Background: Aspergillus fumigatus produces two galactose-containing exopolysaccharides, galactomannan and galactosaminogalactan.
Results: Galactosaminogalactan synthesis requires the UDP-glucose 4-epimerases, Uge5 and Uge3, whereas galactomannan synthesis requires Uge5 alone.
Conclusion: Epimerases in A. fumigatus play both distinct and overlapping roles in exopolysaccharide synthesis.
Significance: Uncovering the biosynthetic pathways of galactosaminogalactan will be crucial in developing therapeutics targeting this exopolysaccharide.
Keywords: Carbohydrate Biosynthesis, Cell Wall, Galactose Metabolism, Glycobiology, Mycology, Polysaccharide
Abstract
The cell wall of Aspergillus fumigatus contains two galactose-containing polysaccharides, galactomannan and galactosaminogalactan, whose biosynthetic pathways are not well understood. The A. fumigatus genome contains three genes encoding putative UDP-glucose 4-epimerases, uge3, uge4, and uge5. We undertook this study to elucidate the function of these epimerases. We found that uge4 is minimally expressed and is not required for the synthesis of galactose-containing exopolysaccharides or galactose metabolism. Uge5 is the dominant UDP-glucose 4-epimerase in A. fumigatus and is essential for normal growth in galactose-based medium. Uge5 is required for synthesis of the galactofuranose (Galf) component of galactomannan and contributes galactose to the synthesis of galactosaminogalactan. Uge3 can mediate production of both UDP-galactose and UDP-N-acetylgalactosamine (GalNAc) and is required for the production of galactosaminogalactan but not galactomannan. In the absence of Uge5, Uge3 activity is sufficient for growth on galactose and the synthesis of galactosaminogalactan containing lower levels of galactose but not the synthesis of Galf. A double deletion of uge5 and uge3 blocked growth on galactose and synthesis of both Galf and galactosaminogalactan. This study is the first survey of glucose epimerases in A. fumigatus and contributes to our understanding of the role of these enzymes in metabolism and cell wall synthesis.
Introduction
In immunosuppressed patients, the mold Aspergillus fumigatus causes an invasive pulmonary infection that can disseminate hematogenously to the brain and other deep organs. In recent years, the incidence of invasive aspergillosis due to A. fumigatus has risen dramatically in patients undergoing immunosuppressive and cytotoxic chemotherapy (1). Although earlier classes of antifungals such as the polyenes and azoles target the fungal plasma membrane, the carbohydrate cell wall is emerging as an effective target for antifungals (2). A better understanding of the composition and biosynthesis of the cell wall and its components will be important in developing new cell wall active antifungals.
The cell wall of A. fumigatus is composed of an inner fibrillar layer and an outer amorphous layer (3). The inner layer is composed of a mesh of chitin, β-1,3-glucans, and associated glycoproteins. The outer layer contains α-1,3-glucans, galactomannan, and galactosaminogalactan. Of note, both galactomannan and galactosaminogalactan contain galactose residues, indicating the importance of fungal galactose metabolism in cell wall biosynthesis.
Galactomannan is a branched glycan consisting of an α-linked mannosyl backbone with branches of 4–5 β-linked galactofuranose (Galf)3 units (4, 5). Galf residues are also found in a variety of other fungal glycoproteins and glycolipids (6, 7). Although the role of galactomannan, and Galf, in virulence remains unclear (6, 8), immunodetection of Galf antigen by EB-A2 antibody is a widely used diagnostic test for invasive aspergillosis (9–11). Galactomannan biosynthesis has been best studied in the nonpathogenic species Aspergillus nidulans. In this species, UDP-glucose is converted to UDP-galactose (UDP-galactopyranose) in the cytoplasm by the UDP-glucose 4-epimerase, UgeA (12). UDP-galactopyranose is in turn modified to UDP-galactofuranose through the activity of the UgmA, an UDP-glucose mutase. UDP-galactofuranose is then transported by the glycosyl transporter, UgtA (13), into the Golgi where the glycosylation of carbohydrates, proteins, and lipids occurs. Although the glycosyltransferases involved in chain formation remain unknown in Aspergillus, galactofuranosyltransferases have been studied in bacteria (14–17) and protozoans (18). Although the A. fumigatus orthologs of ugmA and ugtA, termed ugm1 (glfA) and glfB, respectively, have been studied and found to have a similar function to their A. nidulans orthologs, the A. fumigatus ortholog of the ugeA epimerase has yet to be identified or characterized (5, 8, 19–21).
Galactosaminogalactan is a recently described galactose-containing cell wall polysaccharide that plays an important role in virulence (22, 23). This glycan mediates adhesion to a variety of host substrates and is immunosuppressive both through directly inducing leukocyte apoptosis, as well as by concealing fungal β-1,3-glucan from immune recognition by the pattern recognition receptor Dectin-1 (22, 23). Galactosaminogalactan is a linear heteropolysaccharide composed of varying combinations of α-1,4-linked galactose and N-acetylgalactosamine (GalNAc) (23). The pathways governing synthesis of galactosaminogalactan are largely unknown. Previously, we reported that disruption of uge3 (Afu3g07910), predicted to encode an UDP-glucose 4-epimerase, resulted in a complete absence of cell wall GalNAc and galactosaminogalactan (22). Galf production and the galactose content of the cell wall were not altered in the Δuge3 mutant, suggesting that uge3 encodes a GlcNAc-GalNAc epimerase. The source of the galactose component of galactosaminogalactan remains unknown.
In addition to uge3, the genome of A. fumigatus contains genes predicted to encode two other UDP-glucose 4-epimerases, uge5 (Afu5g10780) and uge4 (Afu4g14090). Although neither of these epimerases has been studied in A. fumigatus, Uge5 is most homologous to the A. nidulans, UgeA, which was previously reported to have UDP-glucose 4-epimerase activity (12). Uge4 shares 55% amino acid homology with Uge3 and 60% with a putative epimerase of Aspergillus niger (An12g10410), but it has not been studied in either organism. We undertook this study to elucidate the role of these three UDP-glucose 4-epimerases in galactose metabolism and the biosynthesis of A. fumigatus cell wall galactose-containing glycans.
MATERIALS AND METHODS
Fungal Strains and Growth Conditions
A. fumigatus strain Af293 (a generous gift from P. Magee, University of Minnesota, St. Paul, MN) was used as the parent wild-type strain for all molecular manipulations. The Δuge3 mutant was described previously (22). Unless otherwise noted, strains were grown and harvested on YPD agar (Fisher) at 37 °C as described previously (24). For growth in liquid medium, Brian medium (23), Aspergillus Minimum Medium (25), and RPMI 1640 medium (Wissent) were used as indicated. For agar plates and liquid medium with galactose as the sole source of carbon, Brian agar and liquid medium were modified to include galactose as the only carbon source. For adherence and apoptosis assays, germinated conidia (germlings) were obtained growing either 1 × 105 or 2 × 105 conidia in either 24-well plates or 1.5-ml microcentrifuge tubes for 9 h at 37 °C, 5% CO2 incubation in phenol-free RPMI 1640 medium.
Molecular and Genetic Manipulations
Deletion of uge4 and uge5 was performed as described previously (22). Briefly, pAN7.1 plasmid was modified for Gateway® (Invitrogen) use by digestion with restriction enzymes BmgBI or NaeI followed by fusion of an attR::ccdB target sequence at the site of each digestion using the Gateway® vector conversion system, to generate plasmids pHY and pYG (22). To generate the disruption constructs, ∼1 kb of the flanking sequences of uge5 was amplified by PCR from Af293 genomic DNA using primers U5-1, U5-2, and U5-3, U5-4 to generate fragments FS1 and FS4, respectively. The resulting PCR products were then cloned into pENTR-D -TOPO® entry plasmid. An LR recombination resulted in recombination of pENTR::FS1 with pHY and pENTR::FS4 with pYG, yielding the fusion of each flanking sequence with the hph cassette in plasmids pHY and pYG. Finally, the DNA fragments for transformation were generated by PCR, using the primers U5–1,HY with pHY::FS1 and U5–4,YG with pYG::FS4. For Δuge4, the same strategy was used as with Δuge5, except that U4-1, U4-2, U4-3, and U4-4 were used as flanking sequence primers to amplify genomic DNA from Af293. A. fumigatus wild-type strain Af293 was then transformed with each pair of disruption cassettes using protoplasting (24). All integrations were confirmed by PCR, and the expected gene expression profile was confirmed by real time RT-PCR for the gene of interest.
For the generation of the Δuge3 Δuge5 double mutant, plasmid p402 was modified for Gateway® use by digestion with BsaBI or BamHI, followed by a mung bean nuclease treatment in the case of BamHI, and then fusion of an attR::ccdB target sequence at the site of each digestion to generate plasmids pBL and pLE. An LR recombination allowed recombination of previously produced pENTR::FS1 with pBL and pENTR::FS4 with pLE, resulting in the fusion of each flanking sequence with the ble cassette in plasmids pBL and pLE. Finally, the DNA fragments for transformation were generated by PCR, using the primers U5-1,BL with pBL::FS1 and U5-4,LE with pLE::FS4. Protoplasts of the A. fumigatus Δuge3 mutant strain (22) were then transformed with each DNA fragment, as described previously (22). Transformants were selected on 0.015% phleomycin-enriched plates. Complete deletion of the uge5 open reading frame was confirmed as described above.
For construction of the Δuge5::uge5 complemented strain, the plasmid pSK485, bearing a pyrithiamine resistance cassette (26) and obtained from the Fungal Genetics Stock Center (27), was modified by PCR using Sbf-pSK and Asc-pSK primers to add unique SbfI and AscI restriction sites upstream of the βrec::trpA cassette. The plasmid was then linearized by SbfI and AscI digestion. A 2.9-kb DNA fragment, corresponding to the restriction site SbfI, 1.2 kb of the uge5 promoter, the entire uge5 ORF, 0.8 kb of the uge5 terminator and the restriction sites AscI, was amplified by PCR using Af293 DNA as a template and the primers U5-1b and U5-5. The resulting PCR product was cloned into the linearized pSK485 plasmid via the Infusion® cloning reaction, following the manufacturer's instructions. The resulting pSK485::uge5 plasmid was linearized at the unique DraI site, located upstream of the uge5 partial promoter, and was used to transform protoplasts of the Δuge5 strain. Transformants were selected on 0.1 μg/ml pyrithiamine-enriched plates, and uge5 expression was confirmed by RT-PCR.
