concentration almost 25 times higher than that available from nucleated blood cells extracted from a similar volume of whole maternal blood. There have now been numerous reports on the use of cell-free DNA (cfDNA) for NIPT for chromosomal aneuploidies—especially trisomy (an extra copy of a chromosome) or monosomy (a missing chromosome)—and a number of commercial products are already being marketed for this indication. This article reviews the various techniques being used to analyze cell-free DNA in the maternal circulation for the prenatal detection of chromosome abnormalities and the evidence in support of each. A number of areas of ongoing controversy are addressed, including the timing of maternal blood sampling, the need for genetic counseling, and the use of confirmatory invasive testing. Future applications for this technology are also reviewed.
Key words: Noninvasive prenatal testing, Prenatal diagnosis, Cell-free fetal DNA, Trisomy 21
In 1959, Lejeune and colleagues6 demonstrated that Down syndrome is caused by an extra copy of chromosome 21 (trisomy 21 [T21]). This finding was of great clinical importance as Down syndrome is the single most common cause of mental retardation and has the highest incidence at birth of any chromosome abnormality.7 Children with Down syndrome have an increased risk for congenital defects and infectious morbidity, and all have some degree of mental retardation, with a mean intelligence quotient of 24. They also have a significantly shortened life span, with a 10- to 20-fold increased risk for leukemia and an increased risk for early-onset dementia. Although most fetuses with chromosomal aneuploidies are nonviable and lead to early miscarriage,8 a handful have the potential to survive to the newborn period. In some cases, as with trisomy 13 (T13) and trisomy 18 (T18), they are associated with significant clinical morbidity and a high rate of mortality shortly after birth. For all these reasons, considerable effort has been expended over the years to identify such fetuses early in pregnancy in order to provide couples with sufficient time to consider their reproductive options. Although patients may choose to decline prenatal testing, a discussion of the various options available for prenatal diagnosis is now considered a standard of care for all pregnant women in developed countries. Options currently recommended by both the American Congress of Obstetricians and Gynecologists (ACOG)9 and the American College of Medical Genetics (ACMG)10 include diagnostic testing, which requires that fetal cells be harvested using one of two invasive procedures (CVS or amniocentesis); or noninvasive screening, which uses a combination of first trimester risk assessment (FTRA) at 11 to 14 weeks and/or maternal serum analyte (quad) screening at 15 to 20 weeks and/or a sonographic fetal structural survey at 18 to 22 weeks to adjust a woman’s a priori risk of having an aneuploid fetus based on her age. The diagnostic accuracy of karyotyping cultured cells obtained by invasive testing has been found to be 97.5% to 99.8%.11–15 However, invasive testing also carries a risk for procedure-related miscarriage.16,17
Aneuploidy screening by ultrasound and/or analysis of various maternal serum biochemical markers is primarily targeted at detecting Down syndrome and, to a lesser extent, T18; it has reported detection rates of 75% to 96% (depending on the screening approach utilized) with false-positive rates ranging from 5% to 10%.9,10,18–29 In addition, ultrasound and maternal serum analysis are considered screening procedures and both require follow-up by CVS or amniocentesis in screen-positive cases for a definitive diagnosis of a chromosome abnormality in the fetus.
The presence of fetal cells in maternal blood was initially reported in 1969,30 and the possibility that these cells could be isolated during pregnancy generated an exciting new noninvasive approach for identifying fetal genetic abnormalities. Several fetal cell types have been reported to exist in the maternal circulation, including fetal trophoblasts, lymphocytes, granulocytes, nucleated erythrocytes, and platelets. Fetal erythroid cells are the most commonly studied cell type as the existence of erythroid progenitors in adult blood is scarce in comparison with its quantitative constitution in fetal blood.31 Nucleated erythrocytes are also fairly well differentiated and likely to have a limited life span in the maternal circulation.32 The detection of certain fetal aneuploidies and triploidy from a maternal blood sample has been accomplished using fluorescence in situ hybridization (FISH) for chromosome-specific DNA probes following various sorting and enrichment procedures.33–37 However, the inconsistent and low yield of fetal cells retrieved after sorting and enrichment has prevented the transition of this approach to routine clinical practice. Although a few commercial entities continue to persevere with aneuploidy testing of intact fetal cells, primarily utilizing newer molecular techniques, the focus of NIPT has shifted toward aneuploidy analysis of cfDNA.
In 1997, Lo and colleagues38 showed that cfDNA could be reliably detected in the maternal circulation during pregnancy. A year later, the same group reported that a surprisingly high mean concentration of fetal DNA (3.4%–6.2%) can be found in total maternal plasma DNA.39 This translates to approximately 20- to 25-times greater concentrations than that in the cellular fraction of maternal blood at the same gestational stage.40 Fetal DNA can also be detected in as little as 10 μL of maternal plasma and serum, in amounts significantly higher than that available from nucleated blood cells extracted from a similar volume of whole blood.39 The approach of using cfDNA instead of fetal cells provided a far easier, less labor intensive, and less time consuming way to work with fetal DNA derived from the maternal circulation, and this opened up new opportunities for NIPT. Since that time, there have been numerous reports on the use of cfDNA for NIPT for fetal chromosome aneuploidies—primarily for T21, but also for T18 (Edwards syndrome), T13 (Patau syndrome), and sex chromosome anomalies—and a number of commercial products are already being marketed for this indication.
Cell/DNA Trafficking in Pregnancy
In his landmark 1953 publication designed to explain immunologic tolerance during pregnancy (and for which he was subsequently awarded the Nobel Prize in 1960), Dr. Peter Brian Medawar proposed that there may be a “true anatomic barrier between the mother and the fetus.”41 He proved to be wrong. Indeed, throughout pregnancy, fetal cells are constantly trafficking across the placenta into the maternal circulation and vice versa42; some of these are undoubtedly fetal stem cells. As such, every woman who has ever been pregnant can be regarded as being a recipient of a stem cell “transplant,” and there is a burgeoning literature on the effect that such microchimerism may have on chronic disease states in women.43 It is estimated that, after the first trimester, there is approximately one fetal cell in the maternal circulation for every 10,000 to 1 million maternal cells. This translates to approximately 20 fetal cells in 20 mL of maternal blood.44 Given their scarcity, efforts to date to isolate and purify these fetal cells for subsequent analysis have been largely unsuccessful38,45–48; for this reason, attention has turned to cfDNA.
The presence of cfDNA in the circulation was documented in adult serum as early as 1947 (see Swaminathan and Butt49 for a review). It likely comes from lysis of cells within the circulation and from physiologic apoptosis occurring within various organ systems, and an extensive scavenging system exists to remove this DNA from the circulation. In 1997, Lo and colleagues38 reported the presence of cfDNA from the Y chromosome of male fetuses in the maternal plasma during pregnancy. It is present as early as 5 to 7 weeks of gestation, released continually by apoptotic cells throughout pregnancy, and is typically cleared from circulation within a matter of hours.48 It is now known that 3% to 10% of the cfDNA in the maternal circulation during pregnancy comes from the fetoplacental unit, most of which is shed from the placenta. Unfortunately, the absolute amount of fetal cfDNA is very small—typically less than 1 μg in 20 mL of whole blood44,50–52—and reliably separating fetal cfDNA from maternal cfDNA is not technically feasible at this time. A number of companies have been spearheading the effort to develop a reliable and accurate commercial NIPT for fetal aneuploidy detection. All analyze the full cfDNA complement in the maternal blood without extracting or enriching the fetal fraction. Although the precise technology used by each of these companies varies, they all rely on the same premise: that existing high-throughput approaches to DNA sequencing coupled with sophisticated data analysis will be able to detect abnormal amounts of chromosome-specific cfDNA in the maternal circulation in those pregnancies with fetal aneuploidy. The expectation is that one or all of these tests will prove to be superior to the currently available ultrasound/biochemical noninvasive tests and may eventually replace invasive prenatal testing altogether.
Current Recommendations for Prenatal Diagnosis of Fetal Aneuploidy
A discussion of the various options available for prenatal diagnosis is now considered a standard of care for all pregnant women in developed countries.9,10,48 Several options are discussed below.
Diagnostic Testing
This approach requires the direct harvesting of fetal cells during pregnancy for subsequent karyotype and/or genetic analysis. Two invasive procedures are commonly used.
CVS, which involves a biopsy of placental cells, can be performed either transcervically or transabdominally. The major benefit of CVS is that it can be performed early in the pregnancy, typically between 10 and 13 weeks of gestation. Preliminary results may be obtained in 1 to 2 days if a short-term direct culture of actively dividing villous cytotrophoblastic tissue is initiated, but in most cases a final karyotype result is issued within 7 to 10 days following long-term culture of mesenchymal cells from the villi. The disadvantage of CVS lies primarily in the source of cells being analyzed (the cells come from the trophectoderm and not the fetus itself), which can lead, in a small number of instances (1%–2% of cases), to diagnostic ambiguity in the setting of confined placental mosaicism.12,53,54 In addition, the procedure-related pregnancy loss rate following CVS may be as high as 1%.10,48,55–57
Amniocentesis, on the other hand, involves placing a needle directly into the uterine cavity and aspirating some amniotic fluid containing fetal cells (amniocytes), which typically originate from fetal urine, pulmonary secretions, and skin. These amniocytes are cultured, harvested, and subjected to karyotype and/or genetic analysis. Compared with CVS, the cells primarily derive from the fetus itself, and the procedure-related pregnancy loss rate is generally regarded as being lower than for CVS (quoted as 1:200 to 1:600).9,10,55–58 Amniocentesis is usually only offered after 15 weeks (earlier amniocentesis is associated with higher pregnancy loss rates)55,59 and fewer viable cells are retrieved, precipitating long culture times and consequently a longer turnaround time (typically 8–14 days).
