Abstract
Here we report a new peptide modified mesoporous silica nanocontainer (PMSN) as a novel controlled release system. The peptides are part of a stimuli responsive nanovalve and ensure an efficient cellular uptake.
Stimuli responsive nanomaterials have attracted attention as nanodevices for the controlled and selective release of drugs as these materials can entrap drug molecules in their porous structure. Furthermore, the payload is protected from enzymatic degradation. At the same time these systems reduce some severe side effects associated with many drugs.1–8 Nanovalve-capped mesoporous silica nanoparticles can protect and carry hydrophobic drugs with high efficacy and without any surface modification. Zink et al. have demonstrated that mesoporous silica nanoparticles without surface modification can act as a nanocontainer of hydrophobic anticancer drug such as camptothecin (CPT), which showed to have a high efficiency against PANC-1 cells.7 In order to increase the cellular uptake and to develop a controlled release system, mesoporous silica nanoparticles have been modified on the surface with different molecules. These systems include nanocarriers with inorganic valves, such as CdS,9 Au10,11 or Fe3O412 which can be cleaved under reducing conditions, and nanovalves activated by light, redox reactions,13,14 temperature15 or as a function of pH.
A common goal in mesoporous silica nanoparticle drug delivery systems is to deliver the cargo in the cells responsible for the disease and only when they arrive at the targeted site, the active compound is released. Several strategies are being pursued to achieve this aim and one of them involves binding the targeting moiety to the silica structure through a disulfide bond cleavable subsequently with reducing conditions.9,16–18 In living cells the reducing condition is localized in the cytosol due to the high concentration of glutathione. Nevertheless this molecule is also present in the blood but with a concentration of ca. 2 μM instead of 10 mM of the cellular cytoplasm.19,20
However, most of the valves previously used do not exploit specific targeting stoppers and these moieties are often hydrophobic which limits their application in vivo due to severe aggregation in buffered solutions and cell-culturing media. Therefore, hydrophilic valves have also been reported making in vivo applications more feasible.21 Examples of hydrophilic surface modifications include polymers,22 DNA/dendrimer complexes23 and lipid bilayers.24 Monoclonal antibodies were used as part of the valve system and targeting moeity.25 Cheng et al. modified the surface of mesoporous nanoparticles with a small peptide for targeting cancer cells.26 However, in this case the peptide was not used as part of a valve system.
Herein, we report the use of oligopeptides as part of the nanovalves system in peptide modified mesoporous silica nanocontainers (PMSN) for the controlled release of drugs. The peptides decorating the surface of the particles have two functions: (I) to act as stimuli responsive nanovalves and (II) to ensure efficient cell uptake. The peptide sequence used in this study has a high selectivity towards cellular membranes and at the same time allows the suspension of the silica nanocarrier in water solutions in order to perform in vivo studies. In addition we studied the effect of the spacer between the silica nanoparticle and the peptide on the quality of the nanovalves (i.e. leakage) and its dispersion in aqueous media.
Spherical silica nanoparticles possessing a hexagonal mesoporosity (2.8 nm) were synthesized, see Fig. S1–S3 in the supporting information (SI).27–29 Previously we developed methods for the functionalization of SNP with rotaxanes as the nanovalves.30 Here we present a drug delivery system consisting of drug loaded mesoporous silica nanoparticles functionalized with different alkoxysilane linkers on which a α-cyclodextrin (α-CD) was tethered to form a rotaxane (Scheme 1).
Scheme 1.
Synthesis of the peptide modified mesoporous silica nanocontainers (PMSN). The modification with APTES is shown as an example.
To ensure that the α-CD remains at the thread we coupled a peptide stopper at the terminus of the tether via a reducible disulfide bond. This 12-mer oligopeptide is a fragment derived from the HIV-1 TAT peptide, which was first discovered in 1988.31 The TAT regulatory protein is an 86 amino-acid long nuclear protein of which 12 amino acid residues, CGRRRQRRKKRG, are responsible for translocation of exogenous molecules across the cell plasma membrane. The high affinity of this peptide to the membrane proteins allows the HIV-1 virus to easily penetrate in the immuno cells; hence this important feature has been exploited in many systems to overcome biological barriers, such as superparamagnetic contrast agents.
