Abstract
INTRODUCTION
ARHI, an imprinted tumor suppressor gene, is expressed in normal immortalized prostate epithelial cells, but is dramatically down-regulated in prostate cancer cell lines. Here we investigated the mechanisms of ARHI silencing in prostate cancer through miRNA and genistein mediated pathways.
EXPERIMENTAL PROCEDURE
We evaluated ARHI mRNA and protein levels by real time PCR and immunostaining of prostate tissue array. Then, ARHI was over-expressed in prostate cancer PC-3 cells followed by functional studies. Finally, miRNA inhibitor studies and dual luciferase pMIR-REPORT assay were performed to prove the direct target of miR-221&222 to ARHI.
RESULTS
Both ARHI mRNA and protein levels were down regulated in prostate cancer tissues compared to adjacent normal tissues. Over-expression of ARHI can inhibit cell proliferation, colony formation, invasion and induced apoptosis. Further studies on a new mechanism of ARHI down regulation showed a significant inverse relationship between ARHI and miR-221 & 222 which were up-regulated in cancer cell lines. Transfection of miR-221 & 222 inhibitors into PC-3 cells caused a significant induction of ARHI expression. A direct interaction of miR-221 or 222 with a target site on the 3’UTR of ARHI was confirmed by a dual luciferase pMIR-REPORT assay.
CONCLUSIONS
ARHI is a tumor suppressor gene down regulated in prostate cancer and over-expression of ARHI can inhibit cell proliferation, colony formation and invasion. This study demonstrates for the first time that prostate cancer cells have decreased level of ARHI which could be caused by direct targeting of 3’UTR of ARHI by miR221/222.
Keywords: ARHI, prostate cancer, miRNA, tumor suppressor gene
Introduction
Prostate cancer is the most commonly diagnosed cancer among males in United States. It is estimated that approximately 192,280 new cases will be reported with 27,360 deaths in 2009. Although the prostate cancer death rate has declined with the improvement of detection and diagnosis since the 1990s, it remains the second most common cancer leading to death in men in the United States. So far, the molecular mechanisms involved in prostate cancer are not fully understood.
ARHI is a maternally imprinted human tumor suppressor gene that maps to chromosome 1p31 and encodes a 26-kDa small G protein with 60% homology to rap and ras (1). ARHI has been reported to be expressed in several normal tissues including ovarian (1), breast (2, 3), liver (4), thyroid (5) and pancreas (6). However its expression is lost or down-regulated in their respective cancer tissues. Several mechanisms for this loss have been studied, for example, loss of heterozygosity contributes up to 40% of ARHI gene loss in ovarian and breast cancers (2, 7). ARHI is also down-regulated by epigenetic mechanisms like DNA methylation, histone methylation and histone acetalyation (8, 9). It has also been reported that ARHI is down-regulated by binding of transcription factor E2F to the ARHI promoter region (10, 11). This kind of negative regulation can be reversed by TSA treatment, E2F binding site mutation and E2F siRNA (11).
Over expression of ARHI in ovarian and breast cancer cells can inhibit cell growth, decrease cell invasiveness and cause caspase-independent apoptosis (12). Meanwhile, expression of ARHI at physiological levels induces autophagy rather than apoptosis (13). Furthermore, low expression of ARHI has been closely related to shorter progression-free survival in pancreatic cancer (14).
However, little is known about ARHI expression in prostate cancer and whether its mis-regulation contributes to prostate cancer tumorigenesis. In this study, we investigated ARHI expression levels in prostate cancer cells and prostate cancer tissues. We also explored the functional role of ARHI in the pathogenesis of prostate cancer and for the first time show a new mechanism of ARHI regulation by miRNAs and genistein, a dietary isoflavone from soy.
Materials and Methods
Cell and Tissue Specimens
Immortalized normal human prostate epithelial cells RWPE-1 and several prostate cancer derived cell lines (PC-3, LNCap, Du145) were employed in this study. RWPE-1 cells were cultured in Keratinocyte Serum Free Medium (K-SFM) (GIBCO Kit Catalog Number 17005-042) which was supplied with bovine pituitary extract (BPE) and human recombinant epidermal growth factor (EGF)). Prostate cancer derived cell lines were cultured in RPMI media (UCSF facility) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (UCSF facility).
Prostate cancer tissue arrays were obtained from US Biomax, Inc., Rockville, MD) which included 95 cores, 48 cases with 36 cancers, 8 matched normal adjacent tissues and 4 cases of bone metastasis.
Reverse transcription and real-time PCR
Total RNA was extracted from cultured cells using an RNeasy mini kit or miRNeasy Mini Kit (for collection of small RNAs) (Qiagen, Valencia, CA). Gene expression was measured by realtime quantitative PCR (RT-QPCR) using an Applied Biosystems 7500 Fast Sequence Detection System and gene-specific Taqman assay kits. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and RNU48 were used as endogenous controls to normalized expression data. The thermal cycling conditions were according to the TaqMan Fast Universal PCR protocol. Each sample was analyzed in quadruplicate and each result was concluded from three independent experiments.
PCR array and Tissue Scan q-PCR
To study the expression profile of genes in a particular pathway, pathway-specific PCR arrays (Cell cycle array, Apoptosis Array) (SABioscience, Frederick, MD) were used according to the manufacturer’s instructions. Data was analyzed by the instrument’s software. Dysregulated genes were further validated by Taqman gene expression assays (Applied Biosystems, Foster city, CA)
Tissue Scan q-PCR array (prostate cancer panel I) (Origene, Rockville, MD) which contains 48 prostate samples (7 adjacent normal, 10 tumor, 31 lesions). Only the data from clearly defined adjacent normal and tumor tissues were compared to each other for ARHI gene expression. The manufacturer’s protocol was followed.
Flow Cytometry Analysis
To prepare single cell suspensions for flow cytometry, cell culture plates were incubated with 0.25% trypsin-EDTA (Invitrogen) at 37°C for 5 min and released by pipetting. The cell pellet was washed three times with ice-cold PBS and resuspended in 1 ml nuclear stain DAPI (Beckman Coulter, Inc. Fullerton, CA) for cell cycle analysis. Stained cells in a volume of 1 ml were analyzed by Flow Cytometer (Cell Lab Quanta SC; Beckman Coulter, Inc). For apoptosis analysis, cells were double stained with viability dye 7-ADD and Annexin-V-FITC using Annexin V-FITC/7-AAD Kit (Beckman Coulter, Inc. Fullerton, CA) and then analyzed.