FISH, using chromosome-specific fluorescence-tagged probes, has provided a rapid way to detect the most common fetal aneuploidies found at the time of prenatal testing. The prenatal FISH panel typically targets chromosomes 13, 18, 21, X, and Y, and results from FISH analysis of interphase nuclei derived from chorionic villi or amniocytes may be available within 24 to 48 hours. The sensitivity and specificity of prenatal FISH for detection of all aneuploidies targeted by the typical FISH chromosome panel is > 99.6% and > 99.98%, respectively, 60 and a large multicenter retrospective study demonstrated an extremely high concordance rate (99.8%) between FISH on interphase amniocytes and standard cytogenetic analysis for the specific chromosome abnormalities that the FISH panel is designed to detect.60 Although prenatal FISH provides a swift way to diagnose common fetal aneuploidies, its diagnostic scope has reduced sensitivity compared with conventional cytogenetic analysis; that is, cases with cytogenetic abnormalities other than the most frequent ones (eg, translocations, inversions, markers) will not be identified by this technique. Furthermore, aneuploidy due to familial Robertsonian translocations cannot be identified by FISH because this requires visual inspection of the G-banded karyotype. For these reasons, FISH results should always be followed by routine chromosome analysis for a complete cytogenetic evaluation of the fetus.
Chromosomal microarray analysis (CMA) has provided a way to detect submicroscopic imbalances that remain undetected by conventional cytogenetic analysis. Unlike FISH, CMA permits high-resolution assessment of the entire genome. CMA can either be performed using array-comparative genomic hybridization (aCGH) or single nucleotide polymorphism (SNP) oligonucleotide microarray analysis (SOMA). Although both techniques offer comparable detection of copy number imbalances (also referred to as copy number variances), SOMA is able to utilize the SNP genotype information to detect triploidy, uniparental isodisomy, and consanguinity. SOMA can also assess zygosity in multiple gestations and, if a maternal sample is concurrently run, SOMA can detect maternal cell contamination. The recent Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) Multicenter Prenatal Microarray Study indicated 100% concordance between CMA (aCGH and SOMA) and conventional cytogenetic analysis for the detection of nonmosaic common aneuploidies.61 In this cohort, CMA detected clinically relevant genomic imbalances in an additional 2.5% of cases that had a normal karyotype by G-band analysis.61
Noninvasive Testing
Noninvasive testing grew out of a desire to avoid direct contact with the growing fetus/placenta and concomitantly risking the health of the fetus. NIPT refers specifically to techniques that evaluate fetal cells or cfDNA in a blood sample drawn from the mother during pregnancy. However, for the purposes of this article, it applies to any form of NIPT for fetal aneuploidy. Importantly, it is a screening test, not a diagnostic test. This is a critical distinction and should be explained carefully to every patient during prenatal counseling. A screening test will determine if an individual patient is at high or low risk of having an aneuploid fetus; it does not confirm or refute the diagnosis. If a patient is assessed as high-risk on the basis of her NIPT results (typically regarded as a risk > 1:200) or if she is not satisfied with a simple risk estimate, then she may choose to proceed with invasive testing (CVS or amniocentesis). Such tests are diagnostic in that, with very rare exception (such as mixing up samples in the laboratory and low level mosaicism), they can definitively confirm or refute the presence of a chromosome abnormality.11–15
Prenatal screening has been defined as “the identification, among apparently normal pregnancies, of those at sufficient risk for a specific fetal disorder to justify subsequent invasive and/or costly prenatal diagnostic tests or procedures.”62 The objective is to develop a test with a high detection rate (ideally > 95%) and low false-positive rate (< 1%). Several NIPT options are currently approved and recommended by both ACOG9 and ACMG.10 Current NIPTs perform just as well in women over the age of 35 (advanced maternal age) as they do in younger women; as such, it is no longer necessary or appropriate to counsel women of advanced maternal age to proceed directly to amniocentesis.9,10,48
FTRA is typically done at 11 to 14 weeks of gestation or, more correctly, at a crown-rump length of 45 to 84 mm.9,10,20,22,24,26,28 It involves two elements, both of which are required to accurately interpret the test: the sonographic nuchal translucency, and maternal serum biochemical testing for two analytes (β-human chorionic gonadotropin [β-hCG] and pregnancy-associated plasma protein-A). Based on these results, as well as her age and a few other minor variables (such as body mass index and singleton or twin pregnancy), the patient can be given an adjusted risk that she may be carrying an aneuploid fetus.
Maternal serum analyte (quad) screening can be offered at 15 to 20 weeks of gestation.9,10,18,20,25,26 It involves measuring circulating levels of four biomarkers in the mother’s blood: maternal serum alpha-fetoprotein, β-hCG, unconjugated estriol, and inhibin-A. Again, based on these results, as well as her age, the patient can be given an adjusted risk that she may be carrying an aneuploid fetus. Such testing has been validated for twins (although the detection rate is lower21), but not for triplets or other higher-order multiple pregnancies.
FTRA can be combined with the quad screen, but this should only be done using an integrated algorithm that does not include both β-hCG measurements.10,23,27,29,56,63,64 A single result should then be given to the patient once the entire integrated screen is completed. Independent assessment of T21 risk followed by second trimester screening (serial testing) is generally discouraged because of the high false-positive rates associated with this approach.20,65
The sonographic fetal structural survey is not part of the second trimester quad screen for fetal aneuploidy. That said, it is commonly offered to women at 18 to 20 weeks of gestation, and allows the obstetric care provider to screen the fetus for two reasons: to identify the presence or absence of a major structural defect, such as a cardiac defect or neural tube defect, which may or may not be associated with a chromosomal abnormality; and to identify the presence or absence of so-called soft markers of fetal aneuploidy. These soft markers are not structural abnormalities per se, and include such findings as short femurs, thickened nuchal fold, renal pyelectasis, or echogenic bowel, but may suggest the presence of an underlying fetal aneuploidy. This is especially true when several of these sonographic markers are seen together. Absence of the fetal nasal bone has been shown to be both sensitive and specific in identifying fetuses with T21.66,67 However, overall, aneuploidy detection by ultrasound is not especially sensitive, and is dependent in large part on the skill and experience of the sonographer. In one landmark study of 15,000 women screened by routine ultrasonography at two separate intervals during pregnancy, only 35% of fetal anatomic abnormalities were detected, and only 17% of these were detected prior to 24 weeks of gestation.68 Moreover, because 40% to 50% of fetuses with T21 have no abnormalities on ultrasound, these sonographic markers have a low sensitivity and specificity for detecting T21.69–71 As such, ultrasonography is not recommended as a primary screening tool for T21.9,10 Although rare (1:20,000 to 1:40,000 pregnancies), ultrasound is more sensitive when it comes to the diagnosis of T18 and T13, because the majority of these fetuses have major structural anomalies with or without fetal growth restriction.69
Weaknesses of Current NIPT Options
Obtaining a fetal genetic test result early in the pregnancy facilitates early reproductive decision making. In this regard, FTRA has a significant advantage over second trimester maternal serum quad screening or integrated screening. However, FTRA requires both a blood test and an ultrasound, which typically entails two prenatal visits. Although these noninvasive screening tests are safe for the pregnancy, they are primarily targeted at detecting T21 (and to a lesser extent T18) and they have poor accuracy with false-negative rates between 12% and 23% and false-positive rates between 1.9% and 5.2%.9,10,18–29,63–65 The performance of these tests for the detection of T21 is summarized in Table 1.
Table 1.
