Abstract
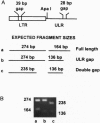
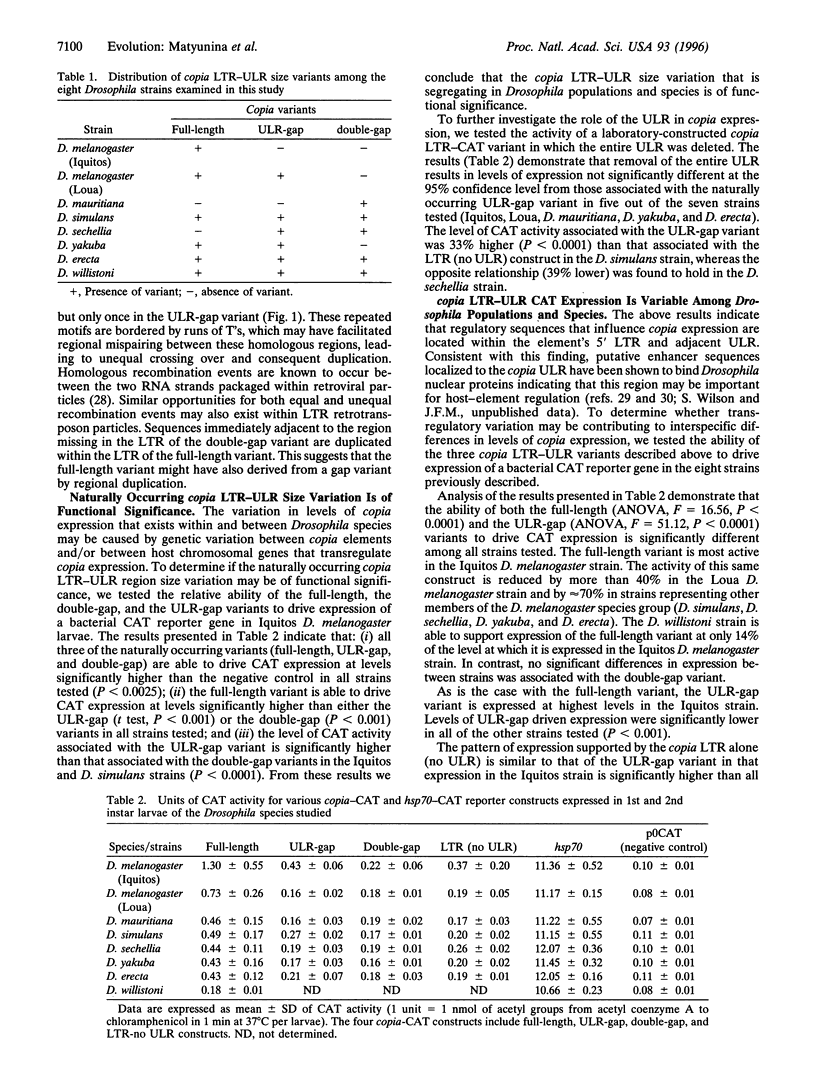
Significant differences in levels of copia [Drosophila long terminal repeat (LTR) retrotransposon] expression exist among six species representing the Drosophila melanogaster species complex (D. melanogaster, Drosophila mauritiana, Drosophila simulans, Drosophila sechellia, Drosophila yakuba, and Drosophila erecta) and a more distantly related species (Drosophila willistoni). These differences in expression are correlated with major size variation mapping to putative regulatory regions of the copia 5' LTR and adjacent untranslated leader region (ULR). Sequence analysis indicates that these size variants were derived from a series of regional duplication events. The ability of the copia LTR-ULR size variants to drive expression of a bacterial chloramphenicol acetyltransferase reporter gene was tested in each of the seven species. The results indicate that both element-encoded (cis) and host-genome-encoded (trans) genetic differences are responsible for the variability in copia expression within and between Drosophila species. This finding indicates that models purporting to explain the dynamics and distribution of retrotransposons in natural populations must consider the potential impact of both element-encoded and host-genome-encoded regulatory variation to be valid. We propose that interelement selection among retrotransposons may provide a molecular drive mechanism for the evolution of eukaryotic enhancers which can be subsequently distributed throughout the genome by retrotransposition.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bender W., Spierer P., Hogness D. S. Chromosomal walking and jumping to isolate DNA from the Ace and rosy loci and the bithorax complex in Drosophila melanogaster. J Mol Biol. 1983 Jul 25;168(1):17–33. doi: 10.1016/s0022-2836(83)80320-9. [DOI] [PubMed] [Google Scholar]
- Bialojan S., Falkenburg D., Renkawitz-Pohl R. Characterization and developmental expression of beta tubulin genes in Drosophila melanogaster. EMBO J. 1984 Nov;3(11):2543–2548. doi: 10.1002/j.1460-2075.1984.tb02170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Corces V. G. Transcription and reverse transcription of retrotransposons. Annu Rev Microbiol. 1989;43:403–434. doi: 10.1146/annurev.mi.43.100189.002155. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Garfinkel D. J., Styles C. A., Fink G. R. Ty elements transpose through an RNA intermediate. Cell. 1985 Mar;40(3):491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- Bryant L. A., Brierley C., Flavell A. J., Sinclair J. H. The retrotransposon copia regulates Drosophila gene expression both positively and negatively. Nucleic Acids Res. 1991 Oct 25;19(20):5533–5536. doi: 10.1093/nar/19.20.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavarec L., Heidmann T. The Drosophila copia retrotransposon contains binding sites for transcriptional regulation by homeoproteins. Nucleic Acids Res. 1993 Nov 11;21(22):5041–5049. doi: 10.1093/nar/21.22.