Abstract
Over the past decade, compelling evidence has emerged from population-based studies to suggest that AF is a heritable disease. More recently, we have begun to elucidate the genetic substrate underlying AF. Genome wide association studies (GWAS) have led to the identification of multiple risk loci that confer increased susceptibility to the arrhythmia. These loci harbor intriguing candidate genes including those encoding ion channels, transcription factors and signaling molecules. Current efforts are ongoing to functionally validate the role of these genes in disease pathogenesis. In the future, novel genotyping technologies such as exome sequencing and whole genome sequencing promise to uncover a greater proportion of the heritability underlying AF. In this article we review recent advances in AF genetics research and discuss future developments in the field.
Keywords: Atrial fibrillation, Genetics, Inherited arrhythmias
Introduction
Over a century after the electrocardiographic descriptions of atrial fibrillation (AF),1 the molecular basis of the arrhythmia remains incompletely understood. In recent years, multiple population-based studies have convincingly demonstrated that AF is a heritable disease.2–4 Initial research in AF genetics focused on rare familial forms of the arrhythmia and identified a number of isolated mutations, many of which were located in ion channel genes.5–11 These discoveries were followed by a wave of candidate gene association studies; however, such candidate gene approaches were associated with significant limitations, including a low pretest probability of the selected variants playing a role in disease pathogenesis. The advent of genome wide association studies (GWAS) has been a major advance in the field as nine risk loci have been identified for the arrhythmia,12–15 yet a number of challenges remain. Firstly, the causal variants at the AF risk loci have not been identified and functionally validated. Secondly, a significant proportion of the heritability of AF remains unaccounted for. In the future, emerging techniques such as exome sequencing and whole genome sequencing promise to uncover a significant proportion of the ´missing heritability´ of AF. Ultimately, the identification of the genetic substrate underlying AF may enable the discovery of novel therapeutic targets and also allow for more accurate risk stratification of this common and morbid arrhythmia. Since monogenic forms of AF and candidate gene association studies in AF have been discussed extensively in previous reviews,16 we will focus on recent GWAS results and future directions in the field of AF genetics.
Genome wide association studies in atrial fibrillation
The 4q25 locus for AF
The first GWAS for AF was performed in an Icelandic population and identified a risk locus on chromosome 4q25.12 The most significant single nucleotide polymorphism (SNP) at this locus, rs2200733 was associated with a p value of 6.1×10−41 and an odds ratio (OR) of 1.71. Multiple subsequent studies have consistently demonstrated a robust association between this locus and AF.14–15, 17–19
The 4q25 locus has been the most comprehensively studied of the AF risk loci. While the mechanism of the association between this locus and AF remains unclear, these studies have provided valuable insights into potential mechanisms underlying the association and also provide a framework for investigating other AF risk loci. The following section, we will discuss in detail the evidence relating to the 4q25 locus and AF followed by a discussion of the other susceptibility loci for AF.
The SNPs at the 4q25 locus are located in an extensive intergenic region within which there are no genes or putative genes. However, approximately 150,000 base pairs downstream lies a strong candidate gene, the paired-like homeodomain transcription factor 2 (PITX2). PITX2 plays a role in asymmetric morphogenesis of the heart and the gut and embryonic rotation.20–21 PITX2 also plays a critical role in suppression of formation of a sinus node in the left atrium and, possibly most importantly in the context of AF, development of the pulmonary myocardium.22–23
SNPs identified by GWAS typically act as a markers for a region of disease susceptibility that harbors the causative variants. These so-called linkage disequilibrium or LD blocks often span several thousands of base pairs. Fine mapping can be performed to refine the signal and to identify additional signals associated with disease.24 Lubitz et al recently performed such an analysis of the 4q25 locus. In addition to the top SNP at the locus, rs2200733, they identified two additional risk SNPs that were independently associated with AF. An interesting finding from the study was that the variants were associated with a graded risk of AF and the presence of all three variants identified a subset of patients with a 6-fold increased risk of AF.25
The potential role of PITX2 in the pathogenesis of AF has subsequently been investigated in a number of studies. Homozygous knockout of PITX2 is lethal in utero and is associated with multiple structural defects of the heart.23 Therefore, functional studies have been performed in either heterozygous or cardiac restricted PITX2 knockout mice. Heterozygote PITX2 knockout mice have an increased susceptibility to AF during programmed stimulation.26–27 Further, loss of PITX2 results in an abbreviated action potential, a reduction in the action potential amplitude and a more depolarized resting membrane potential.26, 28 These changes would be predicted to create a proarrhythmogenic substrate in the atrium. Interestingly, PITX2 expression is also downregulated in atrial tissue from human subjects with AF.28
The association between the AF risk variants at the 4q25 locus has also been investigated in a variety of clinical settings. Husser et al demonstrated that the risk SNPs at the 4q25 locus conferred an increased risk of AF recurrence following pulmonary vein isolation.29 Similarly, in a study of patients undergoing cardiac surgery, the risk variants at the 4q25 locus were independently associated with postoperative AF.30 The 4q25 locus has also previously been associated with ischemic stroke in a GWAS. Amongst stroke patients, the strongest association was observed with cardioembolic stroke.31
The majority of the studies relating to the 4q25 locus discussed thus far have been performed in populations of European descent. It is important to note however that a number of the replication studies have also been extended to non-European populations. For instance, following the initial GWAS in 2007, replication was performed in a cohort of Chinese descent and the top SNP at 4q25 was associated with a p value of 6.4 ×10−4 and an OR of 1.42. More recently, in a population of Japanese ancestry, a strong association was reported at the 4q25 locus with a p value of 3.7×10−17 and an OR of 1.84. These studies provide further corroborating evidence for the robust association between 4q25 and AF.
Other GWAS loci associated with AF
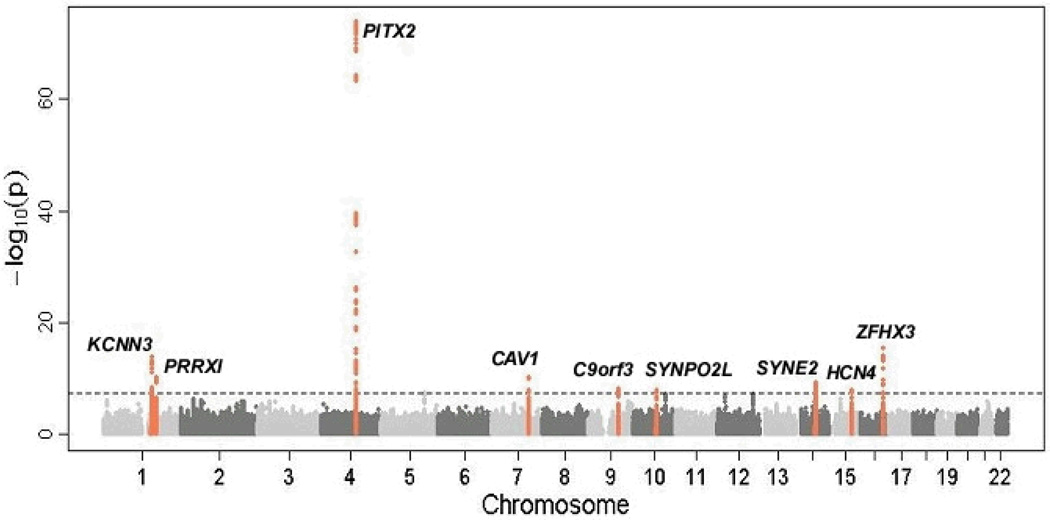
Since the original AF GWAS in 2007, three large-scale meta-analyses of GWAS data have been conducted.13–14, 32 In addition to confirming the association between the 4q25 locus and AF, eight novel risk loci for the arrhythmia have been identified (summarized in table 1 and figure 1). A number of these loci harbor compelling candidate genes for AF.
