Abstract
Oncolytic viruses, which preferentially lyse cancer cells and stimulate an antitumor immune response, represent a promising approach to the treatment of cancer. However, how they evade the antiviral immune response and their selective delivery to, and replication in, tumor over normal tissue has not been investigated in humans. Here,we treated patients with a single cycle of intravenous reovirus before planned surgery to resect colorectal cancer metastases in the liver. Tracking the viral genome in the circulation showed that reovirus could be detected in plasma and blood mononuclear, granulocyte, and platelet cell compartments after infusion. Despite the presence of neutralizing antibodies before viral infusion in all patients, replication-competent reovirus that retained cytotoxicity was recovered from blood cells but not plasma, suggesting that transport by cells could protect virus for potential delivery to tumors. Analysis of surgical specimens demonstrated greater, preferential expression of reovirus protein in malignant cells compared to either tumor stroma or surrounding normal liver tissue. There was evidence of viral factories within tumor, and recovery of replicating virus from tumor (but not normal liver)was achieved in all four patients from whom fresh tissue was available. Hence, reovirus could be protected from neutralizing antibodies after systemic administration by immune cell carriage, which delivered reovirus to tumor.These findings suggest new preclinical and clinical scheduling and treatment combination strategies to enhance in vivo immune evasion and effective intravenous delivery of oncolytic viruses to patients in vivo.
INTRODUCTION
Naturally occurring or genetically modified oncolytic viruses (OVs)specifically target tumor cells for replication and cell death (1). In addition to their direct cytotoxic effects, OVs can also stimulate a therapeutic antitumor immune response (2). A number of OVs have now progressed through preclinical and early clinical testing, with no indication of major toxicity and encouraging evidence of antitumor activity (3). A phase 3 study of a herpes simplex virus (OncoVex) has been completed in melanoma (4), and a randomized trial using a vaccinia virus (JX-594) to treat liver tumors is due to open shortly (5).
The optimal route of administration for clinical application of OVs remains unresolved. Direct intratumoral injection ensures that the virus effectively accesses the tumor microenvironment for immune activation as well as direct cell killing and circumvents the concern of functional inactivation of intravenous virus in the circulation by neutralizing antibodies (NABs) present at baseline and/or induced on repeat administration. However, intratumoral injection is technically challenging and limits application to accessible tumor sites; moreover, systemic delivery remains more acceptable to clinicians.
Reovirus is a genetically unmodified, nonpathogenic double-stranded RNA virus with anticancer activity mediated by both direct targeting of malignant cells with activation of the ras pathway and stimulation of antitumor immunity (6, 7). Clinical-grade reovirus (type 3 Dearing; Reolysin) has been through phase 1/2 trials and is currently being tested intravenously in the phase 3 setting in combination with carboplatin and paclitaxel in squamous cell carcinoma of the head and neck (8). Although all patients carry NABs to reovirus after exposure to the virus in childhood (9), a small number of posttreatment tumor biopsies from early-phase trials have confirmed that reovirus can access tumors after systemic delivery (10, 11). Therefore, although intravenous reovirus is ineffective in mice previously immunized against the virus (12), in humans the presence of circulating NABs does not absolutely preclude successful delivery to tumors. However, how the virus is transported after intravenous injection from blood to tumor in patients has not been explored in humans.
Here, we report a window-of-opportunity clinical study in which a single cycle of intravenous reovirus monotherapy was given to patients before a planned resection of colorectal cancer metastatic to the liver. By analysis of sequential blood samples and resected tumor and normal tissue, we show that reovirus selectively replicates in tumor after protective blood cell carriage in the circulation. These data confirm intravenous reovirus targeting of tumor in patients and demonstrate how the virus evades NABs after systemic administration.
RESULTS
Patients, study design, and toxicity of the trial
Ten patients were recruited into this translational biological endpoint clinical trial. All patients were scheduled to undergo resection of colorectal cancer liver metastases with radical intent as part of standard clinical care. The patients’ clinical characteristics are shown in Table 1, and the trial schema, involving administration of a single cycle of intravenous reovirus before the planned surgery, is illustrated in Fig. 1. Treatment with reovirus was well tolerated, with the most common side effects being flu-like symptoms, consistent with previous clinical experience (8).
Table 1.
Patient demographics and clinical data. NA denotes samplesunavailable for analysis.
| Patient | Age (years) |
Sex | No.of reovirus infusions |
Time between final reovirus infusion and surgery (days) |
ras mutation status |
Toxicity (grade) |
|---|---|---|---|---|---|---|
| 1 | 74 | M | 3 | 7 | 12ASP | Nausea (2) |
| Constipation (2) | ||||||
| Headache (2) | ||||||
| Pyrexia (2) | ||||||
| Myalgia (2) | ||||||
| 2 | 62 | M | 5 | 11 | 13ASP, 12VAL |
None |
| 3 | 73 | M | 5 | 21 | 12VAL | None |
| 4 | 66 | M | 5 | 19 | 12ASP | Pyrexia (2) |
| 5 | 62 | M | 5 | 14 | 12CYS | Rigors (2) |
| 6 | 50 | F | 5 | 6 | None | Myalgia (1) |
| Pyrexia (1) | ||||||
| 7 | 58 | M | 4 | 28 | None | Pyrexia (1) |
| Leukopenia (2) | ||||||
| 8 | 65 | F | 1 | 10 | NA | Insomnia (2) |
| Hallucination (1) | ||||||
| Myalgia (2) | ||||||
| Rigors (2) | ||||||
| 9 | 70 | M | 5 | 6 | None | Pyrexia (1) |
| 10 | 63 | M | 5 | 14 | None | Pyrexia (1) |
| Hypotension (1) |
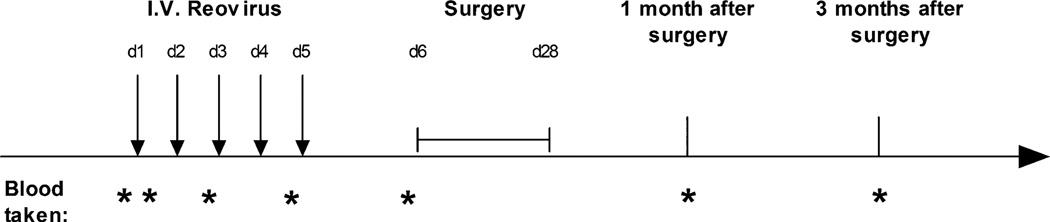
Fig. 1.
Trial schema is presented: timing of reovirus infusion, sample collection, and surgery. I.V., intravenous.
There were no grade 3 or 4 toxicities in any patient (Table 1). In three patients, fewer than the planned five doses of reovirus were given. In one patient, a single infusion was omitted because of clinical concern about a falling white cell count, whereas in the other two cases, only one and three treatments were given, respectively, because of the patients’ own concerns that flu-like symptoms might interfere with the planned surgery, leading them to decline subsequent infusions. Surgery took place between 6 and 28 days after the last reovirus treatment; in no case was surgery delayed because of reovirus toxicity.
The endpoints of this trial were as follows: (i) tracking the development of the NAB response to reovirus after treatment; (ii) detection of the virus in different blood compartments [plasma, peripheral blood mononuclear cells (PBMCs), granulocytes, platelets, and red cells]; (iii) assessment of reovirus within resected tumor and normal liver; and (iv) monitoring of toxicity, particularly in relation to the planned surgery.
