Abstract
The complement system is composed of soluble factors in plasma that enhance or “complement” immune-mediated killing through innate and adaptive mechanisms. Activation of complement causes recruitment of immune cells; opsonization of coated cells; and direct killing of affected cells through a membrane attack complex (MAC). Tumor cells up-regulate complement inhibitory factors -one of several strategies to evade the immune system. In many cases as the tumor progresses, dramatic increases in complement inhibitory factors are found on these cells. This review focuses on the classic complement pathway and the role of major complement inhibitory factors in cancer immune evasion as well as on how current protein engineering efforts are being employed to increase complement fixing or to reverse complement resistance leading to better therapeutic outcomes in oncology. Strategies discussed include engineering of antibodies to enhance complement fixation, antibodies that neutralize complement inhibitory proteins as well as engineered constructs that specifically target inhibition of the complement system.
Keywords: complement inhibition, tumor immune evasion, CD46, CD55, CD59
THE COMPLEMENT SYSTEM AND ITS REGULATION
For over a hundred years, the interaction of adaptive immunity with a heat-labile serum component that “complements” cytotoxic activity has been recognized [1]. In what is now defined as the “classical pathway”, antibody is bound to the surface of a cell and recruits serum components that lead to cell killing and clearance of pathogens [2]. Two other branches of complement are now recognized: The “lectin pathway”, in which signaling is initiated by binding to certain polymeric molecules and carbohydrates; and the “alternate pathway”, where cells that are not host specific are destroyed due to the lack of inhibitory factors. This pathway initially was thought to be constitutive, but recent research suggests it is also triggered through specific binding interactions [3,4]. The separation into these three cascades is somewhat artificial: in response to various signals complement activation is orchestrated by a network of interactions allowing elegant distinction of healthy host cells from debris, foreign intruders, and apoptotic cells. A description of these specific interactions is beyond the scope of this review and the reader is referred to other excellent articles on the complement system [5,6].
Central to the activation of the complement system is the activity of C3 which is cleaved into active forms C3a and C3b by C3 convertases. Deposition of C3b on cell surfaces and its association with either factor B or the C4bC2a complex leads to further activation of complement through C3 conversion as well as initiation of the terminal complement cascade and formation of the membrane attack complex [7–9]. In concert, the other products of C3 and C5 cleavage, the anaphylatoxins C3a and C5a, have numerous other signaling activities. They generate pro-inflammatory signals, increase vascular permeability, and stimulate phagocytosis [3,6]. Through complement receptors, inflammatory cytokines, and in conjunction with TLR pathways, the products of C3 and C5 cleavage influence B cell maturation, antigen presenting cell activation, and T cell influx providing a bridge to adaptive immunity [10–14].
To prevent uncontrolled amplification of the effects of complement there are a variety of complement regulatory proteins (CRPs). These include soluble factors like C1 inhibitor, factor H, factor I, and vitreonectin as well as membrane-bound complement regulatory proteins (mCRPs) like CD35, CD46, CD55, and CD59. Table 1 summarizes major complement regulators and their functions. Because of the high levels of serum complement proteins that range into the high hundreds of milligrams per liter [15], it is unlikely that cancerous growths could influence the soluble complement protein balance. Tumors that evade complement’s action therefore appear to do so by modulating the levels of the membrane bound components. In addition to direct inhibition of the complement system, these inhibitor can also influence the cellular and humoral immune responses [16,17] and eliminating this inhibition can enhance cellular immunity, with key implications for cancer immune therapy [18].
Table 1.
| A: Soluble complement regulatory factors | |
|---|---|
| REGULATOR | FUNCTION |
| C1 inhibitor | Serine protease that targets the C1s/C1r, inhibiting activation of C4 and C2 [19] |
| Factor H | Modulates C3b formation by acting as a co-factor for Factor I and by accelerating the decay of the C3 convertases [20] |
| Factor I (C3b /C4b inactivator) | Decreases complement activation by cleaving C3b and C4b when complexed with co-factors such as CD46 [21,22] |
| C4 Binding Protein (C4BP) | A co-factor for Factor I, binds C4b increasing proteolytic accessibility [23] |
| Vitronectin (S40) | Inhibits the terminal cascade and formation of the MAC; may have other roles in regulation of disease responses [24–26] |
| Clusterin (Apo J) | Similar to vitronectin, inhibits the formation of the MAC and may have other functions [26] |
| B: Membrane-bound complement regulatory factors | |
|---|---|
| REGULATOR | FUNCTION |
| CD35 (Complement receptor 1, CR1) | Decay accelerating factor for C3/C5 convertases, facilitates phagocytosis of cells with complement activated, co-factor for Factor I, fixes complement immune complexes on erythrocytes, has limited tissue distribution in humans [27] |
| CD46 (Membrane cofactor protein, MCP) | Cofactor for factor I, regulator of T-cell differentiation and apoptosis, widely expressed in humans [16,28,29] |
| CD55 (Decay accelerating factor, DAF) | Inhibits formation and accelerates decay of C3 convertases [30–32] |
| CD59 (MAC inhibitory protein, MAC-IP, 20 kDa homologous restriction factor, HRF20) | Inhibits formation of the MAC by binding C5b/8 complex and interfering with insertion of C9 [33,34] |
CANCERS AND COMPLEMENT REGULATORY PROTEINS
In the development of immune therapies for cancer the role of complement has often been neglected, but as more insight is gained into the mechanisms of action of monoclonal antibodies new approaches to improve specific antibody functions are emerging. Monoclonal antibodies have contributed substantially to progress in treating many types of cancers, but tumors evasion mechanisms lead to low complete response rates for many of these therapies [35–38]. For example, rituximab is a humanized IgG1 antibody against the surface protein CD20 which is expressed on the surface of normal B-lymphocytes and B-cell malignancies but not on hematopoietic stem cells and plasma cells. It is currently used for the treatment of B-cell non-Hodgkin lymphoma, mantle cell lymphoma, hairy cell leukemia, and chronic lymphocytic leukemia. Survival rates for B cell non-Hodgkin’s lymphoma have increased significantly since the introduction of rituximab, but only about half of the patients suffering from this disease survive ten years after diagnosis. The cancer recurs and patients often become resistant to rituximab therapy. One of the mechanisms of action of rituximab involves binding to malignant B-cells with subsequent activation of the complement system [39–43]. By inhibiting the action of complement, cancer cells could be able to evade killing by rituximab.