The release of the drugs from the mesoporous silica nanocontainer occurs when the disulfide bond between the tether and the peptide is cleaved under reducing conditions, which are also found in the cytoplasm of a cell, or by a reducing agent such as dithiothreitol (DTT).
An important criterion towards the development of an efficient drug delivery system is that the mesoporous silica nanocontainers should not release any of its drugs before it reaches its desired location. Therefore we also investigated whether the leakage and release profile was influenced by the tether length and the presence of the rotaxane structure. Thus the mesoporous silica nanoparticles were modified with either (3-Aminopropyl) triethoxysilane (APTES), γ-(2-Aminoethyl)-aminopropyl trimethoxysilane (AAPTMS), or (3-[2-(2-Aminoethylamino) ethylamino]propyltrimethoxysilane) (AEPTMS) resulting in amine surface functionalized nanoparticles with variable tether lengths. Next an activated thiol was introduced by coupling N-succinimidyl 3-[2-pyridyldithio]-propionate (SPDP) to the amine groups and the particles were loaded with fluorescein as a model drug. The mesoporous silica nanocontainers were incubated overnight with α-CD at 5 °C and subsequently the TAT-peptide was coupled at pH =9 for 2 days in order to cap the tether resulting in the formation of the rotaxane structure. The loading capacity of the PMSN is typically 0.4 mmol per gram as measured using UV-VIS spectroscopy. The in vitro release profile of fluorescein from the loaded peptide modified mesoporous silica nanocontainers (5 mg) was investigated upon the addition of DTT as the reducing agent (Fig. 1).
Fig. 1.
Release of fluorescein from PMSN as a function of the spacer composition. In all cases cyclodextrin was immobilized on an APTS (■), AAPTMS (●) and AEPTMS (▲) spacer. At t = 0 DTT was added and typically a lag time of 30 min was observed.
A lag time of 30 min was observed which most likely is due to the steric hindrance by the tethered α-cyclodextrins. This hypothesis was confirmed by the investigation of release profile of fluorescein from PMSNs in the absence of α-CD tethered on the linker, which shows an immediate release without any delay (See Fig. S9).
We observed that in the absence of DTT, the uncontrolled leakage of fluorescein is only slightly dependent on the length of the aliphatic tether (See Fig. S8). PMSNs modified with the longest aliphatic (AEPTMS) spacer released the highest amount of dye. Therefore it was concluded that the nanocarrier composed of cyclodextrin rotaxane tethered on the AAPTMS linker represents a good compromise for an efficient drug delivery system with minimal leakage.
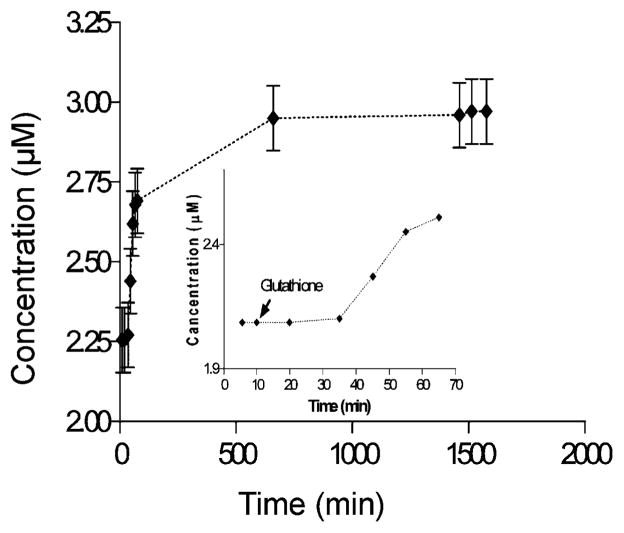
An effective delivery system requires also that it is activated in a cellular environment. The metabolic pathway of glutathione is one of the most common mechanisms exploited by the cell to reduce disulfide bonds in macromolecules. Therefore, we examined the fluorescein release for AAPTMS-PMSNs using glutathione as the reducing agent at a concentration similar to a cellular concentration (Fig. 2). The release profile showed a lag time of circa 30 min reflecting the same trend in fluorescein release compared to the release experiments induced by DTT.