Detection of reovirus genome, but not replication-competent virus, in plasma after systemic delivery, despite the presence of NABs
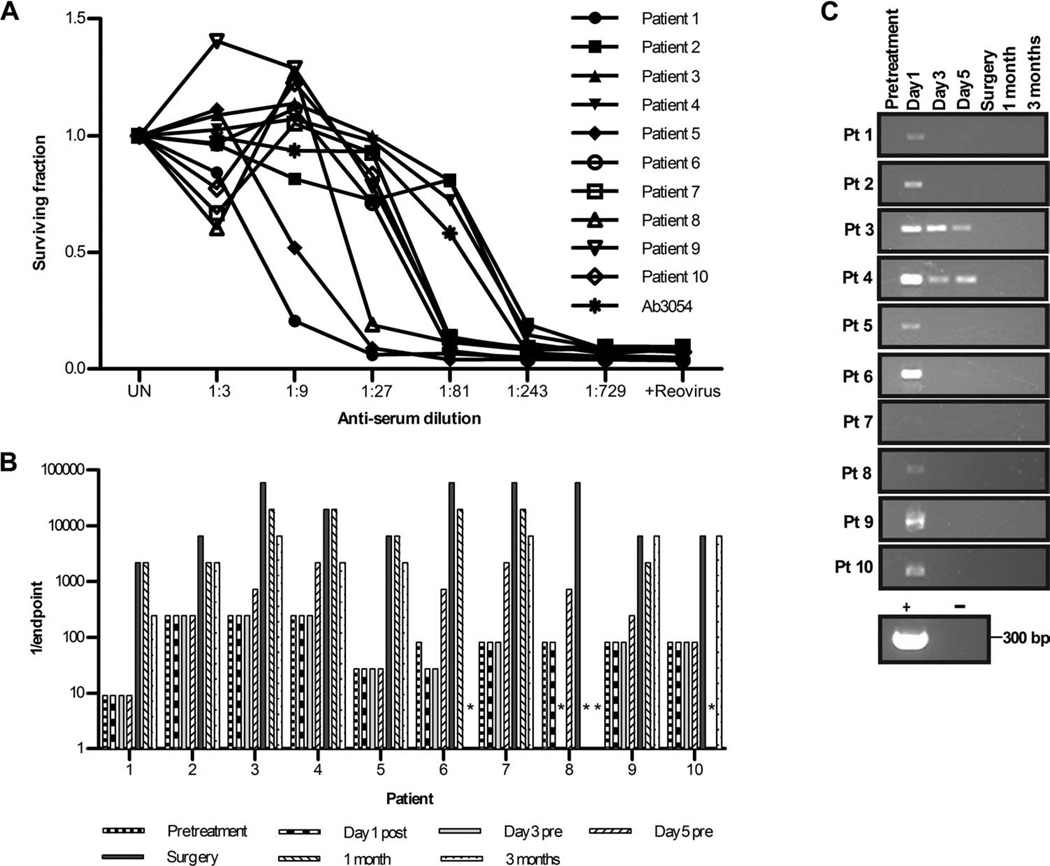
We first confirmed that anti-reovirus NABs were present in our patients at baseline (Fig. 2A) and, consistent with other intravenous monotherapy trials (10, 13), found that titers increased after intravenous treatment, peaking around the time of surgery (Fig. 2B). We then tested for the presence of virus, initially in plasma, using the following assays. First, reverse transcription–polymerase chain reaction (RT-PCR) of RNA was performed directly (“neat” RT-PCR) to look for the viral genome. Next, retrieval of replication-competent virus was assessed by adding plasma to reovirus-sensitive L929 cells in a median tissue culture infectious dose (TCID50) assay. This quantifies viral titer by demon-stration of the cytopathic effect (CPE) and cytotoxicity of serial dilutions of samples against L929 cells (14). Further, to confirm increased genome band intensity consistent with viral replication, we performed an “amplified” RT-PCR on RNA extracted from L929 cells at the end of this TCID50 assay. Clear bands were seen on immediate RT-PCR of plasma at 1 hour after the first infusion, whereas in all but two patients, no reovirus genome was detected at later time points (Fig. 2C). In patients 3 and 4, bands were also seen on days 3 and 5 (in samples taken immediately before the third and fifth infusions, respectively). However, when plasma was tested in virus amplification assays on L929 cells, no replicating reovirus was seen as evidenced by either CPE/cytotoxicity or a positive, amplified RT-PCR signal. Hence, free virus in the circulation is readily detectable in plasma, particularly early during treatment, but is functionally neutralized for productive infection and cell killing, presumably by the NABs present at baseline before the first reovirus infusion.
Fig. 2.
NABs are present at baseline and increase after treatment, whereas the viral genome is only transiently detectable after infusion in plasma. (A) Samples collected before virus infusion were tested for baseline NAB levels. Plot shows neutralization of reovirus-induced killing of L929 cells by serial dilutions of samples as measured by MTT assay at 72 hours. L929 cells were treated with reovirus only (+Reovirus) or left untreated (UN) as positive and negative controls, respectively; anti-reovirus polyclonal goat antibody (Ab3054) was used as a standard curve. (B) Changes in endpoint reovirus NAB titer over time (asterisk denotes samples unavailable for analysis). (C) Patient plasma was assessed for reovirus RNA by RT-PCR over time. Reovirus RNA and RNase-free water were included as positive and negative controls, respectively. bp, base pairs.
Transport of reovirus that is functional for replication and target killing by PBMCs
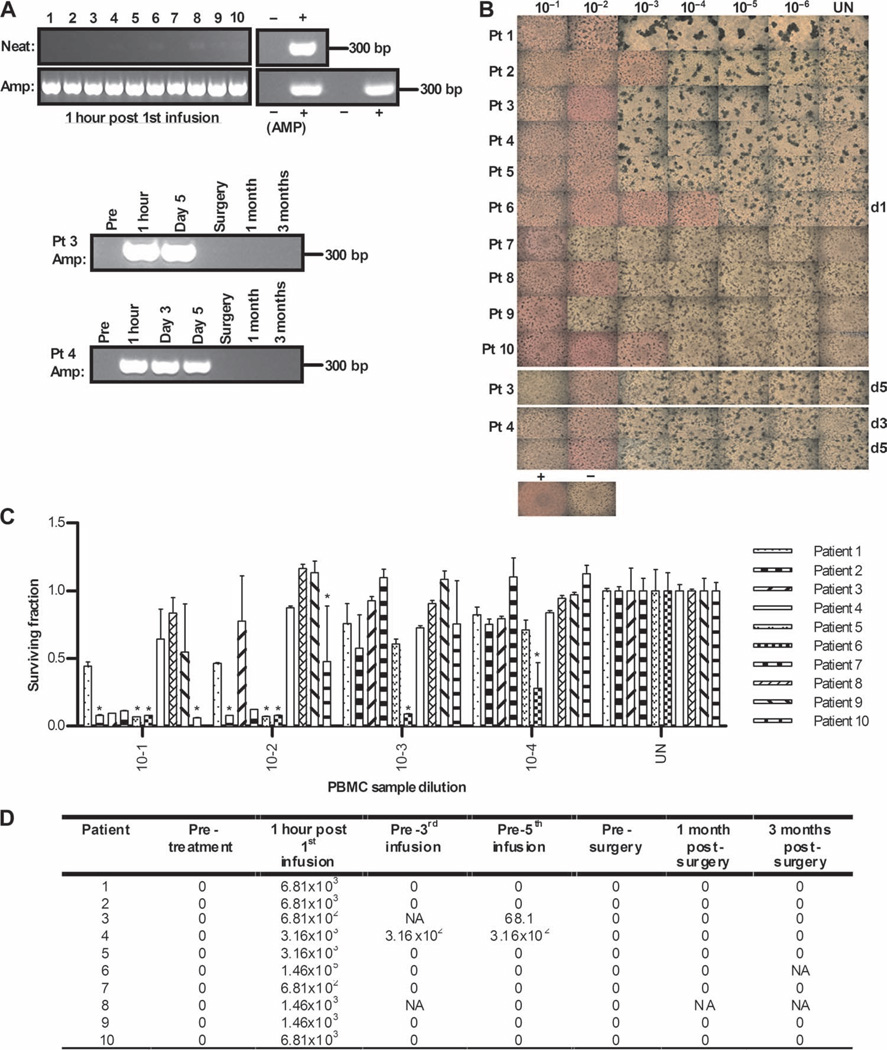
We have previously shown in preclinical models that purified dendritic cells (DCs) and T cells, cultured ex vivo, can act as carrier cells for reovirus and are able to protect the virus from NABs for therapeutic delivery to tumors (12, 15). We therefore hypothesized that PBMCs, which contain both DCs and T cells, may similarly “hitchhike” reovirus in patients after intravenous delivery. PBMCs were isolated from patient blood samples and again tested for input (neat) and amplified (replication-competent) virus. Figure 3A shows by RT-PCR as previously described (10) with a detection limit reovirus copy number of 50 (fig. S1) that neat reovirus could be identified in PBMCs at 1 hour after the first infusion, as evidenced by weak bands visible in patients 4, 6, 8, 9, and 10. Such bands were also apparent in more patients when alternative primers were used (fig. S2). However, in marked contrast to plasma, this signal was markedly amplified in all patients after culture of PBMCs on L929 cells for 7 days, consistent with viral handoff to, and replication in, target L929 cells in vitro. This amplified signal was also detected at later time points in patient 3 (day 5; a day 3 sample was not available) and patient 4 (days 3 and 5); all later time points were negative in all other patients. The presence of replicating, cytotoxic virus on patient PBMCs was further evidenced by CPE of L929 cells (Fig. 3B), as well as L929 cytotoxicity as measured by MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay (Fig. 3C) at the end of the 7-day PBMC/L929 coculture. Finally, the viral titer (TCID50/ml) on PBMCs was calculated (16), as shown in Fig. 3D. Patients 3 and 4 were the only cases in which replicating virus was detected in amplification assays beyond the 1-hour post–first infusion time point (Fig. 3, B to D); these same patients remained plasma-positive by neat PCR at later time points (Fig. 2B). Together, these data show that PBMCs carry virus in patients after intravenous delivery, despite the presence of NABs at baseline (Fig. 2A). In marked contrast to plasma, functional, replicative PBMC-associated virus can be handed off to target L929 cells in vitro, thus providing a potential protective delivery mechanism for systemic reovirus to tumor in patients.