To generate the uge3-gfp construct, the uge3 gene was amplified by PCR using primers uge3-gfp forward and uge3-gfp reverse. This PCR fragment was then cloned into the pGFP plasmid, as described previously, and digested with NcoI and NotI (28, 29). The resulting plasmid, designated puge3-GFP, was used to transform wild-type A. fumigatus Af293 as described previously (30). Transformants were selected by phleomycin resistance, and uge3-gfp expression was verified using confocal microscopy (IX81, Olympus), excitation wavelength 495 nm and emission wavelength 519 nm.
To generate the uge5-rfp construct, uge5 was amplified by PCR using primers uge5-rfp forward and uge5-rfp reverse. This fragment was then cloned in-frame with mRFP1 in the pRFP-HYG plasmid using EcoRV (31). The resulting plasmid pRFP-Uge5-HYG was used to transform wild-type A. fumigatus Af293. Transformants were selected using hygromycin, and uge5-rfp expression was verified using confocal microscopy (IX81, Olympus), excitation 543 nm and emission wavelength 563 nm.
For His6-Uge3, the ORF of uge3 was produced by PCR with an additional CACC at the 5′ end, using the primers U3-start ORF, U3-end ORF, and the Af293 genomic DNA as template. The resulting PCR product was then cloned into pENTR-D-TOPO® entry plasmid. An LR recombination allowed the recombination of pENTR::U3-ORF with pDest17®, a plasmid engineered to produce His6 proteins with a high rate in proper Escherichia coli strains. The resulting pDest17::uge3 plasmid was transformed in E. coli BL21 DE3 strain. All primer sequences are listed in Table 1.
TABLE 1.
List of primers for plasmid construction and gene expression experiments
| Primer name | Target | Sequence |
|---|---|---|
| U5-1 | uge5 5′-flanking sequence | CACCTTCAAGCGGAACTGGAC |
| U5-2 | uge5 5′-flanking sequence | AAAGGAAGCAGAAGAGCAGAAAGAA |
| U5-3 | uge5 3′-flanking sequence | CACCTGTGAGCGAGAAAGACTGGAAA |
| U5-4 | uge5 3′-flanking sequence | ACATGAAGCAATTAGAGGCAGCA |
| U5-ext 1 | Analysis of uge5 locus | CTTAGTGGACAGCAGACCAGGGG |
| U5-ext 4 | Analysis of uge5 locus | GCGAGAGCCTGACCTATTGCATCT |
| uge5 RT-sense | uge5 cDNA | ATGAGGCCGAGAAGTGGAAC |
| uge5 RT-antisense | uge5 cDNA | CGTGAGAGGCATAGTCGTCA |
| U5-1b | uge5 transcription unit | TACCTGCAGGGTGACATTGATGACGGA |
| U5-5 | uge5 transcription unit | TGGCGCGCCGACAAATATCCAAACGGTA |
| U4-1 | uge4 5′-flanking sequence | CACCGTCGGCTACATTCGT |
| U4-2 | uge4 5′-flanking sequence | AAGGGTTCGCAGTCATCCTC |
| U4-3 | uge4 3′-flanking sequence | CACCAATCTCACAGCTAACG |
| U4-4 | uge4 3′-flanking sequence | CGCAGACATACCACTTCTTG |
| U4-ext 1 | Analysis of uge5 locus | GGCCCCAAAATCAGGAGT |
| U4-ext 4 | Analysis of uge5 locus | ATCGCATCTACGCCATGATT |
| uge4 RT-sense | uge4 cDNA | CATCCACACCCCTCTGAAGT |
| uge4 RT-antisense | uge4 cDNA | CGGACGAGGAGAAGATGAAG |
| U3-start ORF | His6-Uge3 production | CACCATGGACAGCTACCAGCAATC |
| U3-end ORF | His6-Uge3 production | AAGGGACATGCGACAACATC |
| HY | hph | CAACCACGGCCTCCAGAAGAAGA |
| YG | hph | GCGAGAGCCTGACCTATTGCATCT |
| tef1-RT sense | tef1 | CCATGTGTGTCGAGTCCTTC |
| tef1-RT antisense | tef1 | GAACGTACAGCAACAGTCTGG |
| Sbf-pSK | pSK485 modification | AGCTTCCTGCAGGTAAATCAAAAGAATAGACCGAGATA |
| Asc-pSK | pSK485 modification | TGGCGCGCCTAAGGGATTTTGCCGATTTC |
| Uge5-RFP forward | Campoli et al. (31) | CATCACCCCATGGATATGTCTGCTGGTTCAGTT |
| Uge5-RFP-reverse | Campoli et al. (31) | GGAGGAGGCCATGATCTTCTTGAGCTGTTCCAG |
| Uge3-gfp forward | Choe et al. (29) | AGACATCACCCCATGGATGGACAGCTACCAGCA |
| Uge3-gfp reverse | Choe et al. (29) | CTCACCATCGCGGCCGCAGTAGATAACCCACTGA |
Real Time RT-PCR
Expression of the genes of interest was quantified by relative real time RT-PCR analysis as described previously (32). The primers used for each of the genes are shown in Table 1 or in Gravelat et al. (22). First strand synthesis was performed from total RNA with the QuantiTec reverse transcription kit (Qiagen) using random primers. Real time PCR was then performed using an ABI 7000 thermocycler (Applied Biosystems). Amplification products were detected with the Maxima® SYBR Green qPCR system (Fermentas). Fungal gene expression was normalized to A. fumigatus tef1 expression. To verify the absence of genomic DNA contamination, negative controls were used for each gene set in which reverse transcriptase was omitted from the mix.
Localization Studies
Conidia from uge3-gfp, uge5-rfp, or A. fumigatus Af293 strains were grown for 9 h at 37 °C on coverslips in a 24-well polystyrene plate in RPMI 1640 medium without phenol. Young hyphae were washed in PBS, stained with Draq® nuclear staining at 1:100 dilution (Cell Signaling, Inc), mounted in Slow Fade® Gold Antifade (Invitrogen), and imaged under a confocal microscope (IX81, Olympus) at excitation 495, 543, and 633 nm, respectively.
Cell Culture Assays
Type II pneumocyte cell line CCL-185 (lung epithelial cells A549) and murine bone marrow-derived dendritic cells were cultured as described previously (22).
Aspergillus adherence to A549 cells was determined by co-incubating germlings of the strain of interest on a monolayer of A549 cells for 30–45 min, as described previously (24). To determine induction of apoptosis via caspase-3 activity, germlings of each strain were co-incubated with bone marrow-derived dendritic cells at a multiplicity of infection of 10:1 for 3 h. Caspase-3 activity was measured using EnzChek® Caspase-3 assay kit following the manufacturer's instructions (Invitrogen). Samples containing fungus or bone marrow-derived dendritic cells alone were included as controls.
Polysaccharide Analysis
Galactosaminogalactan and Galf productions were assayed as described previously, with minor modifications (22, 23). Briefly, 4 × 106 conidia were grown in modified Brian medium for 72 h. Culture supernatants were filtered, and extracellular galactosaminogalactan or galactomannan was precipitated by 2.5 or 4.0 volumes of ethanol, respectively. Galactosaminogalactan composition was determined by gas chromatography after hydrolysis, reduction, and peracetylation with meso-inositol as internal standard. Total neutral hexoses were quantified by the phenol sulfuric assay. Galf quantification was assayed by enzyme immunoassay using the Platelia® Aspergillus kit (Bio-Rad), following the manufacturer's instructions. For quantification of β-1,3-glucan exposure, 1 × 105 conidia were grown in Brian medium for 12 h in a 96-well opaque bottom plate (Nunclon, Inc.), fixed in 4% paraformaldehyde, and then labeled with 10 μg/ml Fc-Dectin-1 (a generous gift from Dr. G. D. Brown, University of Aberdeen, Aberdeen, UK), followed by FITC-labeled AffiniPure F(ab′) fragment donkey anti-human IgG, FCy fragment-specific (Jackson ImmunoResearch). Fluorescence was measured at 495 nm excitation and 515 nm emission using Spectramax® fluorescence microplate reader (Molecular Devices, Inc.). The biofilm adherence assay and scanning electron microscopy were performed as described previously (22, 24).
His-tagged Uge3 Extraction and Purification
Protein expression of His6-Uge3 was performed in E. coli BL21(DES) in autoinduction medium supplemented with 100 μg/ml ampicillin (33). Cells were grown for 20–24 h at 28 °C and harvested by centrifugation. Pellets were flash-frozen and then lysed and filtered through a 0.2-μm nylon membrane filter and incubated for 1.5 h in Ni2+-agarose beads (Qiagen). After successive washing with increasing imidazole concentrations, bound His6-Uge3 was eluted with 250 mm imidazole. Fractions containing His6-Uge3 determined by SDS-PAGE Coomassie Blue staining were pooled, concentrated, and quantified with either Bradford assay or NanoDrop®. Alternatively, after cell lysis, His6-Uge3 was purified through metal affinity chromatography on a POROS-MC20 perfusion chromatography column with Ni2+ as chelating metal. Elution fractions were pooled and further purified on a preparative scale high resolution Superdex-200 gel filtration column. Purification of His6-Uge3 was validated though Western blotting using HRP-tagged anti-His6 (Abcam) and protein mass spectrometry. Enzymes were stored at −20 °C in PBS supplemented with 25% glycerol and 0.5 mm DTT.
Enzyme Activity Assays
Product Formation
Evolution of product was detected using NMR by incubating 5 μg of His6-Uge3 with 1 mm of either UDP-glucose or UDP-GlcNAc (Sigma) in a reaction mix of 250 μl for 1 h at 37 °C. No co-factor was added because preliminary experiments did not show any changes to the reaction rate by adding co-factors such as NAD+, Mg2+, or Ca2+ (data not shown), as similarly reported in other epimerases (34–36). All reagents were in D2O (Calbiochem), and spectra were recorded at 25 °C with acetone internal reference (2.23 ppm) using standard pulse COSY, TOCSY (mixing time 120 ms), and 1H,31P HMQC. NMR experiments were performed on Varian INOVA® 500 MHz spectrometer with a 3-mm gradient probe. Spectra assignment was performed using Bruker Topspin Version 3.1 program for spectra visualization.