Overexpression of complement inhibitory proteins is a well-documented phenomenon in cancer cells and has been proposed as an escape mechanism from monoclonal antibody therapy [44–46]. This up-regulation blocks complement signaling and allows cells with bound antibody to evade killing by the complement system. The pattern of complement inhibition is wide spread for many different types of tumors, stages of tumor and can exceed many orders of magnitude of overexpression versus primary, normal tissue. That being said, the levels of complement inhibitory proteins do not necessarily correlate with functional resistance to lysis. As Golay and Macor have shown with neutralizing antibodies, even low level expression of individual CRPs is sufficient to protect tumors from lysis induced by monoclonal therapeutics [47–49].
Although by no means an exhaustive summary of all literature on mCRPs in cancer, Table 2 shows how widespread enhancement of surface expression of major membrane bound complement inhibitory proteins is in cancers.
Table 2.
Complement regulatory proteins and documented increases in expression1
| TISSUE TYPE | CD46 | CD55 | CD59 | CIT. |
|---|---|---|---|---|
| Lung cancers | Consistent high levels found | Low levels on few tumors | Variable levels detected on majority of cancers | [50] |
| Breast cancer | Expressed in all breast carcinoma and normal tissue examined, increase associated with poor prognosis | No staining on cancer cells in ductal carcinomas | Trend towards increase in staining versus normal tissues; variable on some patient samples | [50–52] |
| Colorectal cancer | Strong increase in staining found on most samples | None found | Increase in staining, variable for some samples | [51] |
| Prostate cancer | Expressed but does not increase | Increase in cancer and further with malignancy | Expressed, but does not increase | [53] |
| Bladder cancer | Upregulation up to around 10 fold in 77% of samples tested | Upregulation up to around 10 fold in 55% of samples tested | Upregulation up to around 10 fold in 59% of samples tested | [54] |
| Malignant endometrial tissue | 2.5 fold rise in optical density on stained image | 2.2 fold rise in optical density on stained image | 1.7 fold rise in optical density on stained image | [55] |
| Head and Neck Cancer | Highly expressed in all forms, low to no staining in normal surrounding tissue | Highly expressed in all forms, low to no staining in normal surrounding tissue | Highly expressed in all forms, low to no staining in normal surrounding tissue | [56] |
| Esophageal cancer | Dramatic increase in staining | Pronounced decrease in staining | Uniform between normal and cancer tissue | [57] |
| Non-Hodgkin’s lymphoma2 | High level expression with possible correlation to outcome | High level expression with possible correlation to outcome | High level expression with possible correlation to outcome | [58,59] |
| Renal cell cancers | Low, scattered staining | No staining detected | High levels on most tumors tested | [50] |
| Primary gynecologic carcinosarcoma | High level expression | High level expression | High level expression | [60] |
| Ovarian cancer | Robust expression on the surface of cells | Robust expression on the surface of cells | Robust expression on the surface of cells | [61] |
No information is usually found on CD35 so this mCRP is not presented
The conclusions in the two papers contradict as to correlation with outcome; high levels were found in both studies
ENGINEERING ANTIBODIES TO MORE EFFICIENTLY FIX COMPLEMENT
Because of the ability of tumor cells to evade complement-dependent killing there have been multiple efforts to engineer antibodies to increase the efficiency of complement fixation and thereby enhance complement dependent cell cytotoxicity (CDC). Examining the mechanism of C1q binding that initiates the deposition of complement in the classical pathway, one can envision that numerous approaches could be successful: (1) Targeting high-density membrane localized epitopes to increase the number of bound antibodies per cell; (2) Positioning the antibody binding site so that it is membrane proximal and increases the efficiency of membrane deposition of complement components; (3) increasing the antibody’s affinity to the target by reducing the off rate so that it remains on the cells for a prolonged time; and (4) increasing the interaction of the antibody with C1q. The first three approaches are embodied in the anti-CD20 antibody, ofatumumab or “Arzerra®” that was intended to be an improvement over rituximab. Ofatumumab was found to have a longer off-rate than the competitor and the unique localization of the epitope near the surface of the cell membrane is thought to enhance deposition of complement on the membrane [62]. These complement enhancing features translate into an enhancement of killing of complement resistant cells from 25% (rituximab) to 75% (ofatumumab) [63].
Other protein engineering efforts focus primarily on the interaction between the first component of complement, C1, and the Fc region of the target antibody. In this approach, either specific isotypes known to fix complement better are chosen as an antibody scaffold or key Fc residues are mutated to promote favorable energetics of binding between the Fc region of the antibody and the complement component C1q. Figure 1 shows key amino acids that have been targeted when modulating antibody /C1q interactions [64,65]. Modifying these specific amino acids can lead to dramatic increases in C1q binding and enhance the potency of antibodies as much as 600 fold [64,66].
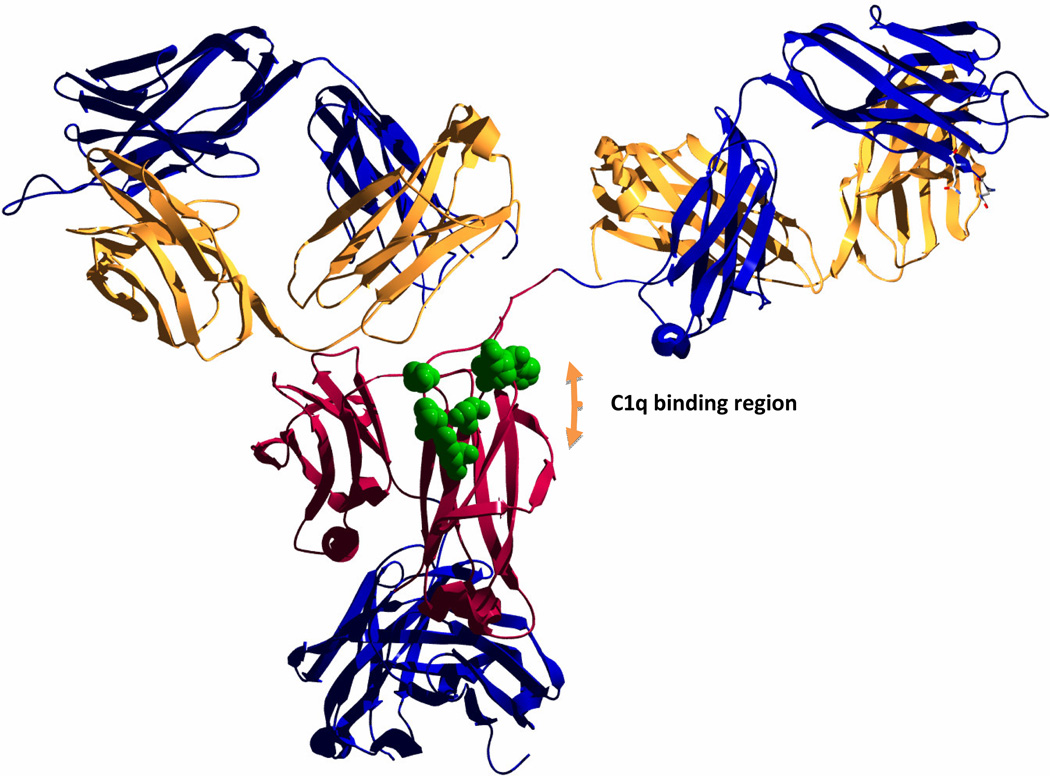
Figure 1. Protein engineering targeting the Fc region to enhance complement fixation.