Fig. 3.
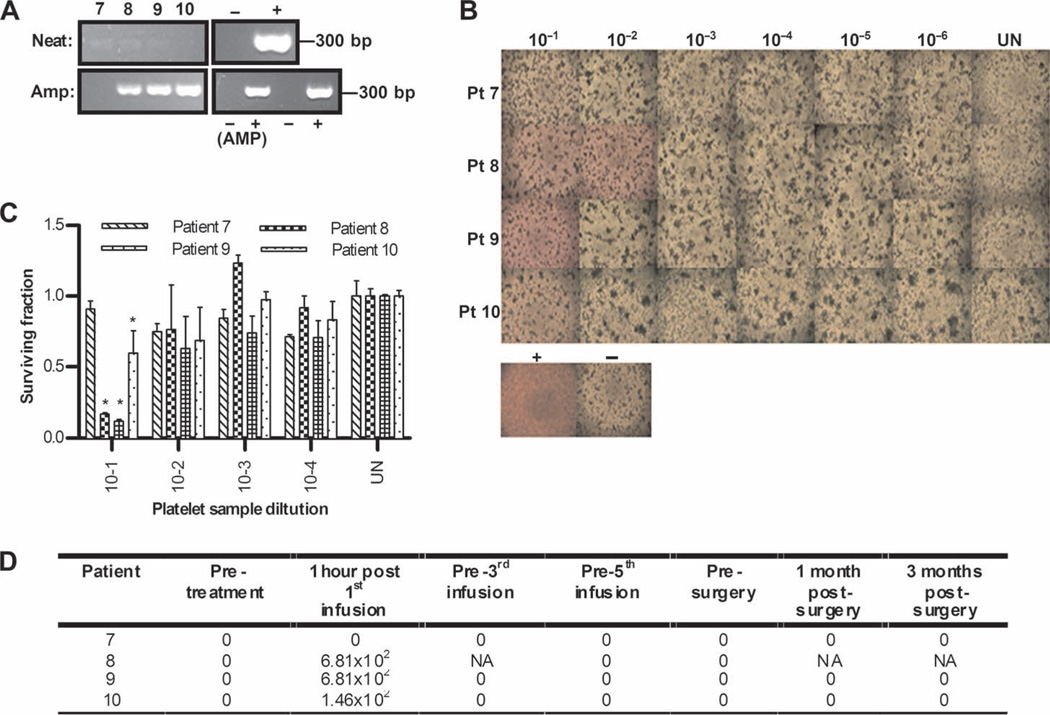
Despite circulating NABs, PBMCs transiently carry reovirus after infusion, which can replicate in and kill target cells in vitro. (A) Day 1 post-infusion PBMCs were assessed directly for reovirus RNA by RT-PCR (neat) or after an additional amplification step on L929 cells for 7 days (amplified). Reovirus RNA and RNase-free water were included as positive and negative controls, respectively, alongside a 1:10 dilution of stock reovirus or 5% DMEM incubated on L929 cells as amplified positive and negative controls (AMP). Later time points for patients 3 and 4 are also shown. (B) PBMCs were assessed for functional reovirus in a TCID50 assay. L929 cells were cultured with serial dilutions of PBMCs and observed 7 days later for CPE. Photomicrographs show results from day 1 post-infusion PBMCs for all samples and later time points for patients 3 and 4. Dilution (1:10) of stock reovirus or 5% DMEM (UN) was incubated on L929 cells as positive and negative controls, respectively. Rounded up cells and unused (red) media signify CPE. (C) Reovirus-induced cell killing by day 1 posttreatment PBMCs was further confirmed by MTT analysis. *P < 0.05 versus untreated control; error bars represent SEM. (D) Viral titers (TCID50/ml) from PBMCs over time (NA denotes samples unavailable for analysis).
Transport of reovirus by granulocytes and platelets, but not red blood cells, in patients
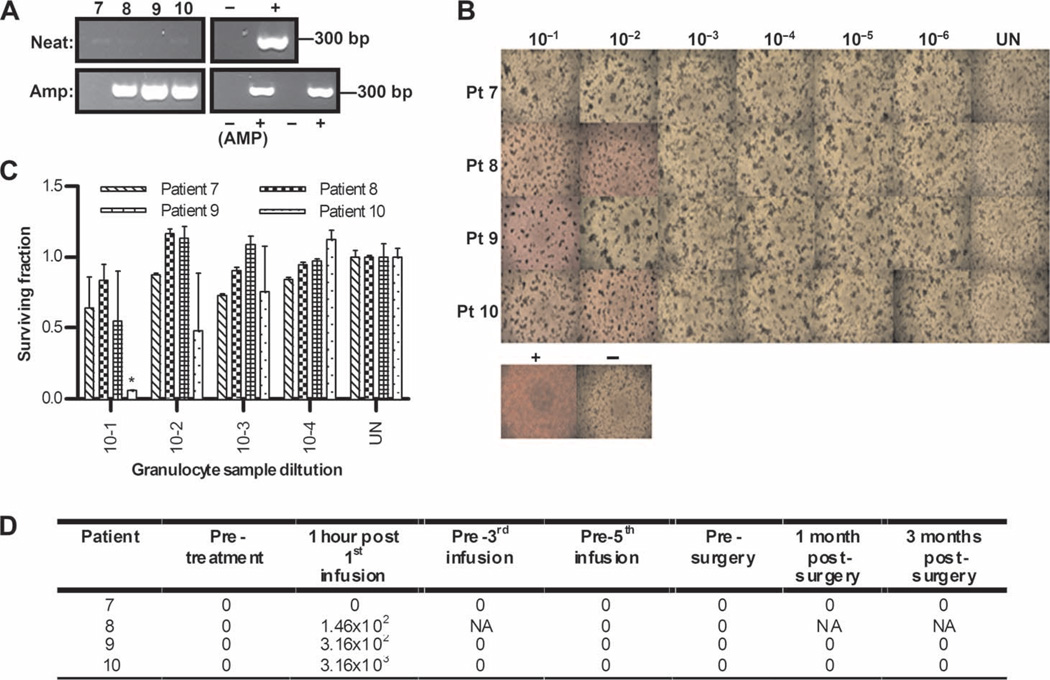
Using the same assays as described for PBMCs, we next explored other fractions of blood cells collected from patients for their ability to hitchhike reovirus. Specifically, we tested granulocytes, platelets, and red blood cells, blood components potentially able to bind to and/or carry virus (granulocytes and platelets were available only from patients 7 to 10). Figures 4 and 5 show that, in three of four patients, both granulocytes and platelets, similar to PBMCs, carried oncolytically functional virus, as evidenced by assays of neat and amplified RT-PCR (Figs. 4 and 5A and fig. S2), CPE (Figs. 4 and 5B), and target killing (Figs. 4 and 5C) of L929 cells. Viral titers are shown in Figs. 4 and 5D, which also show that reovirus was detected only in granulocytes and platelets 1 hour after the first infusion (the corresponding neat and amplified RT-PCR assays were also negative at later time points in all cases for granulocytes and platelets). Moreover, no reovirus was detected in red blood cells at any time point in any sample. However, because immediate RT-PCR was the only assay technically feasible for these samples, we cannot absolutely exclude red cells as capable of functional virus carriage. Hence, granulocytes and platelets, as well as PBMCs, although not apparently red blood cells, can potentially hitchhike reovirus to tumor targets in patients despite the presence of NABs.