Table 1.
Summary of atrial fibrillation susceptibility loci identified by GWAS
| Locus | Top SNP | Nearest gene |
Location of SNP | RR | P-value | Initial report |
Replication |
|---|---|---|---|---|---|---|---|
| 1q21 | rs13376333 | KCNN3 | Intronic | 1.52 | 1.8×10−21 | 13 | 15 |
| 1q24 | rs3903239 | PRRX1 | 46 kb upstream | 1.14 | 8.4×10−14 | 15 | |
| 4q25 | rs2200733 | PITX2 | 150 kb upstream | 1.72 | 3.3×10−41 | 12 | 14–15, 17–19 |
| 7q31 | rs3807989 | CAV1 | Intronic | 0.90 | 3.6×10−12 | 15 | |
| 9q22 | rs10821415 | C9orf3 | Intronic | 1.11 | 4.2×10−11 | 15 | |
| 10q22 | rs10824026 | SYNPO2L | 5 kb upstream | 0.87 | 4.0×10−9 | 15 | |
| 14q23 | rs1152591 | SYNE2 | Intronic | 1.13 | 5.8×10−13 | 15 | |
| 15q24 | rs7164883 | HCN4 | Intronic | 1.19 | 2.8×10−17 | 15 | |
| 16q22 | rs2106261 | ZFHX3 | Intronic | 1.25 | 1.8×10−15 | 14 | 15, 68 |
Abbreviations: SNP, single nucleotide polymorphism; RR, relative risk. Note: The RR and P-values taken from the initial report of each SNP.
Figure 1.
Manhattan plot demonstrating association AF and risk loci identified by GWAS. The physical position of SNPs on each chromosome are depicted on the x axis while the −log10(P value) for each SNP is plotted on the y axis. The dashed line indicates the threshold for genome wide significance (P <5×10−8). The nine loci that exceed the genome wide threshold are indicated in orange. Reprinted by permission from Macmillan Publishers Ltd: Nature Genetics, 2012 Apr 29;44(6):670-5.
In addition to PITX2, two loci containing transcription factors have been associated with AF. The top SNP at the 16q22 locus was located within the ZFHX3 gene.14 ZFHX3 encodes a transcription factor, which has previously been demonstrated to mediate neural and myogenic differentiation.33–34 The risk variants at the 1q24 locus is intronic to PRRX1, a member of a family of homeobox transcription factors which regulate embryonic patterning by modulating transcription of downstream target genes.35–36 Interestingly in a murine model, ablation of PRRX1 expression is associated with abnormal development of the pulmonary vasculature and the great vessels.37–38
At two of the AF susceptibility loci, the risk variants cluster in ion channel genes. The risk SNPs at the 1q21 are located in the KCNN3 gene which encodes a calcium activated potassium channel (SK3 or KCa2.3).13 In neural tissues, this channel reduces excitability by mediating afterhyperpolarization.39 The role of the SK3 channel in cardiac physiology has yet to be characterized; however, the related SK2 channel has been shown to modulate repolarization.40
The most significant risk SNP at the 15q24 locus is located in the HCN4 gene which encodes the hyperpolarization-activated cyclic nucleotide-gated channel.36 The HCN4 channel underlies the funny current (If) that contributes to the generation of spontaneous action potentials in cardiac pacemaker cells. Of note, mutations in HCN4 have been reported to cause dysfunction of the sinus node pacemaker.41–42 While the association between sinus node dysfunction and AF is well recognized, the specific role of this channel in AF pathogenesis has yet to be elucidated.
Other compelling candidate genes at GWAS loci include SYNPO2L and MYOZ1 at the 10q22 locus, and CAV1 at the 7q31 locus.36 SYNPO2L and MYOZ1 encode proteins that localize to the Z disc in cardiac and skeletal muscle and are postulated to play critical roles in cell signaling and muscle function.43–44 CAV1 encodes caveolin-1, which is one of a family of coat proteins of caveolae which also plays an important role in cell signaling .45
Future directions
Despite the success of GWAS in identifying multiple susceptibility loci for AF, a number of questions remain. Specifically:
How do we functionally validate the role of candidate variants in disease pathogenesis?
A number of different strategies can be used to functionally validate the role of GWAS risk loci in disease pathogenesis. One approach is screen AF cohorts for causative mutations in candidate genes at GWAS loci. A second possibility would be to create knockout models in fish or mice of compelling candidate genes to examine the effect of gene loss on atrial structure or function. As described previously, such a strategy has been used for further analysis of the potential role of PITX2 at the 4q25 locus where loss of this gene was demonstrated to increase susceptibility to AF.26–27
A third potential approach would be to examine expression quantitative trait loci (eQTL).46 In this approach the top SNP related to AF at a locus is related to RNA levels of genes in the locus. If expression of a gene is strongly associated with the AF related SNP, then the gene is likely the causative gene at the locus. While eQTL mapping is a powerful approach it is often tissue specific and is thus limited by the availability of large repositories of cardiac tissue for these analyses.
Finally, for many noncoding SNPs it is often assumed that the SNP is regulating the function of an adjacent gene perhaps by altering the function of a promoter or enhancer. One strategy for the identification of these elements is to search for regions of phylogenetic conservation, as regulatory elements are typically highly conserved through evolution.47 Once potential regulatory elements have been identified, the next stage involves determining whether common variants associated with AF alter the function of these elements. Cell-based assays represent a useful tool for initial high-throughput screening for regulatory function.48 Further characterization can subsequently be performed using in vivo models such as fish and mice.49 While powerful, the identification of such regulatory elements is tedious and time-consuming.
How do we uncover the ‘missing heritability’ for AF?