Cell Proliferation Assay and Invasion Assay
PC-3 cells were plated in 96-well microplates at a density of 5×103 cells per well 24 hours before transfection. Cell proliferation was determined by CellTiter 96 Aqueous One Solution Cell Proliferation Assay Kit (Promega, Madison, MI) at 24h, 48h and 72h after transfection. Absorbance was recorded at 490nm with a kinetic microplate reader (Spectra MAX 190; Molecular Devices Co., Sunnyvale, CA). Experiments were performed three times in quadruplicate.
For invasion assay, we used BD Matrigel coated chambers (BD Bioscience, San Jose, CA) according to the manufacture’s protocol. Briefly, 48 hours after cells were transfected with ARHI plasmid or control plasmid; cells were counted and plated on Matrigel inserts or control inserts at an equal number (1×105cells/ml) in serum free medium and allowed to grow for 15 h at 37°C. Cells migrating through the membrane were fixed, stained with a Hema 3 kit (Fisher Chemicals, Pittsburge, PA) and counted manually.
Western Blot Analysis
Whole cell extracts were prepared from RIPA buffer (Thermal Scientific, Pierce). Protein lysate (50 μg) was electrophoresed in 12 % polyacrylamide gels and were transferred to PVDF membrane at 75 mA overnight at 4°C. Next day, the PVDF membrane was blocked and incubated with 1:100 polyclonal goat anti-ARHI IgG (sc-30321, Santa Cruz Biotechnology, Santa Cruz, CA) overnight at 4°C. The secondary antibody was HRP conjugated horse anti-goat IgG (Cell signaling, Beverly, MA) diluted 1:2,500 in fresh blocking solution. ARHI was visualized using the Super Signal West Pico Chemiluminescence Detection Kit (Pierce). Membranes were exposed to X-OMAT AR film (Kodak, Rochester, NY). Antibody of GADD45A (ab54740), CASP1 (ab54932), CD70 (ab96323), P21 (ab16767), HRK (ab84822) were obtained from Abcam, Cambridge, MA.
Immunohistochemistry
Immunohistochemistry was done on formalin-fixed, paraffin-embedded prostate cancer tissue arrays. The slice was deparafinized and antigen retrieval was carried out by microwaving the slices in 10mmol/L sodium citrate buffer. The staining kit was a Goat ImmunoCruz Staining System (sc-2053, Santa Cruz Biotechnology, Santa Cruz, CA). The primary antibody was the same for Western blots. All procedures strictly followed the manufacture’s protocol. The intensity of the staining was graded by two individuals from “−“as no staining to “++++”as strong staining. Figures were showing at 200X magnification in both adjacent normal and cancer tissues.
Sodium Bisulfite Treatment and Bisulfite Sequencing
Genomic DNA from RWPE-1, PC-3, DU145, LNCap cells were treated with bisulfate by using an EpiTect Bisulfite kit (Qiagen, Valencia, CA) as per the manufacturer’s instructions. Treated DNAs were analyzed for methylation status by using the specific methylation sequence. The specific primers were designed for the CpG island of the ARHI gene as followings:
-
CpG island I: Forward, ATAGGTAAGGGAGAAAGAAGTTAGA;
Reverse, AATACTACTATCCTAACAAAACCCTC.
-
CpG island II: Forward,TGTTTGAAGTTTATTTTAATAGGTGAGAA;
Reverse, ACTAAACACCCCAAAAACTCACTC.
-
CpG island III: Forward, GGGGTATTTGAAAATAATAAGTGTTAGATA
Reverse, TCCAAAATACAATTACAAAACCTACC
The sequencing was done by an outside lab (MCLAB, South San Francisco, CA).
Luciferase Activity Assay
According to the miRDB analysis, oligonucleotides containing human ARHI 3’-UTR target sequence were annealed and cloned into the pMIR-REPORTER Luciferase plasmid (Applied Biosystem, Foster city, CA) following the manufacture’s protocol to generate vector pMIR-ARHI-221/222.
ARHI-3’UTR for miR-221:
F: CTAGTATTGGCATTTGAAGAGGAGAACAAAATGTAGCATGATGTATTA
R: ATAACCGTAAACTTCTCCTCTTGTTTTACATCGTACTACATAATTCGA
ARHI-3’UTR for miR-222:
F: CTAGTTGGCATTTGAAGAGGAGAACAAAATGTAGCATGATGTATTA
R: AACCGTAAACTTCTCCTCTTGTTTTACATCGTACTACATAATTCGA
Empty vector pMIR without the inserts was using as a negative control. pMIR-REPOTR β-gal control plasmid was used for transfection normalization. PC-3 cells were cultured in 24 well plates and transfected with 800ng of pMIR-ARHI or pMIR together with 100ng of pMIR-β-gal and 20pmol of miR-221/222(Applied Biosystems, Foster city, CA). Lipofectamine 2000 and OPTI-MEMI reduced serum medium (Invitrogen) were used for transfection. 48 hours after transfection, luciferase and β-gal activity were measured using the Dual-Light System (Applied Biosystems, Foster city, CA).
MicroRNA inhibitors were specifically designed by Applied Biosystems for targeting the respective miRNAs (`miR-221 inhibitor catalog # AM 10337, miR-222 inhibitor catalog # PM11376).
Statistics
Statistical analysis was performed with Statview 5.0 for Windows as needed. P values less than 0.05 were regarded as statistically significant and are represented by * on the bars in the figures.
Results
ARHI is down-regulated in prostate cancer cell lines and prostate cancer tissues
To evaluate ARHI expression levels in prostate cancer cell lines, ARHI mRNA level and protein levels were measured in immortalized prostate cell line RWPE-1 and two prostate cancer cell lines: PC-3, LNCap by real-time PCR and Western blot. We found that both ARHI mRNA and protein levels were dramatically down–regulated in prostate cancer cell lines compared to RWPE-1 cells (Fig 1A).
Figure 1. ARHI is down-regulated in prostate cancer.