Fig. 4.
Granulocytes similarly carry replication-competent reovirus after infusion. (A) Day 1 post-infusion granulocytes were assessed for reovirus RNA by RT-PCR, using both neat and amplified samples as for PBMCs in Fig. 3A. (B) Granulocytes were assessed for functional reovirus in a TCID50 assay as for PBMCs in Fig. 3B. Photomicrographs show day 1 post-infusion granulocyte dilutions; rounded up cells and unused (red) media signify CPE. (C) Reovirus-induced cell killing by day 1 posttreatment granulocytes was further confirmed by MTT analysis. *P < 0.05 versus untreated control; error bars represent SEM. (D) Viral titers (TCID50/ml) from granulocytes over time (NA denotes samples unavailable for analysis).
Fig. 5.
Platelets also carry reovirus after infusion. (A) Day 1 post-infusion platelets were assessed for reovirus RNA by RT-PCR, using both neat and amplified samples as for PBMCs in Fig. 3A. (B) Platelets were assessed for functional reovirus in a TCID50 assay as for PBMCs in Fig. 3B. Photomicrographs show day 1 post-infusion platelet dilutions; rounded up cells and unused (red) media signify CPE. (C) Reovirus-induced cell killing by day 1 posttreatment platelets was further confirmed by MTT analysis. *P < 0.05 versus untreated control; error bars represent SEM. (D) Viral titers (TCID50/ml) in platelets over time (NA denotes samples unavailable for analysis).
Preferential detection of reovirus in tumor cells after systemic delivery
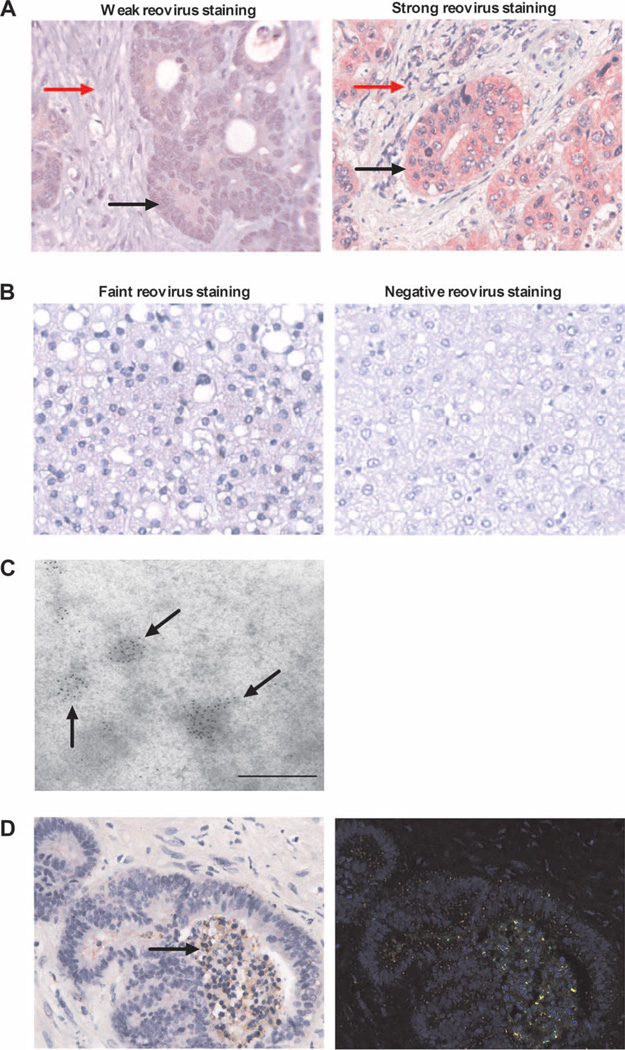
Resected tumor and surrounding normal liver (excised as a margin around metastases) were first tested for reovirus by immunohistochemistry for the σ3 capsid protein (11). In 9 of the 10 patients (but in none of 3 control patients tested who had resections out with the trial), reovirus protein was detected. Tumor staining was scored as absent (1 patient), weak (3 patients), or strong (6 patients). Figure 6A shows representative data from 1 patient with weak and one with strong tumor staining. In all nine cases where tumor was positive for reovirus, the colorectal metastases stained more strongly than tumor stroma (black versus red arrows in Fig. 6A) or surrounding liver (see below, Fig. 6B), consistent with selective reovirus delivery to, and/or replication in, malignant over nonmalignant cells. Some staining of normal liver tissue was seen. In five patients, faint positive staining of hepatocytes was seen (in four of which there was associated strong tumor staining, and in one weak tumor staining), whereas the liver tissue samples of the other five cases were scored as negative (including the single case in which tumor was negative). Figure 6B shows a representative example of faint hepatocyte staining as well as a negative case. The presence of reovirus protein in resected tumor was further confirmed by electron microscopy (EM) (Fig. 6C). Further, in four patients where enough tissue was available for analysis, reovirus was seen to colocalize with caspase-3 (Fig. 6D), indicating tumor cell apoptosis. The changes of nuclear and cytoplasmic degeneration in infected tumor cells (black arrow) are also consistent with reovirus-associated apoptosis in the caspase-3–positive cells. The complete histological and EM data for all patients are summarized in table S1. Hence, systemically administered reovirus was found in resected tissue in 9 of 10 cases, with tumor cells consistently staining more strongly than either tumor-associated stroma or resected, adjacent normal liver.
Fig. 6.
Intravenous reovirus is preferentially detected in metastatic colorectal tumor cells within the liver. (A) Immunohistochemistry images showing expression of reovirus protein (red stain) in resected colorectal liver metastases (magnification, ×400). One representative case each of weak (left; patient 1) and strong (right; patient 6) staining is shown. Malignant cells and tumor stroma are marked by black and red arrows, respectively. (B) Immunohistochemistry images for expression of reovirus protein (red stain) in normal liver (magnification, ×400). One representative case each of faint (left; patient 8) and negative (right; patient 2) staining is shown. (C) Representative EM image (patient 8) showing immunogold staining of reovirus σ3 capsid protein within colorectal liver metastases. Scale bar, 500 nm. (D) RGB image analyses of resected colorectal liver metastases, using the Nuance System (magnification, ×400). Images are representative (patient 9) and show reovirus staining (red) and caspase-3 staining (brown) (left image; arrow indicates changes of nuclear and cytoplasmic degeneration in reovirus-infected tumor cells). Right image shows conversion of RGB image to fluorescent green (caspase-3), fluorescent red (reovirus), and yellow (coexpression).