Fig. 2.
Release profile of fluorescein from AAPTMS-PMSNs with cyclodextrin immobilized on the spacer. At t = 10 min glutathione is added to the system and a lag time of circa 30 min is observed.
It is a prerequisite for efficient cellular uptake that the PMSNs do not aggregate massively or even precipitate when dispersed in cell culturing media. Therefore the hydrodynamic radius of the PMSNs was studied using dynamic light scattering (DLS) in water and PBS buffer (See Fig. S11.) Interestingly, the presence of the α-CD showed to a positive effect on the aggregation of the particles. However, in all cases the aggregation was limited. Due to the high biocompatibility of silica nanoparticles with biological systems, the cellular uptake and internalization was studied using the HeLa cell line. A solution of the PMSN system (50 μg/mL) with the AAPTMS linker and the cyclodextrin [2] rotaxane was added to HeLa cells and incubated at 37 °C for 1 h.
Confocal laser scanning microscopy imaging using z-scans showed that the PMSNs were effectively internalized in all cells (Fig. 3). This behavior is most likely due to the high affinity of the TAT peptide towards cell membrane. The uptake was confirmed and quantified by flow cytometry analysis showing that 98% of the cells contained the particles. In contrast, silica nanoparticles without peptides showed only a 13% uptake.
Fig. 3.
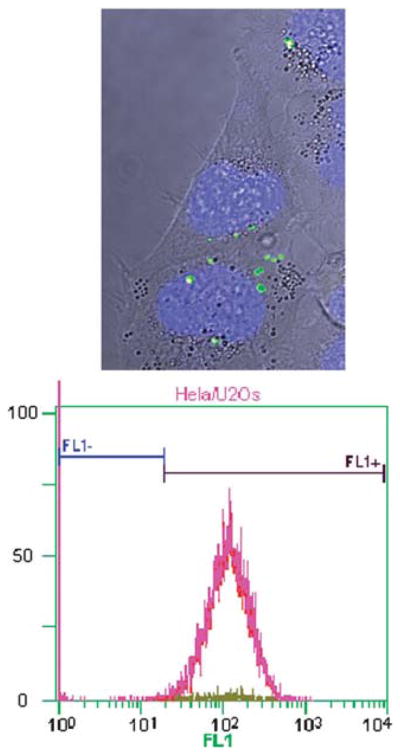
(Top) Bright-field confocal laser scanning microscopy z-scan image with fluorescent overlay of HeLa cells after incubation with PMSN (green) and with nuclei stained with DAPI (blue). (Bottom) Flow cytometry shows an internalization of the fluorescent PMSN of 98%. Fl1+ shows the fluorescent-labeled PMSN internalized cells, Fl1− shows the non-fluorescent cells.
In conclusion, we have shown that the PMSNs are effectively internalized into HeLa cells. Furthermore, the short targeting peptide sequence also acts as part of the reducible stopper for the cyclodextrin rotaxane valve. Current studies are investigating the potential use of these PMSNs as a targeted drug delivery tool will be tested in vivo using the zebrafish model.32 In this study we used the rather generic TAT peptide sequence to demonstrate the concept, but more specific peptide sequences will be used to achieve cell selectivity.
Supplementary Material
Acknowledgments
The authors acknowledge the support of the Smart Mix Programme of the Netherlands Ministry of Economics Affairs and the Netherlands Ministry of Education, Culture and Science. Claude Backendorf and Hans den Dulk are acknowledged for their support in cell culture and flow cytometry and Ke Peng for the synthesis of some intermediates. JIZ acknowledges the support of the US NIH RO1CA133697.