While GWAS have successfully identified multiple risk variants associated with AF, these variants result in relatively subtle increments in disease risk and therefore only explain a small proportion of the observed heritability of the trait.50 Potential sources of the unexplained or ‘missing heritability’ of AF including rare variants, common variants that have not been identified by current GWAS due to stringent significant thresholds and structural variations in the genome such as copy number variations.50
Potential techniques for the identification of rare variants include analyses of data from an exome chip, exome sequencing, and whole genome sequencing. The exome chip is an array designed to cost-effectively capture both common and rare coding variants and is currently being run in many large-scale population studies. Exome sequencing on the other hand involves sequencing the entire coding region of the genome and is therefore a highly effective strategy for the identification of de novo coding variants. Exome sequencing is also increasingly commonly being used in population-based studies. In the longer term, as sequencing costs fall, it will be possible to perform whole genome sequencing in AF populations to identify both rare and common variants underlying the trait.51
What about non-European populations?
GWAS in AF, and indeed in most complex traits, have primarily been performed in cohorts of European descent. This approach is associated with important limitations as populations of European descent only harbor a fraction of human genetic variation.52 Therefore, the results from GWAS in European cohorts may not always be applicable to other ethnic populations.52
Interestingly, despite a higher incidence of traditional risk factors for AF, non-European populations, and in particular populations of African descent, have a lower risk of developing AF.53–54 These observations suggest an inherent difference in the genetic susceptibility to AF between different races. Further evidence to support this notion has recently emerged from a study by Marcus et al who demonstrated that amongst African Americans, the risk of developing AF was predicted by the degree of European ancestry.55 By implication therefore, a higher degree of African ancestry was protective.
Increasing efforts are currently being made to determine whether the results from GWAS of European cohorts are generalizable to other ethnic populations. As discussed previously, in a recent GWAS of cohorts of Japanese descent, the 4q25/PITX2 locus was reported to have a robust association with AF. Of note however, the other risk loci reported in European GWAS did not reach genome wide significance in the Japanese GWAS. These observations underscore the point that not all the risk loci identified in European GWAS confer increased AF risk in other ethnic populations.
Performing GWAS in more non-European populations in the future has a number of potential advantages. Firstly, these studies will lead to the identification of novel loci that confer increased susceptibility to AF in different populations and may provide novel insights into the pathogenesis of the arrhythmia. Secondly, genetic structure varies by race and can be exploited to narrow the boundaries of the known loci and facilitate the identification of causative variants at these loci.
Can we use variants identified by GWAS to predict risk of developing AF in a clinical setting?
Lubitz et al recently demonstrated that a family history of AF is an independent risk factor for developing the arrhythmia.56 Based on these findings, one may predict that risk variants associated with AF will enhance risk prediction. However, the AF risk SNPs identified to date are associated with modest effect sizes.12–14, 36 Therefore, before genotype-based risk prediction algorithms can be developed for AF, a larger proportion of the risk variants underlying the arrhythmia would have to be identified. For some phenotypes, initial attempts have been made to use the most significantly associated SNPs from GWAS as predictors of risk.57 Not surprisingly, most of these studies have had limited success, also due the relatively small effect sizes of the variants.58
The potential role of genetic testing for risk stratification of patients with AF or family members of AF patients has recently been addressed in the HRS/EHRA Expert Consensus Statement on Genetic Testing. Since the SNPs related to AF have a limited role in risk prediction or clinical outcomes, the guidelines do not recommended genetic testing for these AF related variants at this time.59
Interestingly, in addition to GWAS SNPs, biomarkers are emerging as potentially important predictors of AF risk.60 A promising future possibility is the development of clinically applicable risk algorithms that incorporate novel risk markers such as GWAS SNPs and biomarkers with established risk factors such as hypertension and diabetes to accurately determine the individual risk of developing AF.
What are the therapeutic implications of the identification of genetic variants underlying AF?
GWAS in AF have already identified a number of candidate genes that represent attractive therapeutic targets including the ion channel genes KCNN3 and HCN4.
Since the KCNN3 locus was implicated in AF pathogenesis, the effect of pharmacological modulation of this channel has been investigated in a number of studies. The KCNN3 channel is one of a family of three potassium channels, collectively referred to as SK channels. Diness et al recently demonstrated that in animal models of acute AF, two potent SK channel pore blockers, UCL1684 and ICA, terminated AF. They and others have also demonstrated that NS8593, a compound that negatively modulates SK channels, suppresses AF.61–64
Interestingly, drugs that block the HCN4 channel are already in clinical use as heart rate reducing medications.65 While the mechanistic link between AF pathogenesis remains to be elucidated, it is possible that in future such drugs could be also be used to suppress AF.
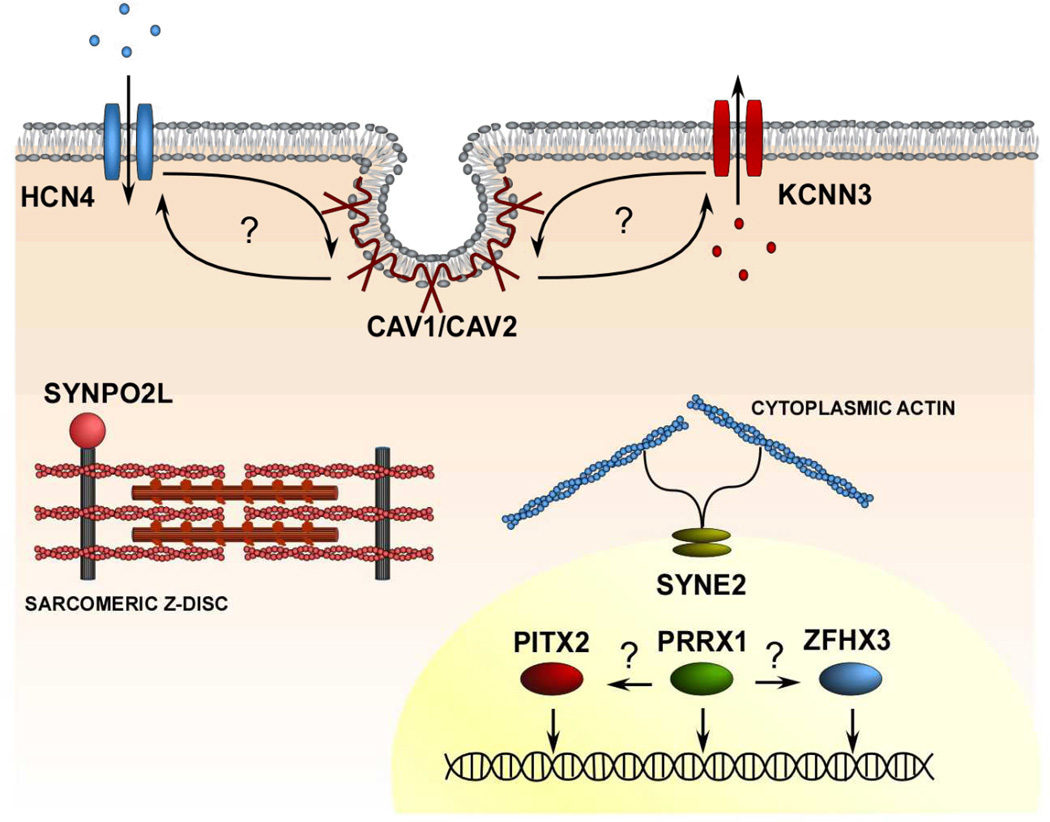
In addition to individual candidate genes, GWAS provide a unique opportunity to identify previously unsuspected molecular pathways involved in disease pathogenesis. An intriguing possibility is that the variants for AF interact with one another and form part of the same functional pathways. Indeed, previous studies have demonstrated that both the HCN4 and KCNN3 channels interact with caveolins.66–67 Further, transcription factors are also known to interact with one another. It is plausible therefore that the PITX2, ZFHX3, and PRRX1 transcription factors identified by GWAS are in a common molecular pathway. Systems biological approaches that integrate genomic, transcriptional, and sequencing data may facilitate the identification of the molecular pathways leading to AF.