Retrieval of replication-competent reovirus from resected tumor but not liver tissue
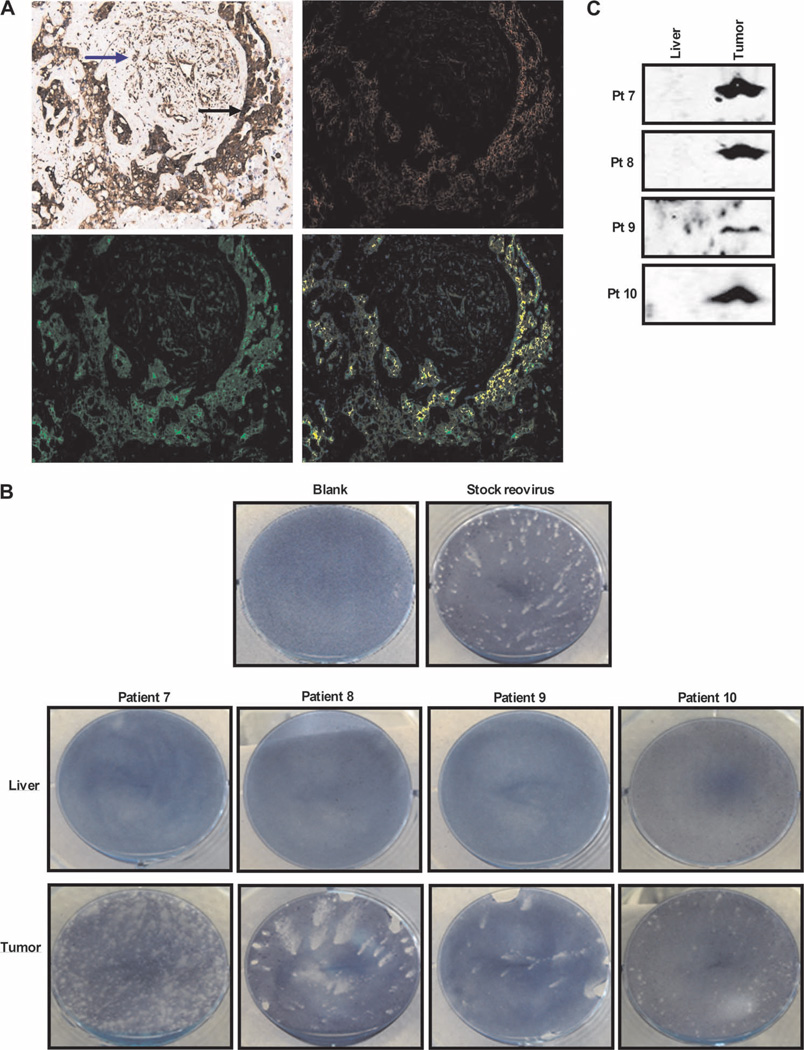
The detection of reovirus capsid protein illustrates successful delivery of virus to target tissue, but does not address whether the virus is, or has been, functional for replication. To further address the question of replication of reovirus specifically in tumor, we first stained sections for colocalization of reovirus and tubulin as an indirect marker of replication within viral factories (11). Colocalization was seen in four of six assessable tumors, with costaining confined to tumor, as opposed to stromal, cells (table S2; a representative patient is shown in Fig. 7A). Next, we sought to directly retrieve replication-competent reovirus from resected specimens. Initially, when freeze-thaw lysates from tumor and liver were pulsed onto L929 cells, no CPE or viral plaques were seen. However, for patients 7 to 10, we were able to perform additional experiments designed to increase the sensitivity of viral detection by taking tissue direct from theater, making single liver or tumor cell suspensions, and processing these directly as described in Materials and Methods. This modified technique avoids any loss of virus during lysate preparation (because of freeze-thaw and/or retention of virus in pelleted cell debris) and also harnesses the enhanced delivery of reovirus to targets from intact infected cells compared to free virus (15). Under these conditions, fresh tumor (but not liver) cells from all four patients tested did yield replicating virus, as demonstrated by plaques on L929 cells as shown in Fig. 7B. Figure 7C confirms these plaques as reovirus by Western blot, whereas liver samples were again negative. Hence, replication-competent reovirus could be retrieved from tumor, but not liver, of patients after intravenous delivery.
Fig. 7.
Replication-competent reovirus can be retrieved from tumor tissue. (A) RGB image analyses of resected colorectal liver metastases, using the Nuance System (magnification, ×200). Images are representative (patient 10) and show (top left) reovirus staining (red) and tubulin staining (brown) (malignant cells and tumor stroma are marked by black and blue arrows, respectively), (top right) conversion of RGB image to fluorescent red (reovirus), (bottom left) conversion of RGB image to fluorescent green (tubulin), and (bottom right) coexpression of reovirus and tubulin (yellow). (B) Plaque assays demonstrating retrieval of reovirus from freshly resected tumor and liver tissue; photographs show representative wells of optimized supernatant dilutions of 1:2500 (patient 7), neat supernatant (patients 8 and 9), and 1:1200 (patient 10). Photographs of all liver samples show representative wells of neat supernatant. (C) Culture supernatants from plaque assays performed in (B) were assessed for reovirus σ3 capsid protein by Western blotting.
Surgical outcome
All 10 patients proceeded to their planned surgery with no delay attributable to reovirus treatment, and there was no unexpected or excessive surgical morbidity in comparison to nontrial patients undergoing similar operations.
DISCUSSION
OVs represent a promising class of anticancer agent that target tumors through both direct cytotoxic and immune-mediated mechanisms (1). Reovirus is one of two OVs that have reached phase 3 testing, and there is currently significant interest and investment in OVs from the commercial as well as academic sectors (17).
The optimal route of OV treatment is unresolved, with the genetically modified herpes simplex virus, OncoVex, given by intratumoral injection (4), unlike clinical-grade reovirus (Reolysin), which has been administered in most studies (including the ongoing phase 3 trial) intravenously (8, 18). Although direct intratumoral delivery ensures viral access to the tumor and avoids potential systemic neutralization, intravenous injection is more practical for clinical use and can readily target multiple tumor sites.
Limited data from posttreatment tumor biopsies in early trials have shown that reovirus can reach tumor in patients, despite the presence of circulating NABs (10, 11). Recently, viral and encoded transgenes from the vaccinia OV JX-594 were shown to be expressed in malignant cells within tumors after systemic delivery, although in only one patient in this single administration phase 1 trial was delivery confirmed in the presence of NABs (19). Although preclinical studies have shown that a variety of purified carrier cells generated ex vivo could shield OVs from neutralization in the circulation (20), intravenous reovirus targeting of tumors and the mechanisms by which OVs in general might evade the systemic immune response have not previously been addressed in patients.
In the biological endpoint study reported here, we used a “window of opportunity” for intravenous reovirus delivery to 10 patients before a planned resection of colorectal cancer metastatic to the liver. Colorectal cancer is an appropriate target for reovirus oncolysis, with a reported ras mutation rate of 37% (21). Within the current study, 5 of the 10 patients’ tumors were ras mutant (Table 1), although the mechanisms determining reovirus sensitivity are complex and incompletely understood. The clinical scenario of tumor spread to the liver may be particularly appropriate for OV therapy because the liver is known to sequestrate viruses after systemic delivery (22). We showed that in 9 of 10 patients, more reovirus protein was seen at the time of surgery in malignant cells compared to tumor stroma or normal liver and that the presence of reovirus was associated with cellular degeneration and apoptosis (Fig. 6 and table S1). Although these data demonstrate targeted viral delivery to, and/or replication in, malignant versus normal cells, it cannot definitively prove causation between the presence of reovirus and tumor cell apoptosis. However, reovirus infection does induce apoptosis in human melanoma and colorectal cancer cell lines in vitro (23).
The higher reovirus levels detected in tumor cells in these patients are more likely due to selective replication than greater infection, firstly because reovirus receptors are ubiquitously expressed, and primary human hepatocytes, unlike colorectal cancer cell lines, are resistant to reovirus replication and killing in vitro. Moreover, EM findings and the colocalization of reovirus and tubulin are consistent with viral reproduction “factories” (11). Most importantly, however, replication-competent reovirus could be retrieved from the tumor in four of four patients tested, when an optimized assay using fresh intact tumor cells without freeze-thaw was used (Fig. 7B). Virus could not be similarly retrieved from the corresponding liver samples [despite the presence of low-level viral capsid protein as evidenced by immunohistochemistry (Fig. 6B)], directly demonstrating selective replication of an OV in malignant versus normal tissue within the same patient. Although the selective retrieval of replicating reovirus from tumor in this monotherapy trial is encouraging, levels may be further increased by combination strategies incorporating immunomodulatory chemotherapy (24), and we are currently developing further studies to specifically address this question.
Despite successful delivery of reovirus to tumor, the effectiveness of the early innate response for viral clearance was apparent from analysis of various blood compartments, in which RT-PCR signals for the reovirus genome were restricted in all but two patients to the 1-hour post–first infusion sample. Most of later day 3 and day 5 samples (taken immediately before the third and fifth infusions, and therefore almost 24 hours after the second and fourth infusions, respectively) were negative, which suggests that the virus was generally cleared from the circulation within hours of administration. Whether blood taken an hour after infusions 2 to 5 would carry virus in the same way as early samples after the first treatment, and how much these later treatments contribute to additional tumor delivery, is unknown and will be a focus of further investigation. Nevertheless, the finding that the tumors of the three patients who received fewer than the five planned infusions all stained positive for reovirus, and that two of these (of two tested) also yielded replicating virus (of which one had only one injection), whereas the only negative case was from a patient who received all five infusions, raises the question of whether a repeated daily reovirus regimen is optimal. Moreover, these clinical findings highlight how biological endpoint studies of this type may help inform protocol development; for example, more widely spaced virus infusions, timed to coincide with a chemotherapy-induced blunted innate immune response, might enhance viral persistence in the circulation and associated delivery to tumors.