Footnotes
Electronic supplementary information (ESI) available: Experimental details; synthesis and characterization of the peptide modified silica nanocontainers. See DOI: 10.1039/c0cp02959a
Notes and references
- 1.Kataoka K, Harada A, Nagasaki Y. Adv Drug Delivery Rev. 2001;47:113–131. doi: 10.1016/s0169-409x(00)00124-1. [DOI] [PubMed] [Google Scholar]
- 2.Lee CC, MacKay JA, Frechet JMJ, Szoka FC. Nat Biotechnol. 2005;23:1517–1526. doi: 10.1038/nbt1171. [DOI] [PubMed] [Google Scholar]
- 3.Denny WA. Cancer Invest. 2004;22:604–619. doi: 10.1081/cnv-200027148. [DOI] [PubMed] [Google Scholar]
- 4.Duncan R, Ringsdorf H, Satchi-Fainaro R. J Drug Targeting. 2006;14:337–341. doi: 10.1080/10611860600833856. [DOI] [PubMed] [Google Scholar]
- 5.Tietze LF, Major F, Schuberth I. Angew Chem, Int Ed. 2006;45:6574–6577. doi: 10.1002/anie.200600936. [DOI] [PubMed] [Google Scholar]
- 6.Sauer AM, Schlossbauer A, Ruthardt N, Cauda V, Bein T, Brauchle C. Nano Lett. 2010;10:3684–3691. doi: 10.1021/nl102180s. [DOI] [PubMed] [Google Scholar]
- 7.Lu J, Liong M, Zink JI, Tamanoi F. Small. 2007;3:1341–1346. doi: 10.1002/smll.200700005. [DOI] [PubMed] [Google Scholar]
- 8.Slowing, Trewyn BG, Giri S, Lin VSY. Adv Funct Mater. 2007;17:1225–1236. [Google Scholar]
- 9.Lai CY, Trewyn BG, Jeftinija DM, Jeftinija K, Xu S, Jeftinija S, Lin VSY. J Am Chem Soc. 2003;125:4451–4459. doi: 10.1021/ja028650l. [DOI] [PubMed] [Google Scholar]
- 10.Vivero-Escoto JL, Slowing, Wu CW, Lin VSY. J Am Chem Soc. 2009;131:3462. doi: 10.1021/ja900025f. [DOI] [PubMed] [Google Scholar]
- 11.Aznar E, Marcos MD, Martinez-Manez R, Sancenon F, Soto J, Amoros P, Guillem C. J Am Chem Soc. 2009;131:6833–6843. doi: 10.1021/ja810011p. [DOI] [PubMed] [Google Scholar]
- 12.Giri S, Trewyn BG, Stellmaker MP, Lin VSY. Angew Chem, Int Ed. 2005;44:5038–5044. doi: 10.1002/anie.200501819. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen TD, Tseng HR, Celestre PC, Flood AH, Liu Y, Stoddart JF, Zink JI. Proc Natl Acad Sci U S A. 2005;102:10029–10034. doi: 10.1073/pnas.0504109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen TD, Liu Y, Saha S, Leung KCF, Stoddart JF, Zink JI. J Am Chem Soc. 2007;129:626–634. doi: 10.1021/ja065485r. [DOI] [PubMed] [Google Scholar]
- 15.Schlossbauer A, Warncke S, Gramlich PME, Kecht J, Manetto A, Carell T, Bein T. Angew Chem, Int Ed. 2010;49:4734–4737. doi: 10.1002/anie.201000827. [DOI] [PubMed] [Google Scholar]
- 16.Liu R, Zhao X, Wu T, Feng PY. J Am Chem Soc. 2008;130:14418–14419. doi: 10.1021/ja8060886. [DOI] [PubMed] [Google Scholar]
- 17.Liu R, Zhang Y, Feng PY. J Am Chem Soc. 2009;131:15128–15129. doi: 10.1021/ja905288m. [DOI] [PubMed] [Google Scholar]