The figure shows the crystal structure of a human IgG1 (PDB: 1HZH, [84]) with the heavy chain constant domains 2 in red. The rest of the heavy chain trace is in blue and light chains are shown in yellow. The long flexible hinge regions allow for contact and binding of the C1q complement components in the region labeled in the figure. Residues that have been reported to be important in the interaction with C1q and that have been targeted by mutagenesis to modulate complement fixation are shown with the full sidechains in green [64,85]. Protein was visualized with the Swiss-PdbViewer version 4.1.0 [86] and rendered using POV-Ray [87].
PROTEIN ENGINEERING TO TARGET COMPLEMENT INHIBITION
Although enhanced fixation of complement is potentially attractive for increasing CDC, complement inhibitors could eliminate any benefit by rapidly quelling complement triggering. For this reason, engineering efforts targeting complement inhibition could dramatically improve therapy and would “complement” antibody engineering efforts described in the previous section. Efforts to target complement inhibition by knocking down mCRP expression using siRNA have been successful [67,68], but we will focus on protein-based engineering strategies here.
The most straightforward engineering approach is to make antibodies that inhibit the function of the complement inhibitory proteins. Ziller and colleagues screened a human antibody phage display library for recognition of CD55 and CD59 and used the positive clones as variable chain templates that were fused to the hinge-CH2-CH3-Fc fragments of human IgG1 to generate the “minibodies” MB55 (anti-CD55) and MB59 (anti-CD59) [69]. Using rituximab combined with MB55 and MB59 enabled the researchers to enhance survival of LCL2 challenged SCID mice from 30% to 70%. Notably in this experiment, the MB55 /MB59 combination without rituximab was as effective as rituximab in enhancing survival [46].
Intermedilysin is a bacterial cytotoxin isolated from Streptococcus intermedius that binds to CD59 and initiates pore formation in human cells resulting in cell death [70]. The protein has a membrane attack complex/perforin fold (MACPF) region that is responsible for pore formation on the surface of the affected cell, and a “domain 4”, an avid CD59 binding region [71]. By removing the pore and linker domains and leaving only domain 4, Qin and Hu et al were able to generate a 114 amino acid, affinity-tagged, recombinant protein that inhibited CD59 function, “rILYd4” [72]. This protein was then further engineered by removing the affinity tag to give a well behaved lead candidate targeting CD59 [73]. When rILYd4 was used in combination with CD20 targeted monoclonal antibodies for cancer, a dose dependent increase in cytolysis was determined with the pretreatment allowing more than double the amount of cell killing of rituximab resistant cells by both rituximab and ofatumumab [74–76].
In another approach, the discovery that certain group B adenoviruses, including serotype 35, can interact with CD46 and lead to its internalization and degradation [77,78] combined with the fact that high levels of CD46 were found on tumors [79] led to the hypothesis that adjuvanting antibody therapy with adenovirus-derived removal of complement inhibition would be a viable clinical path to improve cancer therapy. Simply infusing large amounts of adenovirus would be impractical so that an engineered protein was sought to develop a therapeutic candidate. The ideal candidate would be a relatively small protein that could penetrate tumors and be straightforward to manufacture, as well as having high binding affinities for the inhibitory target. Adenoviruses display fiber proteins that extend out from each vertix of the virus and interact with the virus’ receptor (Figure 2). The receptor binding domain is localized in the C-terminal domain of the trimeric fiber and is called the fiber knob. The adenovirus serotype knob protein has two CD46 contacting loops and due to the trimeric knob protein structure makes high avidity contacts when bound to CD46. This coding region for a functional Ad35knob protein end was inserted into an E. coli expression vector and mutated by error prone PCR [80,81] resulting in high affinity variants; individual mutations at knob protein positions 207, 245, and 256 (Figure 2B) resulted in up to 8 fold increases in affinity and combined mutations resulted in a lead candidate protein, Ad35K++, with a drop from 14 nanomolar dissociation constants down to 630 picomolar KD’s, an increase in affinity of more than 23 fold. This new construct was then tested for its ability to enhance rituximab killing of lymphoma cells in vitro and was found to dramatically enhance complement dependent killing (Figure 3 and [82]). In later in vivo studies the combination of rituximab and Ad35K++ markedly increased responses to lymphoma xenographs in mice and sensitized non-human primate CD20 positive B-cells to the action of rituximab after intravenous injection into macaques [83].