Conclusions
The identification of the genetic substrate underlying AF will significantly enhance our understanding of the molecular basis underlying this common and morbid arrhythmia. GWAS have already led to the identification of multiple, previously unsuspected molecular pathways underlying AF. The advent of novel genotyping technologies such as exome sequencing and whole genome sequencing will likely uncover an even greater proportion of the genetic basis of AF. These advances represent significant steps towards the ultimate goal of AF genetics research, which is personalized, genotype-based medicine.
Figure 2.
Representative illustration of a cardiomyocyte demonstrating protein products of genes implicated in GWAS and the mechanisms of action of these proteins. The arrows indicate hypothetical interactions between the protein products.
BOX 1: GLOSSARY OF TERMS
- Candidate gene association study
A population-based study designed to test the association between a SNP in or near a specific gene and a disease. Candidate genes are typically selected based on their involvement in specific biological pathways.69 Such studies are often limited by a low-pretest probability that the SNP or gene being tested is directly associated with the disease.
- Exome sequencing
A technique which involves sequencing the entire protein coding region of the genome. The exome consists of approximately 1% of the genome and therefore this recently developed technique represents a cost-effective strategy for the identification of mutations and rare variants in the coding region of genes.
- Expression quantitative trait loci (eQTL)
SNPs that influence the quantity of gene expression are referred to as expression quantitative trait loci (eQTL). eQTLs may be located in the vicinity of the genes that they regulate or may exert their effect over larger genetic distances.46 eQTL studies involve an analysis of the correlation between SNPs of interest and the abundance of specific gene transcripts.
- Fine mapping
A strategy designed to refine a GWAS risk locus. Fine mapping involves genotyping densely spaced SNPs in the linkage disequilibrium block tagged by the GWAS marker SNP and associating these SNPs with disease. The identification of SNPs that are more strongly associated with disease allows one to delineate a narrower region within the risk locus which harbors the causative variant.24
- Genome-wide association study (GWAS)
A population-based study aimed at identifying genomic loci that harbor disease-causing variants. GWAS involve assaying numerous SNPs distributed throughout the genome and relating them to disease by comparing the frequency of these SNPs in disease cases as compared to controls. The disease-associated SNPs identified by GWAS typically act as surrogate markers which point to disease causing variants.24
- Genome-wide significance
The association between a SNP and disease is considered to reach genome-wide significance when the p value is less than a prespecified threshold (typical p values are in the order of 10−8). Stringent significance thresholds are set to avoid false positives that may arise due to multiple testing.72
- Linkage disequilibrium (LD)
Refers to the non-random segregation of alleles at different loci. Alleles that are in LD with one another have a higher chance of being inherited together. Thus knowledge of the genotype of a SNP allows one to make predictions about the genotype of adjacent SNPs at the locus. A single SNP can therefore be used as a marker for multiple variants in a linkage disequilibrium block that are predicted to be inherited together.73
- Manhattan plot
A bar chart that is commonly used to display results of GWAS. The positions of marker SNPs on each chromosome are conventionally displayed on the x axis while the −log10 of the p value for each SNP is plotted on the y axis. The height of the signal in the chart is proportional to the strength the association between a given marker SNP and disease. The genome-wide significance threshold is also commonly depicted in Manhattan plots.
- Missing heritability
Multiple genetic variants are likely to underlie the heritability of complex traits like AF. The variants identified by GWAS to date only explain a fraction of the observed heritability of these traits. The proportion of the heritability that remains unexplained is referred to as the ‘missing heritability’.50
- Single-nucleotide polymorphisms (SNP)
-
A single base pair change in the genome. SNPs considered in GWAS typically have a frequency of ≥1% in the population.70
SNPs can be further classified into coding, non-coding, synonymous and nonsynonymous variants. A coding SNPs is located within the translated region or exons of a gene. Such coding variants can either be synonymous or result in no change in the protein sequence or nonsynonymous variants that alter the amino acid sequence.
Noncoding SNPs on the other hand maybe intronic, intergenic or maybe located in untranslated regions of genes.71 Noncoding SNPs may influence disease risk by altering the function of regulatory elements which control gene expression.
REFERENCES
- 1.Snellen H. Two pioneers of electrocardiography. The Correspondence Between Einthoven And Lewis From 1908–1926. Netherlands: Alphen a/d Rijn; 1983. [Google Scholar]
- 2.Fox CS, Parise H, D'Agostino RB, Sr, Lloyd-Jones DM, Vasan RS, Wang TJ, Levy D, Wolf PA, Benjamin EJ. Parental atrial fibrillation as a risk factor for atrial fibrillation in offspring. Jama. 2004;291:2851–2855. doi: 10.1001/jama.291.23.2851. [DOI] [PubMed] [Google Scholar]
- 3.Ellinor PT, Yoerger DM, Ruskin JN, MacRae CA. Familial aggregation in lone atrial fibrillation. Hum Genet. 2005;118:179–184. doi: 10.1007/s00439-005-0034-8. [DOI] [PubMed] [Google Scholar]
