SUMMARY
Genes disrupted in schizophrenia may be revealed by de novo mutations in affected persons from otherwise healthy families. Furthermore, during normal brain development, genes are expressed in patterns specific to developmental stage and neuroanatomical structure. We identified de novo mutations in persons with schizophrenia, then mapped the responsible genes onto transcriptome profiles of normal human brain tissues from age 13 weeks gestation to adulthood. In the dorsolateral and ventrolateral prefrontal cortex during fetal development, genes harboring damaging de novo mutations in schizophrenia formed a network significantly enriched for transcriptional co-expression and protein interaction. The 50 genes in the network function in neuronal migration, synaptic transmission, signaling, transcriptional regulation, and transport. These results suggest that disruptions of fetal prefrontal cortical neurogenesis are critical to the pathophysiology of schizophrenia. These results also support the feasibility of integrating genomic and transcriptome analyses to map critical neurodevelopmental processes in time and space in the brain.
INTRODUCTION
Schizophrenia is a complex brain disorder characterized by aberrant perceptions, thought processes, and behavior. It is one of the leading causes of disability worldwide (Press, 2008), and is both highly heritable and highly genetically heterogeneous (McClellan and King, 2010). Individually rare gene-disrupting copy number variants (CNVs) contribute substantially to the disorder (Walsh et al., 2008; Xu et al., 2008; Stefansson et al., 2008; The International Schizophrenia Consortium 2008), in particular CNVs that disrupt genes involved with signaling, synaptic plasticity and neurodevelopmental processes (Walsh et al., 2008; Kirov et al., 2012). These mutations are de novo or recent in origin, with de novo CNVs specifically enriched in patients with sporadic, rather than familial, illness (Xu et al., 2008; Kirov et al., 2012). Most rare gene-disrupting CNVs detected in affected persons are unique, although some recur independently at genomic hotspots, including on chromosomes 1q21.1, 3q29, 15q11.2, 15q13.1, 16p11.2, and 22q11.2 (McClellan and King, 2010) and at the neuropeptide receptor VIPR2 (Vacic et al., 2011).
Advances in genomics and neurobiology have enabled the next generation of these studies. The analysis of de novo mutations in schizophrenia can now include point mutations and small insertions and deletions (indels) as well as CNVs (Girard et al., 2011; Xu et al., 2011; Xu et al., 2012). In parallel, transcriptome databases of the human brain have been generated (Kang et al. 2011; Hawrylycz et al., 2012). Integrating these approaches has led to new insights into genetic aberrations in mental illness. For example, patterns of gene expression distinguishing frontal and temporal cortex have been shown to be significantly attenuated in brains of persons with autism (Voineagu et al., 2011). Furthermore, genes implicated in schizophrenia, including some harboring de novo mutations, have been shown to cluster in networks that are highly expressed in brain, particularly during prenatal development (Xu et al., 2012; Gilman et al., 2012).
The goal of this project was to identify temporal and spatial processes of brain development critical to the neuropathogenesis of schizophrenia. Our approach was to examine the contribution of de novo mutations to schizophrenia, then to characterize functional networks in brain of the genes harboring these mutations. The study design integrates genomics, transcriptomics, and proteomics. We first identified genes harboring de novo putatively damaging mutations in persons with schizophrenia from otherwise healthy families. We then evaluated the extent to which the proteins encoded by these genes interact, and the extent to which they are transcriptionally co-expressed in different brain regions across developmental stages. The co-expression and protein interaction profiles were then used to generate a network whose interconnectedness was quantifiable by the number of connections (edges) between implicated genes. Across brain regions and developmental stages, we compared the interconnectedness of networks of genes harboring de novo mutations in probands versus networks from 10,000 simulations of genes harboring de novo mutations in unaffected persons.
RESULTS
Identification of de novo mutations by whole exome sequencing
De novo mutations were identified by exome sequencing of quads and trios comprised of a proband with schizophrenia, his/her unaffected parents, and whenever available, an unaffected sibling. Families were selected for negative family history of severe mental illness other than the proband; that is, the probands were sporadic, or singleton, cases. The study design was based on quads because unaffected siblings provide ideal controls for ancestry, for nongenetic familial effects, and for sampling strategy (Sebat et al., 2007). A quad-based study design also optimizes experimental conditions since the affected and unaffected persons are sequenced in the same way and the same variant-assessment filters are applied to all subjects, blind to disease status. In each family, de novo variants were identified by comparing exome sequences of each child to his/her parents. Variants were experimentally validated as present in the child and absent from both parents, then classified as damaging or not damaging to protein function as defined in Experimental Procedures. Exome sequencing of genomic DNA was carried out for 399 persons, including 105 probands with schizophrenia, 84 unaffected sibs, and their 210 unaffected parents (Table S1).
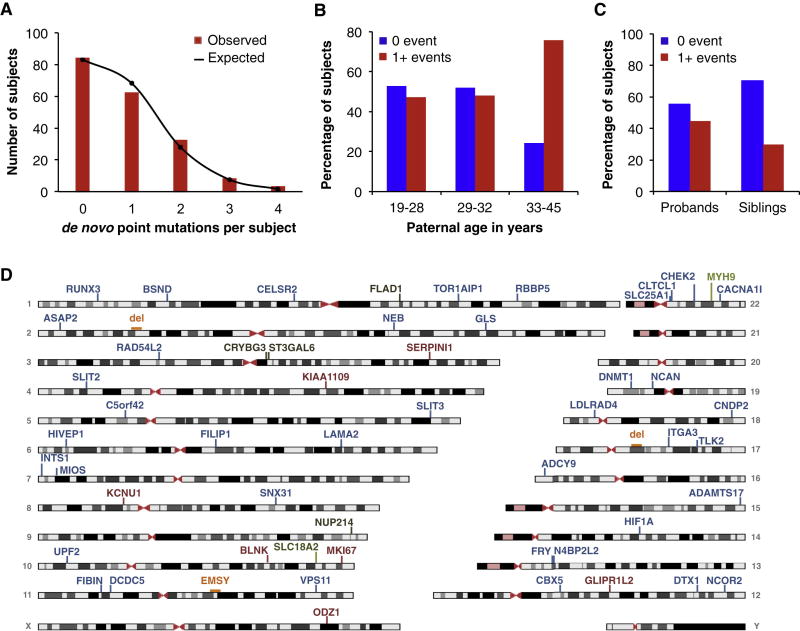
We first considered all de novo point mutations in probands and unaffected sibs, regardless of predicted effect on protein function. Of the 105 probands, 66 probands (63%) carried 96 de novo point mutations, 0.91 mutations per individual; of the 84 unaffected siblings, 39 siblings (46%) carried 66 such mutations, 0.79 mutations per individual (Table 1). The difference reflects a modest enrichment among probands in the likelihood of carrying at least one de novo mutation (X2 = 5.10; P = 0.024). Combining silent, missense, and nonsense mutations yields 155 de novo point mutations in coding sequence, of which 68% (105/155) were non-synonymous, similar to most previous studies of schizophrenia and autism (Xu et al., 2012; Iossifov et al., 2012; O’Roak et al., 2012b: Neale et al., 2012; Sanders et al., 2012) (Table S2). Mutation frequency was distributed as a Poisson distribution with mean 0.82 (Figure 1A).