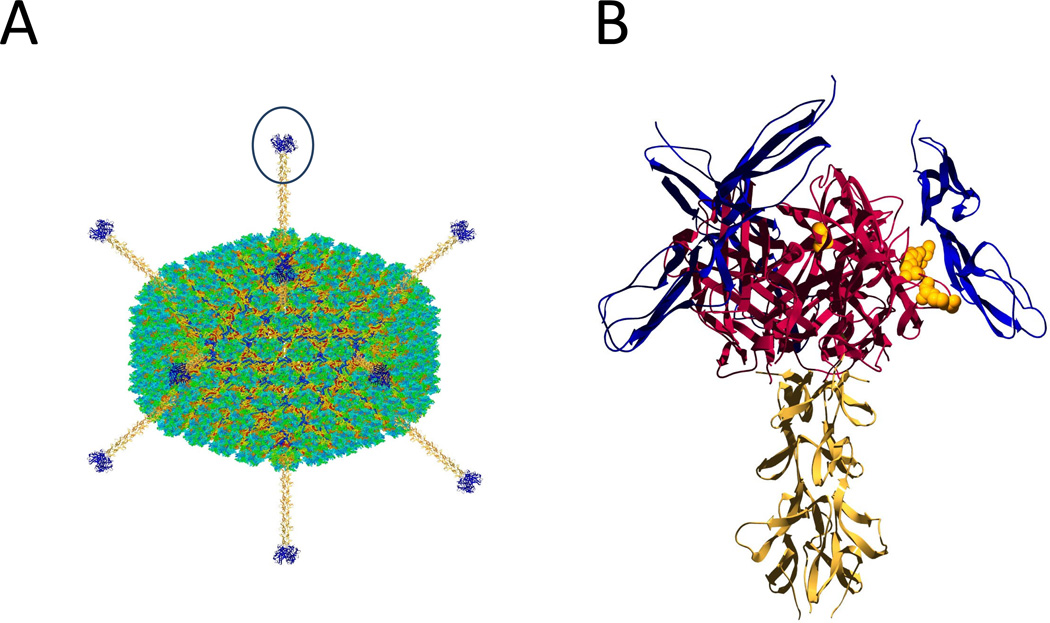
Figure 2. Engineering of the adenoviral fiber knob as an adjunct therapy to remove complement inhibition.
Panel A: Adenoviruses are icosahedral particles with fibers extending from the vertices of the icosahedron. The knob portion of the adenovirus interacts with the virus receptor on the host cell and allows for viral entry. Panel B: Close up of one of the Ad35 fiber knob regions interacting with CD46. The trimeric knob region used to generate Ad35K++ is shown in red; the fiber shaft regions in yellow and the bound CD46 domains are in blue. Residues that were mutated and found to have effects on binding of CD46 [81] are shown with full sidechains in yellow. For Panel A the adenovirus structure from Fabry et al. [88] was used and rendered using Jmol [89] and POV-Ray [87] after adding fiber knobs modeled in the Swiss-PDBViewer [86]. The same software packages were used in Panel B, using the CD46 /knob structure reported as PDB 2O39 [90] and the adenovirus fiber protein in PDB: 1QIU [91].
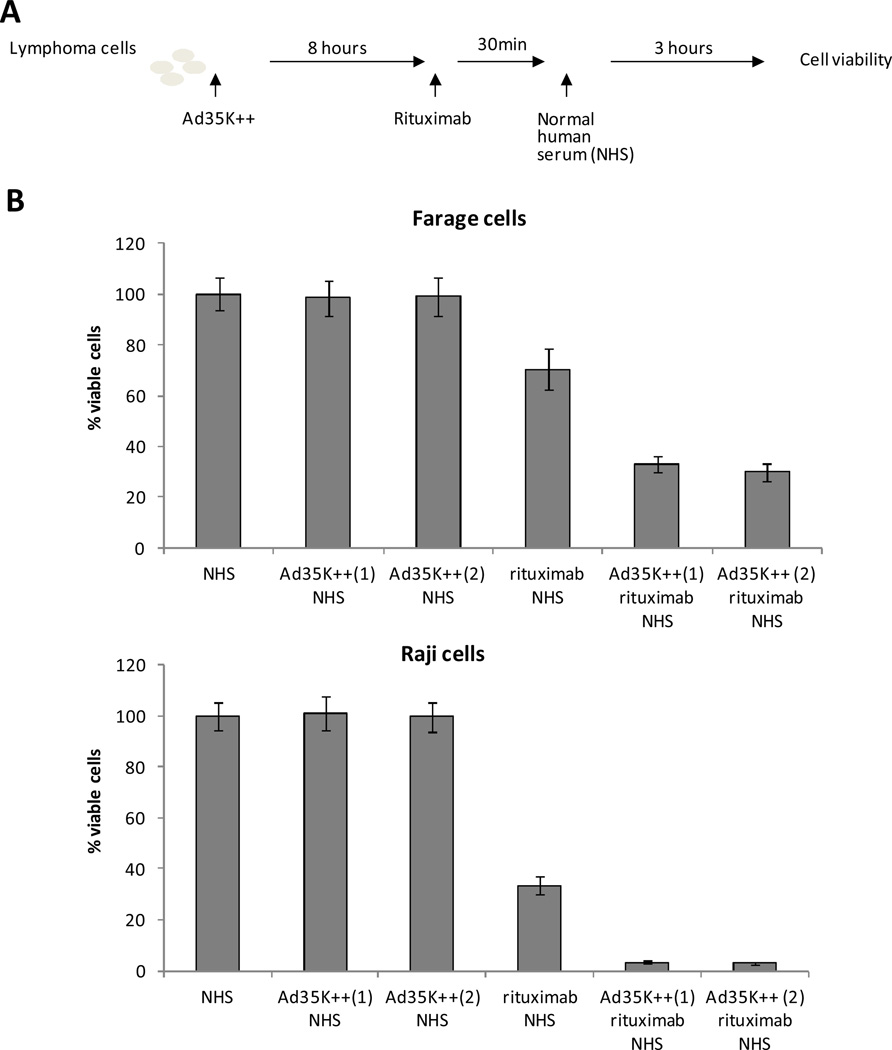
Figure 3. Using engineered knob proteins to enhance killing of lymphoma cells.
Panel A – Experimental setup. Lymphoma cells were incubated with the engineered knob protein Ad35K++ as reported in Wang et al. [82]. NHS is used as source of complement. Panel B – Both research and scaled preparations have equivalent efficacy. N>6. Ad35K++ (1) is a protein lot that was produced previously and used in published studies. Ad35K++ (2) is a protein made with a scalable process. The difference between rituximab + NHS and Ad35K++ + rituximab + NHS is significant (p<0.01). The difference between Ad35K++ (1) + rituximab + NHS and Ad35K++ (2) + rituximab + NHS is not significant.
TRANSLATIONAL DEVELOPMENT OF ANTI-COMPLEMENT THERAPIES
Clinical testing and commercial deployment of new proteins targeting complement inhibition in cancer will require efficient manufacturing processes that comply with regulatory requirements. Once the concept of reversing complement inhibition in cancer is successfully proven in pre-clinical models, the focus should be on how to achieve the same results with a commercializable production process (e.g. as demonstrated in Figure 3). Investigators engineering proteins as adjunct therapies that sensitize to the action of approved drugs such as monoclonals in cancer should keep in mind the development issues that such a product faces: Costs need to be reduced to ensure that the combination product has an attractive commercial profile.