- 4.Darbar D, Herron KJ, Ballew JD, Jahangir A, Gersh BJ, Shen WK, Hammill SC, Packer DL, Olson TM. Familial atrial fibrillation is a genetically heterogeneous disorder. J Am Coll Cardiol. 2003;41:2185–2192. doi: 10.1016/s0735-1097(03)00465-0. [DOI] [PubMed] [Google Scholar]
- 5.Chen YH, Xu SJ, Bendahhou S, Wang XL, Wang Y, Xu WY, Jin HW, Sun H, Su XY, Zhuang QN, Yang YQ, Li YB, Liu Y, Xu HJ, Li XF, Ma N, Mou CP, Chen Z, Barhanin J, Huang W. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299:251–254. doi: 10.1126/science.1077771. [DOI] [PubMed] [Google Scholar]
- 6.Xia M, Jin Q, Bendahhou S, He Y, Larroque MM, Chen Y, Zhou Q, Yang Y, Liu Y, Liu B, Zhu Q, Zhou Y, Lin J, Liang B, Li L, Dong X, Pan Z, Wang R, Wan H, Qiu W, Xu W, Eurlings P, Barhanin J, Chen Y. A Kir2.1 gain-of-function mutation underlies familial atrial fibrillation. Biochem Biophys Res Commun. 2005;332:1012–1019. doi: 10.1016/j.bbrc.2005.05.054. [DOI] [PubMed] [Google Scholar]
- 7.Hong K, Piper DR, Diaz-Valdecantos A, Brugada J, Oliva A, Burashnikov E, Santos-de-Soto J, Grueso-Montero J, Diaz-Enfante E, Brugada P, Sachse F, Sanguinetti MC, Brugada R. De novo KCNQ1 mutation responsible for atrial fibrillation and short QT syndrome in utero. Cardiovasc Res. 2005;68:433–440. doi: 10.1016/j.cardiores.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 8.Das S, Makino S, Melman YF, Shea MA, Goyal SB, Rosenzweig A, Macrae CA, Ellinor PT. Mutation in the S3 segment of KCNQ1 results in familial lone atrial fibrillation. Heart Rhythm. 2009;6:1146–1153. doi: 10.1016/j.hrthm.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olson TM, Alekseev AE, Liu XK, Park S, Zingman LV, Bienengraeber M, Sattiraju S, Ballew JD, Jahangir A, Terzic A. Kv1.5 channelopathy due to KCNA5 loss-of-function mutation causes human atrial fibrillation. Hum Mol Genet. 2006;15:2185–2191. doi: 10.1093/hmg/ddl143. [DOI] [PubMed] [Google Scholar]
- 10.Olson TM, Michels VV, Ballew JD, Reyna SP, Karst ML, Herron KJ, Horton SC, Rodeheffer RJ, Anderson JL. Sodium channel mutations and susceptibility to heart failure and atrial fibrillation. JAMA. 2005;293:447–454. doi: 10.1001/jama.293.4.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe H, Darbar D, Kaiser DW, Jiramongkolchai K, Chopra S, Donahue BS, Kannankeril PJ, Roden DM. Mutations in sodium channel beta1- and beta2-subunits associated with atrial fibrillation. Circ Arrhythm Electrophysiol. 2009;2:268–275. doi: 10.1161/CIRCEP.108.779181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, Jonasdottir A, Baker A, Thorleifsson G, Kristjansson K, Palsson A, Blondal T, Sulem P, Backman VM, Hardarson GA, Palsdottir E, Helgason A, Sigurjonsdottir R, Sverrisson JT, Kostulas K, Ng MC, Baum L, So WY, Wong KS, Chan JC, Furie KL, Greenberg SM, Sale M, Kelly P, MacRae CA, Smith EE, Rosand J, Hillert J, Ma RC, Ellinor PT, Thorgeirsson G, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 13.Ellinor PT, Lunetta KL, Glazer NL, Pfeufer A, Alonso A, Chung MK, Sinner MF, de Bakker PI, Mueller M, Lubitz SA, Fox E, Darbar D, Smith NL, Smith JD, Schnabel RB, Soliman EZ, Rice KM, Van Wagoner DR, Beckmann BM, van Noord C, Wang K, Ehret GB, Rotter JI, Hazen SL, Steinbeck G, Smith AV, Launer LJ, Harris TB, Makino S, Nelis M, Milan DJ, Perz S, Esko T, Kottgen A, Moebus S, Newton-Cheh C, Li M, Mohlenkamp S, Wang TJ, Kao WH, Vasan RS, Nothen MM, MacRae CA, Stricker BH, Hofman A, Uitterlinden AG, Levy D, Boerwinkle E, Metspalu A, Topol EJ, Chakravarti A, Gudnason V, Psaty BM, Roden DM, Meitinger T, Wichmann HE, Witteman JC, Barnard J, Arking DE, Benjamin EJ, Heckbert SR, Kaab S. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet. 2010;42:240–244. doi: 10.1038/ng.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benjamin EJ, Rice KM, Arking DE, Pfeufer A, van Noord C, Smith AV, Schnabel RB, Bis JC, Boerwinkle E, Sinner MF, Dehghan A, Lubitz SA, D'Agostino RB, Sr, Lumley T, Ehret GB, Heeringa J, Aspelund T, Newton-Cheh C, Larson MG, Marciante KD, Soliman EZ, Rivadeneira F, Wang TJ, Eiriksdottir G, Levy D, Psaty BM, Li M, Chamberlain AM, Hofman A, Vasan RS, Harris TB, Rotter JI, Kao WH, Agarwal SK, Stricker BH, Wang K, Launer LJ, Smith NL, Chakravarti A, Uitterlinden AG, Wolf PA, Sotoodehnia N, Kottgen A, van Duijn CM, Meitinger T, Mueller M, Perz S, Steinbeck G, Wichmann HE, Lunetta KL, Heckbert SR, Gudnason V, Alonso A, Kaab S, Ellinor PT, Witteman JC. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet. 2009;41:879–881. doi: 10.1038/ng.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, Arking DE, Muller-Nurasyid M, Krijthe BP, Lubitz SA, Bis JC, Chung MK, Dorr M, Ozaki K, Roberts JD, Smith JG, Pfeufer A, Sinner MF, Lohman K, Ding J, Smith NL, Smith JD, Rienstra M, Rice KM, Van Wagoner DR, Magnani JW, Wakili R, Clauss S, Rotter JI, Steinbeck G, Launer LJ, Davies RW, Borkovich M, Harris TB, Lin H, Volker U, Volzke H, Milan DJ, Hofman A, Boerwinkle E, Chen LY, Soliman EZ, Voight BF, Li G, Chakravarti A, Kubo M, Tedrow UB, Rose LM, Ridker PM, Conen D, Tsunoda T, Furukawa T, Sotoodehnia N, Xu S, Kamatani N, Levy D, Nakamura Y, Parvez B, Mahida S, Furie KL, Rosand J, Muhammad R, Psaty BM, Meitinger T, Perz S, Wichmann HE, Witteman JC, Kao WH, Kathiresan S, Roden DM, Uitterlinden AG, Rivadeneira F, McKnight B, Sjogren M, Newman AB, Liu Y, Gollob MH, Melander O, Tanaka T, Stricker BH, Felix SB, Alonso A, Darbar D, Barnard J, Chasman DI, Heckbert SR, Benjamin EJ, Gudnason V, Kaab S. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012;44:670–675. doi: 10.1038/ng.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahida S, Lubitz SA, Rienstra M, Milan DJ, Ellinor PT. Monogenic atrial fibrillation as pathophysiological paradigms. Cardiovasc Res. 89:692–700. doi: 10.1093/cvr/cvq381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaab S, Darbar D, van Noord C, Dupuis J, Pfeufer A, Newton-Cheh C, Schnabel R, Makino S, Sinner MF, Kannankeril PJ, Beckmann BM, Choudry S, Donahue BS, Heeringa J, Perz S, Lunetta KL, Larson MG, Levy D, Macrae CA, Ruskin JN, Wacker A, Schomig A, Wichmann HE, Steinbeck G, Meitinger T, Uitterlinden AG, Witteman JC, Roden DM, Benjamin EJ, Ellinor PT. Large scale replication and meta-analysis of variants on chromosome 4q25 associated with atrial fibrillation. Eur Heart J. 2009 doi: 10.1093/eurheartj/ehn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viviani Anselmi C, Novelli V, Roncarati R, Malovini A, Bellazzi R, Bronzini R, Marchese G, Condorelli G, Montenero AS, Puca AA. Association of rs2200733 at 4q25 with atrial flutter/fibrillation diseases in an Italian population. Heart. 2008;94:1394–1396. doi: 10.1136/hrt.2008.148544. [DOI] [PubMed] [Google Scholar]