Although immediate RT-PCR bands were seen in plasma, virus competent for handoff, PCR amplification and killing of L929 targets, could not be retrieved from this compartment, demonstrating effective functional neutralization of reovirus in plasma, most likely by NABs. In contrast, PBMCs, granulocytes, and platelets collected from treated, NAB-positive patients effectively hitchhiked and protected reovirus for handoff and killing of target cells ex vivo. However, carriage by different blood cells within one patient was not inevitably linked; PBMCs from patient 7 hitchhiked reovirus effectively, whereas the patient’s granulocytes and platelets did not. Moreover, detection of the viral genome in cells did not always guarantee protective carriage, as illustrated by the neat RT-PCR band seen in patient 7’s granulocytes and platelets, which could not be amplified by culture with L929 cells, and was not associated with L929 killing. Although purified human DCs have previously been shown in vitro to shield reovirus from NABs by internalizing the virus (15), further studies addressing the mechanisms by which different cell populations may carry and protect OVs in the circulation are warranted. Moreover, the current study cannot exclude a role for non–cell-associated delivery of free virus to tumor within the first hour after treatment.
Accepting the small numbers in this study, no obvious correlation was seen in any patient between baseline NAB levels/number of injections/time to surgery and viral titers/prolonged detection in blood, or reovirus detection in/retrieval from tumor. The failure of NABs to block virus delivery is encouraging for clinical application, because it shows that cell carriage allows OVs to evade NABs for delivery to tumor and suggests that concerns that baseline and/or induced NABs will inevitably prevent tumor access in patients have been overstated. Moreover, additional adjustments to scheduling, such as increasing very early dose intensity relative to prolonged repeat administration, may further enhance systemic OV delivery to tumors by maximizing the viral load before significant antiviral immune activation.
Although this trial had primarily translational biological endpoints, it involved clinical systemic delivery of an OV before a planned operation. The absence of any observed surgical toxicity is important because it supports the development of OV neoadjuvant protocols with defined clinical endpoints for colorectal and other cancers.
Although this study does not address the therapeutic potential of reovirus in this neoadjuvant setting and cannot definitively prove a causal link between cell-associated reovirus in the circulation and specific delivery to and/or replication in tumor leading to apoptosis, these data confirm that systemically delivered OVs can access and specifically replicate in tumors in patients despite the presence of NABs. These human data contrast with preclinical mouse models, in which successful therapy in virus preimmune animals is difficult to achieve (12), and show that complex strategies, including ex vivo culture of protective carrier cells, may not be necessary in the clinic. Rather, an improved understanding of how cells can shield OVs for tumor targeting, and which components of the immune response are most relevant for systemic viral delivery as well as therapy, will enlighten how best to manipulate OV application and scheduling to realize the promise of this class of anticancer agent.
MATERIALS AND METHODS
Patients, study procedures, and sample collection
Patients with colorectal cancer metastatic to the liver scheduled to undergo routine, planned resection with radical intent were approached about the study at a single center. Written, informed consent was obtained in accordance with local institutional ethics and review approval. Clinical-grade Dearing type 3 Reolysin was provided by Oncolytics Biotech Inc., but otherwise, the trial was sponsored, run, and funded by the University of Leeds, UK. Eligibility criteria included age 18 to 75 years, completion of any previous therapy at least 4 weeks before entry into the study (2 of 10 patients had had neoadjuvant chemotherapy to reduce disease burden before surgery), Eastern Cooperative Oncology Group (ECOG) performance score of ≤2, life expectancy of at least 3 months, absolute neutrophil count ≥1.5 × 109/liter, platelets ≥100 × 109/liter, hemoglobin ≥9.0 mg/dl, serum creatinine ≤1.5 times the institutional upper limit of normal (ULN), total bilirubin ≤1.5 times the ULN, aspartate transaminase/alanine transaminase ≤2.5 times the ULN, and a negative pregnancy test for females of childbearing potential. Exclusion criteria included extensive liver disease requiring surgery more extensive than an extended hemihepatectomy; known brain metastases; known HIV, hepatitis B, or hepatitis C infections; pregnancy or breast-feeding; clinically significant cardiac disease (New York Heart Association class III or IV); and dementia or altered mental state that would prohibit informed consent. The study was approved by the appropriate ethics and biological safety committees (EUDRACT number 2007/000258-29; MREC08/H1306/73). As described in previous studies (10), a single cycle of 1 × 1010 TCID50 reovirus was prepared and administered as an intravenous infusion over 60 min daily from days 1 to 5, between 6 and 28 days before surgery. Blood samples were taken immediately before and 1 hour after the first reovirus infusion; immediately before infusions on days 3 and 5; within 3 hours before surgery; and at 1 and 3 months after surgery. Tumor and adjacent normal hepatic parenchyma were excised, preserving margins for histological diagnosis and confirmation of the adequacy of resection at the time of surgery.
Blood sample processing
PBMCs were isolated from K3EDTA anticoagulated whole blood by density gradient centrifugation with Lymphoprep (Axis-Shield UK) as per the manufacturer’s instructions. PBMCs were frozen in freezing medium [90% fetal calf serum (FCS); Biosera] containing 10% dimethyl sulfoxide (DMSO) (Sigma-Aldrich Ltd.) at a cell density of 5 × 106/ml. Granulocytes were isolated from the same K3EDTA anticoagulated whole blood with a two-step procedure. Briefly, after PBMC harvest,granulocytes were collected from the lower white cell interface. Granulocytes were then further purified with Polymorphprep (Axis-Shield), as per the manufacturer’s instructions. Purified granulocytes were frozen in freezing medium at a cell density of 1 × 107/ml. Platelets and red blood cells were isolated from K3EDTA anticoagulated whole blood collected in a 6-ml Vacuette (Greiner Bio-One Ltd.). The Vacuette was centrifuged at 210g with no brake for 10 min, and the platelet-rich plasma (PRP) top layer was collected (the red blood cells were retained for processing separately) before a further centrifugation at 210g for 10 min was performed to remove any contaminating white blood cells. PRP was then centrifuged at 800g for 10 min to pellet the platelets. Platelets were then washed twice in 5 ml of MACS buffer [Ca2+/Mg2+-free phosphate-buffered saline (PBS) (Oxoid) + 1% FCS + 0.2% EDTA (Sigma)], centrifuging at 800g for 10 min for each wash. Platelets were frozen in 1 ml of ribonuclease (RNase)–free water (Sigma). The red blood cells retained in the Vacuette were centrifuged at 2000g for 10 min. The upper white blood cell layer was removed before aliquots of red blood cells were collected and frozen. Serum was isolated from whole blood collected in serum clot activator Vacuettes (Greiner), and plasma was isolated from whole blood collected in K3EDTA Vacuettes. Vacuettes were spun at 2000g for 10 min before serum and plasma were collected from the upper interfaces and aliquots were frozen. All samples were stored at −80°C until required.
Cell lines and reovirus
The murine fibroblastic cell line L929 was cultured in Dulbecco’s modified Eagle’s medium (DMEM; Sigma) supplemented with 5% (v/v) FCS, 1% (v/v) glutamine (Sigma), and 0.5% (v/v) penicillin/streptomycin (5% DMEM) (Sigma). Cells were cultured at 37°C in a humidified atmosphere containing 5% CO2 and were routinely tested for, and found to be negative of, Mycoplasma infection. Clinical-grade Dearing type 3 reovirus (Reolysin) was titered with a standard plaque assay protocol and stored in the dark at −80°C for laboratory experiments.
TCID50 experiments
L929 cells were seeded at 1 × 104 in 96-well plates and incubated for 24 hours before experiments. Supernatants were aspirated and 100 µl of 10-fold dilutions (starting at 1:10) of PBMCs, granulocytes, platelets, and plasma samples was added to L929 cells. Additional plating medium was added to the wells 1 to 2 hours after infection/treatment, and cells were then incubated for 6 days. A 1:10 dilution of stock reovirus and 5% DMEM were incubated on L929 cells as positive and negative controls, respectively. CPE (reovirus-induced cell death) was assessed by examination under a light microscope to calculate viral titer in TCID50/ml [with the Spearman-Karber statistical method, as previously described (16)]; photomicrographs were also taken. L929 cell survival/cell death was also confirmed at the end of the assay with an MTT assay, as described below.