Enhanced immune effector functions may increase toxicological concerns and deciding on viable administration doses and timings is a complex pharmacokinetic and pharmacodynamic problem. addition, off-target neutralization of complement inhibition could lead to undesirable triggering of complement responses that damage healthy cells and organ systems. To address this, new ways to target the therapeutic to tumor cells could be required – especially if these cells have the same redundancy in complement inhibitory proteins found on normal cells and requiring neutralization of multiple CRPs. Nonetheless, the broad applicability of a successful co-immunotherapeutic makes this strategy highly relevant for the treatment of cancer patients.
CONCLUSION
The complement pathway may have been underappreciated in terms of how important evasion may be to tumor cell survival. Both direct killing of cells as well as recruitment of cellular effectors can be initiated by complement and there is a strong body of literature supporting the fact that cancers up-regulate complement inhibitory factors. Innovative protein engineering efforts have co-opted natural protein inhibitors of complement inhibition and utilized these to target tumor cells. Genetic engineering strategies made small, commercially-deployable proteins with very high affinity for membrane associated complement regulatory proteins. Preclinical testing of these molecules offers hope that the dramatic effects in animal models may be recapitulated in human trials. Successful completion of these trials may open the gate to a new line of protein therapies and a revolution in cancer immune therapy.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
LITERATURE CITED
- 1.Chaplin H., Jr Review: the burgeoning history of the complement system 1888–2005. Immunohematology. 2005;21:85–93. [PubMed] [Google Scholar]
- 2.Sim RB. Complement, Classical Pathway. In: Peter JD, editor. In Encyclopedia of Immunology. Second Edition. Oxford: Elsevier; 1998. pp. 604–612. [Google Scholar]
- 3.Murphy KTPWMJC. Janeway's immunobiology, Garland Science. New York: 2012. [Google Scholar]
- 4.Spitzer D, Mitchell LM, Atkinson JP, Hourcade DE. Properdin can initiate complement activation by binding specific target surfaces and providing a platform for de novo convertase assembly. J Immunol. 2007;179:2600–2608. doi: 10.4049/jimmunol.179.4.2600. [DOI] [PubMed] [Google Scholar]
- 5.Carroll MV, Sim RB. Complement in health and disease. Adv Drug Deliv Rev. 2011;63:965–975. doi: 10.1016/j.addr.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11:785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Degn SE, Thiel S. Humoral Pattern Recognition and the Complement System. Scandinavian Journal of Immunology. 2013;78:181–193. doi: 10.1111/sji.12070. [DOI] [PubMed] [Google Scholar]
- 8.Dodds AW, Ren XD, Willis AC, Law SK. The reaction mechanism of the internal thioester in the human complement component C4. Nature. 1996;379:177–179. doi: 10.1038/379177a0. [DOI] [PubMed] [Google Scholar]
- 9.Bally I, et al. Expression of recombinant human complement C1q allows identification of the C1r/C1s-binding sites. Proc Natl Acad Sci U S A. 2013;110:8650–8655. doi: 10.1073/pnas.1304894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carroll MC. The complement system in regulation of adaptive immunity. Nat Immunol. 2004;5:981–986. doi: 10.1038/ni1113. [DOI] [PubMed] [Google Scholar]
- 11.Carroll MC. The complement system in B cell regulation. Mol Immunol. 2004;41:141–146. doi: 10.1016/j.molimm.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 12.Carroll MC, Isenman DE. Regulation of humoral immunity by complement. Immunity. 2012;37:199–207. doi: 10.1016/j.immuni.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunkelberger JR, Song WC. Complement and its role in innate and adaptive immune responses. Cell Res. 2010;20:34–50. doi: 10.1038/cr.2009.139. [DOI] [PubMed] [Google Scholar]
- 14.Dunkelberger JR, Song WC. Role and mechanism of action of complement in regulating T cell immunity. Mol Immunol. 2010;47:2176–2186. doi: 10.1016/j.molimm.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pomeroy C, Mitchell J, Eckert E, Raymond N, Crosby R, Dalmasso AP. Effect of body weight and caloric restriction on serum complement proteins, including Factor D/adipsin: studies in anorexia nervosa and obesity. Clinical & Experimental Immunology. 1997;108:507–515. doi: 10.1046/j.1365-2249.1997.3921287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ni Choileain S, Astier AL. CD46 processing: A means of expression. Immunobiology. 2012;217:169–175. doi: 10.1016/j.imbio.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marie JC, Astier AL, Rivailler P, Rabourdin-Combe C, Wild TF, Horvat B. Linking innate and acquired immunity: divergent role of CD46 cytoplasmic domains in T cell induced inflammation. Nat Immunol. 2002;3:659–666. doi: 10.1038/ni810. [DOI] [PubMed] [Google Scholar]
- 18.Sivasankar B, Longhi MP, Gallagher KME, Betts GJ, Morgan BP, Godkin AJ, Gallimore AM. CD59 Blockade Enhances Antigen-Specific CD4+ T Cell Responses in Humans: A New Target for Cancer Immunotherapy? The Journal of Immunology. 2009;182:5203–5207. doi: 10.4049/jimmunol.0804243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeerleder S. C1-inhibitor: more than a serine protease inhibitor. Semin Thromb Hemost. 2011;37:362–374. doi: 10.1055/s-0031-1276585. [DOI] [PubMed] [Google Scholar]
- 20.Makou E, Herbert AP, Barlow PN. Functional anatomy of complement factor H. Biochemistry. 2013;52:3949–3962. doi: 10.1021/bi4003452. [DOI] [PubMed] [Google Scholar]