- 19.Shi L, Li C, Wang C, Xia Y, Wu G, Wang F, Xu C, Wang P, Li X, Wang D, Xiong X, Bai Y, Liu M, Liu J, Ren X, Gao L, Wang B, Zeng Q, Yang B, Ma X, Yang Y, Tu X, Wang QK. Assessment of association of rs2200733 on chromosome 4q25 with atrial fibrillation and ischemic stroke in a Chinese Han population. Hum Genet. 2009;126:843–849. doi: 10.1007/s00439-009-0737-3. [DOI] [PubMed] [Google Scholar]
- 20.Logan M, Pagan-Westphal SM, Smith DM, Paganessi L, Tabin CJ. The transcription factor Pitx2 mediates situs-specific morphogenesis in response to left-right asymmetric signals. Cell. 1998;94:307–317. doi: 10.1016/s0092-8674(00)81474-9. [DOI] [PubMed] [Google Scholar]
- 21.Yoshioka H, Meno C, Koshiba K, Sugihara M, Itoh H, Ishimaru Y, Inoue T, Ohuchi H, Semina EV, Murray JC, Hamada H, Noji S. Pitx2, a bicoid-type homeobox gene, is involved in a lefty-signaling pathway in determination of left-right asymmetry. Cell. 1998;94:299–305. doi: 10.1016/s0092-8674(00)81473-7. [DOI] [PubMed] [Google Scholar]
- 22.Mommersteeg MT, Hoogaars WM, Prall OW, de Gier-de Vries C, Wiese C, Clout DE, Papaioannou VE, Brown NA, Harvey RP, Moorman AF, Christoffels VM. Molecular pathway for the localized formation of the sinoatrial node. Circ Res. 2007;100:354–362. doi: 10.1161/01.RES.0000258019.74591.b3. [DOI] [PubMed] [Google Scholar]
- 23.Mommersteeg MT, Brown NA, Prall OW, de Gier-de Vries C, Harvey RP, Moorman AF, Christoffels VM. Pitx2c and Nkx2–5 are required for the formation and identity of the pulmonary myocardium. Circ Res. 2007;101:902–909. doi: 10.1161/CIRCRESAHA.107.161182. [DOI] [PubMed] [Google Scholar]
- 24.Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 363:166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 25.Lubitz SA, Sinner MF, Lunetta KL, Makino S, Pfeufer A, Rahman R, Veltman CE, Barnard J, Bis JC, Danik SP, Sonni A, Shea MA, Del Monte F, Perz S, Muller M, Peters A, Greenberg SM, Furie KL, van Noord C, Boerwinkle E, Stricker BH, Witteman J, Smith JD, Chung MK, Heckbert SR, Benjamin EJ, Rosand J, Arking DE, Alonso A, Kaab S, Ellinor PT. Independent susceptibility markers for atrial fibrillation on chromosome 4q25. Circulation. 122:976–984. doi: 10.1161/CIRCULATIONAHA.109.886440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirchhof P, Kahr PC, Kaese S, Piccini I, Vokshi I, Scheld HH, Rotering H, Fortmueller L, Laakmann S, Verheule S, Schotten U, Fabritz L, Brown NA. PITX2c is expressed in the adult left atrium, and reducing Pitx2c expression promotes atrial fibrillation inducibility and complex changes in gene expression. Circ Cardiovasc Genet. 4:123–133. doi: 10.1161/CIRCGENETICS.110.958058. [DOI] [PubMed] [Google Scholar]
- 27.Wang J, Klysik E, Sood S, Johnson RL, Wehrens XH, Martin JF. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc Natl Acad Sci U S A. 107:9753–9758. doi: 10.1073/pnas.0912585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chinchilla A, Daimi H, Lozano-Velasco E, Dominguez JN, Caballero R, Delpon E, Tamargo J, Cinca J, Hove-Madsen L, Aranega AE, Franco D. PITX2 insufficiency leads to atrial electrical and structural remodeling linked to arrhythmogenesis. Circ Cardiovasc Genet. 4:269–279. doi: 10.1161/CIRCGENETICS.110.958116. [DOI] [PubMed] [Google Scholar]
- 29.Husser D, Adams V, Piorkowski C, Hindricks G, Bollmann A. Chromosome 4q25 variants and atrial fibrillation recurrence after catheter ablation. J Am Coll Cardiol. 55:747–753. doi: 10.1016/j.jacc.2009.11.041. [DOI] [PubMed] [Google Scholar]
- 30.Body SC, Collard CD, Shernan SK, Fox AA, Liu KY, Ritchie MD, Perry TE, Muehlschlegel JD, Aranki S, Donahue BS, Pretorius M, Estrada JC, Ellinor PT, Newton-Cheh C, Seidman CE, Seidman JG, Herman DS, Lichtner P, Meitinger T, Pfeufer A, Kaab S, Brown NJ, Roden DM, Darbar D. Variation in the 4q25 chromosomal locus predicts atrial fibrillation after coronary artery bypass graft surgery. Circ Cardiovasc Genet. 2009;2:499–506. doi: 10.1161/CIRCGENETICS.109.849075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gretarsdottir S, Thorleifsson G, Manolescu A, Styrkarsdottir U, Helgadottir A, Gschwendtner A, Kostulas K, Kuhlenbaumer G, Bevan S, Jonsdottir T, Bjarnason H, Saemundsdottir J, Palsson S, Arnar DO, Holm H, Thorgeirsson G, Valdimarsson EM, Sveinbjornsdottir S, Gieger C, Berger K, Wichmann HE, Hillert J, Markus H, Gulcher JR, Ringelstein EB, Kong A, Dichgans M, Gudbjartsson DF, Thorsteinsdottir U, Stefansson K. Risk variants for atrial fibrillation on chromosome 4q25 associate with ischemic stroke. Ann Neurol. 2008;64:402–409. doi: 10.1002/ana.21480. [DOI] [PubMed] [Google Scholar]