Performance Parameters of Noninvasive Screening Tests for Fetal Trisomy 21
| Risk Factor/Biomarkera | GA (wks) | Sensitivity (%) | Screen Positive (%) | Specificity | DRb (for a 5% FPR) (%) | FPRc (for an 85% DR) (%) | Study |
|---|---|---|---|---|---|---|---|
| Age ≥ 35 y | — | 30 | 5 | 1 in 100 | 30–50 | 48–55 | ACOG,9 Driscoll DA et al,10 Haddow JE et al,19 Wald NJ et al,25 Wald NJ et al,26 Wapner R et al.28 |
| First trimester risk assessment | 11–14 | 82–87 | 5 | 1 in 25 | 79–90 | 5–9 | ACOG,9 Driscoll DA et al,10 Malone FD et al,20 Nicolaides KH,22 Spencer K et al,24 Wald NJ et al,26 Wapner R et al.28 |
| Second trimester serum analyte (quad) screen | 15–20 | 80 | 5 | 1 in 40 | 75–85 (only ∼ 50% for twins) | 5–10 | ACOG,9 Driscoll DA et al,10 Benn PA et al,18 Malone FD et al,20 Neveux LM et al,21 Wald NJ et al,26 Wald NJ et al.27 |
| Integrated screen | 11–20 | 94–97 | 4–5 | 1 in 30 | 90–96 | 1–5 | Driscoll DA et al,10 Malone FD et al,20 Platt LD et al,23 Wald NJ et al,26 Wald NJ et al,27 Wright D et al,29 Cuckle H et al,63 Palomaki GE et al.64 |
| Cell-free DNA — MPSS | > 10 | 98.6–100 | — | 97.9–99.8 | — | — | Fan HC et al,72 Palomaki GE et al,73 Fan and Quake,74 Chiu RW et al,76 Bianchi DW et al,77 Palomaki GE et al,78 Chen EZ et al,79 Sehnert AJ et al,84 Ehrich M et al,100 Liao GJ et al.101 |
| — Targeted sequencing + DANSR 1 FORTE | > 10 | 100 | — | 99.7–100 | — | — | Sparks AB et al,75 Sparks AB et al,80 Ashoor G et al.86 |
| — Targeted sequencing + PS | > 9 | 100 | — | 99.8–100 | — | — | Zimmermann B et al,85 Nicolaides KH et al.92 |
a All serum biomarker results include maternal age
b DR refers to the percentage of affected pregnancies (those with a Down syndrome fetus) called screen-positive by the test. In this table, DR data are reported for a fixed 5% FPR. cFPR refers to the percentage of unaffected (normal) pregnancies called screen-positive by the test. In this table, FPR data are reported for a fixed 85% DR. DANSR, Digital Analysis of Selected Regions; DR, detection rate; FORTE, Fetalfraction Optimized Risk of Trisomy Evaluation; FPR, false-positive rate; GA, gestational age; MPSS, massively parallel shotgun sequencing; PS, Parental Support™ (Natera; San Carlos, CA).
Next-Generation NIPT Using cfDNA
Given these weaknesses, several companies have focused on the analysis of cfDNA in a sample of maternal blood collected in the first trimester to develop a more accurate and reliable NIPT. There are currently two primary nextgeneration sequencing approaches for gathering genetic data from cfDNA. The first, massively parallel shotgun sequencing (MPSS), sequences DNA fragments from the whole genome, whereas the second, targeted sequencing, selectively sequences specific genomic regions of interest.
MPSS and Counting
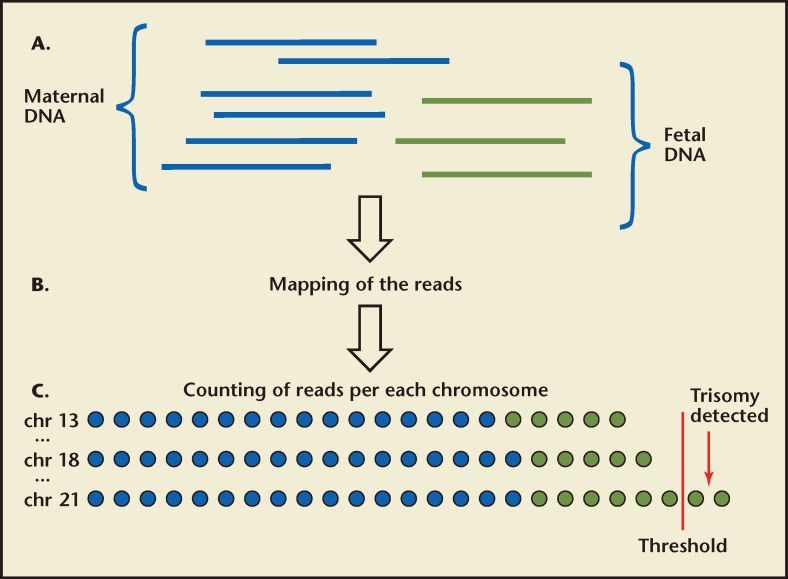
MPSS is a high-throughput technique that uses miniaturized platforms for sequencing large numbers of small DNA sequences called reads from the entire genome. This approach allows for tens of millions of short-sequence DNA tags or fragments (typically 25–36 bp in length) to be sequenced rapidly and simultaneously in a single run. After sequencing the cfDNA present in the maternal plasma, the chromosomal origin of each 25- to 36-bp DNA fragment is obtained by comparison of the sequence data from each DNA fragment with a euploid reference copy of the human genome. Fragments are categorized by chromosome (these include maternal and fetal DNA) and the number of reads mapping to the chromosomes of interest are compared with the number of reads mapping to one or more presumably normal reference chromosomes. This procedure is referred to as counting. If the amount of a chromosome-specific sequence exceeds the threshold that represents a normal (disomic) chromosome, the result is reported as positive for trisomy for that chromosome (Figure 1). A trisomic fetus has 50% more genetic material because of the extra chromosome (3 copies), resulting in an increase in the relative amount of cfDNA from the affected chromosome found in the maternal plasma. It is precisely this difference that the test attempts to detect. This difference is quantitative, not qualitative. In other words, no effort is made to distinguish maternal from fetal DNA. Because maternal DNA is the majority of cfDNA sample, the incremental difference due to fetal trisomy is very small when maternal and fetal DNA measurements are combined. This means that the ability to detect the increased chromosomal dosage resulting from fetal aneuploidy is directly related to the fraction of fetal cfDNA in the maternal circulation. For example, chromosome 21 represents approximately 1.5% of the total genome and an extra copy (T21) would increase the amount of DNA from chromosome 21 from the expected 1.5% to 2.25% (a 50% increase). If the fraction of fetal cfDNA in the maternal circulation is 10%, the relative change in the total cfDNA sample would only increase from 1.5% to 1.575% [1.5% (copy # = 2) × 0.9 (90% maternal)] + [2.25% (copy # = 3) × 0.1 (10% fetal) = 1.575%]. At lower fetal fractions, the increase becomes more marginal. Because fetal fraction tends to rise with increasing gestational age (although there is significant variation from individual to individual72,73), this consideration is particularly important at early gestational ages. The ability to distinguish these minor differences with high degree of confidence requires a large number of reads from chromosome 21, typically > 95,000.72,74 Because MPSS obtains sequence information from all chromosomes and because chromosome 21 represents approximately 1.5% of the human genome, it is estimated that approximately 6.3 million uniquely mapped reads from the entire genome would be required to ensure sufficient chromosome 21 counts to make this distinction.75 The efficiency of MPSS is such that only approximately 25% of reads are uniquely mapped, which requires approximately 25 million raw sequencing reads per sample covering the entire genome in order to return sufficient data for analysis.75 This is especially important if assessment of additional clinically relevant chromosomes (13, 18, X, and Y) is considered. The requirement for a large number of reads necessitates the use of traditional large sequencing machines and represents enormous redundancy especially because the clinically significant chromosomes represent only approximately 14% of the human genome. In addition, throughput is limited, which could potentially affect turnaround times.
Figure 1.
Massively parallel shotgun sequencing for the noninvasive prenatal detection of fetal chromosomal aneuploidy. (A) Fetal DNA (green) circulates in the maternal plasma as a minor population among a high background of maternal DNA (blue). A sample of maternal plasma is obtained. Short fragments of cell-free DNA are then sequenced. (B) The chromosomal origin of each sequence is identified by mapping the reads to the human reference genome. (C) The number of unique sequences mapped to each chromosome is then counted and the number of sequences for the chromosomes of interest is determined in subsequent analysis. Aneuploidy is detected by various statistical techniques based on the number of reads representing the chromosome of interest compared with the other chromosomes. The blue and green circles represent the reads mapped to each chromosome from maternal and fetal DNA fractions, respectively. However, there is no way to distinguish the origin of the individual reads; the only difference is in the total number of reads that are detected. If the reads exceed that expected, the fetus is trisomic for that chromosome (shown for chr 21 above).
An important limitation of counting appears to be the variability with which standard polymerase chain reaction conditions amplify certain chromosomes, and this variability seems to be linked to the GC base content of an individual chromosome.72,76 This limitation has ramifications for clinical testing because it means that the accuracy rate for detection of fetal aneuploidies using MPSS will be different for each chromosome assessed. Indeed, reported detection rates for T13, T18, and T21 all show accuracies to be highest for T21, followed by T18, and are lowest for T13.73,77,78 The issue of GC content has, to some extent, been dealt with using newer bioinformatics algorithms, which has led to improved sensitivity and specificity in the detection of T13 and T18.79
Targeted Sequencing
Instead of sequencing random genomic fragments from all chromosomes, targeted sequencing selectively amplifies specific genomic regions of interest (such as nonpolymorphic loci on chromosomes 21 and 18, or specific polymorphic loci of interest) and then reads and counts only those specific sequences. This means that nearly all sequences are utilized for assigning a diagnosis. This strategy significantly reduces the total number of reads analyzed with a concomitant improvement in efficiency and a 10-fold reduction in overall costs.80 Also, the reduced read requirement enables the use of small bench-top sequencers that affords both financial and practical benefits to the laboratory. The ability to selectively sequence specific regions of the genome in cfDNA allows for a focused analysis of clinically important chromosomes such as 13, 18, 21, X, and Y. Although the capacity to preselect the regions for analysis has resulted in a higher sensitivity and specificity for the detection of fetal T18 and T21 in maternal plasma,81 the detection rates using purely quantitative targeting methods still appear to vary from chromosome to chromosome. Specifically, detection rates using quantitative read counting remain the highest for T21, followed by T18, and then T13.82
Analysis and Interpretation of Sequence Data
Just as important—and perhaps more important—than the technology used for genomic sequencing is the choice of bioinformatics platform for post-hoc data analysis. Several approaches have been taken.