Enzyme Kinetics
The rates of product formation, linearity, and kinetics studies were performed using Beckman gold capillary electrophoresis with a 57-cm bare silica capillary, and 32 Karat software application, as described previously (37). Concentrations of substrates, ranging from 0.2 to 5.0 mm for UDP-GlcNAc and from 0.01 to 1 mm for UDP-galactose, were used to obtain product formation data in the presence of 0.25 or 0.53 pmol of Uge3 for each substrate. For kinetics study, enzyme with varying concentrations of substrates in a 10-μl reaction volume of 0.1 m Tris/HCl buffer at pH 8.0 was incubated at 37 °C for the specified amount of time, quenched by boiling at 95 °C for 5 min, and immediately stored at −80 °C. Prior to incubation, all reagents and enzymes were kept at 4 °C. Electropherogram peaks were integrated using 32 Karat software application to estimate substrate conversion. Vmax values of the reactions were calculated by taking the reciprocal of the Y-intercept, and Km values of the reactions were calculated by taking the slope over the Y-intercept (data not shown).
Bioinformatics Analysis
Annotation and Homology
UDP-glucose 4-epimerase genes and their respective amino acid sequences were retrieved from the Aspergillus Genome Database (38). The amino acid sequences of each of the candidate epimerases were analyzed using Eukaryotic Linear Motif (39), ConSurf (40), Conserved Domain Database (41), and HHpred (42), including SCOP domains. Clusters and protein families by domain predictions were cross-referenced using InterPro (43, 44) and Pfam (45). Furthermore, each of the Aspergillus epimerases were matched with the closest protein entry available in the Protein Data Bank (46) and further annotated.
Homology Structural Modeling
The amino acid sequence of Uge3 was aligned with human GalE PDB code 1HZJ (47), Trypanosoma brucei GalE PDB code 1GY8 (48), or Pseudomonas aeruginosa WbpP PDB code 1SB8 (49) using ClustalW (50). Aligned Uge3 sequences were then modeled against each of the template structures using Modeler Version 9.11 (51). Resulting models were verified using Pairwise Structure Alignment (46). MacPyMOL Version 1.3 (academic license, Schrodinger LLC) was used to align respective structures and identify and analyze key residues in the catalytic site.
Statistical Analysis
All charts and graphs were produced and analyzed using Prism 6 (GraphPad software). All tables were created using MS Excel (Microsoft Inc.).
RESULTS
Uge5 Is the Most Highly Expressed Epimerase Gene in A. fumigatus and Is Required for Normal Galactose Metabolism
We first performed expression analysis of the three epimerase genes in wild-type A. fumigatus grown in Brian medium (Fig. 1). Of the three epimerase-encoding genes, uge5 was the most highly expressed, at a level more than 5-fold higher than that of uge3. Expression of uge4 was minimal, approaching the limits of detection by real time RT-PCR. We therefore hypothesized that Uge5 likely plays an important role in galactose metabolism and cell wall glycan synthesis.
FIGURE 1.
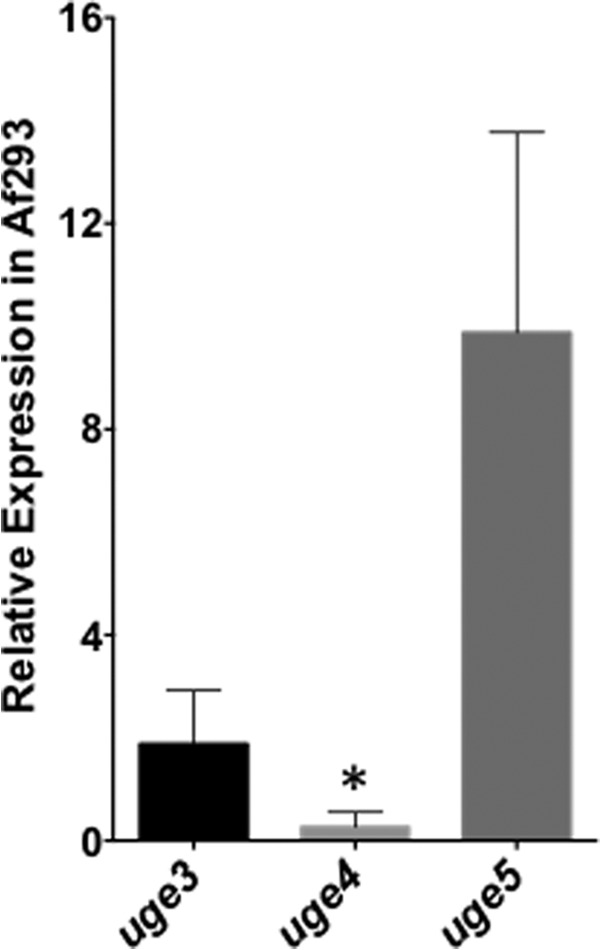
uge5 gene is the most highly expressed of the three epimerase genes in A. fumigatus, whereas uge4 mRNA expression is barely detectable. Strain Af293 was grown in Brian medium for 18 h, and the levels of uge3, uge4, and uge5 mRNA were measured by real time RT-PCR. *, indicates significantly different expression as compared with tef1 reference gene (22).
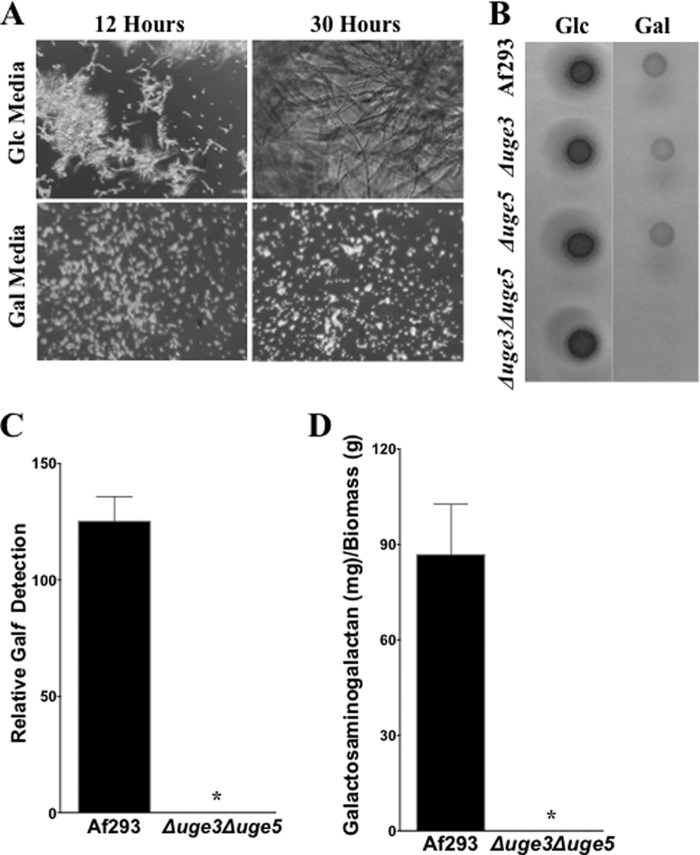
To test the role of each of these three epimerases in galactose metabolism, Δuge4 and Δuge5 A. fumigatus deletion mutants were constructed. The ability of these mutants to utilize glucose and galactose was compared with the previously constructed Δuge3 mutant and wild-type A. fumigatus strains (22). All three mutant strains exhibited wild-type growth on medium with glucose as the sole carbon source (Fig. 2, A–E). Similarly, both the Δuge3 and Δuge4 mutants grew normally on medium with galactose as a sole carbon source (Fig. 2, F–H). Deletion of uge5 resulted in a marked impairment of growth in medium containing galactose as a sole carbon source (Fig. 2, I–J), consistent with the findings reported with the deletion of ugeA, the A. nidulans ortholog of uge5 (12). However, unlike the A. nidulans ΔugeA mutant, the Δuge5 mutant was not completely blocked in hyphal growth under these conditions and was able to grow and form hyphae after 30 h of growth in galactose (Fig. 2N). Collectively, these results suggest that A. fumigatus differs from A. nidulans in that whereas uge5 is the major epimerase responsible for the interconversion of UDP-galactose and UDP-glucose, other pathways or enzymes in A. fumigatus can mediate galactose metabolism in the absence of Uge5.
FIGURE 2.
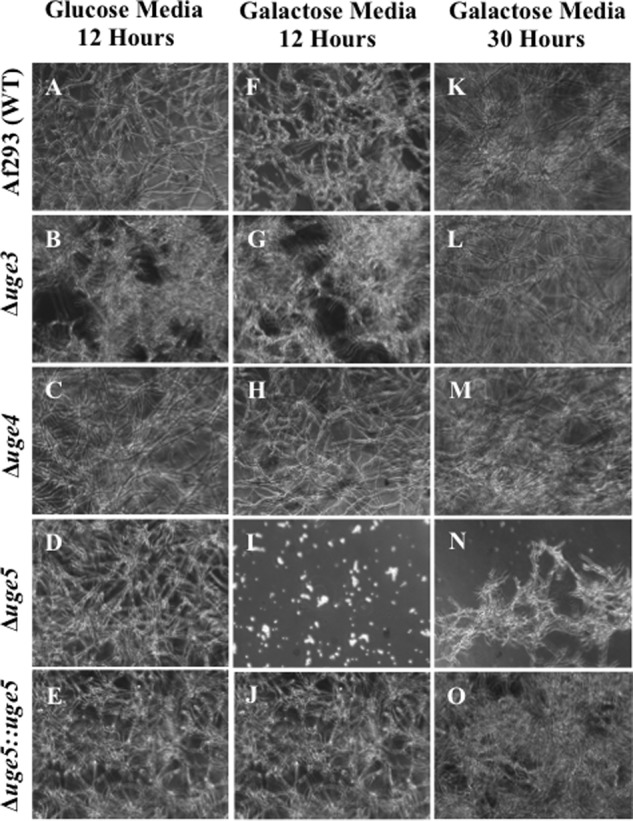
Deletion of uge5 results in a partial galactose auxotrophy. The indicated strains were grown for the indicated time periods in Brian medium with either glucose (A–E) or galactose (F–O) as the sole carbon source. Bright field images at a magnification of ×200 are shown.