- 21.Nilsson SC, Sim RB, Lea SM, Fremeaux-Bacchi V, Blom AM. Complement factor I in health and disease. Mol Immunol. 2011;48:1611–1620. doi: 10.1016/j.molimm.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Masaki T, Matsumoto M, Nakanishi I, Yasuda R, Seya T. Factor I-dependent inactivation of human complement C4b of the classical pathway by C3b/C4b receptor (CR1, CD35) and membrane cofactor protein (MCP, CD46) J Biochem. 1992;111:573–578. doi: 10.1093/oxfordjournals.jbchem.a123799. [DOI] [PubMed] [Google Scholar]
- 23.Hofmeyer T, et al. Arranged sevenfold: structural insights into the C terminal oligomerization domain of human C4b-binding protein. J Mol Biol. 2013;425:1302–1317. doi: 10.1016/j.jmb.2012.12.017. [DOI] [PubMed] [Google Scholar]
- 24.Preissner KT. Structure and biological role of vitronectin. Annu Rev Cell Biol. 1991;7:275–310. doi: 10.1146/annurev.cb.07.110191.001423. [DOI] [PubMed] [Google Scholar]
- 25.Mesnard L, et al. Vitronectin dictates intraglomerular fibrinolysis in immune-mediated glomerulonephritis. FASEB J. 2011;25:3543–3553. doi: 10.1096/fj.11-180752. [DOI] [PubMed] [Google Scholar]
- 26.Chauhan AK, Moore TL. Presence of plasma complement regulatory proteins clusterin (Apo J) and vitronectin (S40) on circulating immune complexes (CIC) Clin Exp Immunol. 2006;145:398–406. doi: 10.1111/j.1365-2249.2006.03135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The structure, genetic polymorphisms, expression and biological functions of complement receptor type 1 (CR1/CD35) Immunopharmacology and Immunotoxicology. 2009;31:524–535. doi: 10.3109/08923970902845768. [DOI] [PubMed] [Google Scholar]
- 28.Barilla-LaBarca ML, Liszewski MK, Lambris JD, Hourcade D, Atkinson JP. Role of Membrane Cofactor Protein (CD46) in Regulation of C4b and C3b Deposited on Cells. The Journal of Immunology. 2002;168:6298–6304. doi: 10.4049/jimmunol.168.12.6298. [DOI] [PubMed] [Google Scholar]
- 29.Cardone J, et al. Complement regulator CD46 temporally regulates cytokine production by conventional and unconventional T cells. Nat Immunol. 2010;11:862–871. doi: 10.1038/ni.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicholson-Weller A, Burge J, Fearon DT, Weller PF, Austen KF. Isolation of a human erythrocyte membrane glycoprotein with decay-accelerating activity for C3 convertases of the complement system. The Journal of Immunology. 1982;129:184–189. [PubMed] [Google Scholar]
- 31.Vainer ED, Meir K, Furman M, Semenenko I, Konikoff F, Vainer GW. Characterization of novel CD55 isoforms expression in normal and neoplastic tissues. Tissue Antigens. 2013;82:26–34. doi: 10.1111/tan.12138. [DOI] [PubMed] [Google Scholar]
- 32.Medof ME, Kinoshita T, Nussenzweig V. Inhibition of complement activation on the surface of cells after incorporation of decay44 accelerating factor (DAF) into their membranes. J Exp Med. 1984;160:1558–1578. doi: 10.1084/jem.160.5.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kimberley FC, Sivasankar B, Paul Morgan B. Alternative roles for CD59. Molecular Immunology. 2007;44:73–81. doi: 10.1016/j.molimm.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 34.Farkas I, et al. CD59 blocks not only the insertion of C9 into MAC but inhibits ion channel formation by homologous C5b-8 as well as C5b-9. The Journal of Physiology. 2002;539:537–545. doi: 10.1113/jphysiol.2001.013381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blick SK, Scott LJ. Cetuximab: a review of its use in squamous cell carcinoma of the head and neck and metastatic colorectal cancer. Drugs. 2007;67:2585–2607. doi: 10.2165/00003495-200767170-00008. [DOI] [PubMed] [Google Scholar]
- 36.Nightingale G. Ofatumumab: a novel anti-CD20 monoclonal antibody for treatment of refractory chronic lymphocytic leukemia. Ann Pharmacother. 2011;45:1248–1255. doi: 10.1345/aph.1P780. [DOI] [PubMed] [Google Scholar]
- 37.Pegram M, Liao J. Trastuzumab treatment in multiple lines: current data and future directions. Clin Breast Cancer. 2012;12:10–18. doi: 10.1016/j.clbc.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 38.Von Minckwitz G, Loibl S, Untch M. What is the current standard of care for anti-HER2 neoadjuvant therapy in breast cancer? Oncology (Williston Park) 2012;26:20–26. [PubMed] [Google Scholar]
- 39.Di Gaetano N, et al. Complement activation determines the therapeutic activity of rituximab in vivo. J Immunol. 2003;171:1581–1587. doi: 10.4049/jimmunol.171.3.1581. [DOI] [PubMed] [Google Scholar]
- 40.Golay J, et al. The role of complement in the therapeutic activity of rituximab in a murine B lymphoma model homing in lymph nodes. Haematologica. 2006;91:176–183. [PubMed] [Google Scholar]
- 41.Reff ME, et al. Depletion of B cells in vivo by a chimeric mouse human monoclonal antibody to CD20. Blood. 1994;83:435–445. [PubMed] [Google Scholar]
- 42.Bellosillo B, Villamor N, Lopez-Guillermo A, Marce S, Esteve J, Campo E, Colomer D, Montserrat E. Complement-mediated cell death induced by rituximab in B-cell lymphoproliferative disorders is mediated in vitro by a caspase-independent mechanism involving the generation of reactive oxygen species. Blood. 2001;98:2771–2777. doi: 10.1182/blood.v98.9.2771. [DOI] [PubMed] [Google Scholar]
- 43.van der Kolk LE, Grillo-Lopez AJ, Baars JW, Hack CE, van Oers MH. Complement activation plays a key role in the side-effects of rituximab treatment. Br J Haematol. 2001;115:807–811. doi: 10.1046/j.1365-2141.2001.03166.x. [DOI] [PubMed] [Google Scholar]
- 44.Gelderman KA, Tomlinson S, Ross GD, Gorter A. Complement function in mAb-mediated cancer immunotherapy. Trends Immunol. 2004;25:158–164. doi: 10.1016/j.it.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 45.Gorter A, Meri S. Immune evasion of tumor cells using membrane-bound complement regulatory proteins. Immunol Today. 1999;20:576–582. doi: 10.1016/s0167-5699(99)01537-6. [DOI] [PubMed] [Google Scholar]