Z-Score
The first approach to determine the difference in the total cfDNA attributed to fetal trisomy was based on a simple z-score. The z-score reflects the number of standard deviations the proportion of reads from a particular chromosome (in relation to the proportion of reads from all other chromosomes) is above the mean. If the amount of a chromosome-specific sequence exceeds a threshold that would be expected if the fetal karyotype was normal (euploid)—typically a z-score of > 2.5—the result is reported as positive for trisomy for that chromosome (Figure 1). The basic z-score method does not account for the variation in GC base content from chromosome to chromosome and, as such, is subject to different aneuploidy detection rates depending on the specific chromosome assessed.
Z-Score With GC Correction
The observation that the GC content of an individual chromosome determines how much of this chromosome is represented in the amplified sample and consequently its detection rate led to a bioinformatics approach to account for these differences. Instead of using read counts to directly calculate the proportion of a specific chromosome of interest, a GC-corrected read count is used with the purpose of eliminating the GC bias.79 As hypothesized, the detection rates for T13 and T18 are indeed improved using z-scores derived after quantitative correction for GC content,79 and additional improvements can be achieved using other modifications such as use of a non-repeat-masked reference genome instead of a repeat-masked reference genome.78
Z-Score With GC Correction Using an Internal Control
A more recent strategy utilizes a specific internal reference chromosome for each chromosome being assessed. The optimal internal reference chromosome is one that has a similar GC content to the chromosome of clinical interest.83 This approach appears to be markedly more adept at detecting aneuploidies other than T21, and a recent proof of concept study using this approach demonstrated 100% accuracy for detection of T13, T18, T21, 45,X, and 47,XXY in a small sample of 32 aneuploid cases.83
Normalized Chromosome Value
The normalized chromosome value (NCV) approach differs in the normalization process that compares the reads from the chromosome of interest with the number of counts from a reference set derived from an unaffected group of samples.77,84 The NCV algorithm helps to minimize the intra- and inter-run sequencing variation.84
Parental Support™
In contrast to the quantitative methods previously discussed in this review, Parental Support™ (PS; Natera, San Carlos, CA) focuses on measuring single nucleotide polymorphisms (SNPs). By measuring polymorphic loci, this approach is able to extract multiple pieces of information (including the number and identity of each allele) from each sequence read. PS then incorporates allelic information from the mother (and from the father, if available) to model a set of hypotheses (viz, monosomy, disomy, or trisomy), corresponding to different genetic inheritance patterns and crossover locations for every possible copy number count. Bayesian statistics then assign a probability to each hypothesis, and a maximum likelihood estimation analysis is performed to select the most likely hypothesis and calculate the probability of that hypothesis being correct.85
Commercial NIPT: Are We There Yet?
A number of companies have been spearheading the effort to develop the next generation of NIPT tests, including Sequenom Center for Molecular Medicine (San Diego, CA), Verinata Health (Redwood City, CA), Ariosa Diagnostics (San Jose, CA), and Natera. These companies all use a sequencing-based approach for gathering the genetic information contained within the cfDNA. In some cases, MPSS is the sequencing methodology of choice, whereas targeted sequencing is utilized by others. Each entity utilizes a unique and proprietary algorithm for interpretation of the genetic data. Although the exact technology may vary, the implications for clinical practice are the same; namely, these are all screening tests performed by analyzing cfDNA in a sample of maternal blood, and all positive test results should be confirmed by amniocentesis or CVS before acting upon the information. Detection rates reported by the commercial entities differ, as does the scope of chromosomal aneuploidies assessed. These, together with the specific analysis technique offered, are described below and summarized in Table 2.
Table 2.
Comparison of Techniques for Prenatal Diagnosis Using Cell-Free DNA
| Accuracy of Detection |
|||||||
|---|---|---|---|---|---|---|---|
| Company | Technical Approach | Anomaly | Sensitivity (%) | Specificity (%) | Depends on Measurement of Specific Loci or Alleles? | Maternal/Paternal Samples Used? | Study |
| Sequenom Center for Molecular Medicine (San Diego, CA) | MPSS | T21 | 98.6 | 99.8 | No | No | Fan HC et al,72 Chiu RW et al,76 Palomaki GE et al,78 Chen EZ et al,79 Ehrich M et al,99 Liao GJ et al,100 Palomaki GE et al.73 a |
| T18 | 100 | 99.7 | |||||
| T13 | 91.7 | 99.0 | |||||
| 45,X | — | — | |||||
| 47,XXY | — | — | |||||
| Verinata Health (Redwood City, CA) | MPSS | T21 | 100 | 100 | No | No | Fan and Quake,74 Sehnert AJ et al,84 Bianchi DW et al.77 a |
| T18 | 97.2 | 99.8 | |||||
| T13 | 78.6 | 99.4 | |||||
| 45,X | 93.8 | 99.8 | |||||
| 47,XXY | — | — | |||||
| Ariosa Diagnostics (San Jose, CA) | Targeted sequencing + DANSR + FORTE | T21 | 100 | 100 | Yes (chromosome 21, 18 and 13) | No | Sparks AB et al,75 Sparks AB et al,80 Ashoor G et al,82 Ashoor G et al.86 |
| T18 | 98 | 100 | |||||
| T13 | — | — | |||||
| 45,X | — | — | |||||
| 47,XXY | — | — | |||||
| Natera (San Carlos, CA) | Targeted sequencing + PS | T21 | 100 | 100 | Yes (SNPs) | Yes | Zimmermann B et al,85 Rabinowitz M et al,91 Nicolaides et al.92 |
| T18 | 100 | 100 | |||||
| T13 | 100 | 100 | |||||
| 45,X | 100 | 100 | |||||
| 47,XXY | 100 | 100 | |||||
a Prospective study.
DANSR, Digital Analysis of Selected Regions; FORTE, Fetal Fraction Optimized Risk of Trisomy Evaluation; MPSS, massively parallel shotgun sequencing; PS, Parental Support™ (Natera); SNPs, single nucleotide polymorphisms; T13, trisomy 13; T18, trisomy 18; T21, trisomy 21.
The Sequenom Center for Molecular Medicine uses the MPSS approach with z-score GC correction. Data from its international collaborative study indicate sensitivity > 91% for T13 and > 99% for both T21 and T18.78 The study reported specificity for all three chromosomes to be > 99.6%.78
Verinata Health uses MPSS with the NCV algorithm. Published in 2012, the Maternal Blood Is Source to Accurately Diagnose Fetal Aneuploidy (MELISSA) study was conducted as a prospective, multicenter, observational study with blinded, nested, case-control analyses, and demonstrated a high sensitivity and specificity for the detection of T21, T18, and T13, as well as sex chromosome anomalies.77
Ariosa Diagnostics uses a targeted sequencing approach, which it calls Digital Analysis of Selected Regions (DANSR), coupled with its post-hoc bioinformatics algorithm, Fetal Fraction Optimized Risk of Trisomy Evaluation (FORTE), which is designed to account for an individual’s age-related risks and fetal fraction of the sample. This approach is closely related to MPSS in that it also uses counting, but includes an initial targeted amplification step in which approximately 400 loci of each chromosome of interest are selectively amplified prior to MPSS analysis. This targeted amplification results in an improvement in sequencing efficiency, with 100,000 to 300,000 reads achieved per target chromosome.75,80,86 DANSR has recently been shown to detect T21, T18, and T13 with good accuracy and greater efficiency than MPSS alone82,86 (Table 2). DANSR permits aneuploidy detection using approximately 1 million raw reads per subject, enabling analysis of 96 subjects per sequencing lane. In contrast, MPSS evaluates the entire genome and requires approximately 25 million raw reads per subject, which limits sequencing throughput to 4 to 6 samples per lane. Thus, DANSR may enjoy an advantage over MPSS in terms of sequencing cost and throughput. As with all purely quantitative methods, however, the approach is dependent on chromosomes having low amplification variability (GC content influenced), which may limit its diagnostic accuracy for some chromosomes.