Deletion of Uge5 Blocks Galf Synthesis and Results in the Production of Galactosaminogalactan with Reduced Galactose Content
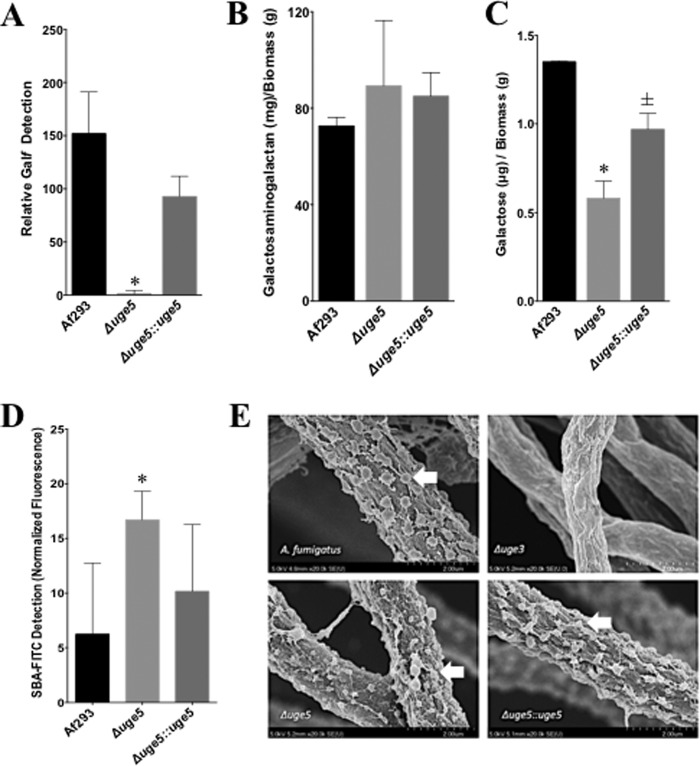
To test the contribution of each of the three epimerases to the synthesis of galactose-containing glycans, we measured the production of galactosaminogalactan and Galf by each of these mutant strains. We previously found that deletion of uge3 results in a complete block in galactosaminogalactan synthesis but had no effect on Galf synthesis (22). Deletion of uge4 had no effect on Galf detection or galactosaminogalactan production (Fig. 3, A and B). Consistent with reports of ugeA deletion in A. nidulans (12), deletion of uge5 resulted in the absence of detectable Galf antigen and, by extension, the absence of galactomannan (Fig. 4A). Unexpectedly, however, multiple assays demonstrated that deletion of uge5 did not block galactosaminogalactan synthesis. Scanning electron microscopy of the Δuge5 mutant identified normal production of the cell wall decorations that have been associated with galactosaminogalactan production (Fig. 4E) (22), and levels of total galactosaminogalactan produced by the Δuge5 mutant were slightly higher than those seen with the wild type, although this was not statistically significant (Fig. 4B). Compositional analysis of galactosaminogalactan from the Δuge5 mutant revealed a significant reduction in the galactose content of this heteropolysaccharide (Fig. 4C). This decrease in galactose was not associated with an increase in other hexose such as glucose or mannose (data not shown). The Δuge5 mutant also exhibited increased staining with the GalNAc-specific soybean agglutinin lectin (Fig. 4D), consistent with the production of GalNAc-rich, galactose-poor galactosaminogalactan. Collectively, these data suggest that although Uge5 activity is required for UDP-Galf synthesis for the production of galactomannan, other enzymes or pathways can also contribute UDP-galactose to the synthesis of galactosaminogalactan in the absence of Uge5 activity.
FIGURE 3.
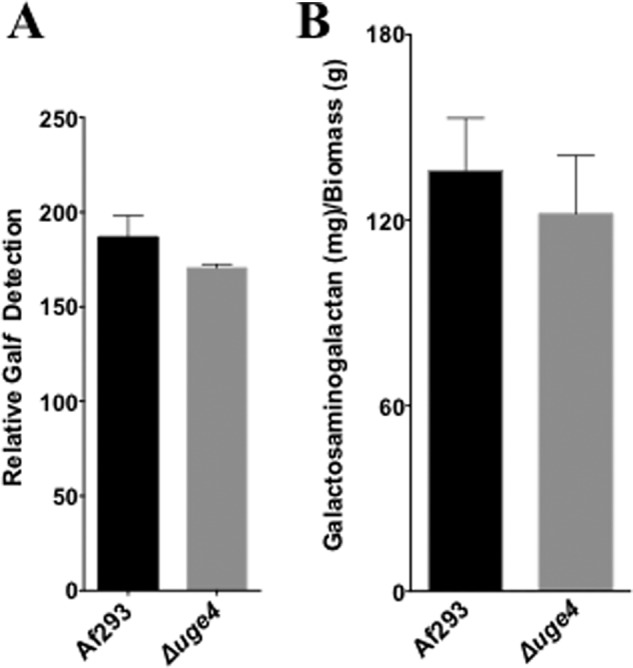
Deletion of uge4 does not alter galactosaminogalactan or Galf production. A, relative Galf detection by ELISA (EB-A2) in culture supernatant of Af293 or Δuge4 mutant after 72 h growth of indicated strains in Brian medium. B, galactosaminogalactan production by biomass from culture supernatant of Af293 or Δuge4 mutant after 72 h growth of the indicated strains in Brian medium.
FIGURE 4.
Deletion of uge5 blocks Galf synthesis and results in production of galactosaminogalactan with a reduced galactose content. A, Galf content of culture supernatants as determined by ELISA. B, galactosaminogalactan content of culture supernatants, normalized to mycelia biomass. C, galactose content of galactosaminogalactan from the indicated strains as detected by gas chromatography followed by hexose and hexosamine quantification. D, FITC-tagged GalNAc-specific soybean agglutinin (SBA) lectin binding on 12-h, Brian medium-grown hyphae of indicated strains. Total fluorescence was measured with Spectramax® fluorescence microplate reader. E, scanning electron micrograph of hyphae of indicated strains after 24 h of growth at 37 °C, 5% CO2 in phenol-free RPMI 1640. Hyphae were fixed, sequentially dehydrated in ethanol, dried in CO2, coated in Pd-Au, and imaged under scanning electron microscope (Hitachi). Arrows indicate surface decorations associated with galactosaminogalactan production. *, significant reduction compared with Af293 wild-type, analysis of variance with pairwise comparison p < 0.05. ±, not statistically significant compared with Af293 but statistically significant compared with the Δuge5 mutant, p < 0.05. A–C, indicated strains were grown for 72 h in Brian medium.
Uge3 Activity May Compensate for the Lack of Uge5
The Δuge5 mutant was able to utilize galactose as a carbon source and produced galactosaminogalactan that still contained galactose. These data suggest that this strain retained some glucose/galactose epimerase activity, possibly mediated by Uge4 or Uge3. Although disruption of uge4 had no effect on galactose metabolism, galactosaminogalactan or Galf production, uge4 was only expressed at very low levels in the wild-type strain of A. fumigatus, and therefore, it could be up-regulated in the absence of Uge5. To test for compensatory up-regulation of uge4 in the absence of Uge5, we performed real time RT-PCR analysis of uge4 expression in this mutant. The expression of uge4 remained minimally detectable in the Δuge5 mutant under galactosaminogalactan-inducing conditions (Fig. 5). In contrast, not only was uge3 expression detectable in wild-type A. fumigatus, but a trend toward increased uge3 expression was observed in the absence of Uge5 (Fig. 5). Collectively, these data suggest that Uge3 may have dual substrate specificity and can mediate the interconversion of both UDP-glucose to UDP-galactose and UDP-GlcNAc to UDP-GalNAc. Consistent with this model, expression of uge3-gfp or uge5-rfp in wild-type A. fumigatus demonstrated that both of these epimerases are located in the cytoplasm (Fig. 6), and thus could provide galactose to the same downstream glycosyltransferases or mediate conversion of galactose to glucose for metabolic use.
FIGURE 5.
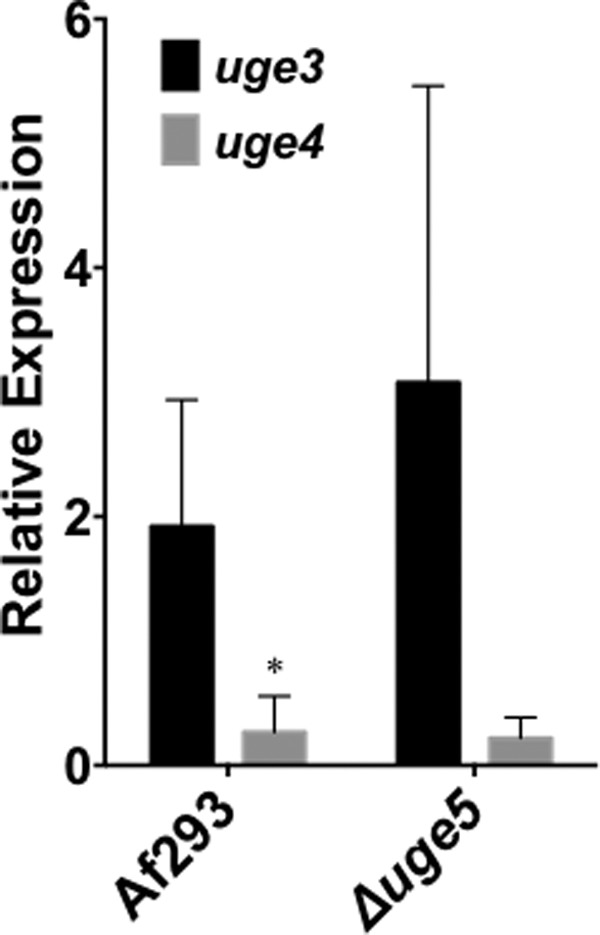
Deletion of uge5 is not associated with significant up-regulation of expression of uge3 or uge4. The indicated strains were grown in Brian medium for 18 h, and the levels of uge3 (black bar) and uge4 (gray bar) mRNA from indicated strains were measured by real time RT-PCR. *, significant difference between uge3 and uge4 expression in the indicated strain as compared with tef1 reference gene, factorial analysis of variance with pairwise comparison, p < 0.05.
FIGURE 6.
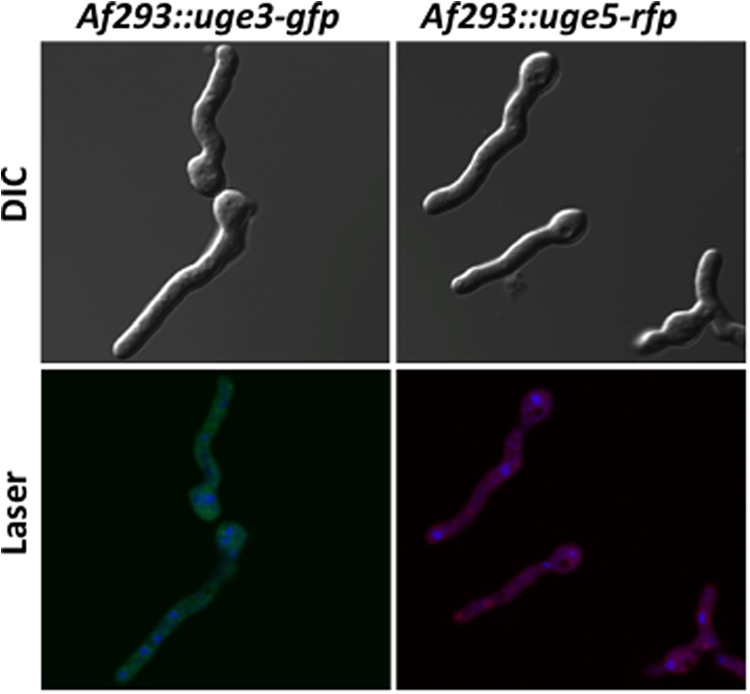
Uge3 and Uge5 are cytoplasmic. Af293 strains expressing uge3-gfp or uge5-rfp were grown in Brian medium for 12 h and imaged by confocal microscopy. For nuclear staining, Draq5® stain was used, and pseudocolor blue was red fluorescent protein at 543 nm, and Draq5® at 633 nm.