- 32.Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, Arking DE, Muller-Nurasyid M, Krijthe BP, Lubitz SA, Bis JC, Chung MK, Dorr M, Ozaki K, Roberts JD, Smith JG, Pfeufer A, Sinner MF, Lohman K, Ding J, Smith NL, Smith JD, Rienstra M, Rice KM, Van Wagoner DR, Magnani JW, Wakili R, Clauss S, Rotter JI, Steinbeck G, Launer LJ, Davies RW, Borkovich M, Harris TB, Lin H, Volker U, Volzke H, Milan DJ, Hofman A, Boerwinkle E, Chen LY, Soliman EZ, Voight BF, Li G, Chakravarti A, Kubo M, Tedrow UB, Rose LM, Ridker PM, Conen D, Tsunoda T, Furukawa T, Sotoodehnia N, Xu S, Kamatani N, Levy D, Nakamura Y, Parvez B, Mahida S, Furie KL, Rosand J, Muhammad R, Psaty BM, Meitinger T, Perz S, Wichmann HE, Witteman JC, Kao WH, Kathiresan S, Roden DM, Uitterlinden AG, Rivadeneira F, McKnight B, Sjogren M, Newman AB, Liu Y, Gollob MH, Melander O, Tanaka T, Stricker BH, Felix SB, Alonso A, Darbar D, Barnard J, Chasman DI, Heckbert SR, Benjamin EJ, Gudnason V, Kaab S. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. doi: 10.1038/ng.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berry FB, Miura Y, Mihara K, Kaspar P, Sakata N, Hashimoto-Tamaoki T, Tamaoki T. Positive and negative regulation of myogenic differentiation of C2C12 cells by isoforms of the multiple homeodomain zinc finger transcription factor ATBF1. J Biol Chem. 2001;276:25057–25065. doi: 10.1074/jbc.M010378200. [DOI] [PubMed] [Google Scholar]
- 34.Jung CG, Kim HJ, Kawaguchi M, Khanna KK, Hida H, Asai K, Nishino H, Miura Y. Homeotic factor ATBF1 induces the cell cycle arrest associated with neuronal differentiation. Development. 2005;132:5137–5145. doi: 10.1242/dev.02098. [DOI] [PubMed] [Google Scholar]
- 35.Chesterman ES, Gainey GD, Varn AC, Peterson RE, Jr, Kern MJ. Investigation of Prx1 protein expression provides evidence for conservation of cardiac-specific posttranscriptional regulation in vertebrates. Dev Dyn. 2001;222:459–470. doi: 10.1002/dvdy.1198. [DOI] [PubMed] [Google Scholar]
- 36.Schnabel RB, Sullivan LM, Levy D, Pencina MJ, Massaro JM, D'Agostino RB, Sr, Newton-Cheh C, Yamamoto JF, Magnani JW, Tadros TM, Kannel WB, Wang TJ, Ellinor PT, Wolf PA, Vasan RS, Benjamin EJ. Development of a risk score for atrial fibrillation (Framingham Heart Study): a community-based cohort study. Lancet. 2009;373:739–745. doi: 10.1016/S0140-6736(09)60443-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ihida-Stansbury K, McKean DM, Gebb SA, Martin JF, Stevens T, Nemenoff R, Akeson A, Vaughn J, Jones PL. Paired-related homeobox gene Prx1 is required for pulmonary vascular development. Circ Res. 2004;94:1507–1514. doi: 10.1161/01.RES.0000130656.72424.20. [DOI] [PubMed] [Google Scholar]
- 38.Bergwerff M, Gittenberger-de Groot AC, Wisse LJ, DeRuiter MC, Wessels A, Martin JF, Olson EN, Kern MJ. Loss of function of the Prx1 and Prx2 homeobox genes alters architecture of the great elastic arteries and ductus arteriosus. Virchows Arch. 2000;436:12–19. doi: 10.1007/pl00008193. [DOI] [PubMed] [Google Scholar]
- 39.Kohler M, Hirschberg B, Bond CT, Kinzie JM, Marrion NV, Maylie J, Adelman JP. Small-conductance, calcium-activated potassium channels from mammalian brain. Science. 1996;273:1709–1714. doi: 10.1126/science.273.5282.1709. [DOI] [PubMed] [Google Scholar]
- 40.Li N, Timofeyev V, Tuteja D, Xu D, Lu L, Zhang Q, Zhang Z, Singapuri A, Albert TR, Rajagopal AV, Bond CT, Periasamy M, Adelman J, Chiamvimonvat N. Ablation of a Ca2+-activated K+ channel (SK2 channel) results in action potential prolongation in atrial myocytes and atrial fibrillation. J Physiol. 2009;587:1087–1100. doi: 10.1113/jphysiol.2008.167718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schulze-Bahr E, Neu A, Friederich P, Kaupp UB, Breithardt G, Pongs O, Isbrandt D. Pacemaker channel dysfunction in a patient with sinus node disease. J Clin Invest. 2003;111:1537–1545. doi: 10.1172/JCI16387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milanesi R, Baruscotti M, Gnecchi-Ruscone T, DiFrancesco D. Familial sinus bradycardia associated with a mutation in the cardiac pacemaker channel. N Engl J Med. 2006;354:151–157. doi: 10.1056/NEJMoa052475. [DOI] [PubMed] [Google Scholar]
- 43.Frey N, Olson EN. Calsarcin-3, a novel skeletal muscle-specific member of the calsarcin family, interacts with multiple Z-disc proteins. J Biol Chem. 2002;277:13998–14004. doi: 10.1074/jbc.M200712200. [DOI] [PubMed] [Google Scholar]
- 44.Beqqali A, Monshouwer-Kloots J, Monteiro R, Welling M, Bakkers J, Ehler E, Verkleij A, Mummery C, Passier R. CHAP is a newly identified Z-disc protein essential for heart and skeletal muscle function. J Cell Sci. 2010;123:1141–1150. doi: 10.1242/jcs.063859. [DOI] [PubMed] [Google Scholar]
- 45.Sowa G. Caveolae, caveolins, cavins, and endothelial cell function: new insights. Front Physiol. 2:120. doi: 10.3389/fphys.2011.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y, Liu XS, Wei L, Altman RB, Batzoglou S. Eukaryotic regulatory element conservation analysis and identification using comparative genomics. Genome Res. 2004;14:451–458. doi: 10.1101/gr.1327604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carey M. Transcriptional Regulation in Eukaryotes: Concepts, Strategies, and Techniques. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 49.Loots GG. Genomic identification of regulatory elements by evolutionary sequence comparison and functional analysis. Adv Genet. 2008;61:269–293. doi: 10.1016/S0065-2660(07)00010-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 52.Rosenberg NA, Huang L, Jewett EM, Szpiech ZA, Jankovic I, Boehnke M. Genome-wide association studies in diverse populations. Nat Rev Genet. 2010;11:356–366. doi: 10.1038/nrg2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Go AS, Hylek EM, Phillips KA, Chang Y, Henault LE, Selby JV, Singer DE. Prevalence of diagnosed atrial fibrillation in adults: national implications for rhythm management and stroke prevention: the AnTicoagulation and Risk Factors in Atrial Fibrillation (ATRIA) Study. JAMA. 2001;285:2370–2375. doi: 10.1001/jama.285.18.2370. [DOI] [PubMed] [Google Scholar]