Natera has been working in the field of genetic testing since 2009 with a focus on preimplantation genetic diagnosis, molecular karyotyping of products of conception, and noninvasive paternity testing using cfDNA.87–89 More recently, the company has become interested in NIPT and uses a novel approach called PS, which combines a targeted amplification step (that targets approximately 2000 polymorphic loci per chromosome under investigation chosen from cross-ethnic databases to avoid interethnic bias) with sophisticated statistical analysis. PS utilizes between 500,000 and 2 million sequence reads per chromosome, a number that falls between MPSS and DANSR.85 The PS approach has a number of distinct advantages. First, it is based on allelic distribution patterns rather than quantifying chromosome-specific read counts and, as such, does not suffer from chromosome amplification variability or require the use of a reference chromosome. PS is therefore able to determine fetal copy number at chromosomes 21, 18, 13, X, and Y with similar accuracy, and it has the same accuracy across all tested chromosomes regardless of fetal fraction.85 Owing to the lack of chromosome specificity, and because PS does not require comparison against a reference chromosome, the technology has the capacity to detect sex chromosome aneuploidies (45,X; 47,XXY, and 47,XYY), uniparental isodisomy, and triploidy. The ability to extend NIPT to the sex chromosomes is important, because these variants represent almost half of the at-birth chromosomal abnormalities90 and an even higher fraction in the mid-trimester when prenatal screens are typically offered. Second, the PS methodology calculates a per-test, per-chromosome accuracy for each sample, thereby providing an individualized risk score for each patient and each aneuploidy under investigation, taking into account the major variables (maternal age and fetal fraction of cfDNA). This is especially important when testing women at early gestational ages (9–12 weeks instead of 15–20 weeks as in many of the published trials), because the fetal fraction of cfDNA is likely to be lower in these cases. Such an approach may, at least in theory, increase the likelihood of a “no-call” for any given aneuploidy, but this is generally preferred over a “wrong call” (false-positive or false-negative). If a patient has a no-call at 10 weeks of gestation, it is likely that a second test could simply be performed 1 or 2 weeks later. And third, the PS targeted sequencing technique under development will enable testing to be carried out on a small desktop sequencing machine, thereby improving efficiency and significantly shortening turnaround time. Recent data have shown that the PS informatics-based method can noninvasively detect fetuses with T13, T18, T21, 45,X and 47,XXY with high sample-specific calculated accuracies for each individual chromosome and across all five chromosomes,85 and preliminary data from an improved version of the PS method, termed Next-Generation Aneuploidy Test Using SNPs (NATUS), show that it delivers similar performance but produces a no-call rate similar to other reported techniques.91 In a recent independent, prospective, blinded study of 242 singleton pregnancies in the United Kingdom, maternal blood was collected prior to clinically indicated CVS at 11 to 13 weeks of gestation. cfDNA was isolated from maternal plasma, subjected to targeted multiplex PCR amplification followed by sequencing of 19,488 SNPs covering five chromosomes (13, 18, 21, X, and Y), and analyzed using the NATUS algorithm.92 Results were reported in 94.6% (229/242) of cases and compared with the fetal karyotype obtained from CVS. In subjects for whom a result was reported, all fetuses with aneuploidy (including 25 with T21, 3 with T18, 1 with T13, 2 with Turner syndrome, and a case of triploidy) were accurately detected with no false-positive or false-negative results. Fetal gender was also correctly determined in all cases. The “no call” rate was 5.4% (13/242), which included two cases of T21. The authors conclude that “… cfDNA testing in maternal blood using targeted sequencing of SNPs at chromosomes 13, 18, 21, X, and Y and use of the NATUS algorithm holds promise as an accurate method for detecting fetal autosomal aneuploidies, sex chromosome aneuploidies, and triploidy in the first trimester of pregnancy.”
In summary, these four approaches have more in common than they differ. The major differences appear to be in the post-hoc bioinformatics and how the data are analyzed once the DNA fragments are sequenced. They all give a result within 2 weeks from a single maternal blood sample drawn early in pregnancy, and all appear to have a better sensitivity (> 99%) for T21 detection (Table 1) than current recommended screening options. The major advantage of these tests is in their specificity. Using routine serum analyte screening, the likelihood that a positive screening test result actually represents a true positive (ie, a T21 karyotype on amniocentesis) is only approximately 2% to 4%. In contrast, using cfDNA technology, the vast majority (> 99%) of screen positives are likely to be true positives.93
Future Applications for the Technology
In addition to prenatal diagnosis, analysis of cfDNA in maternal blood also has the potential to establish the Rh genotype of the fetuses and identify fetuses with congenital adrenal hyperplasia.48,94 Fetal sex determination is also possible from an early gestational age,48,95,96 although this raises a number of ethical concerns and, at least for now, is best reserved for clear medical indications. The advent of chromosomal microarray technology has uncovered multiple new genomic regions that lead to a clinical phenotype when deleted or duplicated. These types of aberrations are commonly referred to as microdeletions and microduplications, and their clinical consequences can be observed in pediatric populations. Examples include the 22q11.2 microdeletion syndrome, which causes DiGeorge syndrome and occurs with a frequency of 1 case in 4000 births,97 and Prader-Willi syndrome, which can result from a microdeletion of the proximal 15q11.2 region. It seems obvious that future efforts in NIPT will be directed at detecting these smaller regions of genomic imbalance. However, the clinical consequences of microdeletions and microduplications are not always certain, they may be extremely variable, and in some cases may be unknown. Initial microdeletion/microduplication targets are sure to be restricted to regions with well characterized clinical outcomes. NIPT techniques using targeted amplification (DANSR and PS/NATUS) could be expanded to target detection of subchromosomal genomic imbalances (microdeletions/microduplications), as well as single gene disorders and/or carrier status for paternally inherited mutations.48,98,99
Conclusions
Although safe for the pregnancy, currently available biochemical/ultrasound-based noninvasive screen ing tests for fetal aneuploidy have poor accuracy with false-negative rates between 12% and 23% and false-positive rates between 1.9% and 5.2%.9,10,18–27,29,61,63–65 Indeed, using current NIPT methodology, one out of every six cases of Down syndrome will go undetected. And, of all women deemed to be at high risk of having an aneuploid fetus, more than 95% are actually carrying a healthy baby, leading to unnecessary anxiety and miscarriage risk from amniocentesis or CVS. NIPT of cfDNA in a sample of the mother’s blood can significantly increase the detection rate of fetuses with T21 and decrease false-positive results. The accuracy of NIPT depends on the precise technology used and on the percentage of fetal DNA in a pregnant woman’s blood. NIPT has not been sufficiently validated in women at low risk for fetal aneuploidy. As such, it is not currently recommended for all pregnant women. Indications for the use of cfDNA for fetal aneuploidy are summarized in Table 3. If NIPT is ever to completely replace conventional cytogenetic analysis following CVS or amniocentesis, it will need to match the diagnostic accuracy as well as the scope of anomalies that can be detected. The diagnostic scope of traditional prenatal cytogenetic analysis has recently been extended to include detection of microdeletions and microduplications using new genomic technologies such as microarray analysis. The recently completed NICHD Multicenter Prenatal Microarray Study indicated that 1.7% of routine low-risk cases (for such indications as advanced maternal age, abnormal FTRA, and parental concern) and 6% of cases referred for structural ultrasound abnormalities had a clinically significant copy number change that was not detected by routine cytogenetic analysis.61 As such, there is a much higher risk for these submicroscopic imbalances than there is for Down syndrome. NIPT still has a way to go before it can replace procedures such as CVS and amniocentesis.
Table 3.
Indications for Considering the Use of Cell-Free Fetal DNA Analysis for Fetal Aneuploidy
|
Data from Obstet Gynecol. 2012;120:1532—1534.93
Main Points.
Prenatal detection of chromosome abnormalities has been offered for more than 40 years; however, invasive testing also carries a risk for procedure-related miscarriage. For this reason, a number of noninvasive tests have been developed.
Fetal DNA can be detected in as little as 10 μL of maternal plasma and serum, in amounts significantly higher than that available from nucleated blood cells extracted from a similar volume of whole blood. The approach of using cell-free fetal DNA instead of fetal cells provided a far easier, less labor intensive, and less time consuming way to work with fetal DNA derived from the maternal circulation, and this opened up new opportunities for noninvasive prenatal testing (NIPT).
Noninvasive testing grew out of a desire to avoid direct contact with the growing fetus/placenta and concomitantly risking the health of the fetus. NIPT refers specifically to techniques that evaluate fetal cells or cfDNA in a blood sample drawn from the mother during pregnancy.
The diagnostic scope of traditional prenatal cytogenetic analysis has recently been extended to include detection of microdeletions and microduplications using new genomic technologies such as microarray analysis.
There are currently two primary next-generation sequencing approaches for gathering genetic data from cellfree DNA: massively parallel shotgun sequencing and targeted sequencing.
A number of companies have been spearheading the effort to develop a reliable and accurate commercial NIPT for fetal aneuploidy detection. Although the precise technology used by each of these companies varies, they all rely on the same premise: that existing high-throughput approaches to DNA sequencing coupled with sophisticated data analysis will be able to detect abnormal amounts of chromosome-specific cfDNA in the maternal circulation in those pregnancies with fetal aneuploidy.
Footnotes
Errol Norwitz, MD, PhD, is an unpaid member of the Advisory Board of Natera (San Carlos, CA); Brynn Levy, MSc (Med), PhD, is a consulting Laboratory Director of Natera.