- 18.Ambrogio MW, Pecorelli TA, Patel K, Khashab NM, Trabolsi A, Khatib HA, Botros YY, Zink JI, Stoddarrt JF. Org Lett. 2010;12:3304–3307. doi: 10.1021/ol101286a. [DOI] [PubMed] [Google Scholar]
- 19.Jones DP, Carlson JL, Samiec PS, Sternberg P, Mody VC, Reed RL, Brown LAS. Clin Chim Acta. 1998;275:175–184. doi: 10.1016/s0009-8981(98)00089-8. [DOI] [PubMed] [Google Scholar]
- 20.Koo AN, Lee HJ, Kim SE, Chang JH, Park C, Kim C, Park JH, Lee SC. Chem Commun. 2008:6570–6572. doi: 10.1039/b815918a. [DOI] [PubMed] [Google Scholar]
- 21.Coti KK, Belowich ME, Liong M, Ambrogio MW, Lau YA, Khatib HA, Zink JI, Khashab NM, Stoddart JF. Nanoscale. 2009;1:16–39. doi: 10.1039/b9nr00162j. [DOI] [PubMed] [Google Scholar]
- 22.Casasus R, Marcos MD, Martinez-Manez R, Ros-Lis JV, Soto J, Villaescusa LA, Amoros P, Beltran D, Guillem C, Latorre J. J Am Chem Soc. 2004;126:8612–8613. doi: 10.1021/ja048095i. [DOI] [PubMed] [Google Scholar]
- 23.Radu DR, Lai CY, Jeftinija K, Rowe EW, Jeftinija S, Lin VSY. J Am Chem Soc. 2004;126:13216–13217. doi: 10.1021/ja046275m. [DOI] [PubMed] [Google Scholar]
- 24.Liu JW, Jiang XM, Ashley C, Brinker CJ. J Am Chem Soc. 2009;131:7567. doi: 10.1021/ja902039y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai CP, Chen CY, Hung Y, Chang FH, Mou CY. J Mater Chem. 2009;19:5737–5743. [Google Scholar]
- 26.Cheng SH, Lee CH, Chen MC, Souris JS, Tseng FG, Yang CS, Mou CY, Chen CT, Lo LW. J Mater Chem. 2010;20:6149–6157. [Google Scholar]
- 27.Patel K, Angelos S, Dichtel WR, Coskun A, Yang YW, Zink JI, Stoddart JF. J Am Chem Soc. 2008;130:2382–2383. doi: 10.1021/ja0772086. [DOI] [PubMed] [Google Scholar]
- 28.Kresge CT, Leonowicz ME, Roth WJ, Vartuli JC, Beck JS. Nature. 1992;359:710–712. [Google Scholar]
- 29.Huh S, Wiench JW, Yoo JC, Pruski M, Lin VSY. Chem Mater. 2003;15:4247–4256. [Google Scholar]
- 30.Liong M, Angelos S, Choi E, Patel K, Stoddart JF, Zink JI. J Mater Chem. 2009;19:6251–6257. [Google Scholar]
- 31.Josephson L, Tung CH, Moore A, Weissleder R. Bioconjugate Chem. 1999;10:186–191. doi: 10.1021/bc980125h. [DOI] [PubMed] [Google Scholar]
- 32.Brittijn SA, Duivesteijn SJ, Belmamoune M, Bertens LFM, Bitter W, De Bruijn JD, Champagne DL, Cuppen E, Flik G, Vandenbroucke-Grauls CM, Janssen RAJ, De Jong IML, De Kloet ER, Kros A, Meijer AH, Metz JR, Van der Sar AM, Schaaf MJM, Schulte-Merker S, Spaink HP, Tak PP, Verbeek FJ, Vervoordeldonk MJ, Vonk FJ, Witte F, Yuan HP, Richardson MK. Int J Dev Biol. 2009;53:835–850. doi: 10.1387/ijdb.082615sb. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.