Homology Modeling Suggests Uge3 Is a Group 2 Epimerase with Dual Substrate Specificity
To examine the possibility that Uge3 could exhibit dual substrate binding and catalysis, the structure of Uge3 was compared with those of other UDP-glucose 4-epimerases by homology modeling. UDP-glucose 4-epimerases are composed of three groups based on substrate specificity (52) as follows: group 1 enzymes have specificity for hexoses, group 2 for both hexoses and hexosamines, and group 3 for hexosamines. Uge3 was therefore modeled using T. brucei tGalE (PDB code 1GY8) as a group 1 template, human hGalE (PDB code 1HZJ) as a group 2 template, and P. aeruginosa WbpP (PDB code 1SB8) as a group 3 template. The N-terminal portion of Uge3 encoding a predicted signal peptide was omitted from modeling. Inspection of amino acid residues in the catalytic sites of the Uge3 model and templates revealed similarities and differences in the predicted substrate-binding pocket region that may provide further insights to Uge3's catalytic activity. Similar to tGalE, hGalE, and WbpP, the SYK triad required for catalytic activity is conserved in the Uge3 model (Ser191, Tyr238, and Lys242) (49, 53). Previously, six amino acid residues forming the hexagonal substrate-binding pocket were identified to be important in enzymatic function (49, 52). Identification of the corresponding residues on Uge3 revealed that these residues are not only present in Uge3, but that they are identical to hGalE, which belongs to the bifunctional group 2 (Table 2). In fact, these six residues in Uge3 and hGalE align almost in a complete overlap around the cofactor and substrate (Fig. 7). Thus, the in silico analysis suggests that Uge3 is structurally more closely related to the group 2 epimerase hGALE and may play a role in the interconversion of both UDP-Glc/UDP-Gal, as well as UDP-GlcNAc/UDP-GalNAc.
TABLE 2.
Alignment of the six amino acid residues within the catalytic fold of Uge3, hGalE, tGalE, and WbpP shows highest alignment between Uge3 and hGalE
Six amino acid residues in the catalytic pocket were identified on the Uge3 model. These residues were then compared with those in hGalE, tGalE, and WbpP. Visualization was performed using MacPyMOL Version 1.3.
| Species | Enzyme | Residues | Group | |||||
|---|---|---|---|---|---|---|---|---|
| A. fumigatus | Uge3 | Lys151 | Ser191 | Tyr238 | Asn268 | Asn288 | Cys391 | 2 |
| H. sapiens | hGalE | Lys92 | Ser132 | Tyr157 | Asn187 | Asn206 | Cys307 | 2 |
| T. brucei | tGalE | Leu102 | Ser142 | Tyr173 | Asn202 | His202 | Leu342 | 1 |
| P. aeruginosa | WbpP | Gly102 | Ser142 | Tyr166 | Asn195 | Ala209 | Ser306 | 3 |
FIGURE 7.
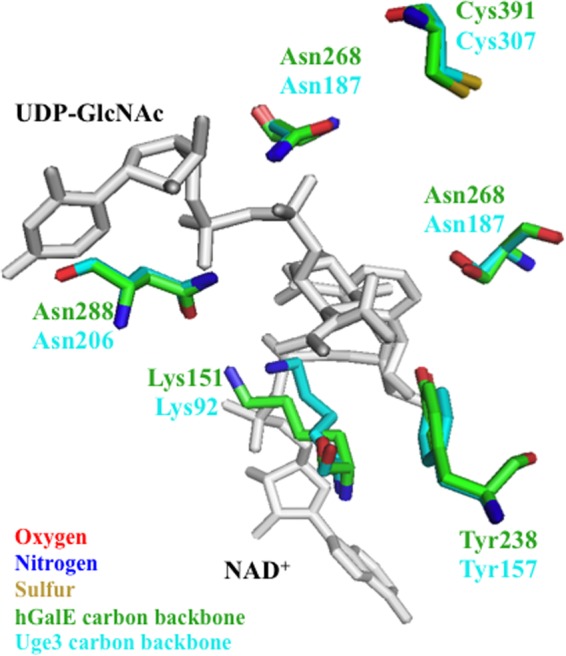
Substrate-binding pocket of Uge3 model aligns with hGalE. The hexagonal substrate-binding pocket of Uge3 model (green) was aligned to hGalE (cyan), with the six residues required for catalytic activity annotated. For Uge3 homology modeling, the N-terminal nonalignment regions were discarded, and resulting aligned sequences were modeled using Modeler Version 9.11. Bound UDP-GalNAc and NAD+ are indicated in gray at top and bottom, respectively.
Uge3 Is a Dual Substrate Epimerase That Can Utilize Both UDP-glucose and UDP-N-acetylglucosamine as Substrates
To test if Uge3 has dual substrate activity, recombinant Uge3 was produced (Fig. 8, A and B) and its enzymatic activity measured. Using 1H NMR, product formation of both UDP-galactose and UDP-GalNAc was detected in the presence of Uge3 when either UDP-glucose or UDP-GlcNAc was provided as a substrate, respectively (Fig. 8, C and D, and Table 3). To obtain better resolution of the peaks, COSY and TOCSY two-dimensional 1H NMR experiments were performed and demonstrated that the cross-peaks of coupled protons show both substrate and product chemical shifts in the respective reaction mixes (data not shown). To further validate that the products retained their UDP moieties and were not derivatives or different species of galactose or GalNAc, 31P NMR was performed and detected the same phosphorus chemical shifts in both respective substrate and product proton peaks (data not shown). Uge3 activity was further characterized using capillary electrophoresis. At steady-state equilibrium, Uge3 converted 30% of UDP-GlcNAc to UDP-GalNAc and 15% of UDP-glucose to UDP-galactose (Fig. 8E). The range of reaction time that resulted in linearity of product formation of less than 10% for Michaelis-Menten analysis was determined to be between 0 and 30 min for both substrates (data not shown). Conversion from UDP-GlcNAc to UDP-GalNAc had Km of 3.2 mm and Vmax of 417 pmol/min (data not shown). Epimerization from UDP-glucose to UDP-galactose could not be quantified due to a strong reverse reaction; however, conversion of UDP-galactose to UDP-glucose had a Km of 0.9 mm and Vmax of 146 pmol/min (data not shown). Collectively, these results support the hypothesis that the galactose component of galactosaminogalactan in the Δuge5 mutant strain likely originates from activity of Uge3.
FIGURE 8.
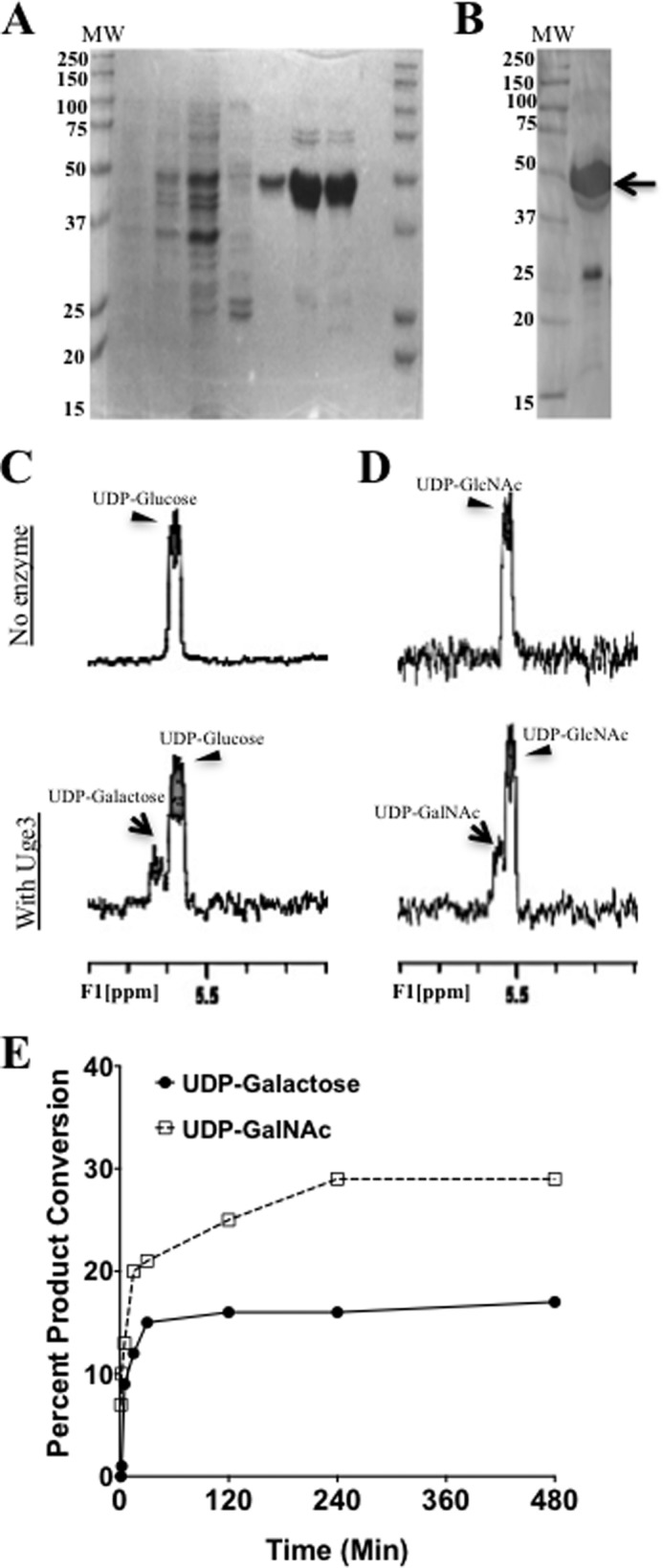
Uge3 exhibits bifunctional UDP-Glc/UDP-Gal and UDP-GlcNAc/UDP-GalNAc epimerase activity. A, SDS-PAGE of lysate and purified His6-Uge3 stained with Commassie Blue from His6-Uge3 expressing BL21(DE) E. coli strain. Lanes are as follows: noninduced cells; cells grown in auto-induction medium; crude lysate; wash fraction eluted using 20 mm imidazole; and four fractions of His6-Uge3 eluted with 250 mm imidazole. B, Western blot of purified lysates for detection of His6-Uge3 using HRP-tagged anti-His6 antibody. Arrow indicates expected band for His6-Uge3. C and D, 1H NMR spectra were measured in a reaction mix containing either UDP-glucose (C) or UDP-GlcNAc (D) in phosphate buffer, in the presence or absence of Uge3. For all NMR experiments, products were detected after 1 h of co-incubation of Uge3 and respective substrates at 37 °C using Varian 500 MHz NMR spectroscopy. E, rate of product formation was measured using capillary electrophoresis over time in a reaction mix containing 20 pmol of Uge3 and 0.1 mm of either UDP-glucose or UDP-GlcNAc as substrate. Reactions took place at 37 °C in a total volume of 10 μl. Products were detected at 254 nm (UV) measuring UDP-moiety endogenous fluorescence. UDP-linked sugars were separated by borate adduct formation.