MTT assay
Cell survival was quantified with an MTT (Sigma) assay. Twenty microliters per well of MTT at 5 mg/ml was added to treated cells. After 4 hours of incubation at 37°C, crystals were solubilized in DMSO and absorbance was measured at 550 nm.
RNA detection
RNA was extracted from 7 × 105 PBMCs, granulocytes, platelets, red blood cells, and plasma samples with the QIAamp Viral Mini Kit and amplified with the OneStep RT-PCR Kit (both Qiagen). Reovirus σ3 complementary DNA targeted primers (Sigma) used were 5′-GGGCTGCACATTACCACTGA (forward) and 5′-CTCCTCGCAATACAACTCGT (reverse), and a detection limit of 35 cycles was used for evaluation, as described previously (10). Samples were run on a 2% agarose gel and analyzed for reovirus RNA by the presence of a 300–base pair PCR product. Positive (reovirus RNA) and negative (RNase-free water) controls were included. For viral detection after amplification (to assess whether functional, replication-competent virus was present in samples, but only at low levels), a 1:10 dilution of samples (alongside positive and negative controls) was incubated on L929 cells as described above for the TCID50 assay before cells and supernatants were harvested and tested for reovirus RNA as described.
Neutralizing anti-reovirus antibody detection
Patient antibody titers were detected in samples with a modified NAB assay as described previously (14).
Immunohistochemistry and fluorescence for reovirus, caspase, and tubulin
Immunohistochemistry was performed as previously described (11) with the Benchmark LT automated slide stainer (Ventana Medical Systems) according to the manufacturer’s instructions. Caspase-3 and tubulin antibodies were purchased from Abcam, and reovirus antibody was supplied by Oncolytics Biotech Inc. Optimized dilutions used for detection of antibodies were as follows: 1:6000 (reovirus), 1:50 (caspase- 3), and 1:100 (tubulin). All antigen retrievals were performed for 30 min. The antigens were detected with the ultraView Universal DAB or Fast Red system (Ventana), with a counterstain of hematoxylin. Negative controls included omission of primary antibody and internal controls of cells/tissues known to be negative for the targets. Colocalization signal was interpreted with the Nuance microscope/computer-based interface system (Cambridge Research Instrumentation Inc.), using coexpression analyses as previously described (25, 26). In brief, the Nuance system dissects the colorimetric-based signal for different chromogens and converts these color-based signals to fluorescence-based signals. This allows performance of “fluorescence-mixing” combinations to determine whether a given cell has zero, one, or two or more signals.
Electron microscopy
Tissue specimens were fixed in 3% glutaraldehyde (Sigma) for a minimum of 4 hours and stored in 70% ethanol until further processing. Specimens were then dehydrated and embedded by a 1-hour incubation step at −20°C in a 2:1 mixture of LR White (Sigma) and ethanol followed by three 1-hour incubations at −20°C in LR White alone. Finally, a polymerization step (in gelatin capsules) was performed at 37°C for 5 days. Sections (80 to 100 nm) were then cut and set on nickel grids. After blocking in PBS/2% bovine serum albumin (BSA), sections were incubated for 1 hour with 25-µl drops of anti-reovirus σ3 antibody (Developmental Studies Hybridoma Bank), diluted 1:800, followed by a 1-hour incubation with 25-µl drops of rabbit anti-mouse immunoglobulin G (IgG) antibody (DakoCytomation), diluted 1:40. Finally, a 1-hour incubation was performed with 25-µl drops of protein A–immunogold particles (10 nm; Aurion), diluted 1:20, and then a final 20-min stain with uranyl acetate. Specimens were viewed with a Phillips/FEI CM10 transmission electron microscope running at 80 kV. Using an exposure time of 2 s, images were captured onto Kodak Electron Image Film (Type SO-163).
Retrieval of replication-competent virus from tissue
Tissue specimens were dissected into 5-mm cubes before disaggregation into a single-cell suspension with a Cell Dissociation Sieve & Tissue Grinder Kit (Sigma). Cells were then passed through a 70-µm cell strainer (BD Biosciences), and debris was removed by two washes in PBS. The single-cell suspension was then added to semiconfluent L929 cells for 24 to 48 hours before being removed and replaced with 5% DMEM. After a further 5 to 7 days of culture, supernatant was collected and reovirus replication was determined by standard plaque assay with L929 cells. Briefly, samples (either neat or diluted in serumfree medium) were added to L929 cells and incubated at 37°C for hours. Samples were gently removed before overlay medium comprising DMEM supplemented with 5% (v/v) FCS and 1.6% (w/v) carboxymethylcellulose (Sigma) was added to each well. After 72 hours, supernatants were collected (and stored for analysis by Western blotting; see below) before cells were gently washed three times with PBS and fixed using 0.1% (v/v) glutaraldehyde (Sigma) for 10 min. Plaques were visualized with 1% methylene blue (Sigma), and images were taken with a Canon IXUS 100 digital camera.
Western blotting
Samples (collected from plaque assays as described above) were prepared in 2× Laemmli buffer and denatured at 95°C for 5 min. Proteins were separated on 10% SDS–polyacrylamide gels by electrophoresis and transferred to a Hybond-C Super nitrocellulose membrane (Amersham Bio Sciences) before being blocked in Odyssey blocking buffer (Li-Cor Biosciences UK Ltd.) overnight at 4°C. Membranes were probed with anti-reovirus σ3 antibody (1:200 dilution) and then secondary IgG–Alexa Fluor 680 (1:5000 dilution; Invitrogen). Nitrocellulose membranes were visualized on a Li-Cor Odyssey Infrared Imager at 700 nm and analyzed with Odyssey Application Software (v1.2). The presence of reovirus in the samples was confirmed by a band at 41 kD.
Statistical analysis
P values were calculated with two-way analysis of variance (ANOVA), and statistical significance is denoted by *P < 0.05.
Supplementary Material
Acknowledgments
Funding: Cancer Research UK, Leeds Experimental Cancer Medicine Centre, Leeds Cancer Research UK Centre, Leeds Cancer Vaccine Appeal, and Rays of Hope Appeal. This work is supported by the National Institute of Health (R01CA107082 and R01CA130878).
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencetranslationalmedicine.org/cgi/content/full/4/138/138ra77/DC1
Materials and Methods
Fig. S1. Reoviral RNA is detectable when ~50 copies enter the reaction, but not ~5 copies or fewer.
Fig. S2. Reoviral RNA can also be detected in patient PBMCs, granulocytes, and platelets using an alternative, second set of primers.
Table S1. Histological data summary for all patients.
Table S2. Coexpression of reovirus and tubulin in tumor.
Author contributions: R.A.A., S.F., A.A., A.M., R.P., O.D., H.P., K.H., and A.A.M. designed the study/protocols; R.A.A., D.B., D.A.A., R.D., and G.T. contributed to patient recruitment and care; V.R., R.A.A., K.J.S., R.M., G.J.N., M.F., A.R., V.A.J., E.J.W., and J.K. performed research and data analysis; M.C., P.S., R.V., K.H., and A.A.M. wrote the manuscript; and A.A.M. was the principal investigator.