References
- 1.Hassold T, Hunt PA, Sherman S. Trisomy in humans: incidence, origin and etiology. Curr Opin Genet Dev. 1993;3:398–403. doi: 10.1016/0959-437x(93)90111-2. [DOI] [PubMed] [Google Scholar]
- 2.Penrose LS. The relative aetiological importance of birth order and maternal age in mongolism. Proc Roy Soc Lond [Biol] 1934;115:431–450. [Google Scholar]
- 3.Sherman SL, Petersen MB, Freeman SB, et al. Nondisjunction of chromosome 21 in maternal meiosis I: evidence for a maternal age-dependent mechanism involving reduced recombination. Hum Mol Genet. 1994;3:1529–1535. doi: 10.1093/hmg/3.9.1529. [DOI] [PubMed] [Google Scholar]
- 4.Hook EB. Chromosome abnormalities: prevalence, risks and recurrence. In: Brock DJH, Rodeck CH, Ferguson-Smith MA, editors. Prenatal Diagnosis and Screening. Edinburgh, Scotland: Churchill Livingstone; 1992. pp. 351–392. [Google Scholar]
- 5.Gardner RJM, Sutherland GR. Chromosome Abnormalities and Genetic Counseling. New York, NY: Oxford University Press; 1996. p. 478. [Google Scholar]
- 6.Lejeune J, Gautier M, Turpin R. Etude des chromosomes somatiques de neuf enfants mongoliens. C R Hebd Seances Acad Sci. 1959;248:1721–1722. [PubMed] [Google Scholar]
- 7.Gardner RJM, Sutherland GR, Shaffer LG. Chromosome Abnormalities and Genetic Counseling. New York, NY: Oxford University Press; 2012. p. 634. [Google Scholar]
- 8.Menasha J, Levy B, Hirschhorn K, Kardon NB. Incidence and spectrum of chromosome abnormalities in spontaneous abortions: new insights from a 12-year study. Genet Med. 2005;7:251–263. doi: 10.1097/01.gim.0000160075.96707.04. [DOI] [PubMed] [Google Scholar]
- 9.ACOG Committee on Practice Bulletins, authors. ACOG Practice Bulletin No. 77: screening for fetal chromosomal abnormalities. Obstet Gynecol. 2007;109:217–227. doi: 10.1097/00006250-200701000-00054. [DOI] [PubMed] [Google Scholar]
- 10.Driscoll DA, Gross SJ Professional Practice Guidelines Committee, authors. Screening for fetal aneuploidy and neural tube defects. Genet Med. 2009;11:818–821. doi: 10.1097/GIM.0b013e3181bb267b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hahnemann JM, Vejerslev LO. Accuracy of cytogenetic findings on chorionic villus sampling (CVS)—diagnostic consequences of CVS mosaicism and non-mosaic discrepancy in centres contributing to EUCROMIC 1986–1992. Prenat Diagn. 1997;17:801–820. doi: 10.1002/(sici)1097-0223(199709)17:9<801::aid-pd153>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 12.Ledbetter DH, Zachary JM, Simpson JL, et al. Cytogenetic results from the U.S. Collaborative Study on CVS. Prenat Diagn. 1992;12:317–345. doi: 10.1002/pd.1970120503. [DOI] [PubMed] [Google Scholar]
- 13.Lippman A, Tomkins DJ, Shime J, Hamerton JL. Canadian multicentre randomized clinical trial of chorion villus sampling and amniocentesis. Final report. Prenat Diagn. 1992;12:385–408. doi: 10.1002/pd.1970120508. [DOI] [PubMed] [Google Scholar]
- 14.Midtrimester amniocentesis for prenatal diagnosis Safety and accuracy. JAMA. 1976;236:1471–1476. doi: 10.1001/jama.1976.03270140023016. [DOI] [PubMed] [Google Scholar]
- 15.Rhoads GG, Jackson LG, Schlesselman SE, et al. The safety and efficacy of chorionic villus sampling for early prenatal diagnosis of cytogenetic abnormalities. N Engl J Med. 1989;320:609–617. doi: 10.1056/NEJM198903093201001. [DOI] [PubMed] [Google Scholar]
- 16.Kuliev A, Jackson L, Froster U, et al. Chorionic villus sampling safety. Am J Obstet Gynecol; Report of World Health Organization/ EURO meeting in association with the Seventh International Conference on Early Prenatal Diagnosis of Genetic Diseases; May 21, 1994; Tel-Aviv, Israel. 1996. pp. 807–811. [DOI] [PubMed] [Google Scholar]
- 17.Tabor A, Philip J, Madsen M, et al. Randomised controlled trial of genetic amniocentesis in 4606 low-risk women. Lancet. 1986;1:1287–1293. doi: 10.1016/s0140-6736(86)91218-3. [DOI] [PubMed] [Google Scholar]
- 18.Benn PA, Fang M, Egan JF, et al. Incorporation of inhibin-A in second-trimester screening for Down syndrome. Obstet Gynecol. 2003;101:451–454. doi: 10.1016/s0029-7844(02)03159-9. [DOI] [PubMed] [Google Scholar]
- 19.Haddow JE, Palomaki GE, Knight GJ, et al. Reducing the need for amniocentesis in women 35 years of age or older with serum markers for screening. N Engl J Med. 1994;330:1114–1118. doi: 10.1056/NEJM199404213301603. [DOI] [PubMed] [Google Scholar]
- 20.Malone FD, Canick JA, Ball RH, et al. First- and Second-Trimester Evaluation of Risk (FASTER) Research Consortium, authors. First-trimester or secondtrimester screening, or both, for Down’s syndrome. N Engl J Med. 2005;353:2001–2011. doi: 10.1056/NEJMoa043693. [DOI] [PubMed] [Google Scholar]
- 21.Neveux LM, Palomaki GE, Knight GJ, Haddow JE. Multiple marker screening for Down syndrome in twin pregnancies. Prenat Diagn. 1996;16:29–34. doi: 10.1002/(SICI)1097-0223(199601)16:1<29::AID-PD801>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 22.Nicolaides KH. Nuchal translucency and other first-trimester sonographic markers of chromosomal abnormalities. Am J Obstet Gynecol. 2004;191:45–67. doi: 10.1016/j.ajog.2004.03.090. [DOI] [PubMed] [Google Scholar]
- 23.Platt LD, Greene N, Johnson A, et al. First Trimester Maternal Serum Biochemistry and Fetal Nuchal Translucency Screening (BUN) Study Group, authors. Sequential pathways of testing after first-trimester screening for trisomy 21. Obstet Gynecol. 2004;104:661–666. doi: 10.1097/01.AOG.0000139832.79658.b9. [DOI] [PubMed] [Google Scholar]
- 24.Spencer K, Spencer CE, Power M, et al. Screening for chromosomal abnormalities in the first trimester using ultrasound and maternal serum biochemistry in a one-stop clinic: a review of three years prospective experience. BJOG. 2003;110:281–286. [PubMed] [Google Scholar]
- 25.Wald NJ, Kennard A, Hackshaw A, McGuire A. Antenatal screening for Down’s syndrome. J Med Screen. 1997;4:181–246. doi: 10.1177/096914139700400402. [DOI] [PubMed] [Google Scholar]
- 26.Wald NJ, Rodeck C, Hackshaw AK, et al. First and second trimester antenatal screening for Down’s syndrome: the results of the Serum, Urine and Ultrasound Screening Study (SURUSS) J Med Screen. 2003;10:56–104. doi: 10.1258/096914103321824133. [DOI] [PubMed] [Google Scholar]
- 27.Wald NJ, Watt HC, Hackshaw AK. Integrated screening for Down’s syndrome on the basis of tests performed during the first and second trimesters. N Engl J Med. 1999;341:461–467. doi: 10.1056/NEJM199908123410701. [DOI] [PubMed] [Google Scholar]
- 28.Wapner R, Thom E, Simpson JL, et al. First Trimester Maternal Serum Biochemistry and Fetal Nuchal Translucency Screening (BUN) Study Group, authors. Firsttrimester screening for trisomies 21 and 18. N Engl J Med. 2003;349:1405–1413. doi: 10.1056/NEJMoa025273. [DOI] [PubMed] [Google Scholar]
- 29.Wright D, Bradbury I, Benn P, et al. Contingent screening for Down syndrome is an efficient alternative to non-disclosure sequential screening. Prenat Diagn. 2004;24:762–766. doi: 10.1002/pd.974. [DOI] [PubMed] [Google Scholar]
- 30.Walknowska J, Conte FA, Grumbach MM. Practical and theoretical implications of fetal-maternal lymphocyte transfer. Lancet. 1969;1:1119–1122. doi: 10.1016/s0140-6736(69)91642-0. [DOI] [PubMed] [Google Scholar]
- 31.Forestier F, Daffos F, Catherine N, et al. Developmental hematopoiesis in normal human fetal blood. Blood. 1991;77:2360–2363. [PubMed] [Google Scholar]
- 32.Simpson JL, Elias S. Isolating fetal cells in maternal circulation for prenatal diagnosis. Prenat Diagn. 1994;14:1229–1242. doi: 10.1002/pd.1970141308. [DOI] [PubMed] [Google Scholar]
- 33.Elias S, Price J, Dockter M, et al. First trimester prenatal diagnosis of trisomy 21 in fetal cells from maternal blood. Lancet. 1992;340:1033. doi: 10.1016/0140-6736(92)93041-k. [DOI] [PubMed] [Google Scholar]