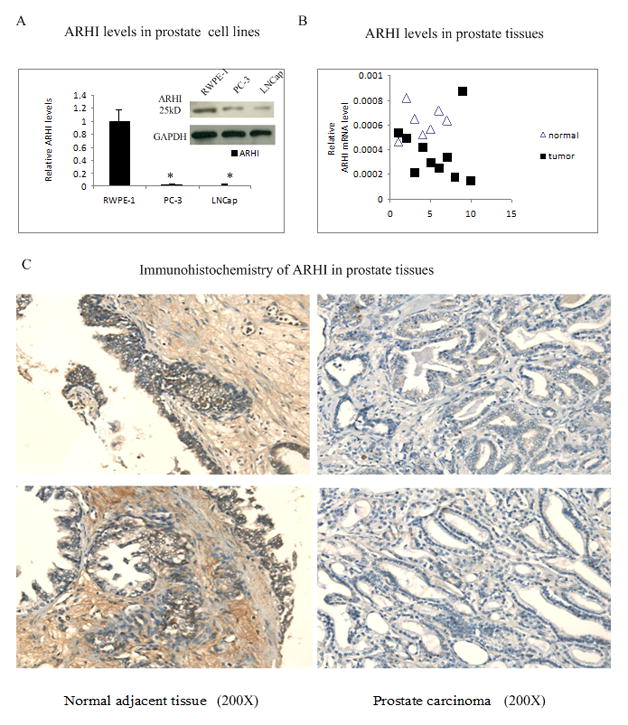
A) ARHI mRNA and protein levels in immortalized RWPE-1 prostate cells, and prostate cancer PC-3 and LNCap cells by realtime PCR and western bolt. Data was normalized to GAPDH signal.
B) ARHI mRNA level in 7 adjacent normal prostate tissues and 10 prostate tumor tissues by Q-PCR plates. Data was normalized to GAPDH signal.
C) Immunohistochemical staining of ARHI in prostate tissues. A prostate tissue microarray was immunostained with ARHI antibody to examine ARHI in matched adjacent prostate normal tissues and carcinoma tissues. Left panels show ARHI staining in adjacent normal tissues and right panels represent the matched carcinoma tissues (×200).
We also examined ARHI mRNA levels in prostate tissues by Q-PCR which consisted of 7 normal adjacent prostate tissues, 10 stage II–III prostate cancers and 31 prostate samples with various levels of lesions. Compared to the normal adjacent controls, over 80% of the marked tumor samples had decreased expression of ARHI (Fig 1B); suggesting that ARHI is frequently down-regulated in prostate cancers at the mRNA level. To further confirm whether expression of ARHI was also reduced in prostate cancer tissues at the protein level, a prostate cancer tissue array (36 cancers, 8 of which were matched adjacent normal tissue and 4 cases of bone metastasis) was immunostained with anti-ARHI antibody. ARHI showed expression in both epithelial cytoplasm and stoma, but the staining intensity was much higher in the epithelial cells. ARHI protein levels were observed to be down-regulated in over 70% of the prostate cancer tissues as shown in representative samples of ARHI immunohistochemistry (Fig 1C).
Over-expression of ARHI causes morphology changes in prostate cancer PC-3 cells
To understand the function of ARHI in prostate cancer, we transiently transfected PC-3 cells with a TrueORF pCMV6 expression vector containing the ARHI ORF. We chose PC-3 cells due to its low endogenous expression of ARHI. The empty vector of pCMV6 was used as a control. Real-time PCR showed a 20000 fold over-expression of ARHI mRNA compared to the controls (Fig 2A). We also performed Western analysis for ARHI protein levels to confirm the successful transfection (Fig 2B). Forty-eight hours after transient transfection, cells showed various morphological changes including smaller size, nuclear fragmentation and increased cell debris, suggesting apoptosis in cells over-expressing ARHI (Fig 2C).
Figure 2. Over-expression of ARHI causes morphological changes in PC-3 cells.
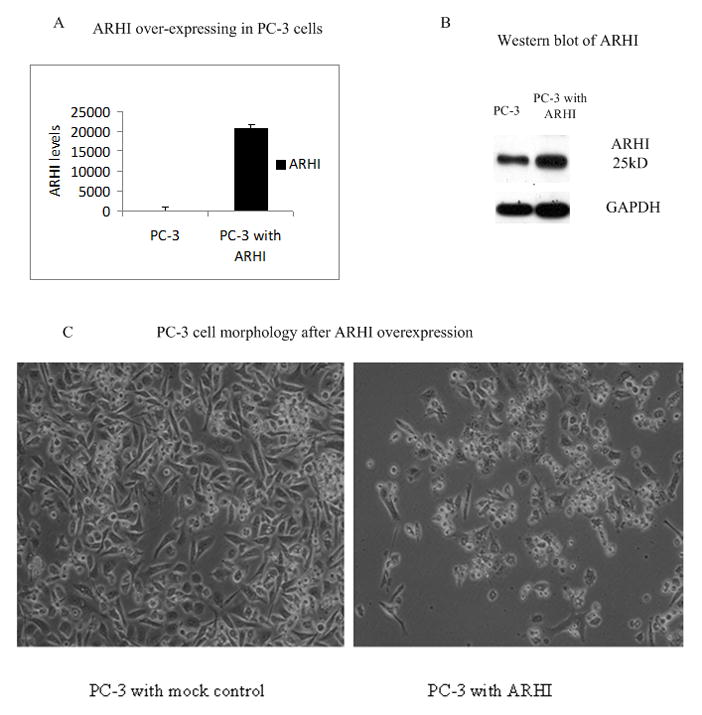
A) and B) Over-expression of ARHI by transfection of PC-3 cells with ARHI plasmid. PC-3 cells were transfected with 2μg of pCMV6-ARHI plasmid or pCMV6 empty vector for 48 hours. Cells were harvested for RNA (A) and protein (B) analysis. Whole cell lysates were analyzed and GAPDH was used as an internal loading control.
(C) PC-3 cell morphology after over-expression of ARHI. PC-3 cell were transfected with either pCMV6 empty vector (mock control) or pCMV6-ARHI plasmid. Pictures were taken after 72 hours (×100).
ARHI affects cellular proliferation, colony formation and invasion
We measured cell proliferation, colony formation and invasion ability after over-expression of ARHI in PC-3 cells. As shown in Fig 3A, cell growth was significantly decreased in a time-dependent manner in ARHI over-expressing cells compared to the control cells, suggesting that the gene ARHI has an anti-proliferative effect.
Figure 3. ARHI affects cellular proliferation, colony formation and invasion.