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavarec L., Jensen S., Heidmann T. Identification of a strong transcriptional activator for the copia retrotransposon responsible for its differential expression in Drosophila hydei and melanogaster cell lines. Biochem Biophys Res Commun. 1994 Aug 30;203(1):392–399. doi: 10.1006/bbrc.1994.2195. [DOI] [PubMed] [Google Scholar]
- Cirera S., Martín-Campos J. M., Segarra C., Aguadé M. Molecular characterization of the breakpoints of an inversion fixed between Drosophila melanogaster and D. subobscura. Genetics. 1995 Jan;139(1):321–326. doi: 10.1093/genetics/139.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J. B., Maddison W. P., Kidwell M. G. Phylogenetic analysis supports horizontal transfer of P transposable elements. Mol Biol Evol. 1994 Jan;11(1):40–50. doi: 10.1093/oxfordjournals.molbev.a040091. [DOI] [PubMed] [Google Scholar]
- Corces V. G., Geyer P. K. Interactions of retrotransposons with the host genome: the case of the gypsy element of Drosophila. Trends Genet. 1991 Mar;7(3):86–90. doi: 10.1016/0168-9525(91)90277-W. [DOI] [PubMed] [Google Scholar]
- Csink A. K., McDonald J. F. Analysis of copia sequence variation within and between Drosophila species. Mol Biol Evol. 1995 Jan;12(1):83–93. doi: 10.1093/oxfordjournals.molbev.a040193. [DOI] [PubMed] [Google Scholar]
- Csink A. K., McDonald J. F. copia expression is variable among natural populations of Drosophila. Genetics. 1990 Oct;126(2):375–385. doi: 10.1093/genetics/126.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle W. F., Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980 Apr 17;284(5757):601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- Flavell A. J., Pearce S. R., Kumar A. Plant transposable elements and the genome. Curr Opin Genet Dev. 1994 Dec;4(6):838–844. doi: 10.1016/0959-437x(94)90068-x. [DOI] [PubMed] [Google Scholar]
- Gerasimova T. I., Gdula D. A., Gerasimov D. V., Simonova O., Corces V. G. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell. 1995 Aug 25;82(4):587–597. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- Kidwell M. G. The Wilhelmine E. Key 1991 Invitational Lecture. The evolutionary history of the P family of transposable elements. J Hered. 1994 Sep-Oct;85(5):339–346. doi: 10.1093/oxfordjournals.jhered.a111478. [DOI] [PubMed] [Google Scholar]
- Levis R., Dunsmuir P., Rubin G. M. Terminal repeats of the Drosophila transposable element copia: nucleotide sequence and genomic organization. Cell. 1980 Sep;21(2):581–588. doi: 10.1016/0092-8674(80)90496-1. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Goodbourn S., Fischer J. A. Regulation of inducible and tissue-specific gene expression. Science. 1987 Jun 5;236(4806):1237–1245. doi: 10.1126/science.3296191. [DOI] [PubMed] [Google Scholar]