Table 1.
De novo mutations in probands with schizophrenia and their unaffected siblings
| Probands (N = 105) | Unaffected siblings (N = 84) | Total (N = 189) | |||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Mutation type | Damaging* | Benign | All de novo | Damaging* | Benign | All de novo | All de novo |
| Frameshift | 4 | 0 | 4 | 1 | 0 | 1 | 5 |
| Nonsense | 7 | 0 | 7 | 5 | 1 | 6 | 13 |
| Missense | 41 | 15 | 56 | 24 | 12 | 36 | 92 |
| Silent | 0 | 31 | 31 | 0 | 19 | 19 | 50 |
| Splice | 2 | 0 | 2 | 5 | 0 | 5 | 7 |
| CNV | 3 | 0 | 3 | 0 | 0 | 0 | 3 |
| Total | 57 | 46 | 103 | 35 | 32 | 67 | 170 |
Predicted to be damaging based on criteria described in text.
Figure 1. De novo mutations in persons with schizophrenia and unaffected siblings.
(A) All de novo point mutations in 189 persons. Observed numbers of events fit a Poisson distribution with m = 0.87 events per person. R2 = 0.97 for goodness of fit of observed to expected values under Poisson assumption.
(B) De novo point mutations in 189 offspring by terciles of paternal age. Presence of at least one de novo point mutation is associated with older paternal age (X2 = 13.96, p = 0.0009).
(C). Presence of at least one de novo putatively damaging mutation in schizophrenia probands and unaffected siblings. 45% (47/105) of the probands and 30% (25/84) of the unaffected siblings carried at least one damaging de novo mutation (X2=4.45, P=0.035).
(D) Genes harboring de novo damaging mutations in persons with schizophrenia. Colors represent types of mutations: blue are missense, red are nonsense, black are frameshift, green are splice, and orange are CNVs.
The likelihood of carrying at least one de novo mutation was related to paternal age at conception (Kong et al. 2012). Among offspring of the youngest third of fathers, ages 19–28 years, 47% (33/70) carried at least one de novo mutation; among offspring of the oldest third of fathers, ages 33–45 years, 76% (50/66) carried at least one de novo mutation (X2= 13.96, p = 0.0009; Table S1, Figure 1B). The relationship between paternal age and de novo mutations was the same for affected and unaffected siblings.
We next considered de novo mutations of all classes (point mutations, indels, and CNVs) predicted to damage protein function. Of the probands, 47/105 (45%) carried 57 de novo damaging mutations, 0.54 damaging mutations per individual; of the unaffected siblings, 25/84 (30%) carried 35 such mutations, 0.42 per individual (Table S3). This difference reflects a modest enrichment among probands in the likelihood of carrying at least one de novo damaging mutation (Figure 1C, OR = 1.91, X2 = 4.45; P = 0.035). Based on this sample, the proportion of schizophrenia attributable to de novo damaging mutations is 21% (see Experimental Procedures).
Schizophrenia gene networks
We next examined how genes harboring de novo putatively damaging mutations might reveal networks critical to development of schizophrenia. The analysis addressed three questions. First, do genes with de novo damaging mutations in schizophrenia form larger and/or more interconnected protein interaction networks than do genes with de novo damaging mutations in controls? Second, is there evidence for transcriptional co-expression of genes harboring these mutations in any brain regions across different stages of development? Third, combining protein interaction and transcriptome data, what networks are generated by the genes harboring de novo damaging mutations in schizophrenia, and which genes are included in these networks?
Overall, 54 different genes harbored de novo damaging point mutations, indels, or single-gene-disrupting CNVs in probands (Table S3A). Each of these genes was included in protein interaction and co-expression analyses. For de novo CNVs, genes were included if disrupted by breakpoints, but not if completely deleted or duplicated.
For comparison, we considered genes carrying de novo damaging mutations in healthy siblings from this study (Table S3B) and from four previous exome studies of schizophrenia and autism (Xu et al., 2012; Iossifov et al., 2012; O’Roak et al., 2012b; Sanders et al., 2012). From the five studies combined, 264 genes in unaffected siblings harbored de novo mutations predicted to be damaging, based on the same criteria used to define de novo damaging events in our probands (Experimental Procedures and Table S4). We carried out the same analyses on 10,000 sets of 54 control genes, each randomly selected from the pool of 264 genes. We constructed a network for each set of 54 genes, yielding 10,001 networks including the network based on case genes, and counted the numbers of nodes and edges for each.
Protein-protein interaction networks of genes with de novo mutations
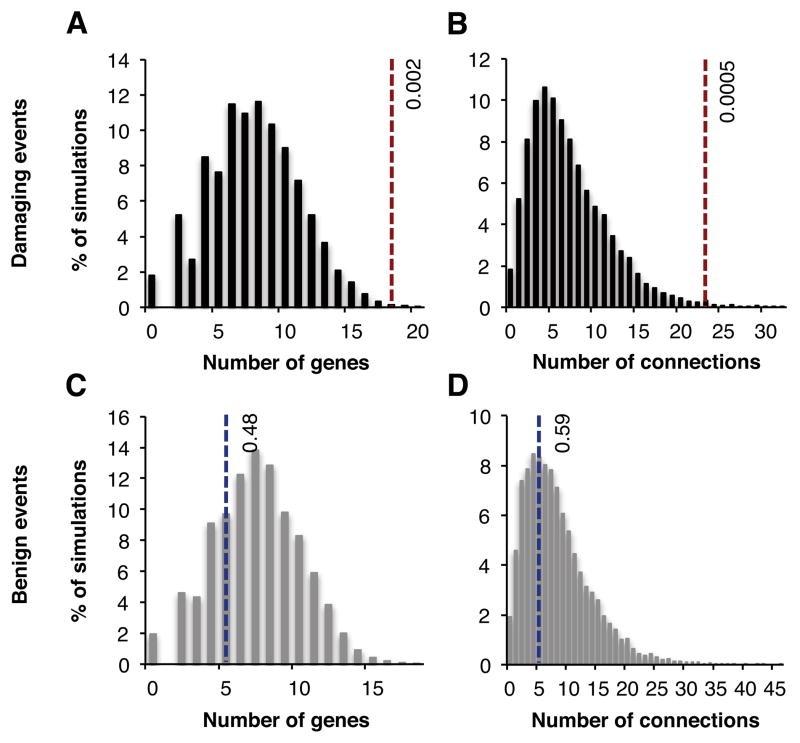
We used the GeneMania physical interaction dataset (Mostafavi et al., 2008) to examine whether genes disrupted by de novo damaging events in probands clustered in protein-protein interaction (PPI) networks. Of the 54 proband genes with de novo damaging mutations, 18 genes mapped to an interconnected PPI network with 23 edges. We evaluated this network by comparing its numbers of nodes and edges to the distribution of nodes and edges of 10,000 simulated networks of genes carrying de novo damaging events in healthy siblings (Figures 2A and 2B). The network of genes harboring de novo damaging mutations in schizophrenia had significantly more nodes (P=0.002) and edges (P=0.0005) than networks of genes harboring de novo damaging mutations in siblings. In contrast, the network of genes harboring de novo benign events in the probands had no more nodes or edges than networks of genes harboring de novo benign events in siblings (Figures 2C and 2D).