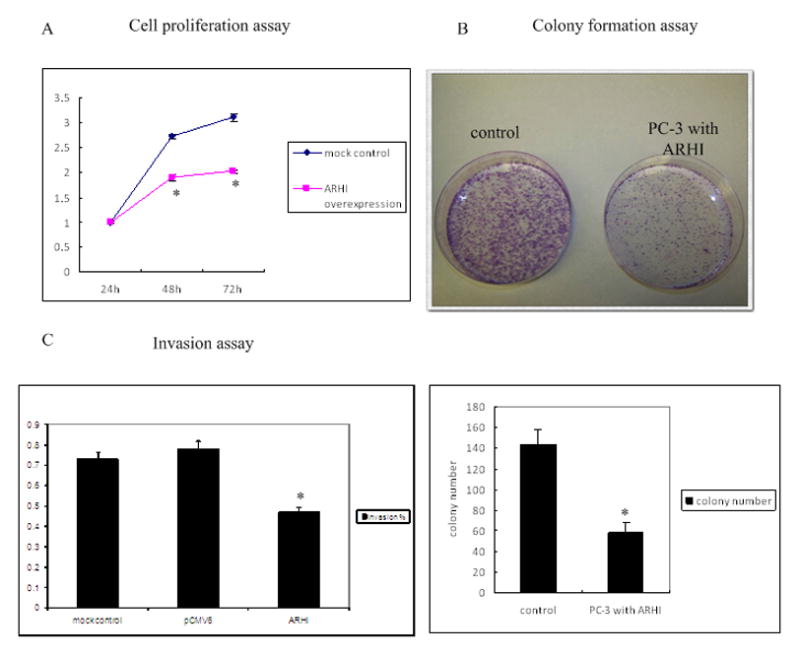
A) Cell proliferation assay. PC-3 cells were transfected with either pCMV6 empty vector (mock control) or pCMV6-ARHI plasmid. Cell proliferation was analyzed at the indicated time points.
B) Colony formation assay. 24 h after PC-3 cells were transfected with either pCMV6 empty vector or pCMV6-ARHI plasmid, cells were placed on 100mm culture dishes and cultured with G418 (1000mg/ml) for 3 weeks. Then the dishes were stained with crystal violet and average colony numbers were taken from three different fields.
C) Matrigel invasion assay. Invasion was analyzed in PC-3 cells with mock control or ARHI plasmid. 1×105 cells were plated on Matrigel or control inserts and were allowed to migrate at 37°C for 15 h. Ten randomly selected fields of migrated cells were counted and the data is presented as percentage of invasion relative to control inserts.
We also conducted colony formation assay to assess the long term effects of ARHI on cell proliferation (Fig 3B). After 21 days of selection in G418 media, cells transfected with ARHI formed significantly fewer colonies than the control cells, consistent with the cell proliferation assay, further suggesting that ARHI inhibited cell proliferation in prostate cancer cells.
We also examined the effect of ARHI on the invasion ability of prostate cancer cells. As shown in Fig 3C, cells over-expressing ARHI had 60% of the invasiveness compared to the control cells, indicating ARHI not only affects prostate cancer cell proliferation, but also modulates cell invasion properties. This suggests that down regulation of ARHI may contribute to the metastatic behavior of prostate cancer cells.
Over-expression of ARHI regulates cell cycle progression and induces apoptosis
Over-expression of ARHI inhibited prostate cancer cell growth that could be caused by regulation of the cell cycle, increased apoptosis or both mechanisms. To better understand how ARHI effects cancer cell growth, we conducted flow cytometry in PC-3 cells 48 hours after ARHI transfection. As shown in Fig 4A, cell cycle analysis showed that cells over-expressing ARHI had a significant increase in cells in the G0/G1 phase while those in the S phase and G2/M phase decreased. This cell cycle distribution suggests that ARHI causes G0/G1 arrest in cell cycle progression. To further uncover the molecular mechanism of this kind of growth arrest, we profiled the expression of key cell cycle related genes in ARHI over-expressing cells and mock controls by using cell cycle PCR arrays from SABioscience. Several genes which were regulated by ARHI had been identified as shown in Fig 4B. Hect domain and RLD5 (HERC5), Cyclin-dependent kinase inhibitor 1A (CDNK1A), growth arrest and DNA-damage-inducible, alpha (GADD45A) are up-regulated more than 5 fold. Protein expression levels of these genes were further confirmed by Western blot (Fig 4E). Most importantly, CDKN1A and GADD45A function as regulators in the G1 phase of cell cycle progression.
Figure 4. Over-expression of ARHI regulates cell cycle progression and induces apoptosis.
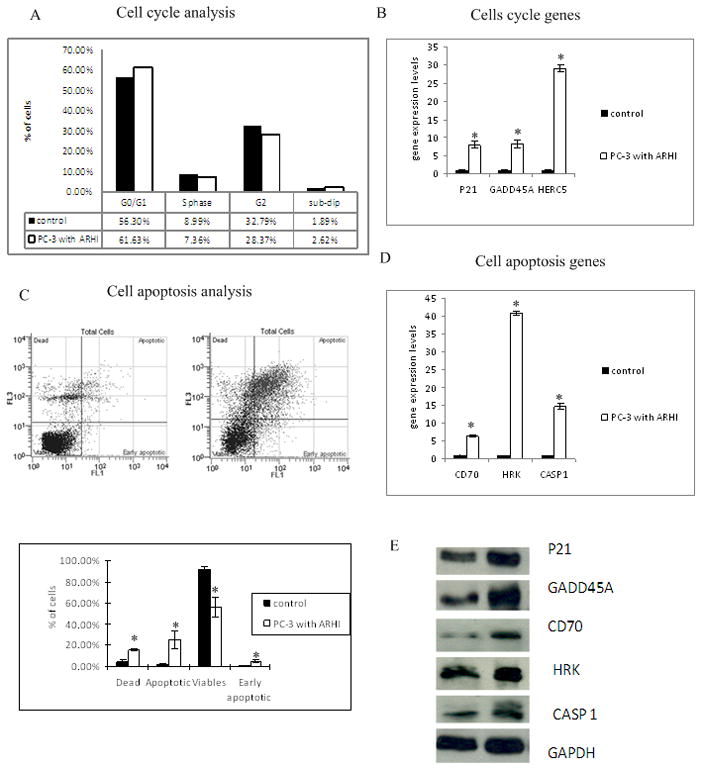
A) Flow cytometry analysis of cell cycle distribution in PC-3 cells over-expressing ARHI. PC-3 cells were harvested 48 hours after transfecting with either empty vector or ARHI plasmid. Cell nuclei were stained with DAPI and analyzed by flow cytometry.
B) Cell cycle genes altered by over-expression of ARHI in PC-3 cells. ARHI regulated genes were identified by cDNA expression profiling of cell cycle related genes. The relative expression of genes was confirmed by RT-PCR by using Taqman gene expression assays. Data were normalized by GAPDH signals.