- 34.Pezzolo A, Santi F, Pistoia V, De Biasio P. Prenatal diagnosis of triploidy using fetal cells in the maternal circulation. Prenat Diagn. 1997;17:389. doi: 10.1002/(sici)1097-0223(199704)17:4<389::aid-pd84>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 35.Gänshirt-Ahlert D, Börjesson-Stoll R, Burschyk M, et al. Detection of fetal trisomies 21 and 18 from maternal blood using triple gradient and magnetic cell sorting. Am J Reprod Immunol. 1993;30:194–201. doi: 10.1111/j.1600-0897.1993.tb00620.x. [DOI] [PubMed] [Google Scholar]
- 36.Price JO, Elias S, Wachtel SS, et al. Prenatal diagnosis with fetal cells isolated from maternal blood by multiparameter flow cytometry. Am J Obstet Gynecol. 1991;165:1731–1737. doi: 10.1016/0002-9378(91)90024-l. [DOI] [PubMed] [Google Scholar]
- 37.Simpson JL, Elias S. Isolating fetal cells from maternal blood. Advances in prenatal diagnosis through molecular technology. JAMA. 1993;270:2357–2361. [PubMed] [Google Scholar]
- 38.Lo YM, Corbetta N, Chamberlain PF, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–487. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 39.Dennis Lo, Tein MS, Lau TK, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998;62:768–775. doi: 10.1086/301800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bianchi DW. Fetal DNA in maternal plasma: the plot thickens and the placental barrier thins. Am J Hum Genet. 1998;62:763–764. doi: 10.1086/301809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Billingham RE, Brent L, Medawar PB. Actively acquired tolerance of foreign cells. Nature. 1953;172:603–606. doi: 10.1038/172603a0. [DOI] [PubMed] [Google Scholar]
- 42.Desai RG, Creger WP. Maternofetal passage of leukocytes and platelets in man. Blood. 1963;21:665–673. [PubMed] [Google Scholar]
- 43.Adams Waldorf, Nelson JL. Autoimmune disease during pregnancy and the microchimerism legacy of pregnancy. Immunol Invest. 2008;37:631–644. doi: 10.1080/08820130802205886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sekizawa A, Samura O, Zhen DK, et al. Apoptosis in fetal nucleated erythrocytes circulating in maternal blood. Prenat Diagn. 2000;20:886–889. doi: 10.1002/1097-0223(200011)20:11<886::aid-pd942>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 45.Bianchi DW, Simpson JL, Jackson LG, et al. Fetal gender and aneuploidy detection using fetal cells in maternal blood: analysis of NIFTY I data. National Institute of Child Health and Development Fetal Cell Isolation Study. Prenat Diagn. 2002;22:609–615. doi: 10.1002/pd.347. [DOI] [PubMed] [Google Scholar]
- 46.Dennis Lo YM, Chiu RW. Prenatal diagnosis: progress through plasma nucleic acids. Nat Rev Genet. 2007;8:71–77. doi: 10.1038/nrg1982. [DOI] [PubMed] [Google Scholar]
- 47.Guetta E, Simchen MJ, Mammon-Daviko K, et al. Analysis of fetal blood cells in the maternal circulation: challenges, ongoing efforts, and potential solutions. Stem Cells Dev. 2004;13:93–99. doi: 10.1089/154732804773099290. [DOI] [PubMed] [Google Scholar]
- 48.Wright CF, Burton H. The use of cell-free fetal nucleic acids in maternal blood for non-invasive prenatal diagnosis. Hum Reprod Update. 2009;15:139–151. doi: 10.1093/humupd/dmn047. [DOI] [PubMed] [Google Scholar]
- 49.Swaminathan R, Butt AN. Circulating nucleic acids in plasma and serum: recent developments. Ann N Y Acad Sci. 2006;1075:1–9. doi: 10.1196/annals.1368.001. [DOI] [PubMed] [Google Scholar]
- 50.Ariga H, Ohto H, Busch MP, et al. Kinetics of fetal cellular and cell-free DNA in the maternal circulation during and after pregnancy: implications for noninvasive prenatal diagnosis. Transfusion. 2001;41:1524–1530. doi: 10.1046/j.1537-2995.2001.41121524.x. [DOI] [PubMed] [Google Scholar]
- 51.Invernizzi P, Biondi ML, Battezzati PM, et al. Presence of fetal DNA in maternal plasma decades after pregnancy. Hum Genet. 2002;110:587–591. doi: 10.1007/s00439-002-0725-3. [DOI] [PubMed] [Google Scholar]
- 52.Lo YM. Fetal DNA in maternal plasma: biology and diagnostic applications. Clin Chem. 2000;46:1903–1906. [PubMed] [Google Scholar]
- 53.Goldberg JD, Wohlferd MM. Incidence and outcome of chromosomal mosaicism found at the time of chorionic villus sampling. Am J Obstet Gynecol. 1997;176:1349–1352. doi: 10.1016/s0002-9378(97)70356-9. discussion 1352–1353. [DOI] [PubMed] [Google Scholar]
- 54.Kalousek DK, Dill FJ, Pantzar T, et al. Confined chorionic mosaicism in prenatal diagnosis. Hum Genet. 1987;77:163–167. doi: 10.1007/BF00272385. [DOI] [PubMed] [Google Scholar]
- 55.Alfirevic Z, Sundberg K, Brigham S. Amniocentesis and chorionic villus sampling for prenatal diagnosis. Cochrane Database Syst Rev. 2003 doi: 10.1002/14651858.CD003252. CD003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.American College of Obstetricians and Gynecologists, authors. ACOG Practice Bulletin No. 88, December 2007. Invasive prenatal testing for aneuploidy. Obstet Gynecol. 2007;110:1459–1467. doi: 10.1097/01.AOG.0000291570.63450.44. [DOI] [PubMed] [Google Scholar]
- 57.Mujezinovic F, Alfirevic Z. Procedure-related complications of amniocentesis and chorionic villous sampling: a systematic review. Obstet Gynecol. 2007;110:687–694. doi: 10.1097/01.AOG.0000278820.54029.e3. [DOI] [PubMed] [Google Scholar]
- 58.Odibo AO, Gray DL, Dicke JM, et al. Revisiting the fetal loss rate after second-trimester genetic amniocentesis: a single center’s 16-year experience. Obstet Gynecol. 2008;111:589–595. doi: 10.1097/AOG.0b013e318162eb53. [DOI] [PubMed] [Google Scholar]
- 59.Randomised trial to assess safety and fetal outcome of early and midtrimester amniocentesis. The Canadian Early and Mid-trimester Amniocentesis Trial (CEMAT) Group. Lancet. 1998;351:242–247. [PubMed] [Google Scholar]
- 60.Tepperberg J, Pettenati MJ, Rao PN, et al. Prenatal diagnosis using interphase fluorescence in situ hybridization (FISH): 2-year multi-center retrospective study and review of the literature. Prenat Diagn. 2001;21:293–301. doi: 10.1002/pd.57. [DOI] [PubMed] [Google Scholar]
- 61.Wapner R. A multicenter, prospective, masked comparison of chromosomal microarray with standard karyotyping for routine and high risk prenatal diagnosis. Am J Obstet Gynecol. 2012;206(suppl):S2. [Google Scholar]
- 62.Cuckle HS, Wald NJ, Lindenbaum RH. Maternal serum alpha-fetoprotein measurement: a screening test for Down syndrome. Lancet. 1984;1:926–929. doi: 10.1016/s0140-6736(84)92389-4. [DOI] [PubMed] [Google Scholar]
- 63.Cuckle H, Benn P, Wright D. Down syndrome screening in the first and/or second trimester: model predicted performance using meta-analysis parameters. Semin Perinatol. 2005;29:252–257. doi: 10.1053/j.semperi.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 64.Palomaki GE, Steinort K, Knight GJ, Haddow JE. Comparing three screening strategies for combining first- and second-trimester Down syndrome markers. Obstet Gynecol. 2006;107:367–375. doi: 10.1097/01.AOG.0000195061.48747.f4. [DOI] [PubMed] [Google Scholar]
- 65.Kadir RA, Economides DL. The effect of nuchal translucency measurement on second-trimester biochemical screening for Down’s syndrome. Ultrasound Obstet Gynecol. 1997;9:244–247. doi: 10.1046/j.1469-0705.1997.09040244.x. [DOI] [PubMed] [Google Scholar]
- 66.Cicero S, Curcio P, Papageorghiou A, et al. Absence of nasal bone in fetuses with trisomy 21 at 11-14 weeks of gestation: an observational study. Lancet. 2001;358:1665–1667. doi: 10.1016/S0140-6736(01)06709-5. [DOI] [PubMed] [Google Scholar]
- 67.Malone FD, Ball RH, Nyberg DA, et al. FASTER Research Consortium, authors. First-trimester nasal bone evaluation for aneuploidy in the general population. Obstet Gynecol. 2004;104:1222–1228. doi: 10.1097/01.AOG.0000143255.46196.7a. [DOI] [PubMed] [Google Scholar]
- 68.Ewigman BG, Crane JP, Frigoletto FD, et al. Effect of prenatal ultrasound screening on perinatal outcome. RADIUS Study Group. N Engl J Med. 1993;329:821–827. doi: 10.1056/NEJM199309163291201. [DOI] [PubMed] [Google Scholar]
- 69.American College of Obstetricians and Gynecologists, authors. ACOG Practice Bulletin No. 101: Ultrasonography in pregnancy. Obstet Gynecol. 2009;113:451–461. doi: 10.1097/AOG.0b013e31819930b0. [DOI] [PubMed] [Google Scholar]