- Mason J. M., Biessmann H. The unusual telomeres of Drosophila. Trends Genet. 1995 Feb;11(2):58–62. doi: 10.1016/s0168-9525(00)88998-2. [DOI] [PubMed] [Google Scholar]
- Mattick J. S. Introns: evolution and function. Curr Opin Genet Dev. 1994 Dec;4(6):823–831. doi: 10.1016/0959-437x(94)90066-3. [DOI] [PubMed] [Google Scholar]
- Mazo A. M., Mizrokhi L. J., Karavanov A. A., Sedkov Y. A., Krichevskaja A. A., Ilyin Y. V. Suppression in Drosophila: su(Hw) and su(f) gene products interact with a region of gypsy (mdg4) regulating its transcriptional activity. EMBO J. 1989 Mar;8(3):903–911. doi: 10.1002/j.1460-2075.1989.tb03451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J. F. Evolution and consequences of transposable elements. Curr Opin Genet Dev. 1993 Dec;3(6):855–864. doi: 10.1016/0959-437x(93)90005-a. [DOI] [PubMed] [Google Scholar]
- Müller M. M., Gerster T., Schaffner W. Enhancer sequences and the regulation of gene transcription. Eur J Biochem. 1988 Oct 1;176(3):485–495. doi: 10.1111/j.1432-1033.1988.tb14306.x. [DOI] [PubMed] [Google Scholar]
- Orgel L. E., Crick F. H. Selfish DNA: the ultimate parasite. Nature. 1980 Apr 17;284(5757):604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- Pennie W. D., Hager G. L., Smith C. L. Nucleoprotein structure influences the response of the mouse mammary tumor virus promoter to activation of the cyclic AMP signalling pathway. Mol Cell Biol. 1995 Apr;15(4):2125–2134. doi: 10.1128/mcb.15.4.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpinelli S., Berloco M., Fanti L., Dimitri P., Bonaccorsi S., Marchetti E., Caizzi R., Caggese C., Gatti M. Transposable elements are stable structural components of Drosophila melanogaster heterochromatin. Proc Natl Acad Sci U S A. 1995 Apr 25;92(9):3804–3808. doi: 10.1073/pnas.92.9.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. M., Lampe D. J. Distribution of transposable elements in arthropods. Annu Rev Entomol. 1995;40:333–357. doi: 10.1146/annurev.en.40.010195.002001. [DOI] [PubMed] [Google Scholar]
- Sneddon A., Flavell A. J. The transcriptional control regions of the copia retrotransposon. Nucleic Acids Res. 1989 Jun 12;17(11):4025–4035. doi: 10.1093/nar/17.11.4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A. C., Rubin G. M. Transposition of cloned P elements into Drosophila germ line chromosomes. Science. 1982 Oct 22;218(4570):341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- Temin H. M. Sex and recombination in retroviruses. Trends Genet. 1991 Mar;7(3):71–74. doi: 10.1016/0168-9525(91)90272-R. [DOI] [PubMed] [Google Scholar]
- Tjian R., Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994 Apr 8;77(1):5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- White S. E., Habera L. F., Wessler S. R. Retrotransposons in the flanking regions of normal plant genes: a role for copia-like elements in the evolution of gene structure and expression. Proc Natl Acad Sci U S A. 1994 Dec 6;91(25):11792–11796. doi: 10.1073/pnas.91.25.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]