C) Flow cytometry analysis of cell apoptosis in PC-3 cells over-expressing ARHI. PC-3 cells were harvested 48 hours after transfection with either pCMV6 empty vector or pCMV6-ARHI plasmid followed by dual staining with viability dye 7-AAD and AnnexinV-FITC. A typical dual staining is represented in the biparametric histograms and shows cells in early (lower right quadrant) and late apoptotic states (upper right quandrant). Viable cells are double negative (Lower left quandrant).
D) Apoptotic genes affected by over-expression of ARHI. Genes were identified by cDNA expression profiling of cell apoptosis related genes and confirmed by RT-PCR using Taqman primers. Data were normalized by GAPDH signals.
E) Western blot of ARHI regulated cell cycle and apoptosis pathway genes. Data were normalized to GAPDH.
We also examined apoptosis in ARHI over-expressing PC-3 cells by flow cytometry analysis after Annexin-V-FITC-7-AAD staining (Fig 4C). There was a dramatic decrease in the viable cell population. The apoptotic population went up from 0.03% to 8.16% and early apoptotic cells increased from 0.71% to 1.69% of the cell population after ARHI transfection. We also noticed a large number of dead cells which could be the effects of apoptosis induced by ARHI. The flow cytometry results confirmed the observation of morphological changes in PC-3 cells after ARHI over-expression showing that ARHI induces apoptosis in prostate cancer cells. Additional studies about gene profiling of apoptosis related genes were also done. CASP1, harakiri, BCL2 interacting protein (HRK) and tumor necrosis factor superfamily, member7 (CD70) were found to be up-regulated in both mRNA levels and protein levels (Fig 4D and 4E). These genes are either involved in apoptosis or play a role in the regulation of apoptosis pathways.
ARHI is promoter methylated and methylation is increased in cancer cell lines PC-3 and Du-145
To clarify the mechanism of ARHI down-regulation in prostate cancer cells, we investigated the epigenetic regulation of this gene. According to Methprimer software, there are three CpG islands in the ARHI promoter and exon region. CpG I is about 300 bp upstream of the transcriptional start site and CpG II and III are downstream of the start site (Supplemental Fig 1A). The methylation status of these ARHI CpG islands was analyzed by using bisulfate DNA sequencing. These data showed ARHI methylation level of approximately 50%–65% in CpG island I and II in immortalized prostate epithelial RWPE-1 cells. However, hypermethylation was observed in CpG island I (about 90%–100%) in prostate cancer cell lines PC-3 (Supplemental Fig 1B) and Du145 (Supplemental Fig 1C). PC-3, but not Du145 cells also had hypermethylation in CpG island II (about 65% to 90%). Another typical prostate cancer cell line LNCap did not have increased methylation of the ARHI promoter region (Supplemental Fig 1D). Furthermore, ARHI expression levels were significantly increased after 5-Aza-2’ dioxycitidine (5-aza, 5uM) treatment for 72 hours in almost all the cell lines except LNCap cells (Supplemental Fig 1). These findings suggest that frequent hypermethylation of the ARHI promoter region may be a critical event associated with the down-regulation of ARHI.
The ARHI 3’-UTR is a target of miR-221 and miR-222, which are up-regulated in prostate cancer PC-3 cells
To investigate the new mechanism of ARHI silencing in PC-3 cells, we focused our attention to microRNAs which are a wide class of small, noncoding RNAs that can regulate gene expression by inhibiting translation or inducing cleavage of target mRNAs.. We performed a bioinformatic analysis by using miRDB, TargetScan and PicTar algorithms and predicted two microRNAs (miR-221 and miR-222) that might target ARHI in 3’UTR region.
To prove that the ARHI 3’UTR is target of miR-221 and miR-222, we first analyzed their expression in PC-3 and RWPE-1 cells (Fig 5A). As shown, miR-221 was increased about 20 fold in PC-3 cells compared to RWPE-1 while miR-222 was increased 5 fold. This data showed an inverse relationship between ARHI and miR-221&222 levels, which was further confirmed by introducing miR-221 and miR-222 inhibitors into PC-3 cells. As shown in Fig 5B, miR-221 and miR-222 levels were significantly down-regulated by the inhibitors, while ARHI was significantly increased in both mRNA and protein levels (Fig 5C). These data strongly support the hypothesis that miR-221 and miR-222 are involved in the regulation of ARHI levels in PC-3 cells.
Figure 5. The ARHI 3’-UTR is a target of miR-221 and miR-222, which are up-regulated in PC-3 cells.
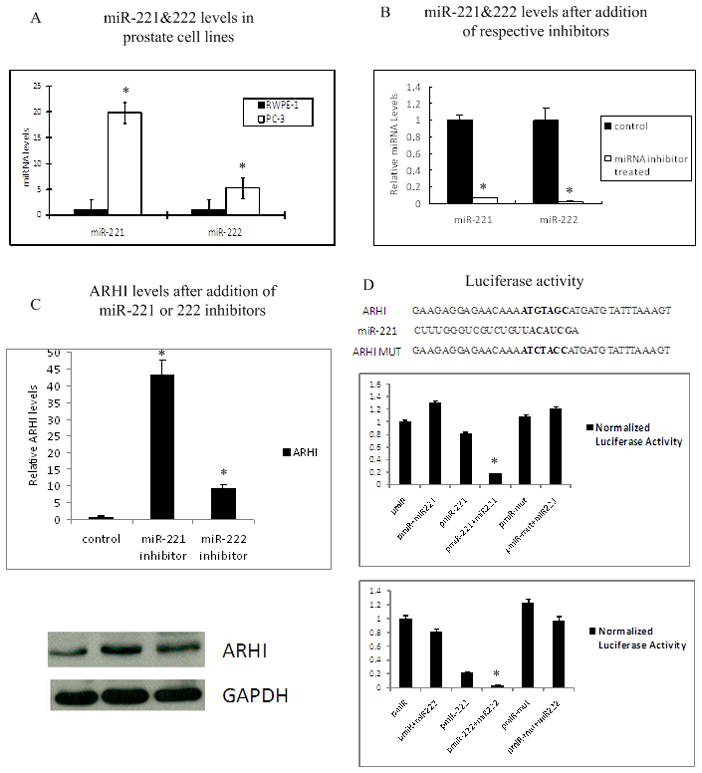
A) This panel displays miR-221 and miR-222 levels in prostate RWPE-1 cells (black) and prostate cancer PC-3 cells (grey) by RT-PCR. Data were normalized to RNU48 signals.