Figure 2. Interconnectedness of protein-protein interaction (PPI) networks based on genes harboring de novo mutations.
(A–B) The 54 genes harboring de novo damaging mutations in schizophrenia cases yield a PPI network of 18 genes (A) and 23 interactions (B), indicated by the vertical dotted lines. For comparison, 10,000 sets of 54 genes were selected at random from 264 genes harboring de novo damaging mutations in unaffected sibs (controls). Each of these 10,000 gene sets also yields a PPI network. The numbers of genes and connecting links for each of these 10,000 control networks was plotted in panels A and B, respectively. P-values for enrichment of genes and connections in the case network were estimated by the proportion of control networks with more genes or interactions than the case network. The network based on cases is enriched both for number of genes (P=0.002) and for number of connections between genes (P=0.0005).
(C–D) Analogous networks based on genes harboring de novo benign events in cases and in controls. The networks based on genes with de novo benign events in cases do not differ from networks based on genes with de novo benign mutations in controls.
Transcriptional co-expression networks of genes with de novo mutations
Next, we evaluated transcriptional co-expression, across different brain regions at different developmental periods, for genes harboring de novo mutations in probands with schizophrenia and for genes harboring de novo mutations in unaffected siblings. Using publicly available RNASeq-based gene expression levels from the BrainSpan Atlas of Developing Human Brain (2013), we generated networks reflecting profiles of co-expressed genes.
Tissues from the BrainSpan Atlas were divided into four anatomic regions: frontal cortex (FC), temporal and parietal regions (TP), sensory-motor regions (SM), and sub-cortical regions (SC); and three developmental stages: fetal (13 – 26 post-conception weeks), early infancy to late childhood (4 months – 11 years) and adolescence to adulthood (13 – 23 years) (Table S5). The number of tissues included by the BrainSpan Atlas ranged from 5–6 for children and young adults to 11–12 for fetal tissues. For each pair of proband genes and each pair of control genes, we calculated Pearson correlation coefficients for RNASeq expression levels across tissues within each brain region and each developmental stage. For each set of tissues, we therefore calculated correlation coefficients for (54×53)/2 = 2860 gene pairs. Gene pairs with |R| ≥ 0.8 were defined as “connected” in subsequent network constructions. Network interconnectedness was measured by the number of connections, or edges (Barabasi and Oltvai, 2004).
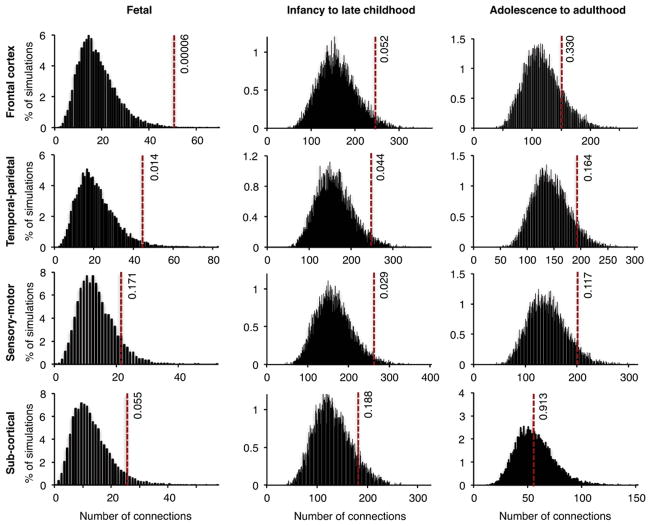
Numbers of edges in the networks of schizophrenia genes and control genes for each brain region by developmental stage are shown in Figure 3. The most striking result is that in fetal frontal cortex, co-expression of genes harboring de novo damaging mutations in probands yields a network with significantly greater connectedness than networks derived from genes harboring de novo damaging mutations in controls (nominal P = 0.00006; corrected for multiple comparisons, P = 0.0007) (Figure 3, Table S6). Connectedness was not significantly greater for case genes than for control genes at other developmental periods. There was no significant enrichment for connectedness in networks based on case genes harboring de novo benign mutations (Figure S2).
Figure 3. Interconnectedness of transcriptional co-expression networks, at various developmental stages and in different brain regions, based on genes harboring de novo damaging mutations.
Co-expression of genes harboring de novo damaging mutations in cases and in controls was evaluated using RNASeq data from the BrainSpan Atlas. Gene pairs were defined as co-expressed if |R| > 0.8 for their RNASeq expression levels across all tissues from a given brain region and a given developmental stage. Networks were created for co-expressed gene pairs as described for Figure 2B. Dotted lines indicate numbers of connections (edges) in networks created using genes with de novo damaging mutations in cases. Histograms represent distributions of the numbers of edges in 10,000 simulated networks using genes with de novo damaging mutations in controls. The most significant enrichment for co-expression of genes mutant in schizophrenia was observed in frontal cortex during fetal development (P=0.00006; Table S6). There was no enrichment for co-expression of genes with de novo benign mutations in schizophrenia compared to controls (Figure S2).
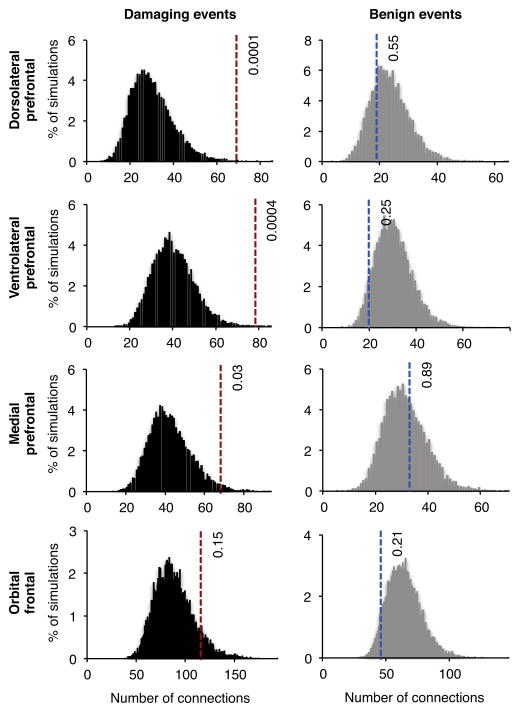
Given the significant interconnectedness of co-expressed case genes in fetal frontal cortex, we carried out the same analyses for four anatomical subregions of fetal frontal cortex: dorsolateral prefrontal cortex, ventrolateral prefrontal contex, medial prefrontal cortex, and orbital frontal cortex. Co-expression profiles of genes mutant in schizophrenia differed most significantly from those of unaffected siblings in tissues from fetal dorsolateral prefrontal cortex (nominal P = 0.0001; corrected P = 0.0005) and fetal ventrolateral prefrontal cortex (nominal P = 0.0004; corrected P = 0.002) (Figure 4, Table S7).