TABLE 3.
1H NMR chemical shift and constant for product and substrate
| Unit | H/C 1 | H/C 2 | H/C 3 | H/C 4 | H/C 5 | H/C 6 |
|---|---|---|---|---|---|---|
| UDP-Glc | 5.6 | 3.53 | 3.78 | 3.47 | 3.9 | 3.78; 3.86 |
| JHn,Hn+1 | 3 | 9 | 9 | 9 | ||
| UDP-Gal | 5.64 | 3.8 | 3.92 | 4.03 | 4.17 | 3.74; 3.77 |
| JHn,Hn+1 | 3 | 9 | <2 | <2 | ||
| UDP-GlcNAc | 5.51 | 3.99 | 3.81 | 3.55 | 3.93 | 3.87; 3.93 |
| JHn,Hn+1 | 3 | 9 | 9 | 9 | ||
| UDP-GalNAc | 5.55 | 4.26 | 3.97 | 4.05 | 4.19 | 3.76; 3.78 |
| JHn,Hn+1 | 3 | 9 | <2 | <2 |
Mutant Deficient in Both Uge3 and Uge5 Is Completely Auxotrophic for Galactose and Produces No Galactomannan or Galactosaminogalactan
To verify that Uge3 activity is responsible for the residual galactose metabolism and galactose content of galactosaminogalactan in the Δuge5 mutant strain, we constructed a mutant deficient in both Uge3 and Uge5. The Δuge3Δuge5 double mutant was unable to grow in medium containing galactose as the sole carbon source (Fig. 9, A and B). Furthermore, the Δuge3Δuge5 double mutant was found to have undetectable levels of both Galf and galactosaminogalactan (Fig. 9, C and D). Collectively these data confirm that Uge3 and Uge5 are the only functional UDP-glucose/galactose epimerases in A. fumigatus.
FIGURE 9.
Deletion of uge3 and uge5 renders the resulting Δuge3Δuge5 double mutant deficient in Galf and galactosaminogalactan. A, indicated strains were grown for the indicated time periods in Brian medium using either glucose or galactose as the sole carbon source. Brightfield images at a magnification of 200x are shown. Glucose is abbreviated as Glc, and galactose as Gal. B, indicated strains were grown for 2 days on either glucose or galactose based Brian medium agar plates. C, relative Galf detection by ELISA in culture supernatant after 72 h growth in Brian medium of the indicated strains. D, galactosaminogalactan production after 72 h growth in Brian medium of the indicated strains. *, significant reduction compared with Af293 wild type and analysis of variance with pairwise comparison, p < 0.05.
Reducing the Galactose Component of Galactosaminogalactan Does Not Impair Its Function
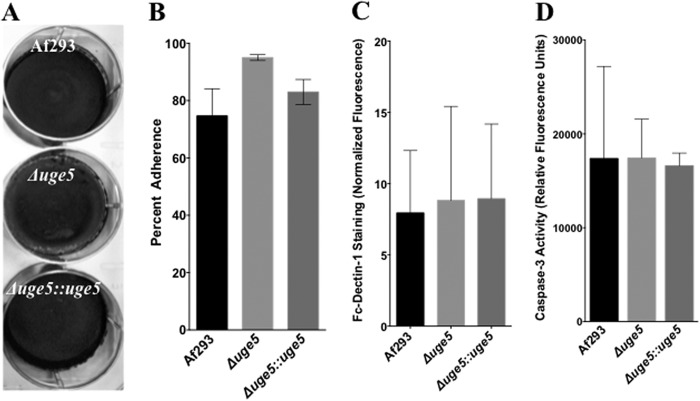
To assess whether the galactose-poor galactosaminogalactan produced by the Δuge5 mutant is altered in function, we characterized the effects of uge5 deletion on the reported functions of galactosaminogalactan (22, 23). Deletion of uge5 did not alter biofilm adherence to the polystyrene surface (Fig. 10A). Furthermore, Δuge5 mutant displayed slightly increased adherence to A549 epithelial cells as compared with wild-type A. fumigatus and the uge5-complemented strain, possibly reflecting the slight increase in total galactosaminogalactan production noted in this strain (Fig. 10B). Similarly the reduced galactose content of galactosaminogalactan did not impair β-(1,3)-glucan masking, as there was no difference between these three strains in the immunodetection of β-(1,3)-glucan by recombinant Fc-dectin-1 (Fig. 10C). Finally, because leukocyte apoptosis has been reported as a mechanism of galactosaminogalactan immunosuppression (23), the ability of the Δuge5 mutant to induce apoptosis of bone marrow-derived dendritic cells was quantified by measuring cellular caspase-3 activity. No difference in the induction of apoptosis by the Δuge5 mutant as compared with wild-type A. fumigatus (Fig. 10D) was observed. Collectively, lowering the galactose content of galactosaminogalactan did not seem to significantly impair adherence, β-(1,3)-glucan masking, or the induction of apoptosis by this glycan.
FIGURE 10.
Galactose-poor Δuge5 galactosaminogalactan retains normal virulence-associated functions. A, biofilm adherence of indicated strains after 24 h of growth on polystyrene plates, after several washes, and visualized by staining with crystal violet. B, adherence of germinated hyphae of the indicated strains to A549 epithelial cells after 30 min. C, detection of Fc-dectin-1 binding using immune staining. Conidia from respective strains were grown for 12 h in Brian medium, fixed, blocked, stained with Fc-dectin-1, and FITC-labeled F(ab) fragment, and total fluorescence was measured with Spectramax® fluorescence microplate reader. D, induction of bone marrow-derived dendritic cell apoptosis as determined by caspase-3 activity. Conidia from indicated strains were grown for 9 h in RPMI 1640 medium and then co-incubated with mouse bone marrow-derived dendritic cells at a multiplicity of infection of 10:1. Caspase-3 activity was measured by commercial assay following the manufacturer's instructions (Invitrogen).
DISCUSSION
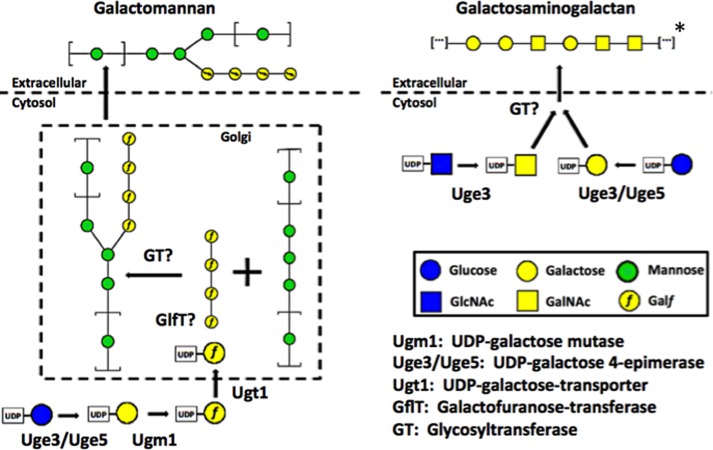
The results of our studies highlight important differences in galactose metabolism, glycan synthesis, and epimerase function between A. fumigatus and A. nidulans. Although deletion of ugeA in A. nidulans resulted in a strain that was completely auxotrophic for galactose, the A. fumigatus Δuge5 mutant exhibited only a partial growth defect in galactose-containing medium. In light of our findings that Uge3 has dual substrate specificity and that other putative epimerases are silent in the Δuge5 mutant, it is likely that Uge3 mediates the interconversion between UDP-glucose and UDP-galactose in the absence of Uge5 (Fig. 11). Although the genome of A. nidulans contains ugeB, an ortholog of uge3, this gene has been reported to be silent (55) and as a result likely does not contribute to UDP-galactose/UDP-glucose interconversion in this species.
FIGURE 11.
Schematic of galactosaminogalactan and galactomannan pathways. Pathway diagrams showing common and distinct components in the biosynthesis of galactosaminogalactan and galactomannan. For Galf symbol representation, the “f” has been inserted to the galactose pyranose symbol. All other symbols follow common nomenclature convention (54). *, note that the depiction of galactosaminogalactan structure is representative of multiple potential combinations of galactose and GalNAc residues.
Disruption of uge5 was associated with normal to increased galactosaminogalactan production and adherence to host cells. These data are consistent with those from mutations in other components of the galactomannan biosynthetic pathways. For example, deletion of ugm1 in A. fumigatus resulted not only in an absence of galactomannan but also an increase in galactosaminogalactan synthesis and biofilm adherence (6). Although galactosaminogalactan production has not been studied in A. nidulans, it was reported that the deletion of ugmA and ugeA was associated with an increase in adherence (55), suggesting a similar phenomenon may also occur in this organism. One possibility is that impaired galactomannan synthesis results in accumulation of precursors that are then redirected to the galactosaminogalactan pathway. If this is true, then such substrate flux must occur at the level of UDP-glucose or earlier given that the Δuge5 mutant retained the ability to produce galactosaminogalactan. Alternatively, the increase in galactosaminogalactan could reflect activation of a regulatory response to alterations in cell wall integrity as has been observed to occur with other mutations or perturbing agents that alter cell wall composition (56–59).