B) MiRNA levels after treatment with miRNA inhibitors. Left two bars are miR-221 levels detected by RT-PCR before and after miR-221 inhibitor treatment. Right two bars are miR-222 levels before and after miR-222 inhibitor treatment. Data were normalized to RNU48 signals.
C) RT-PCR and western blot of ARHI mRNA and protein levels in PC-3 cells treated with control (left), miRNA-221 inhibitor (middle) and miRNA-222 inhibitor (right). Data were normalized to GAPDH signals.
D) Schematic representation of the miR221/222 target sequence with the 3’-UTR of ARHI. The top graph shows relative luciferase activity after PC-3 cells were transfected with pMIR-empty vector (pMIR), pMIR containing the miR-221 target sequence (pMIR-miR221) and pMIR with miR-221 mutated target sequence (pMIR-mut). Data also shows co-transfection of miR-221 precursor. The bottom graph shows luciferase activity after PC-3 cells were transfected with pMIR, pMIR containing miR-222 targeting sequence (pMIR-miR222) and pMIR with miR-222 mutated target sequence (pMIR-mut) with or without miR-222 precursor.
To demonstrate a direct interaction between miR-221 and miR-222 with ARHI, we cloned a 58bp (miR-221) and 56bp (miR-222) ARHI 3’-UTR segment, containing potential target sites for these two microRNAs, into the pMIR vector luciferase open reading frame and transfected these reporter construct into PC-3 cells. Empty vectors and vectors contained mutated target sequence were used as control for comparison. Vectors were co-transfected with miR-221 or miR-222 precursor and the β-gal vector which was used for normalization of transfection differences. As shown in Fig 5D, luciferase activity in PC-3 cells co-transfected with pMIR-ARHI and miR-221 was decreased about 80% compared to the empty vector, while co-transfection with pMIR-ARHI and miR-222 decreased luciferase activity over 90%. No significant changes were observed in PC-3 cells transfected with mutant target sequence groups. Taken together, these results provide strong evidence that the ARHI 3’-UTR is a direct target of miR-221 and miR-222 in prostate cancer PC-3 cells.
Genistein can up-regulate ARHI levels by regulating miR-221 and miR-222
Genistein, a dietary isoflavone from soy, has been investigated as a potential chemopreventive drug. Here we examined whether genistein could regulate tumor suppressor gene ARHI by altering microRNA levels in prostate cancer cells. We treated PC-3 cells with 50μM genistein for 4 days and measured miR-221 and miR-222 levels. As shown in Fig 6A, miR-221 and miR-222 levels decreased about 30% and 55% respectively after genistein treatment. Meanwhile, ARHI mRNA levels were induced 2 fold after genistein treatment (Fig 6B). Western blot confirmed that protein levels of ARHI also increased after genistein treatment (Fig 6B). This data suggests that genistein can partially restore ARHI by decreasing miR-221 and miR-222 levels.
Figure 6. Genistein up-regulates ARHI levels by down regulating miR-221 and miR-222.
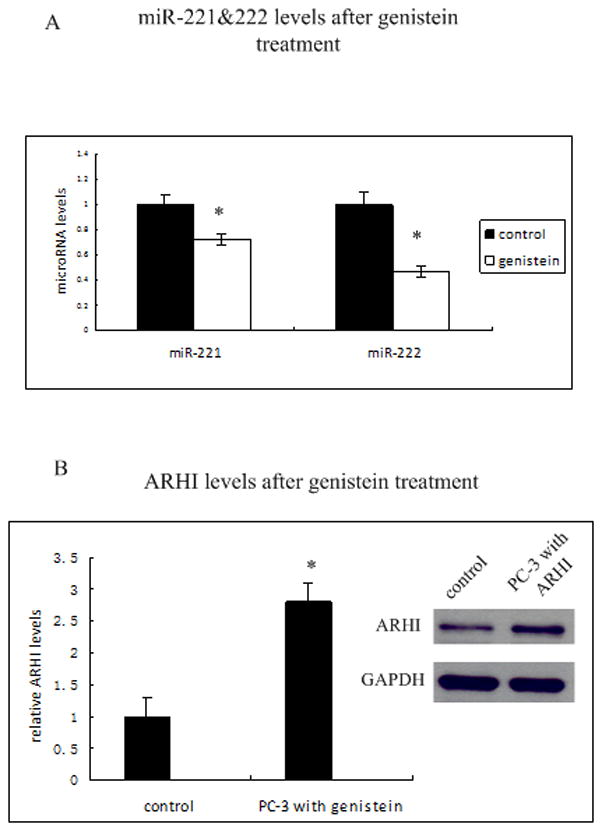
A) MiR-221 and miR-222 levels after genistein treatment. PC-3 cells were treated with 50μM genistein for 96 h, and miR-221 (left) and miR-222 levels (right ) were analyzed by RT-PCR. Data were normalized to RNU48 signals.
B) Relative ARHI mRNA and protein levels in PC-3 cells before and after 50μM genistein treatment for 96 h by RT-PCR and western blot. Data were normalized to GAPDH.
Discussion
ARHI, a tumor suppressor gene, functions by negative regulation of cell proliferation in several different kinds of tissues (3–6). However, due to its special feature of being a maternally imprinted gene, loss or down-regulation of the paternal allele’s expression can lead to the unlimited cell growth, which may contribute to tumorigenesis in certain cancers (3–6). Previous studies have showed that ARHI loss or down-regulation could be caused by loss of heterozygosity (7) and/or epigenetic events such as DNA methylation, histone methylation and histone acetylation (9, 10, 15). However, ARHI expression and its function have not been studied in prostate cancer.
In this study, we evaluated ARHI levels in prostate cancer cell lines and prostate cancer tissues. Both RT-PCR and immunohistochemistry showed that ARHI is down regulated in prostate cancer cells and tissues compared to the adjacent normal. And we also anticipated that ARHI staining would be even stronger in normal tissue (non-malignant tissue) compared to the adjacent normal prostate tissue due to the cancer “field effect”(16). It has been confirmed that multiple genetic and epigenetic changes already occurs in normal adjacent cancers. Difference of ARHI expression between adjacent normal and cancer tissue is consistent with the possibility that this gene might play an important role in regulating cell proliferation in the prostate. We investigated this possibility by over-expression of ARHI in prostate cancer PC-3 cells followed by cell proliferation, invasion and colony formation assays. Our data showed that in PC-3 cells, ARHI negatively regulates cell growth by causing cell cycle G1/G0 arrest and inducing apoptosis.