Figure 4. Interconnectedness of transcriptional co-expression networks from four subregions of the fetal frontal cortex.
Tissues from the fetal frontal cortex with RNASeq data in the BrainSpan Atlas were classified by subregion. Networks were then created, as described for Figure 3, for genes with de novo damaging mutations in cases and in controls (left panels) and for de novo benign mutations in cases and in controls (right panels). The greatest enrichments for co-expression of genes with de novo damaging events were observed in the dorsolateral and ventrolateral prefrontal cortex (Table S7). There were no regions enriched for co-expression of genes harboring de novo benign mutations in cases.
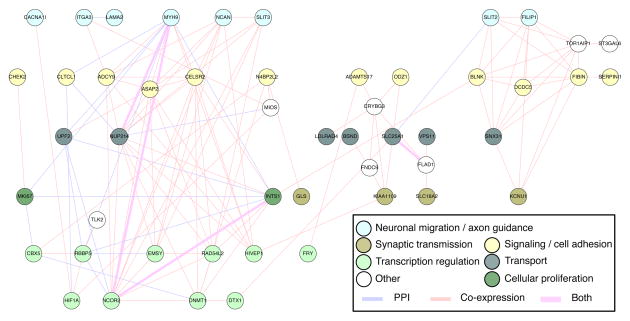
A merged network of protein interaction and transcriptional co-expression in the dorsolateral and ventrolateral prefrontal cortex during fetal development
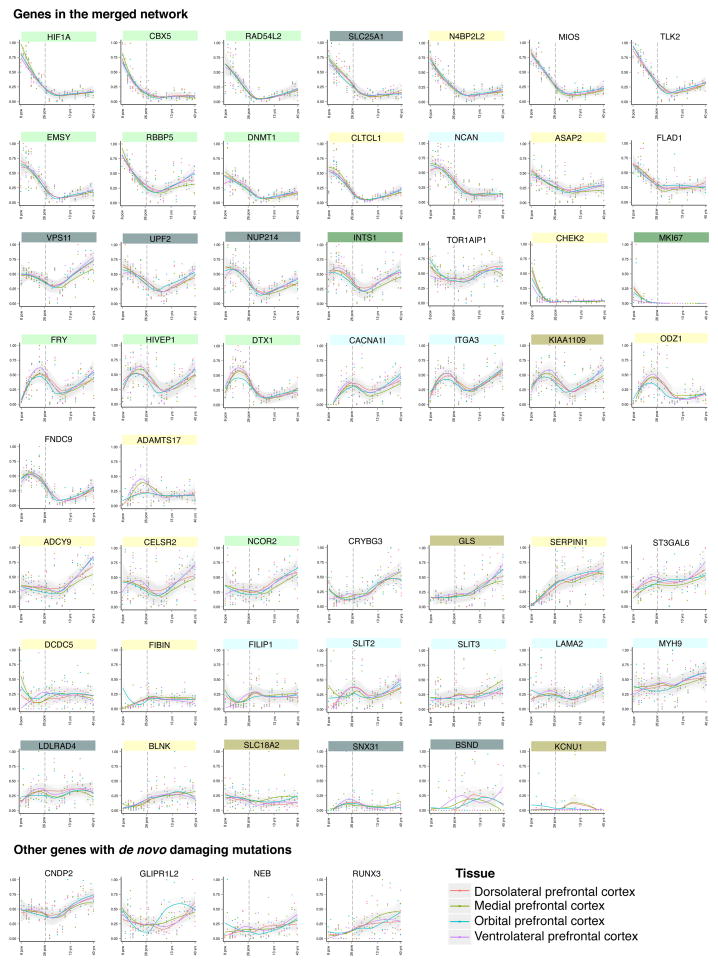
We then merged the co-expression network derived for schizophrenia genes in fetal dorsolateral and ventrolateral prefrontal cortex with the PPI network derived for the same genes. The merged network encompasses 50 of the 54 genes harboring damaging de novo mutations in subjects with schizophrenia, with 126 connections among these genes (Figure 5). Genes in the network function in biological processes critical to neurogenesis and synaptic integrity, including axon guidance and neuronal migration, signaling, cell proliferation, transport, synaptic transmission and transcriptional regulation. Expression levels in fetal frontal cortex of individual network genes are provided in Figure 6. Expression levels of most of the genes are high early in fetal development, then decline at the end of fetal development and in childhood, and gradually increase again in early adulthood. Expression of other genes either increases throughout fetal and adult development or remains at stable levels over time (Figure 6).
Figure 5. Network of genes harboring damaging de novo mutations in schizophrenia that are co-expressed in fetal dorsolateral (DFC) and ventrolateral (VFC) prefrontal cortex.
Merging data from networks of protein-protein interaction (PPI) network and of co-expression in fetal DFC and fetal VFC yields a network of 50 genes (nodes) with 126 connections (edges). Red lines represent co-expressed genes; blue lines represent physical connections between protein products of the genes.
Figure 6. Expression patterns in frontal cortex of genes with de novo damaging mutations in schizophrenia.
Gene expression across all periods of development (13 post-conception weeks to 23 years) for each gene harboring a de novo damaging mutation in schizophrenia is depicted for four brain regions: dorsolateral prefrontal cortex (DFC), anterior (rostral) cingulate (medial prefrontal) cortex (MFC), orbital frontal cortex (OFC), and ventrolateral prefrontal cortex (VFC). Expression levels were obtained from BrainSpan Atlas of Developing Human Brain (2013). RPKM values were max-min normalized and Loess smoothed (Cleveland et al., 1992) across time points.
DISCUSSION
The prefrontal cortex and its role in schizophrenia
Damaging de novo mutations in persons with schizophrenia converge in a network of genes co-expressed in the dorsolateral and ventrolateral prefrontal cortex during fetal development. These results support the neurodevelopmental hypothesis, proposed 25 years ago, that disruptions in fetal prefrontal cortical development are core mechanisms underlying schizophrenia (Weinberger, 1987; Marin, 2012). The specific timing and anatomic locale of the gene network, and the biological functions of its component genes, suggest that mutations leading to schizophrenia disrupt the orchestration of cortical neurogenesis.