It is surprising that, in the absence of Uge5, UDP-galactose production by Uge3 was sufficient to permit production of galactosaminogalactan but not Galf. Although compartmental sequestration of the UDP-galactose produced by these two epimerases could account for this observation, our localization studies suggest that both enzymes are cytoplasmic. Alternatively, differences in the transport of UDP-galactose or the substrate affinity of downstream enzymes specific for each of the two pathways could mediate this preferential funneling of UDP-galactose into galactosaminogalactan synthesis. It is not known if either epimerase complexes with other elements in their respective biosynthetic pathways; however, this explanation could account for differences in accessibility of UDP-galactose between pathways. Finally, there may be different regulatory controls at various levels of gene expression and protein synthesis of other pathway components. Although a deficiency in galactosaminogalactan is not lethal, it renders the strain nonadherent, which is not the case for galactomannan deficiency (5, 6, 8, 12, 22). Thus, in the absence of a specific requirement for galactomannan, it is possible that the fungus preferentially diverts its resources to preserve galactosaminogalactan synthesis for adherence to surfaces or biofilm homeostasis. Testing of these hypotheses will require identification of other elements of the galactosaminogalactan biosynthetic pathways, an area under active research by our group.
We initially hypothesized that enzymes upstream of Uge3 or Uge5 in the Leloir pathway, such as galactokinase or UDP-galactose-1-phosphate uridylyltransferase, may contribute to the residual galactose found in the Δuge5 mutant. Although much of the Leloir pathway is uncharacterized in A. fumigatus, it is well characterized in A. nidulans. In this species, deletion of galactokinase gene galE or UDP-galactose-1-phosphate uridylyltransferase galD results in mutants that are viable, albeit with partial growth defects, in a galactose-based carbon source (60). However, the complete lack of growth of the Δuge3Δuge5 double mutant in galactose-based medium strongly suggests that these epimerases are solely responsible for galactose metabolism under the conditions studied in this report, and other salvage enzymes are insufficient to provide adequate glucose from galactose under the conditions of growth that we tested.
Our findings demonstrate that Uge3 is dispensable for galactose metabolism and Galf synthesis. This observation is likely due, at least in part, to the much lower expression levels of uge3 as compared with uge5. Indeed, it is possible that interconversion of UDP-galactose and UDP-glucose by Uge3 does not occur in the presence of physiologic levels of Uge5 and is only unmasked upon Uge5 deletion. Interestingly, we found that, in vitro, Uge3 produced twice as much UDP-GalNAc compared with UDP-galactose from respective substrates at steady-state equilibrium, thus suggesting a possible preference toward N-acetylated hexosamines as substrates. Nonetheless, this redundancy in UDP-galactose and UDP-glucose interconversion by two epimerases suggests that interconversion of hexoses may be more critical to A. fumigatus than the interconversion of UDP-GlcNAc-UDP-GalNAc, which is mediated by Uge3 alone (22). Indeed, sugar epimerase have important functions outside of cell wall polysaccharide synthesis, such as metabolism. Redundancy in hexose interconversion would ensure that the organism can adapt to different carbon sources and maintain glycolysis and generate derivatives required for other cellular activities (12, 13, 61–63). In contrast, GalNAc seems to be required by A. fumigatus primarily for the synthesis of galactosaminogalactan and is not required for normal growth (22). The need for redundant hexose epimerase activity is unlikely related to galactosaminogalactan synthesis, because production of galactose-poor galactosaminogalactan by the Δuge5 mutant did not impair adherence, biofilm formation, or other virulence associated properties. Indeed, our studies failed to identify a functional role for the galactose component of galactosaminogalactan and suggest that the adherence, apoptosis-inducing, and pathogen-associated molecular pattern-masking phenotypes of galactosaminogalactan are mediated by the GalNAc fraction of this glycan. One possibility is that altering the galactose content of galactosaminogalactan may provide a mechanism for the organism to modulate galactosaminogalactan activity through secondarily changing the relative GalNAc content of the resulting glycan. We are currently investigating the effects of lowering the GalNAc content of galactosaminogalactan to better understand the mechanism of action of galactosaminogalactan in these phenotypes.
Although we found three genes annotated as UDP-glucose 4-epimerases in A. fumigatus, only uge3 and uge5 seem to be active. One explanation could be that the in silico annotation is not correct and that, in fact, uge4 is not a UDP-glucose 4-epimerase. However, based on the close homology of uge4 to the other two epimerases, and the fact that all important domains are predicted with high certainty to be intact in the uge4 sequence, it is likely that uge4 is a UDP-glucose 4-epimerase. It is possible that uge4 is expressed under conditions that were not tested; however, an alternative explanation is that uge4 is a product of gene duplication that could have served a purpose earlier in evolution but has now become silenced. Similar silent gene duplications have been reported in other cell wall-related genes in A. fumigatus (64, 65). Interestingly, silencing of epimerases in Aspergillus species may play a role in virulence because in the nonpathogenic species A. nidulans, ugeB, the ortholog of A. fumigatus uge3, has been reported to be silent (55).
This study broadens our understanding of Aspergillus epimerases and their role in metabolism and carbohydrate synthesis, and it also begins to identify some of the critical steps in the biosynthesis of galactosaminogalactan. These results suggest a model of galactose-containing cell wall polysaccharide synthesis in which Uge5 activity alone mediates production of UDP-galactose as a precursor to Galf and subsequent galactomannan synthesis. Conversely, Uge3 activity is required for the synthesis of UDP-GalNAc for the production of galactosaminogalactan, and both epimerases can contribute to the pool of UDP-galactose used in the synthesis of galactosaminogalactan. Furthermore, Uge5 is responsible for the majority of the epimerase activity within the Leloir pathway, as deletion of uge5, but not uge3, was associated with a defect in growth on galactose-containing medium. A deeper understanding of the biochemical pathways underlying galactose metabolism and the biosynthesis of cell wall glycans in this pathogenic fungus may provide the basis for the development of future antifungal therapies.
Acknowledgments
We are thankful to Marianne Ngure and Moheshwarnath Issur for their guidance on protein purification.
This work was supported, in whole or in part, by National Institutes of Health Grant R01AI073829. This work was also supported by operating funds from the Canadian Institutes of Health Research.
- Galf
- galactofuranose
- PDB
- Protein Data Bank.
REFERENCES
- 1. Garcia-Vidal C., Upton A., Kirby K. A., Marr K. A. (2008) Epidemiology of invasive mold infections in allogeneic stem cell transplant recipients: biological risk factors for infection according to time after transplantation. Clin. Infect. Dis. 47, 1041–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ostrosky-Zeichner L., Casadevall A., Galgiani J. N., Odds F. C., Rex J. H. (2010) An insight into the antifungal pipeline: selected new molecules and beyond. Nat. Rev. Drug Discov. 9, 719–727 [DOI] [PubMed] [Google Scholar]
- 3. Latgé J.-P. (2010) Tasting the fungal cell wall. Cell. Microbiol. 12, 863–872 [DOI] [PubMed] [Google Scholar]
- 4. Latge J. P. (2009) Galactofuranose containing molecules in Aspergillus fumigatus. Med. Mycol. 47, S104–S109 [DOI] [PubMed] [Google Scholar]
- 5. Latgé J. P., Kobayashi H., Debeaupuis J. P., Diaquin M., Sarfati J., Wieruszeski J. M., Parra E., Bouchara J. P., Fournet B. (1994) Chemical and immunological characterization of the extracellular galactomannan of Aspergillus fumigatus. Infect. Immun. 62, 5424–5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lamarre C., Beau R., Balloy V., Fontaine T., Wong Sak Hoi J., Guadagnini S., Berkova N., Chignard M., Beauvais A., Latgé J.-P. (2009) Galactofuranose attenuates cellular adhesion of Aspergillus fumigatus. Cell. Microbiol. 11, 1612–1623 [DOI] [PubMed] [Google Scholar]
- 7. Tefsen B., Ram A. F., van Die I., Routier F. H. (2012) Galactofuranose in eukaryotes: aspects of biosynthesis and functional impact. Glycobiology 22, 456–469 [DOI] [PubMed] [Google Scholar]
- 8. Schmalhorst P. S., Krappmann S., Vervecken W., Rohde M., Müller M., Braus G. H., Contreras R., Braun A., Bakker H., Routier F. H. (2008) Contribution of galactofuranose to the virulence of the opportunistic pathogen Aspergillus fumigatus. Eukaryot. Cell 7, 1268–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Luong M.-L., Filion C., Labbé A.-C., Roy J., Pépin J., Cadrin-Tourigny J., Carignan S., Sheppard D. C., Laverdière M. (2010) Clinical utility and prognostic value of bronchoalveolar lavage galactomannan in patients with hematologic malignancies. Diagn. Microbiol. Infect. Dis. 68, 132–139 [DOI] [PubMed] [Google Scholar]
- 10. Walsh T. J., Anaissie E. J., Denning D. W., Herbrecht R., Kontoyiannis D. P., Marr K. A., Morrison V. A., Segal B. H., Steinbach W. J., Stevens D. A., van Burik J.-A., Wingard J. R., Patterson T. F., Infectious Diseases Society of America (2008) Treatment of aspergillosis: clinical practice guidelines of the Infectious Diseases Society of America. Clin. Infect. Dis. 46, 327–360 [DOI] [PubMed] [Google Scholar]