We also profiled the expression of key cell cycle and apoptosis pathway genes and identified several components that were unregulated. In the cell cycle pathway this included up-regulation of CDKN1A (also known as P21), GADD45A and HERC5. The protein that P21 encodes can bind to and inhibit the activity of cyclin-CDK2 or -CDK4 complexes, and thus functions as a regulator of cell cycle progression at G1. Previously, it has also been reported that ARHI over expression in ovarian cancer is associated with increased expression of p21WAF1/CIP1 (17). GADD45A is a member of a group of genes whose transcript levels are increased following stressful growth arrest conditions and stimulates DNA excision repair, inhibiting cell entry into S phase. HERC5 is a member of the HERC family of ubiquitin ligases and functions as an interferon-induced E3 protein ligase that mediates ISGylation of protein targets. In the apoptosis pathway, CASP1, HRK and CD70 were up-regulated. CASP1 encodes a protease that cleaves IL-1 beta, releasing the mature cytokine which is involved in a variety of inflammatory processes. This gene has been shown to induce cell apoptosis. HRK activates apoptosis and interacts selectively with survival-promoting proteins Bcl-2 and Bcl-X. CD70 binds to CD27 and plays a role in T-cell activation. Taken together, ARHI-mediated regulation of these multiple components of the cell cycle and apoptosis pathways is a novel finding and suggests that ARHI influences cell cycle and apoptosis genes that regulate prostate cancer tumorigenesis.
It is known that aberrant methylation of CpG islands, especially in the promoter regions of genes, serves as an important mechanism for the inactivation of tumor suppressor genes in cancers. Previous reports have shown that ARHI is inactivated by promoter methylation in follicular thyroid carcinoma (5), breast cancers (8), and ovarian cancers (18). Therefore we studied the ARHI promoter and exon CpG island methylation status in several prostate cell lines. We found that all three CpG islands were partially methylated in RWPE-1 cells. In contrast, aberrant methylation of ARHI was observed in prostate cancer cell lines PC-3 and Du145. Especially in CpG island I, hypermethylation (90%–100%) was found in both PC-3 and Du145 cell lines, suggesting methylation in this region could play a critical role in ARHI transcription. Although ARHI expression is down-regulated in several different kinds of cancer cell lines, differences were observed between different cell lines in methylation of the CpG island. LNCap cells had a similar methylation status compared to RWPE-1 cells but markedly lower levels of ARHI, indicating methylation is not the only mechanism causing down-regulation of ARHI.
We also investigated the mechanism of ARHI down-regulation by miRNA regulated pathways. Aberrantly expressed miRNAs are found in many cancer tissues and may be involved in carcinogenesis. Thus, the identification of cancer-specific miRNA targets is critical for understanding the functional role and mechanism. We screened potential miRNAs targeting ARHI with computer software and found more than 10 miRNAs that could potentially target the ARHI gene. Among these miRNAs, miR-221 and miR-222 have been reported to have altered expression in prostate cancer (19, 20). Therefore, we measured their expression levels in PC-3 cells and found increased levels of both miRNAs. Thus these two miRNAs represented attractive candidates for our target study. After introducing of miR-221 and miR-222 inhibitors into PC-3 cells there was a large increase in ARHI level. To confirm this finding, we performed a luciferase reporter assay with a vector containing the ARHI 3’-UTR target site. The luciferase activity in PC-3 cell transfected with ARHI 3’-UTR was significantly lower than those cells transfected with empty control vector. After transfecting the miR-221 or miR-222 precursor, luciferase activity decreased even more, while cells with the control vector showed no measurable difference. Taken together, these results confirmed that miR-221 and miR-222 which are up-regulated in prostate cancer cell line can directly target ARHI at the 3’-UTR region, thereby contributing to prostate cancer tumorigenesis.
Genistein, an isoflavone from soy, has been reported to suppress the growth of several prostate cancer cell lines in vitro including PC-3, Du145 and LNCap (21–23) and in TRAMP mice model (24). This suppression has been attributed to regulation of cell cycle and apoptosis pathways (25–29). In this study we found that genistein regulates ARHI through a novel pathway by regulating miR-221 and 222, thereby affecting ARHI expression levels.
This is the first report to show that ARHI is down-regulated in prostate cancer. We also have shown that ARHI is a direct target of miR-221 and 222 which are up-regulated in prostate cancer, documenting a new mechanism responsible for ARHI down-regulation in prostate cancer. Genistein, a potential non-toxic chemopreventive agent restores expression of ARHI by down-regulation of miR-221& miR-222 and may be an important dietary therapeutic agent for treating prostate cancer.
Acknowledgments
We thank Dr. Roger Erickson for his support and assistance with the preparation of the manuscript. This study was supported by NIH grant R01CA111470, T32DK007790, VA Research Enhancement Award Program (REAP) and VA Merit Review grants.