- 70.Caughey AB, Lyell DJ, Washington AE, et al. Ultrasound screening of fetuses at increased risk for Down syndrome: how many missed diagnoses? Prenat Diagn. 2006;26:22–27. doi: 10.1002/pd.1319. [DOI] [PubMed] [Google Scholar]
- 71.Smith-Bindman R, Chu P, Goldberg JD. Second trimester prenatal ultrasound for the detection of pregnancies at increased risk of Down syndrome. Prenat Diagn. 2007;27:535–544. doi: 10.1002/pd.1725. [DOI] [PubMed] [Google Scholar]
- 72.Fan HC, Blumenfeld YJ, Chitkara U, et al. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proc Natl Acad Sci U S A. 2008;105:16266–16271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Palomaki GE, Kloza EM, Lambert-Messerlian GM, et al. DNA sequencing of maternal plasma to detect Down syndrome: an international clinical validation study. Genet Med. 2011;13:913–920. doi: 10.1097/GIM.0b013e3182368a0e. [DOI] [PubMed] [Google Scholar]
- 74.Fan HC, Quake SR. Sensitivity of noninvasive prenatal detection of fetal aneuploidy from maternal plasma using shotgun sequencing is limited only by counting statistics. PLoS One. 2010;5:e10439. doi: 10.1371/journal.pone.0010439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sparks AB, Struble CA, Wang ET, et al. Noninvasive prenatal detection and selective analysis of cell-free DNA obtained from maternal blood: evaluation for trisomy 21 and trisomy 18. Am J Obstet Gynecol. 2012;206:319. doi: 10.1016/j.ajog.2012.01.030. e1–9. [DOI] [PubMed] [Google Scholar]
- 76.Chiu RW, Chan KC, Gao Y, et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proc Natl Acad Sci U S A. 2008;105:20458–20463. doi: 10.1073/pnas.0810641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bianchi DW, Platt LD, Goldberg JD, et al. MatErnal BLood IS Source to Accurately diagnose fetal aneuploidy (MELISSA) Study Group, authors. Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol. 2012;119:890–901. doi: 10.1097/AOG.0b013e31824fb482. [DOI] [PubMed] [Google Scholar]
- 78.Palomaki GE, Deciu C, Kloza EM, et al. DNA sequencing of maternal plasma reliably identifies trisomy 18 and trisomy 13 as well as Down syndrome: an international collaborative study. Genet Med. 2012;14:296–305. doi: 10.1038/gim.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen EZ, Chiu RW, Sun H, et al. Noninvasive prenatal diagnosis of fetal trisomy 18 and trisomy 13 by maternal plasma DNA sequencing. PLoS One. 2011;6:e21791. doi: 10.1371/journal.pone.0021791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sparks AB, Wang ET, Struble CA, et al. Selective analysis of cell-free DNA in maternal blood for evaluation of fetal trisomy. Prenat Diagn. 2012;32:3–9. doi: 10.1002/pd.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Norton ME, Brar H, Weiss J, et al. Non-Invasive Chromosomal Evaluation (NICE) Study: results of a multicenter prospective cohort study for detection of fetal trisomy 21 and trisomy 18. Am J Obstet Gynecol. 2012;207:137. doi: 10.1016/j.ajog.2012.05.021. e1–8. [DOI] [PubMed] [Google Scholar]
- 82.Ashoor G, Syngelaki A, Wang E, et al. Trisomy 13 detection in the first trimester of pregnancy using a chromosome-selective cell-free DNA analysis method. Ultrasound Obstet Gynecol. 2013;41:21–25. doi: 10.1002/uog.12299. [DOI] [PubMed] [Google Scholar]
- 83.Lau TK, Chen F, Pan X, et al. Noninvasive prenatal diagnosis of common fetal chromosomal aneuploidies by maternal plasma DNA sequencing. J Matern Fetal Neonatal Med. 2012;25:1370–1374. doi: 10.3109/14767058.2011.635730. [DOI] [PubMed] [Google Scholar]
- 84.Sehnert AJ, Rhees B, Comstock D, et al. Optimal detection of fetal chromosomal abnormalities by massively parallel DNA sequencing of cell-free fetal DNA from maternal blood. Clin Chem. 2011;57:1042–1049. doi: 10.1373/clinchem.2011.165910. [DOI] [PubMed] [Google Scholar]
- 85.Zimmermann B, Hill M, Gemelos G, et al. Noninvasive prenatal aneuploidy testing at chromosomes 13, 18, 21, X, and Y, using targeted sequencing at polymorphic loci. Prenat Diagn. 2012;32:1233–1241. doi: 10.1002/pd.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ashoor G, Syngelaki A, Wagner M, et al. Chromosomeselective sequencing of maternal plasma cell-free DNA for first-trimester detection of trisomy 21 and trisomy 18. Am J Obstet Gynecol. 2012;206:322. doi: 10.1016/j.ajog.2012.01.029. e1–5. [DOI] [PubMed] [Google Scholar]
- 87.Johnson DS, Cinnioglu C, Ross R, et al. Comprehensive analysis of karyotypic mosaicism between trophectoderm and inner cell mass. Mol Hum Reprod. 2010;16:944–949. doi: 10.1093/molehr/gaq062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lathi RB, Massie JA, Loring M, et al. Informatics enhanced SNP microarray analysis of 30 miscarriage samples compared to routine cytogenetics. PLoS One. 2012;7:e31282. doi: 10.1371/journal.pone.0031282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rabinowitz M, Ryan A, Gemelos G, et al. Origins and rates of aneuploidy in human blastomeres. Fertil Steril. 2012;97:395–401. doi: 10.1016/j.fertnstert.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 90.Grati FR, Barlocco A, Grimi B, et al. Chromosome abnormalities investigated by non-invasive prenatal testing account for approximately 50% of fetal unbalances associated with relevant clinical phenotypes. Am J Med Genet A. 2010;152A:1434–1442. doi: 10.1002/ajmg.a.33370. [DOI] [PubMed] [Google Scholar]
- 91.Rabinowitz M, Gemelos G, Hill M, et al. Highly accurate non-invasive detection of fetal aneuploidy of chromosomes 13, 18, 21, X and Y by targeted sequencing; Paper presented at: 62nd Annual Meeting of the American Society of Human Genetics; November 6–10, 2012; San Francisco, CA. [Google Scholar]
- 92.Nicolaides KH, Syngelaki A, Gil M, et al. Validation of targeted sequencing of single-nucleotide polymorphisms for non-invasive prenatal detection of aneuploidy of chromosomes 13, 18, 21, X, and Y. Prenat Diagn. doi: 10.1002/pd.4103. [DOI] [PubMed]
- 93.Committee Opinion No. 545: noninvasive prenatal testing for fetal aneuploidy. Obstet Gynecol. 2012;120:1532–1534. doi: 10.1097/01.AOG.0000423819.85283.f4. [DOI] [PubMed] [Google Scholar]
- 94.Maron JL, Bianchi DW. Prenatal diagnosis using cellfree nucleic acids in maternal body fluids: a decade of progress. Am J Med Genet C Semin Med Genet. 2007;145C:5–17. doi: 10.1002/ajmg.c.30115. [DOI] [PubMed] [Google Scholar]
- 95.Devaney SA, Palomaki GE, Scott JA, Bianchi DW. Noninvasive fetal sex determination using cell-free fetal DNA: a systematic review and meta-analysis. JAMA. 2011;306:627–636. doi: 10.1001/jama.2011.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sekizawa A, Kondo T, Iwasaki M, et al. Accuracy of fetal gender determination by analysis of DNA in maternal plasma. Clin Chem. 2001;47:1856–1858. [PubMed] [Google Scholar]
- 97.McDonald-McGinn DM, Sullivan KE. Chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/ velocar diofacial syndrome) Medicine (Baltimore) 2011;90:1–18. doi: 10.1097/MD.0b013e3182060469. [DOI] [PubMed] [Google Scholar]
- 98.Bianchi DW. From prenatal genomic diagnosis to fetal personalized medicine: progress and challenges. Nat Med. 2012;18:1041–1051. doi: 10.1038/nm.2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sekizawa A, Saito H. Prenatal screening of single-gene disorders from maternal blood. Am J Pharmacogenomics. 2001;1:111–117. doi: 10.2165/00129785-200101020-00004. [DOI] [PubMed] [Google Scholar]
- 100.Ehrich M, Deciu C, Zwiefelhofer T, et al. Noninvasive detection of fetal trisomy 21 by sequencing of DNA in maternal blood: a study in a clinical setting. Am J Obstet Gynecol. 2011;204:205. doi: 10.1016/j.ajog.2010.12.060. e1–11. [DOI] [PubMed] [Google Scholar]
- 101.Liao GJ, Lun FM, Zheng YW, et al. Targeted massively parallel sequencing of maternal plasma DNA permits efficient and unbiased detection of fetal alleles. Clin Chem. 2011;57:92–101. doi: 10.1373/clinchem.2010.154336. [DOI] [PubMed] [Google Scholar]