- 46.Macor P, Tedesco F. Complement as effector system in cancer immunotherapy. Immunol Lett. 2007;111:6–13. doi: 10.1016/j.imlet.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 47.Golay J, Lazzari M, Facchinetti V, Bernasconi S, Borleri G, Barbui T, Rambaldi A, Introna M. CD20 levels determine the in vitro susceptibility to rituximab and complement of B-cell chronic lymphocytic leukemia: further regulation by CD55 and CD59. Blood. 2001;98:3383–3389. doi: 10.1182/blood.v98.12.3383. [DOI] [PubMed] [Google Scholar]
- 48.Macor P, Tripodo C, Zorzet S, Piovan E, Bossi F, Marzari R, Amadori A, Tedesco F. In vivo Targeting of Human Neutralizing Antibodies against CD55 and CD59 to Lymphoma Cells Increases the Antitumor Activity of Rituximab. Cancer Research. 2007;67:10556–10563. doi: 10.1158/0008-5472.CAN-07-1811. [DOI] [PubMed] [Google Scholar]
- 49.Macor P, Mezzanzanica D, Cossetti C, Alberti P, Figini M, Canevari S, Tedesco F. Complement Activated by Chimeric Anti–Folate Receptor Antibodies Is an Efficient Effector System to Control Ovarian Carcinoma. Cancer Research. 2006;66:3876–3883. doi: 10.1158/0008-5472.CAN-05-3434. [DOI] [PubMed] [Google Scholar]
- 50.Niehans GA, Cherwitz DL, Staley NA, Knapp DJ, Dalmasso AP. Human carcinomas variably express the complement inhibitory proteins CD46 (membrane cofactor protein), CD55 (decay-accelerating factor), and CD59 (protectin) Am J Pathol. 1996;149:129–142. [PMC free article] [PubMed] [Google Scholar]
- 51.Thorsteinsson L, O'Dowd GM, Harrington PM, Johnson PM. The complement regulatory proteins CD46 and CD59, but not CD55, are highly expressed by glandular epithelium of human breast and colorectal tumour tissues. Apmis. 1998;106:869–878. doi: 10.1111/j.1699-0463.1998.tb00233.x. [DOI] [PubMed] [Google Scholar]
- 52.Madjd Z, Durrant L, Pinder S, Ellis I, Ronan J, Lewis S, Rushmere N, Spendlove I. Do poor-prognosis breast tumours express membrane cofactor proteins (CD46)? Cancer Immunology, Immunotherapy. 2005;54:149–156. doi: 10.1007/s00262-004-0590-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loberg RD, Wojno KJ, Day LL, Pienta KJ. Analysis of membrane-bound complement regulatory proteins in prostate cancer. Urology. 2005;66:1321–1326. doi: 10.1016/j.urology.2005.06.094. [DOI] [PubMed] [Google Scholar]
- 54.Varela JC, Atkinson C, Woolson R, Keane TE, Tomlinson S. Upregulated expression of complement inhibitory proteins on bladder cancer cells and anti-MUC1 antibody immune selection. Int J Cancer. 2008;123:1357–1363. doi: 10.1002/ijc.23676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Murray KP, Mathure S, Kaul R, Khan S, Carson LF, Twiggs LB, Martens MG, Kaul A. Expression of Complement Regulatory Proteins— CD 35, CD 46, CD 55, and CD 59—in Benign and Malignant Endometrial Tissue. Gynecologic Oncology. 2000;76:176–182. doi: 10.1006/gyno.1999.5614. [DOI] [PubMed] [Google Scholar]
- 56.Ravindranath NMH, Shuler C. Expression of complement restriction factors (CD46, CD55 & CD59) in head and neck squamous cell carcinomas. Journal of Oral Pathology & Medicine. 2006;35:560–567. doi: 10.1111/j.1600-0714.2006.00466.x. [DOI] [PubMed] [Google Scholar]
- 57.Shimo K, et al. Complement regulatory proteins in normal human esophagus and esophageal squamous cell carcinoma. Journal of Gastroenterology and Hepatology. 2004;19:643–647. doi: 10.1111/j.1440-1746.2003.03328.x. [DOI] [PubMed] [Google Scholar]
- 58.Dzietczenia J, Wróbel T, Mazur G, Poręba R, Jaźwiec B, Kuliczkowski K. Expression of complement regulatory proteins: CD46, CD55, and CD59 and response to rituximab in patients with CD20(+) non-Hodgkin’s lymphoma. Medical Oncology. 2010;27:743–746. doi: 10.1007/s12032-009-9278-9. [DOI] [PubMed] [Google Scholar]
- 59.Weng W-K, Levy R. Expression of complement inhibitors CD46, CD55, and CD59 on tumor cells does not predict clinical outcome after rituximab treatment in follicular non-Hodgkin lymphoma. Blood. 2001;98:1352–1357. doi: 10.1182/blood.v98.5.1352. [DOI] [PubMed] [Google Scholar]
- 60.Guzzo F, et al. HER2/neu as a potential target for immunotherapy in gynecologic carcinosarcomas. Int J Gynecol Pathol. 2012;31:211–221. doi: 10.1097/PGP.0b013e31823bb24d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cocco E, et al. Tissue factor expression in ovarian cancer: implications for immunotherapy with hI-con1, a factor VII-IgGFc chimeric protein targeting tissue factor. Clinical & Experimental Metastasis. 2011;28:689–700. doi: 10.1007/s10585-011-9401-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teeling JL, et al. The Biological Activity of Human CD20 Monoclonal Antibodies Is Linked to Unique Epitopes on CD20. The Journal of Immunology. 2006;177:362–371. doi: 10.4049/jimmunol.177.1.362. [DOI] [PubMed] [Google Scholar]
- 63.Zhang B. Ofatumumab. MAbs. 2009;1:326–331. doi: 10.4161/mabs.1.4.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moore GL, Chen H, Karki S, Lazar GA. Engineered Fc variant antibodies with enhanced ability to recruit complement and mediate effector functions. MAbs. 2010;2:181–189. doi: 10.4161/mabs.2.2.11158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Strohl WR. Optimization of Fc-mediated effector functions of monoclonal antibodies. Current Opinion in Biotechnology. 2009;20:685–691. doi: 10.1016/j.copbio.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 66.Idusogie EE, Wong PY, Presta LG, Gazzano-Santoro H, Totpal K, Ultsch M, Mulkerrin MG. Engineered Antibodies with Increased Activity to Recruit Complement. The Journal of Immunology. 2001;166:2571–2575. doi: 10.4049/jimmunol.166.4.2571. [DOI] [PubMed] [Google Scholar]
- 67.Geis N, Zell S, Rutz R, Li W, Giese T, Mamidi S, Schultz S, Kirschfink M. Inhibition of membrane complement inhibitor expression (CD46, CD55, CD59) by siRNA sensitizes tumor cells to complement attack in vitro. Curr Cancer Drug Targets. 2010;10:922–931. doi: 10.2174/156800910793357952. [DOI] [PubMed] [Google Scholar]