References
- 1.Yu Y, Xu F, Peng H, et al. NOEY2 (ARHI), an imprinted putative tumor suppressor gene in ovarian and breast carcinomas. Proc Natl Acad Sci U S A. 1999;96:214–9. doi: 10.1073/pnas.96.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peng H, Xu F, Pershad R, et al. ARHI is the center of allelic deletion on chromosome 1p31 in ovarian and breast cancers. Int J Cancer. 2000;86:690–4. doi: 10.1002/(sici)1097-0215(20000601)86:5<690::aid-ijc14>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 3.Wang L, Hoque A, Luo RZ, et al. Loss of the expression of the tumor suppressor gene ARHI is associated with progression of breast cancer. Clin Cancer Res. 2003;9:3660–6. [PubMed] [Google Scholar]
- 4.Huang J, Lin Y, Li L, et al. ARHI, as a novel suppressor of cell growth and downregulated in human hepatocellular carcinoma, could contribute to hepatocarcinogenesis. Mol Carcinog. 2009;48:130–40. doi: 10.1002/mc.20461. [DOI] [PubMed] [Google Scholar]
- 5.Weber F, Aldred MA, Morrison CD, et al. Silencing of the maternally imprinted tumor suppressor ARHI contributes to follicular thyroid carcinogenesis. J Clin Endocrinol Metab. 2005;90:1149–55. doi: 10.1210/jc.2004-1447. [DOI] [PubMed] [Google Scholar]
- 6.Lu X, Qian J, Yu Y, Yang H, Li J. Expression of the tumor suppressor ARHI inhibits the growth of pancreatic cancer cells by inducing G1 cell cycle arrest. Oncol Rep. 2009;22:635–40. [PubMed] [Google Scholar]
- 7.Janssen EA, Ovestad IT, Skaland I, et al. LOH at 1p31 (ARHI) and proliferation in lymph node-negative breast cancer. Cell Oncol. 2009;31:335–43. doi: 10.3233/CLO-2009-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan J, Luo RZ, Fujii S, et al. Aberrant methylation and silencing of ARHI, an imprinted tumor suppressor gene in which the function is lost in breast cancers. Cancer Res. 2003;63:4174–80. [PubMed] [Google Scholar]
- 9.Fujii S, Luo RZ, Yuan J, et al. Reactivation of the silenced and imprinted alleles of ARHI is associated with increased histone H3 acetylation and decreased histone H3 lysine 9 methylation. Hum Mol Genet. 2003;12:1791–800. doi: 10.1093/hmg/ddg204. [DOI] [PubMed] [Google Scholar]
- 10.Luo RZ, Peng H, Xu F, et al. Genomic structure and promoter characterization of an imprinted tumor suppressor gene ARHI. Biochim Biophys Acta. 2001;1519:216–22. doi: 10.1016/s0167-4781(01)00226-3. [DOI] [PubMed] [Google Scholar]
- 11.Lu Z, Luo RZ, Peng H, et al. E2F-HDAC complexes negatively regulate the tumor suppressor gene ARHI in breast cancer. Oncogene. 2006;25:230–9. doi: 10.1038/sj.onc.1209025. [DOI] [PubMed] [Google Scholar]
- 12.Bao JJ, Le XF, Wang RY, et al. Reexpression of the tumor suppressor gene ARHI induces apoptosis in ovarian and breast cancer cells through a caspase-independent calpain-dependent pathway. Cancer Res. 2002;62:7264–72. [PubMed] [Google Scholar]
- 13.Lu Z, Luo RZ, Lu Y, et al. The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells. J Clin Invest. 2008;118:3917–29. doi: 10.1172/JCI35512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalai I, Missiaglia E, Barbi S, et al. Low expression of ARHI is associated with shorter progression-free survival in pancreatic endocrine tumors. Neoplasia. 2007;9:181–3. doi: 10.1593/neo.06838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu Y, Fujii S, Yuan J, et al. Epigenetic regulation of ARHI in breast and ovarian cancer cells. Ann N Y Acad Sci. 2003;983:268–77. doi: 10.1111/j.1749-6632.2003.tb05981.x. [DOI] [PubMed] [Google Scholar]
- 16.Chandran UR, Dhir R, Ma C, Michalopoulos G, Becich M, Gilbertson J. Differences in gene expression in prostate cancer, normal appearing prostate tissue adjacent to cancer and prostate tissue from cancer free organ donors. BMC Cancer. 2005;5:45. doi: 10.1186/1471-2407-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosen DG, Wang L, Jain AN, et al. Expression of the tumor suppressor gene ARHI in epithelial ovarian cancer is associated with increased expression of p21WAF1/CIP1 and prolonged progression-free survival. Clin Cancer Res. 2004;10:6559–66. doi: 10.1158/1078-0432.CCR-04-0698. [DOI] [PubMed] [Google Scholar]
- 18.Luo RZ, Fang X, Marquez R, et al. ARHI is a Ras-related small G-protein with a novel N-terminal extension that inhibits growth of ovarian and breast cancers. Oncogene. 2003;22:2897–909. doi: 10.1038/sj.onc.1206380. [DOI] [PubMed] [Google Scholar]
- 19.Spahn M, Kneitz S, Scholz CJ, et al. Expression of microRNA-221 is progressively reduced in aggressive prostate cancer and metastasis and predicts clinical recurrence. Int J Cancer. 2009 doi: 10.1002/ijc.24715. [DOI] [PubMed] [Google Scholar]
- 20.Sun T, Wang Q, Balk S, Brown M, Lee GS, Kantoff P. The role of microRNA-221 and microRNA-222 in androgen-independent prostate cancer cell lines. Cancer Res. 2009;69:3356–63. doi: 10.1158/0008-5472.CAN-08-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ouchi H, Ishiguro H, Ikeda N, Hori M, Kubota Y, Uemura H. Genistein induces cell growth inhibition in prostate cancer through the suppression of telomerase activity. Int J Urol. 2005;12:73–80. doi: 10.1111/j.1442-2042.2004.00973.x. [DOI] [PubMed] [Google Scholar]
- 22.Rao A, Woodruff RD, Wade WN, Kute TE, Cramer SD. Genistein and vitamin D synergistically inhibit human prostatic epithelial cell growth. J Nutr. 2002;132:3191–4. doi: 10.1093/jn/131.10.3191. [DOI] [PubMed] [Google Scholar]
- 23.Sakamoto K. Synergistic effects of thearubigin and genistein on human prostate tumor cell (PC-3) growth via cell cycle arrest. Cancer Lett. 2000;151:103–9. doi: 10.1016/s0304-3835(99)00423-1. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, Eltoum IE, Lamartiniere CA. Genistein chemoprevention of prostate cancer in TRAMP mice. J Carcinog. 2007;6:3. doi: 10.1186/1477-3163-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao R, Xiang N, Domann FE, Zhong W. Effects of selenite and genistein on G2/M cell cycle arrest and apoptosis in human prostate cancer cells. Nutr Cancer. 2009;61:397–407. doi: 10.1080/01635580802582751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen JC, Klein RD, Wei Q, et al. Low-dose genistein induces cyclin-dependent kinase inhibitors and G(1) cell-cycle arrest in human prostate cancer cells. Mol Carcinog. 2000;29:92–102. doi: 10.1002/1098-2744(200010)29:2<92::aid-mc6>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 27.Constantinou AI, Kamath N, Murley JS. Genistein inactivates bcl-2, delays the G2/M phase of the cell cycle, and induces apoptosis of human breast adenocarcinoma MCF-7 cells. Eur J Cancer. 1998;34:1927–34. doi: 10.1016/s0959-8049(98)00198-1. [DOI] [PubMed] [Google Scholar]
- 28.Spinozzi F, Pagliacci MC, Migliorati G, et al. The natural tyrosine kinase inhibitor genistein produces cell cycle arrest and apoptosis in Jurkat T-leukemia cells. Leuk Res. 1994;18:431–9. doi: 10.1016/0145-2126(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 29.Traganos F, Ardelt B, Halko N, Bruno S, Darzynkiewicz Z. Effects of genistein on the growth and cell cycle progression of normal human lymphocytes and human leukemic MOLT-4 and HL-60 cells. Cancer Res. 1992;52:6200–8. [PubMed] [Google Scholar]