Competing interests: M.C. is an employee of Oncolytics Biotech Inc. M.C. is an author on a patent for the use of reovirus for the treatment of Ras-mediated cellular proliferative disorders. A.A.M., K.H., R.V., H.P., and G.J.N. have received commercial research grants from Oncolytics Biotech Inc. The other authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Garcia-Aragoncillo E, Hernandez-Alcoceba R. Design of virotherapy for effective tumor treatment. Curr. Opin. Mol. Ther. 2010;12:403–411. [PubMed] [Google Scholar]
- 2.Prestwich RJ, Harrington KJ, Pandha HS, Vile RG, Melcher AA, Errington F. Oncolytic viruses: A novel form of immunotherapy. Expert Rev. Anticancer Ther. 2008;8:1581–1588. doi: 10.1586/14737140.8.10.1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eager RM, Nemunaitis J. Clinical development directions in oncolytic viral therapy. Cancer Gene Ther. 2011;18:305–317. doi: 10.1038/cgt.2011.7. [DOI] [PubMed] [Google Scholar]
- 4.Kaufman HL, Bines SD. OPTIM trial: A phase III trial of an oncolytic herpes virus encoding GM-CSF for unresectable stage III or IV melanoma. Future Oncol. 2010;6:941–949. doi: 10.2217/fon.10.66. [DOI] [PubMed] [Google Scholar]
- 5.Breitbach CJ, Thorne SH, Bell JC, Kirn DH. Targeted and armed oncolytic poxviruses for cancer: The lead example of JX-594. Curr. Pharm. Biotechnol. 2011 doi: 10.2174/138920112800958922. [DOI] [PubMed] [Google Scholar]
- 6.Prestwich RJ, Ilett EJ, Errington F, Diaz RM, Steele LP, Kottke T, Thompson J, Galivo F, Harrington KJ, Pandha HS, Selby PJ, Vile RG, Melcher AA. Immune-mediated antitumor activity of reovirus is required for therapy and is independent of direct viral oncolysis and replication. Clin. Cancer Res. 2009;15:4374–4381. doi: 10.1158/1078-0432.CCR-09-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly K, Nawrocki S, Mita A, Coffey M, Giles FJ, Mita M. Reovirus-based therapy forcancer. Expert Opin. Biol. Ther. 2009;9:817–830. doi: 10.1517/14712590903002039. [DOI] [PubMed] [Google Scholar]
- 8.Harrington KJ, Vile RG, Melcher A, Chester J, Pandha HS. Clinical trials with oncolytic reovirus: Moving beyond phase I into combinations with standard therapeutics. Cytokine Growth Factor Rev. 2010;21:91–98. doi: 10.1016/j.cytogfr.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Selb B, Weber B. A study of human reovirus IgG and IgA antibodies by ELISA and Western blot. J. Virol. Methods. 1994;47:15–25. doi: 10.1016/0166-0934(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 10.Vidal L, Pandha HS, Yap TA, White CM, Twigger K, Vile RG, Melcher A, Coffey M, Harrington KJ, DeBono JS. A phase I study of intravenous oncolytic reovirus type 3 Dearing in patients with advanced cancer. Clin. Cancer Res. 2008;14:7127–7137. doi: 10.1158/1078-0432.CCR-08-0524. [DOI] [PubMed] [Google Scholar]
- 11.Comins C, Spicer J, Protheroe A, Roulstone V, Twigger K, White CM, Vile R, Melcher A, Coffey MC, Mettinger KL, Nuovo G, Cohn DE, Phelps M, Harrington KJ, Pandha HS. REO-10: A phase I study of intravenous reovirus and docetaxel in patients with advanced cancer. Clin. Cancer Res. 2010;16:5564–5572. doi: 10.1158/1078-0432.CCR-10-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ilett EJ, Prestwich RJ, Kottke T, Errington F, Thompson JM, Harrington KJ, Pandha HS, Coffey M, Selby PJ, Vile RG, Melcher AA. Dendritic cells and T cells deliver oncolytic reovirus for tumour killing despite pre-existing anti-viral immunity. Gene Ther. 2009;16:689–699. doi: 10.1038/gt.2009.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gollamudi R, Ghalib MH, Desai KK, Chaudhary I, Wong B, Einstein M, Coffey M, Gill GM, Mettinger K, Mariadason JM, Mani S, Goel S. Intravenous administration of Reolysin, a live replication competent RNA virus is safe in patients with advanced solid tumors. Invest. New Drugs. 2010;28:641–649. doi: 10.1007/s10637-009-9279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White CL, Twigger KR, Vidal L, De Bono JS, Coffey M, Heinemann L, Morgan R, Merrick A, Errington F, Vile RG, Melcher AA, Pandha HS, Harrington KJ. Characterization of the adaptive and innate immune response to intravenous oncolytic reovirus (Dearing type 3) during a phase I clinical trial. Gene Ther. 2008;15:911–920. doi: 10.1038/gt.2008.21. [DOI] [PubMed] [Google Scholar]
- 15.Ilett EJ, Bárcena M, Errington-Mais F, Griffin S, Harrington KJ, Pandha HS, Coffey M, Selby PJ, Limpens RW, Mommaas M, Hoeben RC, Vile RG, Melcher AA. Internalization of oncolytic reovirus by human dendritic cell carriers protects the virus from neutralization. Clin. Cancer Res. 2011;17:2767–2776. doi: 10.1158/1078-0432.CCR-10-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Twigger K, Vidal L, White CL, De Bono JS, Bhide S, Coffey M, Thompson B, Vile RG, Heinemann L, Pandha HS, Errington F, Melcher AA, Harrington KJ. Enhanced in vitro and in vivo cytotoxicity of combined reovirus and radiotherapy. Clin. Cancer Res. 2008;14:912–923. doi: 10.1158/1078-0432.CCR-07-1400. [DOI] [PubMed] [Google Scholar]
- 17.Evans J. Recent deal highlights hopes for cancer-killing viruses. Nat. Med. 2011;17:268–269. doi: 10.1038/nm0311-268b. [DOI] [PubMed] [Google Scholar]
- 18.Lal R, Harris D, Postel-Vinay S, de Bono J. Reovirus: Rationale and clinical trial update. Curr. Opin. Mol. Ther. 2009;11:532–539. [PubMed] [Google Scholar]
- 19.Breitbach CJ, Burke J, Jonker D, Stephenson J, Haas AR, Chow LQ, Nieva J, Hwang TH, Moon A, Patt R, Pelusio A, Le Boeuf F, Burns J, Evgin L, De Silva N, Cvancic S, Robertson T, Je JE, Lee YS, Parato K, Diallo JS, Fenster A, Daneshmand M, Bell JC, Kirn DH. Intravenous delivery of a multi-mechanistic cancer-targeted oncolytic poxvirus in humans. Nature. 2011;477:99–102. doi: 10.1038/nature10358. [DOI] [PubMed] [Google Scholar]
- 20.Willmon C, Harrington K, Kottke T, Prestwich R, Melcher A, Vile R. Cell carriers for oncolytic viruses: Fed Ex for cancer therapy. Mol. Ther. 2009;17:1667–1676. doi: 10.1038/mt.2009.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brink M, de Goeij AF, Weijenberg MP, Roemen GM, Lentjes MH, Pachen MM, Smits KM, de Bruine AP, Goldbohm RA, van den Brandt PA. K-ras oncogene mutations in sporadic colorectal cancer in The Netherlands Cohort Study. Carcinogenesis. 2003;24:703–710. doi: 10.1093/carcin/bgg009. [DOI] [PubMed] [Google Scholar]
- 22.Di Paolo NC, van Rooijen N, Shayakhmetov DM. Redundant and synergistic mechanisms control the sequestration of blood-born adenovirus in the liver. Mol. Ther. 2009;17:675–684. doi: 10.1038/mt.2008.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Errington F, White CL, Twigger KR, Rose A, Scott K, Steele L, Ilett LJ, Prestwich R, Pandha HS, Coffey M, Selby P, Vile R, Harrington KJ, Melcher AA. Inflammatory tumour cell killing by oncolytic reovirus for the treatment of melanoma. Gene Ther. 2008;15:1257–1270. doi: 10.1038/gt.2008.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lolkema MP, Arkenau HT, Harrington K, Roxburgh P, Morrison R, Roulstone V, Twigger K, Coffey M, Mettinger K, Gill G, Evans TR, de Bono JS. A phase I study of the combination of intravenous reovirus type 3 Dearing and gemcitabine in patients with advanced cancer. Clin. Cancer Res. 2011;17:581–588. doi: 10.1158/1078-0432.CCR-10-2159. [DOI] [PubMed] [Google Scholar]
- 25.Nuovo GJ, Elton TS, Nana-Sinkam P, Volinia S, Croce CM, Schmittgen TD. A methodology for the combined in situ analyses of the precursor and mature forms of microRNAs and correlation with their putative targets. Nat. Protoc. 2009;4:107–115. doi: 10.1038/nprot.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nuovo GJ. In situ detection of microRNAs in paraffin embedded, formalin fixed tissues and the co-localization of their putative targets. Methods. 2010;52:307–315. doi: 10.1016/j.ymeth.2010.08.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.