- 54.Ruo B, Capra AM, Jensvold NG, Go AS. Racial variation in the prevalence of atrial fibrillation among patients with heart failure: the Epidemiology, Practice, Outcomes, and Costs of Heart Failure (EPOCH) study. J Am Coll Cardiol. 2004;43:429–435. doi: 10.1016/j.jacc.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 55.Marcus GM, Alonso A, Peralta CA, Lettre G, Vittinghoff E, Lubitz SA, Fox ER, Levitzky YS, Mehra R, Kerr KF, Deo R, Sotoodehnia N, Akylbekova M, Ellinor PT, Paltoo DN, Soliman EZ, Benjamin EJ, Heckbert SR. European ancestry as a risk factor for atrial fibrillation in African Americans. Circulation. 122:2009–2015. doi: 10.1161/CIRCULATIONAHA.110.958306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lubitz SA, Yin X, Fontes JD, Magnani JW, Rienstra M, Pai M, Villalon ML, Vasan RS, Pencina MJ, Levy D, Larson MG, Ellinor PT, Benjamin EJ. Association between familial atrial fibrillation and risk of new-onset atrial fibrillation. JAMA. 304:2263–2269. doi: 10.1001/jama.2010.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kooperberg C, LeBlanc M, Obenchain V. Risk prediction using genome-wide association studies. Genet Epidemiol. 2010;34:643–652. doi: 10.1002/gepi.20509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wray NR, Goddard ME, Visscher PM. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 2007;17:1520–1528. doi: 10.1101/gr.6665407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ackerman MJ, Priori SG, Willems S, Berul C, Brugada R, Calkins H, Camm AJ, Ellinor PT, Gollob M, Hamilton R, Hershberger RE, Judge DP, Le Marec H, McKenna WJ, Schulze-Bahr E, Semsarian C, Towbin JA, Watkins H, Wilde A, Wolpert C, Zipes DP. HRS/EHRA Expert Consensus Statement on the State of Genetic Testing for the Channelopathies and Cardiomyopathies This document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) Heart Rhythm. 2011;8:1308–1339. doi: 10.1016/j.hrthm.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 60.Smith JG, Newton-Cheh C, Almgren P, Struck J, Morgenthaler NG, Bergmann A, Platonov PG, Hedblad B, Engstrom G, Wang TJ, Melander O. Assessment of conventional cardiovascular risk factors and multiple biomarkers for the prediction of incident heart failure and atrial fibrillation. J Am Coll Cardiol. 56:1712–1719. doi: 10.1016/j.jacc.2010.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Strobaek D, Hougaard C, Johansen TH, Sorensen US, Nielsen EO, Nielsen KS, Taylor RD, Pedarzani P, Christophersen P. Inhibitory gating modulation of small conductance Ca2+-activated K+ channels by the synthetic compound (R)-N-(benzimidazol-2-yl)-1,2,3,4-tetrahydro-1-naphtylamine (NS8593) reduces afterhyperpolarizing current in hippocampal CA1 neurons. Mol Pharmacol. 2006;70:1771–1782. doi: 10.1124/mol.106.027110. [DOI] [PubMed] [Google Scholar]
- 62.Strobaek D, Jorgensen TD, Christophersen P, Ahring PK, Olesen SP. Pharmacological characterization of small-conductance Ca(2+)-activated K(+) channels stably expressed in HEK 293 cells. Br J Pharmacol. 2000;129:991–999. doi: 10.1038/sj.bjp.0703120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Diness JG, Sorensen US, Nissen JD, Al-Shahib B, Jespersen T, Grunnet M, Hansen RS. Inhibition of small-conductance Ca2+-activated K+ channels terminates and protects against atrial fibrillation. Circ Arrhythm Electrophysiol. 2010;3:380–390. doi: 10.1161/CIRCEP.110.957407. [DOI] [PubMed] [Google Scholar]
- 64.Skibsbye L, Diness JG, Sorensen US, Hansen RS, Grunnet M. The duration of pacing-induced atrial fibrillation is reduced in vivo by inhibition of small conductance Ca(2+)-activated K(+) channels. J Cardiovasc Pharmacol. 2011;57:672–681. doi: 10.1097/FJC.0b013e318217943d. [DOI] [PubMed] [Google Scholar]
- 65.Bucchi A, Tognati A, Milanesi R, Baruscotti M, DiFrancesco D. Properties of ivabradine-induced block of HCN1 and HCN4 pacemaker channels. J Physiol. 2006;572:335–346. doi: 10.1113/jphysiol.2005.100776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barbuti A, Scavone A, Mazzocchi N, Terragni B, Baruscotti M, Difrancesco D. A caveolin-binding domain in the HCN4 channels mediates functional interaction with caveolin proteins. J Mol Cell Cardiol. doi: 10.1016/j.yjmcc.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 67.Lin MT, Adelman JP, Maylie J. Modulation of endothelial SK3 channel activity by Ca2+- dependent caveolar trafficking. Am J Physiol Cell Physiol. doi: 10.1152/ajpcell.00058.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gudbjartsson DF, Holm H, Gretarsdottir S, Thorleifsson G, Walters GB, Thorgeirsson G, Gulcher J, Mathiesen EB, Njolstad I, Nyrnes A, Wilsgaard T, Hald EM, Hveem K, Stoltenberg C, Kucera G, Stubblefield T, Carter S, Roden D, Ng MC, Baum L, So WY, Wong KS, Chan JC, Gieger C, Wichmann HE, Gschwendtner A, Dichgans M, Kuhlenbaumer G, Berger K, Ringelstein EB, Bevan S, Markus HS, Kostulas K, Hillert J, Sveinbjornsdottir S, Valdimarsson EM, Lochen ML, Ma RC, Darbar D, Kong A, Arnar DO, Thorsteinsdottir U, Stefansson K. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41:876–878. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jorgensen TJ, Ruczinski I, Kessing B, Smith MW, Shugart YY, Alberg AJ. Hypothesis-driven candidate gene association studies: practical design and analytical considerations. Am J Epidemiol. 2009;170:986–993. doi: 10.1093/aje/kwp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vignal A, Milan D, SanCristobal M, Eggen A. A review on SNP and other types of molecular markers and their use in animal genetics. Genet Sel Evol. 2002;34:275–305. doi: 10.1186/1297-9686-34-3-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reumers J, Conde L, Medina I, Maurer-Stroh S, Van Durme J, Dopazo J, Rousseau F, Schymkowitz J. Joint annotation of coding and non-coding single nucleotide polymorphisms and mutations in the SNPeffect and PupaSuite databases. Nucleic Acids Res. 2008;36:D825–D829. doi: 10.1093/nar/gkm979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roeder K, Wasserman L. Genome-Wide Significance Levels and Weighted Hypothesis Testing. Stat Sci. 2009;24:398–413. doi: 10.1214/09-STS289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ambrosone CB. The promise and limitations of genome-wide association studies to elucidate the causes of breast cancer. Breast Cancer Res. 2007;9:114. doi: 10.1186/bcr1787. [DOI] [PMC free article] [PubMed] [Google Scholar]