- 68.Loberg RD, Day LL, Dunn R, Kalikin LM, Pienta KJ. Inhibition of decay-accelerating factor (CD55) attenuates prostate cancer growth and survival in vivo. Neoplasia. 2006;8:69–78. doi: 10.1593/neo.05679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ziller F, Macor P, Bulla R, Sblattero D, Marzari R, Tedesco F. Controlling complement resistance in cancer by using human monoclonal antibodies that neutralize complement-regulatory proteins CD55 and CD59. Eur J Immunol. 2005;35:2175–2183. doi: 10.1002/eji.200425920. [DOI] [PubMed] [Google Scholar]
- 70.Nagamune H, Ohnishi C, Katsuura A, Fushitani K, Whiley RA, Tsuji A, Matsuda Y. Intermedilysin, a novel cytotoxin specific for human cells secreted by Streptococcus intermedius UNS46 isolated from a human liver abscess. Infect Immun. 1996;64:3093–4100. doi: 10.1128/iai.64.8.3093-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnson S, Brooks NJ, Smith RA, Lea SM, Bubeck D. Structural basis for recognition of the pore-forming toxin intermedilysin by human complement receptor CD59. Cell Rep. 2013;3:1369–1377. doi: 10.1016/j.celrep.2013.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hu W, et al. A high-affinity inhibitor of human CD59 enhances complement-mediated virolysis of HIV-1: implications for treatment of HIV-1/AIDS. J Immunol. 2010;184:359–368. doi: 10.4049/jimmunol.0902278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu L, et al. Removal of the tag from His-tagged ILYd4, a human CD59 inhibitor, significantly improves its physical properties and its activity. Curr Pharm Des. 2012;18:4187–4196. doi: 10.2174/138161212802430486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hu W, et al. Human CD59 inhibitor sensitizes rituximab-resistant lymphoma cells to complement-mediated cytolysis. Cancer Res. 2011;71:2298–2307. doi: 10.1158/0008-5472.CAN-10-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ge X, Wu L, Hu W, Fernandes S, Wang C, Li X, Brown JR, Qin X. rILYd4, a human CD59 inhibitor, enhances complement-dependent cytotoxicity of ofatumumab against rituximab-resistant B-cell lymphoma cells and chronic lymphocytic leukemia. Clin Cancer Res. 2011;17:6702–6711. doi: 10.1158/1078-0432.CCR-11-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ge X, Wu L, Hu W, Fernandes S, Wang C, Li X, Brown JR, Qin X. rILYd4, a Human CD59 Inhibitor, Enhances Complement-Dependent Cytotoxicity of Ofatumumab against Rituximab-Resistant B-cell Lymphoma Cells and Chronic Lymphocytic Leukemia. Clinical Cancer Research. 2011;17:6702–6711. doi: 10.1158/1078-0432.CCR-11-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gaggar A, Shayakhmetov D, Lieber A. CD46 is a cellular receptor for group B adenoviruses. Nature Medicine. 2003;9:1408–1412. doi: 10.1038/nm952. [DOI] [PubMed] [Google Scholar]
- 78.Wang H, et al. Identification of CD46 binding sites within the adenovirus serotype 35 fiber knob. J Virol. 2007;81:12785–12792. doi: 10.1128/JVI.01732-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tuve S, et al. A new group B adenovirus receptor is expressed at high levels on human stem and tumor cells. J Virol. 2006;80:12109–12120. doi: 10.1128/JVI.01370-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang Z, Kuhr CS, Allen JM, Blankinship M, Gregorevic P, Chamberlain JS, Tapscott SJ, Storb R. Sustained AAV-mediated dystrophin expression in a canine model of Duchenne muscular dystrophy with a brief course of immunosuppression. Mol Ther. 2007;15:1160–1166. doi: 10.1038/sj.mt.6300161. [DOI] [PubMed] [Google Scholar]
- 81.Wang H, et al. In vitro and in vivo properties of adenovirus vectors with increased affinity to CD46. J Virol. 2008;82:10567–10579. doi: 10.1128/JVI.01308-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang H, Liu Y, Li ZY, Fan X, Hemminki A, Lieber A. A recombinant adenovirus type 35 fiber knob protein sensitizes lymphoma cells to rituximab therapy. Blood. 2010;115:592–600. doi: 10.1182/blood-2009-05-222463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Beyer I, et al. Transient Removal of CD46 Is Safe and Increases B-cell Depletion by Rituximab in CD46 Transgenic Mice and Macaques. Mol Ther. 2013;21:291–299. doi: 10.1038/mt.2012.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Saphire EO, Stanfield RL, Max Crispin MD, Parren PWHI, Rudd PM, Dwek RA, Burton DR, Wilson IA. Contrasting IgG Structures Reveal Extreme Asymmetry and Flexibility. Journal of Molecular Biology. 2002;319:9–18. doi: 10.1016/S0022-2836(02)00244-9. [DOI] [PubMed] [Google Scholar]
- 85.Idusogie EE, Presta LG, Gazzano-Santoro H, Totpal K, Wong PY, Ultsch M, Meng YG, Mulkerrin MG. Mapping of the C1q Binding Site on Rituxan, a Chimeric Antibody with a Human IgG1 Fc. The Journal of Immunology. 2000;164:4178–4184. doi: 10.4049/jimmunol.164.8.4178. [DOI] [PubMed] [Google Scholar]
- 86.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 87.Ltd., P.o.V.P., editor. Persistence of Vision Raytracer (Version 3.6) 2004 [Google Scholar]
- 88.Fabry CM, Rosa-Calatrava M, Conway JF, Zubieta C, Cusack S, Ruigrok RW, Schoehn G. A quasi-atomic model of human adenovirus type 5 capsid. Embo J. 2005;24:1645–1654. doi: 10.1038/sj.emboj.7600653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hanson R. Jmol -a paradigm shift in crystallographic visualization. Journal of Applied Crystallography. 2010;43:1250–1260. [Google Scholar]
- 90.Persson BD, Reiter DM, Marttila M, Mei YF, Casasnovas JM, Arnberg N, Stehle T. Adenovirus type 11 binding alters the conformation of its receptor CD46. Nat Struct Mol Biol. 2007;14:164–166. doi: 10.1038/nsmb1190. [DOI] [PubMed] [Google Scholar]
- 91.van Raaij MJ, Mitraki A, Lavigne G, Cusack S. A triple beta spiral in the adenovirus fibre shaft reveals a new structural motif for a fibrous protein. Nature. 1999;401:935–938. doi: 10.1038/44880. [DOI] [PubMed] [Google Scholar]