Human brain evolution is distinguished by the remarkable expansion of the surface area and cytoarchitectural complexity of the neocortex, particularly in the prefrontal cortex. Healthy adult cortical function emerges from synchronized spatiotemporal neuronal migration during fetal development, which depends upon the complex regulation of cellular proliferation, signaling and transcription pathways (Rakic, 2009). Many genes orchestrate these processes, the disruption of any one of which may result in loss of cortical integrity and neuronal homeostasis, leading to neuropsychiatric disease (Ramocki and Zoghbi, 2008).
Prefrontal cortical networks organize input from other cortical and subcortical brain regions to plan and direct motor, cognitive, affective and social behaviors (Kolb et al., 2012). The dorsolateral and ventrolateral prefrontal cortices are distinct neuroanatomic structures, with complementary neurocognitive functions. The dorsolateral prefrontal cortex is linked to cognitive control, which requires the coordination of attention and working memory to address changing environments and task demands. The ventrolateral prefrontal cortex is associated with reward learning and decision-making, which depend upon the ability to gauge potential outcomes in the context of incoming stimuli as a guide to making choices (Glasher et al., 2012). These executive functions endow humans with enormous capacity for problem-solving and adaptation to a wide range of environments.
Several lines of evidence support the role of the prefrontal cortex in schizophrenia. Deficits in executive functions are characteristic of the illness and often predate its onset (Kalkstein et al., 2010). The prefrontal cortex matures late in brain development, with gray matter consolidation and myelination extending into early adulthood—the timing of which is consistent with ages of onset of schizophrenia (Gogtay et al., 2004). Neuroimaging studies of schizophrenia have demonstrated anatomical and functional deficits in the region (Shepherd et al., 2012). Postmortem brain studies have found a variety of cellular pathological findings in prefrontal cortical tissue (Eisenberg and Berman, 2010). Further, a substantial portion of prefrontal cortical neurons express monoaminergic receptors that are targets of antipsychotic medications (Masana et al., 2012). Our results suggest that deficits in prefrontal cortex executive functions are manifestations of schizophrenia, with full presentation occurring as key brain regions mature. Integrating genomic data with transcriptome network analyses can help pinpoint anatomical and developmental mechanisms that are disrupted in this process.
A key feature of schizophrenia appears to be brain disconnectivity, i.e., aberrant connections with and between different brain regions (Fitzsimmons et al., 2013). The prefrontal cortex is normally highly interconnected with other cortical, basal ganglia and limbic regions. Human brain development depends upon the coordinated temporal and spatial regulation of neuronal migration in order to establish integrated cortical networks (Uhlhaas and Singer 2012). Disruptions of individual components within developing neuronal networks may impact the functional integrity of larger dynamic brain systems (Vidal et al., 2011). Therefore, our results do not preclude the involvement of other brain regions in schizophrenia, nor imply that damaging mutations have an impact only during fetal development. Genes harboring de novo damaging mutations in schizophrenia are expressed in multiple brain regions (and other tissues) across the lifespan. Neuroimaging studies of schizophrenia describe multiple anatomical abnormalities, including decreased total brain volumes and losses of grey matter in the anterior cingulate, frontal and temporal lobes, hippocampus, amygdala, thalamus, and insula (Shepherd et al., 2012).
Roles in brain development of genes with de novo mutations in schizophrenia
Genes implicated in schizophrenia and autism function in processes important to fetal brain development, including axon guidance, neuronal cell mobility, synaptic function and chromosomal remodeling (Gilman et al., 2012; Xu et al., 2012). Most of the genes with damaging de novo mutations in probands are known to play critical roles in developing brain, including neuronal migration, (ITGA3, LAMA2; Anton et al., 1999; Relucio et al., 2009), axon guidance (SLIT2, SLIT3; Zhang et al., 2012), signal transduction and cell adhesion (CELSR2; Shima et al., 2007), cellular proliferation (MKI67; Duchrow et al., 1995), transcriptional regulation of glial and neuronal development (DTX1, HIF1A, NCOR2; Patten et al., 2006; Pacary et al., 2007; Jepsen et al., 2007), and neurotransmitter signaling and synaptic transmission (ADCY9, CACNA1I, GLS, SLC18A2; Martin et al., 2007; Simons and van Winkel, 2013).
Several network genes with de novo mutations in probands function in neurotransmitter pathways. ADCY9 is involved in neuronal signaling in glutamate and GABA pathways (Martin et al., 2007). SLC18A2 encodes the vesicular monoamine transporter VMAT2, which is responsible for pumping serotonin, dopamine, norepinephrine, epinephrine, and histamine into vesicles for use in synaptic transmission (Simons and van Winkel, 2013). GLS catalyzes the hydrolysis of glutamine to glutamate. Studies of GLS knockout mice suggest that dysregulation of glutaminase function could play a role in schizophrenia; and that medications inhibiting glutaminase may have therapeutic benefit for the illness (Gaisler-Salomon et al., 2012).
CACNA1I, the only gene disrupted by damaging de novo mutations in more than one patient, is a brain-specific, T-type calcium channel involved in regulation of differential patterns of neuronal firing in the thalamus, striatum, nucleus accumbens and prefrontal cortex (Chemin et al., 2002; Talley et al., 1999; Yunker et al., 2003). In rats, T-type calcium channel antagonists reduced amphetamine-induced psychomotor activity and glutamate release, patterns predictive of antipsychotic effects (Uslaner et al., 2012). The therapeutic activity of antipsychotic medications, including clozapine, may stem in part from the inhibition of T-type calcium channels (Choi and Rhim, 2010).
Our results provide further evidence for the genetic complexity and heterogeneity of schizophrenia. Several network genes have been implicated previously in schizophrenia and other neuropsychiatric disorders, including LAMA2 (Xu et al., 2012), DNTM1 (Costa et al., 2007), and SLC25A1 and CLTCL1, both of which are located in the chromosome 22q11.2 deletion region that confers substantial risk for schizophrenia. Expression profiles of CELSR2, HIVEP1, ITGA3, SERPINI1, and SLIT2 differed between neurons induced from stem cells of persons with schizophrenia versus controls (Brennand et al., 2011). Nine patients carried de novo damaging mutations in more than one gene (Table S8), suggesting the possibility of oligogenic influences on disease (Girirajan et al., 2012).
Causality
Given the degree of genetic heterogeneity of mental illness, establishing causal links between individual genes and schizophrenia will depend on both biological and statistical evidence. The sample size of this study both limits the power of network analyses and is not sufficient to prove causality of any one gene. Resequencing studies of candidate genes in very large cohorts may provide such statistical evidence, as has been demonstrated for autism (O’Roak et al., 2012b).
The goal of modern genetic studies of mental illness is to identify key pathways that, when disrupted, lead to disease, realizing that mutation of any of multiple genes may disrupt the same pathway (McClellan and King, 2010). Our goal for this project has been to use computational tools to define gene networks based on mutations in affected individuals, thereby providing empiric evidence that the networks are related to schizophrenia. The next step is to discover the biological pathways that underlie the computational networks (Konopka et al., 2012). The principal limitation in doing so is incomplete understanding of normal brain development. As pathways critical to neurodevelopment are revealed by mutations that disrupt them, it will become possible to carry out statistical testing for mutations leading to loss of integrity of a pathway rather than of a single gene.
