Abstract
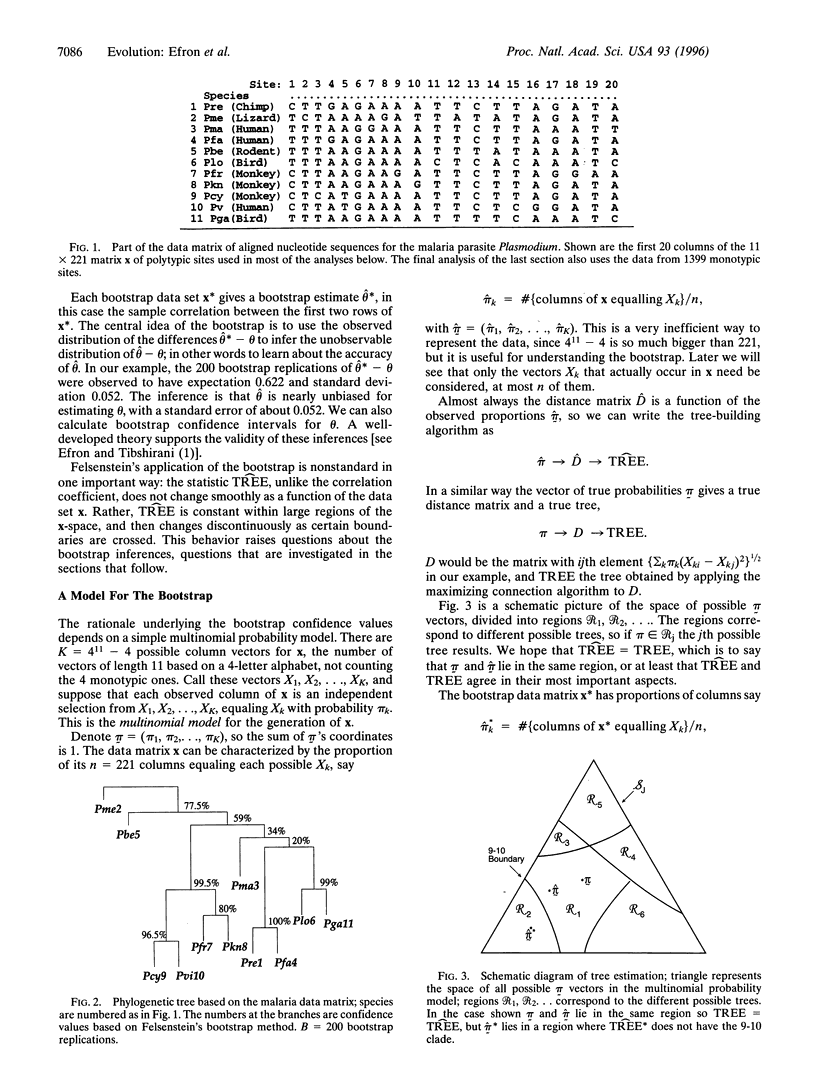
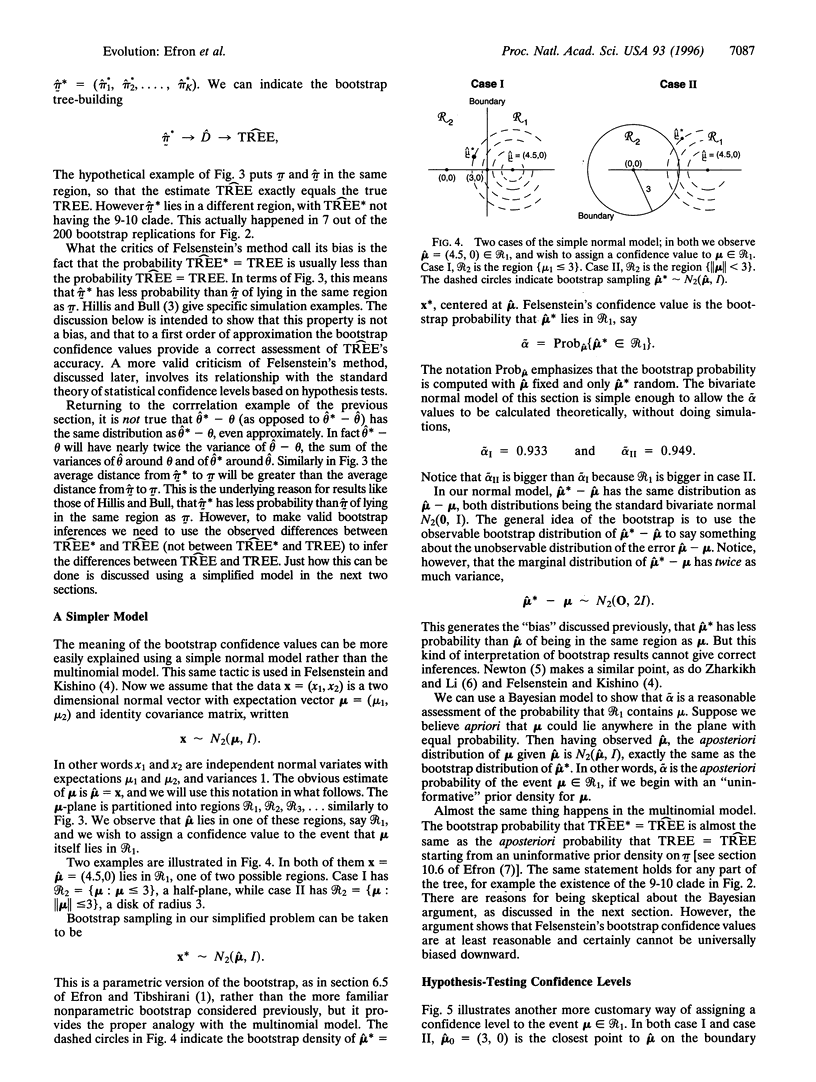
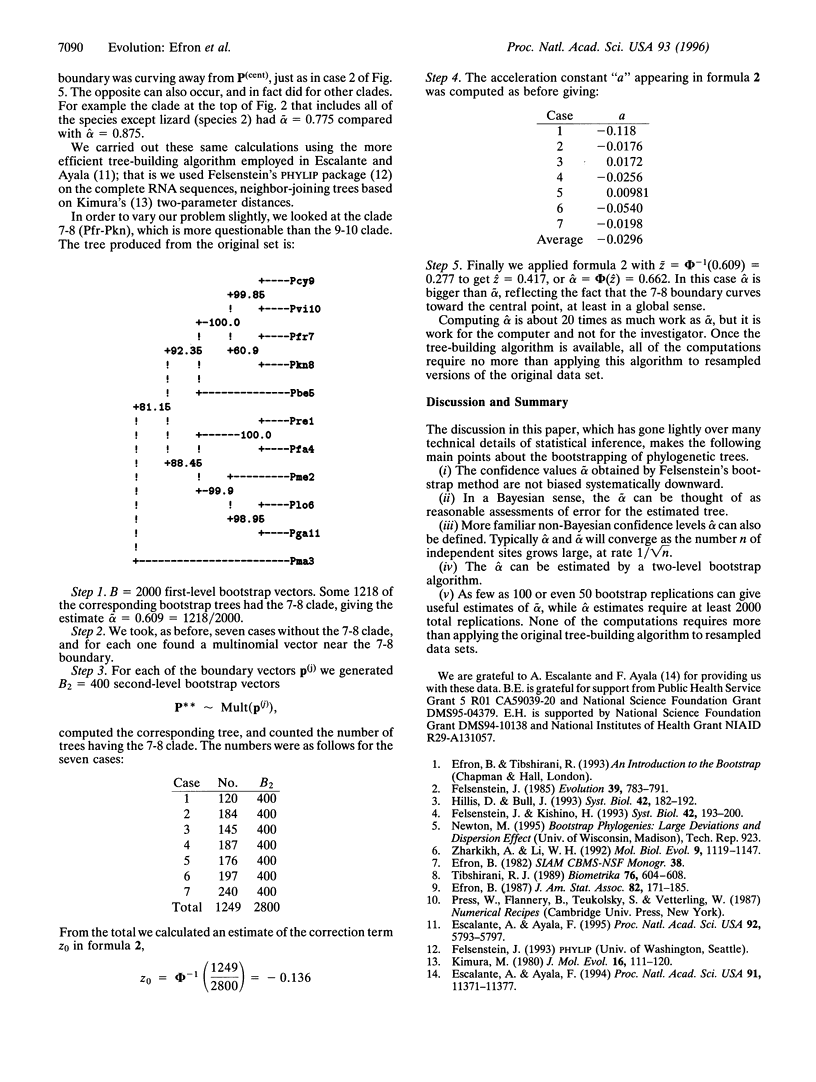
Evolutionary trees are often estimated from DNA or RNA sequence data. How much confidence should we have in the estimated trees? In 1985, Felsenstein [Felsenstein, J. (1985) Evolution 39, 783-791] suggested the use of the bootstrap to answer this question. Felsenstein's method, which in concept is a straightforward application of the bootstrap, is widely used, but has been criticized as biased in the genetics literature. This paper concerns the use of the bootstrap in the tree problem. We show that Felsenstein's method is not biased, but that it can be corrected to better agree with standard ideas of confidence levels and hypothesis testing. These corrections can be made by using the more elaborate bootstrap method presented here, at the expense of considerably more computation.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Escalante A. A., Ayala F. J. Evolutionary origin of Plasmodium and other Apicomplexa based on rRNA genes. Proc Natl Acad Sci U S A. 1995 Jun 20;92(13):5793–5797. doi: 10.1073/pnas.92.13.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante A. A., Ayala F. J. Phylogeny of the malarial genus Plasmodium, derived from rRNA gene sequences. Proc Natl Acad Sci U S A. 1994 Nov 22;91(24):11373–11377. doi: 10.1073/pnas.91.24.11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980 Dec;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Zharkikh A., Li W. H. Statistical properties of bootstrap estimation of phylogenetic variability from nucleotide sequences. I. Four taxa with a molecular clock. Mol Biol Evol. 1992 Nov;9(6):1119–1147. doi: 10.1093/oxfordjournals.molbev.a040782. [DOI] [PubMed] [Google Scholar]