Our results suggest that disruption of genes critical to neurogenesis of the prefrontal cortex are also critical to pathogenesis of schizophrenia. These findings have implications for the development of more biologically specific treatments of the illness (Insel, 2009). Given ethical, clinical, and neurobiological limitations to in utero interventions, treatment research inevitably focuses on compensatory strategies that address downstream effects on brain development. Medications targeting glutamate and T-type calcium channel pathways are potential therapeutic mechanisms suggested by mutant genes in our patients. Large-scale collaborative efforts in genomics and neurobiology are needed to reveal additional therapeutic targets based on the genes and biological mechanisms underlying brain circuitry (Akil et al., 2010).
EXPERIMENTAL PROCEDURES
Exome Sequencing
From genomic DNA of the 399 participants, library construction, exome capture, and sequencing were carried out as previously described (Walsh et al., 2010) with some modifications. Genomic DNA libraries were captured by SeqCap EZ Exome v2 (Nimblegen) pools, then hybridized to biotynylated capture probes and sequenced to median depth of coverage of at least 100x. Paired-end sequence reads (2x101bp) were collected, de-multiplexed, filtered for quality, aligned to exome targets using BWA v0.6.1-r104, realigned with GATK v1.5-21, then genotypes called with MAQ v0.7.1, SAM tools v0.1.18, and GATK v1.5-21. For individual samples, on average 93% of targeted coding regions were covered at ≥10X and 84% at ≥20X. For joint coverage of trios (Iossifov et al., 2012), on average 89% of targeted coding regions were covered at ≥10X and 77% at ≥20X (Figure S1).
Artifacts were excluded by comparison with 800 exomes previously sequenced on the same instrument in our lab. Common variants, with MAF>0.001 in dbSNP v137 or in the NHLBI Exome Sequencing Project (2012), were also excluded. To detect point mutations and small insertions and deletions (1–23 bp in length), variants were retained at sites with at least 30% of total reads representing the variant, after adjusting by individual inspection for genomic complexities (e.g. multiple pseudogenes of CHEK2). No constraint was put on the proportion of variant reads in parental samples. Candidate de novo variants were tested by diagnostic PCR and Sanger sequencing. To detect CNVs, exome-based calls were made by CoNIFER (Krumm et al., 2012), then validated by TaqMan in DNA from the proband or sib and from the parents. For de novo CNVs not flanked by segmental duplications, exact genomic breakpoints were determined with diagnostic PCR primers and Sanger sequencing. De novo variants included in subsequent analyses were defined as variants of any type demonstrated by both exome sequencing and Sanger sequence validation to be present in a proband or a sib and absent from both parents.
The predicted functional impact of each candidate de novo missense variant was assessed with in silico tools. Variants that met any of the following criteria were considered potentially damaging: frameshift; nonsense (unless the introduced nonsense was polymorphic or the natural stop in other species); missense mutation with Polyphen score ≥0.90 and/or SIFT P < 0.05 and/or Grantham score >100 (Adzhubei et al., 2010; Kumar et al., 2009; Grantham, 1974); CNV disrupting a gene or completely deleting a gene or genes; splice or enhancer alteration predicted by one or more algorithms and with GERP score >5.0.
Proportions of probands and unaffected siblings with point mutations, and with damaging mutations of all classes, were compared by chisquare tests using standard procedures. Attributable risk was estimated by p(OR − 1)/[p(OR − 1) + 1] (Schlesselman, 1982), where p is the proportion of controls (unaffected sibs) with a de novo damaging mutations and OR is the odds ratio for harboring a de novo damaging mutation in cases vs controls.
Network construction
Physical interactions from the Homo sapiens database were collected from GeneMANIA 2011-08-03 release of 150 studies (Mostafavi et al., 2008). There were 753,875 physical interactions in 12,010 proteins after removing duplicates. A physical interaction network was created for the schizophrenia genes and for each simulation gene set. Degree is defined as the number of neighbors of each node (Barabasi and Oltvai, 2004; Watts and Strogatz, 1998).
Co-expression based network analyses were conducted using existing data-sets in the BrainSpan Atlas of Developing Human Brain (2013). Normalized gene expression levels for 26 different brain tissues in 10 different developmental periods were obtained from the BrainSpan RNAseq dataset v3 of developing human brain. Tissue qualification, processing and dissection, and experimental and bioinformatics procedures are described in the technical white paper of the dataset (http://help.brain-map.org/display/devhumanbrain/Documentation) (Supplemental materials). Network figures were created using Cytoscape 2.8.3 (Smoot et al., 2011).
To estimate the statistical significance of networks created from PPI and co-expression data, we generated a dataset of de novo predicted damaging mutations among unaffected siblings by pooling results from unaffected siblings in this study with results from unaffected siblings reported in autism and schizophrenia exome studies (Xu et al., 2012; Iossifov et al., 2012; O’Roak et al., 2012b; Sanders et al., 2012). We excluded variants observed in both probands and unaffected siblings, variants without a defined nucleotide change, and variants reported by the authors to have failed validation. From all unaffected siblings, 547 de novo events were included, for which functional consequences were predicted using the same criteria applied to probands in this study (Supplementary Materials). A total of 276 events (2 frameshift, 21 nonsense, 248 missense, and 5 splice) in 267 genes were predicted to be damaging. Three genes not represented in either the GeneMania database or the BrainSpan Atlas were removed from analysis. Using the remaining 264 genes with damaging de novo events in the pooled group of unaffected siblings, we created 10,000 random gene sets that each contained 54 different genes; corresponding to the number of genes harboring predicted damaging events in probands.
PPI and co-expression networks were constructed for each gene set. The mean numbers and standard deviations of nodes and edges in the simulated networks were used to generate normal distributions for PPI networks in each of the four anatomic regions and three developmental stages. Significance levels of networks based on genes mutant in probands were calculated using Z-scores and corrected as appropriate for numbers of tests.
Supplementary Material
Highlights.
De novo damaging mutations are enriched in schizophrenia
Schizophrenia genes are highly co-expressed in fetal prefrontal cortex
Schizophrenia genes operate in pathways important for brain development
Disruptions of fetal prefrontal cortical neurogenesis are critical to schizophrenia
Acknowledgments
We thank Jim Watson for advice and support throughout the project. This work was supported by a gift from Steve and Connie Lieber, by NARSAD grants to M.C.K. and to T.W., and by NIMH grants R01MH083989, R01MH083849, R01MH083756, R01MH084071, and R03TW008696. Grants supporting the collection of clinical material for the NIMH repository are indicated in Supplementary Materials.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akil H, Brenner S, Kandel E, Kendler KS, King MC, Scolnick E, Watson JD, Zoghbi HY. The future of psychiatric research: genomes and neural circuits. Science. 2010;327:1580–1581. doi: 10.1126/science.1188654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton ES, Kreidberg JA, Rakic P. Distinct functions of alpha3 and alpha(v) integrin receptors in neuronal migration and laminar organization of the cerebral cortex. Neuron. 1999;22:277–289. doi: 10.1016/s0896-6273(00)81089-2. [DOI] [PubMed] [Google Scholar]
- Barabási AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- BrainSpan: Atlas of the Developing Human Brain. 2013 www.brainspan.org.
- Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, Li Y, Mu Y, Chen G, Yu D, et al. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemin J, Monteil A, Perez-Reyes E, Bourinet E, Nargeot J, Lory P. Specific contribution of human T-type calcium channel isotypes (alpha1G, alpha1H and alpha1I) to neuronal excitability. J Physiol. 2002;540:3–14. doi: 10.1113/jphysiol.2001.013269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KH, Rhim H. Inhibition of recombinant Ca(v)3.1 (alpha1G) T-type calcium channels by the antipsychotic drug clozapine. Eur J Pharmacol. 2010;626:123–130. doi: 10.1016/j.ejphar.2009.09.035. [DOI] [PubMed] [Google Scholar]
- Cleveland WS, Grosse E, Shyu MJ. Local Regression Models. In: Chambers SJM, Hastie T, editors. Statistical Models. Chapman and Hall; New York: 1992. pp. 309–376. [Google Scholar]
- Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A NISC Comparative Sequencing Program. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa E, Dong E, Grayson DR, Guidotti A, Ruzicka W, Veldic M. Reviewing the role of DNA (cytosine-5) methyltransferase overexpression in the cortical GABAergic dysfunction associated with psychosis vulnerability. Epigenetics. 2007;2:29–36. doi: 10.4161/epi.2.1.4063. [DOI] [PubMed] [Google Scholar]
- Duchrow M, Schlüter C, Key G, Kubbutat MH, Wohlenberg C, Flad HD, Gerdes J. Cell proliferation-associated nuclear antigen defined by antibody Ki-67: a new kind of cell cycle-maintaining proteins. Arch Immunol Ther Exp. 1995;43:117–121. [PubMed] [Google Scholar]
- Eisenberg DP, Berman KF. Executive function, neural circuitry, and genetic mechanisms in schizophrenia. Neuropsychopharmacol. 2010;35:258–277. doi: 10.1038/npp.2009.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaisler-Salomon I, Wang Y, Chuhma N, Zhang H, Golumbic YN, Mihali A, Arancio O, Sibille E, Rayport S. Synaptic underpinnings of altered hippocampal function in glutaminase-deficient mice during maturation. Hippocampus. 2012;22:1027–1039. doi: 10.1002/hipo.22014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman SR, Chang J, Xu B, Bawa TS, Gogos JA, Karayiorgou M, Vitkup D. Diverse types of genetic variation converge on functional gene networks involved in schizophrenia. Nat Neurosci. 2012;15:1723–1728. doi: 10.1038/nn.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard SL, Gauthier J, Noreau A, Xiong L, Zhou S, Jouan L, Dionne-Laporte A, Spiegelman D, Henrion E, Diallo O, et al. Increased exonic de novo mutation rate in individuals with schizophrenia. Nat Genet. 2011;43:860–863. doi: 10.1038/ng.886. [DOI] [PubMed] [Google Scholar]
- Girirajan S, Rosenfeld JA, Coe BP, Parikh S, Friedman N, Goldstein A, Filipink RA, McConnell JS, Angle B, Meschino WS, et al. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N Engl J Med. 2012;367:1321–1331. doi: 10.1056/NEJMoa1200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gläscher J, Adolphs R, Damasio H, Bechara A, Rudrauf D, Calamia M, Paul LK, Tranel D. Lesion mapping of cognitive control and value-based decision making in the prefrontal cortex. Proc Natl Acad Sci USA. 2012;109:14681–14686. doi: 10.1073/pnas.1206608109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogtay N, Giedd JN, Lusk L, Hayashi KM, Greenstein D, Vaituzis AC, Nugent TF, 3rd, Herman DH, Clasen LS, Toga AW, et al. Dynamic mapping of human cortical development during childhood through early adulthood. Proc Natl Acad Sci USA. 2004;101:8174–8179. doi: 10.1073/pnas.0402680101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974;185:862–864. doi: 10.1126/science.185.4154.862. [DOI] [PubMed] [Google Scholar]
- Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, van de Lagemaat LN, Smith KA, Ebbert A, Riley ZL, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391–399. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee YH, Narzisi G, Leotta A, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepsen K, Solum D, Zhou T, McEvilly RJ, Kim HJ, Glass CK, Hermanson O, Rosenfeld MG. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450:415–419. doi: 10.1038/nature06270. [DOI] [PubMed] [Google Scholar]
- Kalkstein S, Hurford I, Gur RC. Neurocognition in schizophrenia. Curr Top Behav Neurosci. 2010;4:373–390. doi: 10.1007/7854_2010_42. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, Moran J, Chambert K, Toncheva D, Georgieva L, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17:142–153. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb B, Mychasiuk R, Muhammad A, Li Y, Frost DO, Gibb R. Experience and the developing prefrontal cortex. Proc Natl Acad Sci USA. 2012;109(Suppl 2):17186–17193. doi: 10.1073/pnas.1121251109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, Gudjonsson SA, Sigurdsson A, Jonasdottir A, Jonasdottir A, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka G, Friedrich T, Davis-Turak J, Winden K, Oldham MC, Gao F, Chen L, Wang GZ, Luo R, Preuss TM, et al. Human-specific transcriptional networks in the brain. Neuron. 2012;75:601–617. doi: 10.1016/j.neuron.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm N, Sudmant PH, Ko A, O’Roak BJ, Malig M, Coe BP, Quinlan AR, Nickerson DA, Eichler EE NHLBI Exome Sequencing Project. Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012;22:1525–1532. doi: 10.1101/gr.138115.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protocols. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Lin M, Hrabovsky A, Pedrosa E, Wang T, Zheng D, Lachman HM. Allele-biased expression in differentiating human neurons: implications for neuropsychiatric disorders. PLoS One. 2012;7:e44017. doi: 10.1371/journal.pone.0044017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín O. Interneuron dysfunction in psychiatric disorders. Nat Rev Neurosci. 2012;13:107–120. doi: 10.1038/nrn3155. [DOI] [PubMed] [Google Scholar]
- Martin C, Jacobi JS, Nava G, Jeziorski MC, Clapp C, Martínez de la Escalera G. GABA inhibition of cyclic AMP production in immortalized GnRH neurons is mediated by calcineurin-dependent dephosphorylation of adenylyl cyclase 9. Neuroendocrinol. 2007;85:257–266. doi: 10.1159/000103557. [DOI] [PubMed] [Google Scholar]
- Masana M, Santana N, Artigas F, Bortolozzi A. Dopamine neurotransmission and atypical antipsychotics in prefrontal cortex: A critical review. Curr Top Med Chem. 2012;12:2357–2374. doi: 10.2174/156802612805289872. [DOI] [PubMed] [Google Scholar]
- McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141:210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9:S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma’ayan A, Samocha KE, Sabo A, Lin CF, Stevens C, Wang LS, Makarov V, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NHLBI Exome Sequencing Project. 2012 http://evs.gs.washington.edu/EVS/
- O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, Carvill G, Kumar A, Lee C, Ankenman K, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacary E, Tixier E, Coulet F, Roussel S, Petit E, Bernaudin M. Crosstalk between HIF-1 and ROCK pathways in neuronal differentiation of mesenchymal stem cells, neurospheres and in PC12 neurite outgrowth. Mol Cell Neurosci. 2007;35:409–423. doi: 10.1016/j.mcn.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Patten BA, Sardi SP, Koirala S, Nakafuku M, Corfas G. Notch1 signaling regulates radial glia differentiation through multiple transcriptional mechanisms. J Neurosci. 2006;26:3102–3108. doi: 10.1523/JNEUROSCI.4829-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press W. Global Burden of Disease. World Health Organization; Geneva, Switzerland: 2008. [Google Scholar]
- Rakic P. Evolution of the neocortex: a perspective from developmental biology. Nat Rev Neurosci. 2009;10:724–735. doi: 10.1038/nrn2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramocki MB, Zoghbi HY. Failure of neuronal homeostasis results in common neuropsychiatric phenotypes. Nature. 2008;455:912–918. doi: 10.1038/nature07457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relucio J, Tzvetanova ID, Ao W, Lindquist S, Colognato H. Laminin alters Fyn regulatory mechanisms and promotes oligodendrocyte development. J Neurosci. 2009;29:11794–11806. doi: 10.1523/JNEUROSCI.0888-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesselman JJ. Case-Control Studies: Design, Conduct, Analysis. Oxford Univ Press; 1982. p. 354. [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd AM, Laurens KR, Matheson SL, Carr VJ, Green MJ. Systematic meta-review and quality assessment of the structural brain alterations in schizophrenia. Neurosci Biobehav Rev. 2012;36:1342–1356. doi: 10.1016/j.neubiorev.2011.12.015. [DOI] [PubMed] [Google Scholar]
- Shima Y, Kawaguchi SY, Kosaka K, Nakayama M, Hoshino M, Nabeshima Y, Hirano T, Uemura T. Opposing roles in neurite growth control by two seven-pass transmembrane cadherins. Nat Neurosci. 2007;10:963–969. doi: 10.1038/nn1933. [DOI] [PubMed] [Google Scholar]
- Simons CJ, van Winkel R. Intermediate phenotype analysis of patients, unaffected siblings, and healthy controls identifies VMAT2 as a candidate gene for psychotic disorder and neurocognition. Schizophr Bull. 2013;39:848–856. doi: 10.1093/schbul/sbs067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data ntegration and network visualization. Bioinformatics. 2011;27:431–2. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, Pietiläinen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talley EM, Cribbs LL, Lee JH, Daud A, Perez-Reyes E, Bayliss DA. Differential distribution of three members of a gene family encoding low voltage-activated (T-type) calcium channels. J Neurosci. 1999;19:1895–1911. doi: 10.1523/JNEUROSCI.19-06-01895.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlhaas PJ, Singer W. Neuronal dynamics and neuropsychiatric disorders: toward a translational paradigm for dysfunctional large-scale networks. Neuron. 2012;75:963–80. doi: 10.1016/j.neuron.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Uslaner JM, Smith SM, Huszar SL, Pachmerhiwala R, Hinchliffe RM, Vardigan JD, Nguyen SJ, Surles NO, Yao L, Barrow JC, et al. T-type calcium channel antagonism produces antipsychotic-like effects and reduces stimulant-induced glutamate release in the nucleus accumbens of rats. Neuropharmacol. 2012;62:1413–1421. doi: 10.1016/j.neuropharm.2010.11.015. [DOI] [PubMed] [Google Scholar]
- Vacic V, McCarthy S, Malhotra D, Murray F, Chou HH, Peoples A, Makarov V, Yoon S, Bhandari A, Corominas R, et al. Duplications of the neuropeptide receptor gene VIPR2 confer significant risk for schizophrenia. Nature. 2011;471:499–503. doi: 10.1038/nature09884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Cusick ME, Barabási AL. Interactome networks and human disease. Cell. 2011;144:986–98. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, Shahin H, Elkan-Miller T, Lee MK, Thornton AM, Roeb W, Abu Rayyan A, Loulus S, Avraham KB, King MC, et al. Whole exome sequencing and homozygosity mapping identify mutation in the cell polarity protein GPSM2 as the cause of nonsyndromic hearing loss DFNB82. Am J Hum Genet. 2010;87:90–94. doi: 10.1016/j.ajhg.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, McClellan JM, McCarthy SE, Addington AM, Pierce SB, Cooper GM, Nord AS, Kusenda M, Malhotra D, Bhandari A, et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science. 2008;320:539–543. doi: 10.1126/science.1155174. [DOI] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- Weinberger DR. Implications of normal brain development for the pathogenesis of schizophrenia. Arch Gen Psychiatry. 1987;44:660–669. doi: 10.1001/archpsyc.1987.01800190080012. [DOI] [PubMed] [Google Scholar]
- Xu B, Ionita-Laza I, Roos JL, Boone B, Woodrick S, Sun Y, Levy S, Gogos JA, Karayiorgou M. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat Genet. 2012;44:1365–1369. doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Roos JL, Dexheimer P, Boone B, Plummer B, Levy S, Gogos JA, Karayiorgou M. Exome sequencing supports a de novo mutational paradigm for schizophrenia. Nat Genet. 2011;43:864–868. doi: 10.1038/ng.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Roos JL, Levy S, van Rensburg EJ, Gogos JA, Karayiorgou M. Strong association of de novo copy number mutations with sporadic schizophrenia. Nat Genet. 2008;40:880–885. doi: 10.1038/ng.162. [DOI] [PubMed] [Google Scholar]
- Yunker AM, Sharp AH, Sundarraj S, Ranganathan V, Copeland TD, McEnery MW. Immunological characterization of T-type voltage-dependent calcium channel CaV3.1 (alpha 1G) and CaV3.3 (alpha 1I) isoforms reveal differences in their localization, expression, and neural development. Neurosci. 2003;117:321–335. doi: 10.1016/s0306-4522(02)00936-3. [DOI] [PubMed] [Google Scholar]
- Zhang C, Gao J, Zhang H, Sun L, Peng G. Robo2-Slit and Dcc-Netrin1 coordinate neuron axonal pathfinding within the embryonic axon tracts. J Neurosci. 2012;32:12589–12602. doi: 10.1523/JNEUROSCI.6518-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.