- 11. Stynen D., Sarfati J., Goris A., Prévost M. C., Lesourd M., Kamphuis H., Darras V., Latgé J. P. (1992) Rat monoclonal antibodies against Aspergillus galactomannan. Infect. Immun. 60, 2237–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. El-Ganiny A. M., Sheoran I., Sanders D. A., Kaminskyj S. G. (2010) Aspergillus nidulans UDP-glucose 4-epimerase UgeA has multiple roles in wall architecture, hyphal morphogenesis, and asexual development. Fungal Genet. Biol. 47, 629–635 [DOI] [PubMed] [Google Scholar]
- 13. Afroz S., El-Ganiny A. M., Sanders D. A., Kaminskyj S. G. (2011) Roles of the Aspergillus nidulans UDP-galactofuranose transporter, UgtA in hyphal morphogenesis, cell wall architecture, conidiation, and drug sensitivity. Fungal Genet. Biol. 48, 896–903 [DOI] [PubMed] [Google Scholar]
- 14. Kremer L., Dover L. G., Morehouse C., Hitchin P., Everett M., Morris H. R., Dell A., Brennan P. J., McNeil M. R., Flaherty C., Duncan K., Besra G. S. (2001) Galactan biosynthesis in Mycobacterium tuberculosis. Identification of a bifunctional UDP-galactofuranosyltransferase. J. Biol. Chem. 276, 26430–26440 [DOI] [PubMed] [Google Scholar]
- 15. Guan S., Clarke A. J., Whitfield C. (2001) Functional analysis of the galactosyltransferases required for biosynthesis of d-galactan I, a component of the lipopolysaccharide O1 antigen of Klebsiella pneumoniae. J. Bacteriol. 183, 3318–3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mikusová K., Belánová M., Korduláková J., Honda K., McNeil M. R., Mahapatra S., Crick D. C., Brennan P. J. (2006) Identification of a novel galactosyltransferase involved in biosynthesis of the mycobacterial cell wall. J. Bacteriol. 188, 6592–6598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wing C., Errey J. C., Mukhopadhyay B., Blanchard J. S., Field R. A. (2006) Expression and initial characterization of WbbI, a putative d-Galf:α-d-Glcβ-1,6-galactofuranosyltransferase from Escherichia coli K-12. Org. Biomol. Chem. 4, 3945–3950 [DOI] [PubMed] [Google Scholar]
- 18. Stoco P. H., Aresi C., Lückemeyer D. D., Sperandio M. M., Sincero T. C., Steindel M., Miletti L. C., Grisard E. C. (2012) Trypanosoma rangeli expresses a β-galactofuranosyltransferase. Exp. Parasitol. 130, 246–252 [DOI] [PubMed] [Google Scholar]
- 19. Bakker H., Kleczka B., Gerardy-Schahn R., Routier F. H. (2005) Identification and partial characterization of two eukaryotic UDP-galactopyranose mutases. Biol. Chem. 386, 657–661 [DOI] [PubMed] [Google Scholar]
- 20. Engel J., Schmalhorst P. S., Dörk-Bousset T., Ferrières V., Routier F. H. (2009) A single UDP-galactofuranose transporter is required for galactofuranosylation in Aspergillus fumigatus. J. Biol. Chem. 284, 33859–33868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oppenheimer M., Poulin M. B., Lowary T. L., Helm R. F., Sobrado P. (2010) Characterization of recombinant UDP-galactopyranose mutase from Aspergillus fumigatus. Arch. Biochem. Biophys. 502, 31–38 [DOI] [PubMed] [Google Scholar]
- 22. Gravelat F. N., Beauvais A., Liu H., Lee M. J., Snarr B. D., Chen D., Xu W., Kravtsov I., Hoareau C. M., Vanier G., Urb M., Campoli P., Al Abdallah Q., Lehoux M., Chabot J. C., Ouimet M.-C., Baptista S. D., Fritz J. H., Nierman W. C., Latgé J. P., Mitchell A. P., Filler S. G., Fontaine T., Sheppard D. C. (2013) Aspergillus galactosaminogalactan mediates adherence to host constituents and conceals hyphal β-glucan from the immune system. PLoS Pathog. 10.1371/journal.ppat.1003575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fontaine T., Delangle A., Simenel C., Coddeville B., van Vliet S. J., van Kooyk Y., Bozza S., Moretti S., Schwarz F., Trichot C., Aebi M., Delepierre M., Elbim C., Romani L., Latge J. P. (2011) Galactosaminogalactan, a new immunosuppressive polysaccharide of Aspergillus fumigatus. PLoS Pathog. 10.1371/journal.ppat.1002372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gravelat F. N., Ejzykowicz D. E., Chiang L. Y., Chabot J. C., Urb M., Macdonald K. D., al-Bader N., Filler S. G., Sheppard D. C. (2010) Aspergillus fumigatus MedA governs adherence, host cell interactions, and virulence. Cell. Microbiol. 12, 473–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gravelat F. N., Askew D. S., Sheppard D. C. (2012) Targeted gene deletion in Aspergillus fumigatus using the hygromycin-resistance split-marker approach. Methods Mol. Biol. 845, 119–130 [DOI] [PubMed] [Google Scholar]
- 26. Hartmann T., Dümig M., Jaber B. M., Szewczyk E., Olbermann P., Morschhäuser J., Krappmann S. (2010) Validation of a self-excising marker in the human pathogen Aspergillus fumigatus by employing the β-rec/six site-specific recombination system. Appl. Environ. Microbiol. 76, 6313–6317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McCluskey K., Wiest A., Plamann M. (2010) The Fungal Genetics Stock Center: a repository for 50 years of fungal genetics research. J. Biosci. 35, 119–126 [DOI] [PubMed] [Google Scholar]
- 28. Al Abdallah Q., Choe S. I., Campoli P., Baptista S., Gravelat F. N., Lee M. J., Sheppard D. C. (2012) A conserved C-terminal domain of the Aspergillus fumigatus developmental regulator MedA is required for nuclear localization, adhesion and virulence. PLoS One, 10.1371/journal.pone.0049959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Choe S. I., Gravelat F. N., Al Abdallah Q., Lee M. J., Gibbs B. F., Sheppard D. C. (2012) Role of Aspergillus niger acrA in arsenic resistance and its use as the basis for an arsenic biosensor. Appl. Environ. Microbiol. 78, 3855–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Twumasi-Boateng K., Yu Y., Chen D., Gravelat F. N., Nierman W. C., Sheppard D. C. (2009) Transcriptional profiling identifies a role for BrlA in the response to nitrogen depletion and for StuA in the regulation of secondary metabolite clusters in Aspergillus fumigatus. Eukaryot. Cell 8, 104–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Campoli P., Perlin D. S., Kristof A. S., White T. C., Filler S. G., Sheppard D. C. (2013) Pharmacokinetics of posaconazole within epithelial cells and fungi: Insights into potential mechanisms of action during treatment and prophylaxis. J. Infect. Dis. 10.1093/infdis/jit358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gravelat F. N., Doedt T., Chiang L. Y., Liu H., Filler S. G., Patterson T. F., Sheppard D. C. (2008) In vivo analysis of Aspergillus fumigatus developmental gene expression determined by real-time reverse transcription-PCR. Infect. Immun. 76, 3632–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Studier F. W. (2005) Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 34. Gu X., Lee S. G., Bar-Peled M. (2011) Biosynthesis of UDP-xylose and UDP-arabinose in Sinorhizobium meliloti 1021: first characterization of a bacterial UDP-xylose synthase, and UDP-xylose 4-epimerase. Microbiology 157, 260–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Creuzenet C., Belanger M., Wakarchuk W. W., Lam J. S. (2000) Expression, purification, and biochemical characterization of WbpP, a new UDP-GlcNAc C4 epimerase from Pseudomonas aeruginosa serotype O6. J. Biol. Chem. 275, 19060–19067 [DOI] [PubMed] [Google Scholar]
- 36. Johnson A. E., Tanner M. E. (1998) Epimerization via carbon-carbon bond cleavage. l- Ribulose-5-phosphate 4-epimerase as a masked class II aldolase. Biochemistry 37, 5746–5754 [DOI] [PubMed] [Google Scholar]
- 37. McCallum M., Shaw G. S., Creuzenet C. (2011) Characterization of the dehydratase WcbK and the reductase WcaG involved in GDP-6-deoxy-manno-heptose biosynthesis in Campylobacter jejuni. Biochem. J. 439, 235–248 [DOI] [PubMed] [Google Scholar]
- 38. Arnaud M. B., Cerqueira G. C., Inglis D. O., Skrzypek M. S., Binkley J., Chibucos M. C., Crabtree J., Howarth C., Orvis J., Shah P., Wymore F., Binkley G., Miyasato S. R., Simison M., Sherlock G., Wortman J. R. (2012) The Aspergillus Genome Database (AspGD): recent developments in comprehensive multispecies curation, comparative genomics and community resources. Nucleic Acids Res. 40, D653–D659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dinkel H., Michael S., Weatheritt R. J., Davey N. E., Van Roey K., Altenberg B., Toedt G., Uyar B., Seiler M., Budd A., Jödicke L., Dammert M. A., Schroeter C., Hammer M., Schmidt T., Jehl P., McGuigan C., Dymecka M., Chica C., Luck K., Via A., Chatr-Aryamontri A., Haslam N., Grebnev G., Edwards R. J., Steinmetz M. O., Meiselbach H., Diella F., Gibson T. J. (2012) ELM–the database of eukaryotic linear motifs. Nucleic Acids Res. 40, D242–D251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Celniker G., Nimrod G., Ashkenazy H., Glaser F., Martz E., Mayrose I., Pupko T., Ben-Tal N. (2013) ConSurf: using evolutionary data to raise testable hypotheses about protein function. Isr. J. Chem. 53, 199–206 [Google Scholar]
- 41. Marchler-Bauer A., Lu S., Anderson J. B., Chitsaz F., Derbyshire M. K., DeWeese-Scott C., Fong J. H., Geer L. Y., Geer R. C., Gonzales N. R., Gwadz M., Hurwitz D. I., Jackson J. D., Ke Z., Lanczycki C. J., Lu F., Marchler G. H., Mullokandov M., Omelchenko M. V., Robertson C. L., Song J. S., Thanki N., Yamashita R. A., Zhang D., Zhang N., Zheng C., Bryant S. H. (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Biegert A., Mayer C., Remmert M., Söding J., Lupas A. N. (2006) The MPI Bioinformatics toolkit for protein sequence analysis. Nucleic Acids Res. 34, W335–W339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Apweiler R., Attwood T. K., Bairoch A., Bateman A., Birney E., Biswas M., Bucher P., Cerutti L., Corpet F., Croning M. D., Durbin R., Falquet L., Fleischmann W., Gouzy J., Hermjakob H., Hulo N., Jonassen I., Kahn D., Kanapin A., Karavidopoulou Y., Lopez R., Marx B., Mulder N. J., Oinn T. M., Pagni M., Servant F., Sigrist C. J., Zdobnov E. M. (2001) The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29, 37–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hunter S., Jones P., Mitchell A., Apweiler R., Attwood T. K., Bateman A., Bernard T., Binns D., Bork P., Burge S., de Castro E., Coggill P., Corbett M., Das U., Daugherty L., Duquenne L., Finn R. D., Fraser M., Gough J., Haft D., Hulo N., Kahn D., Kelly E., Letunic I., Lonsdale D., Lopez R., Madera M., Maslen J., McAnulla C., McDowall J., McMenamin C., Mi H., Mutowo-Muellenet P., Mulder N., Natale D., Orengo C., Pesseat S., Punta M., Quinn A. F., Rivoire C., Sangrador-Vegas A., Selengut J. D., Sigrist C. J., Scheremetjew M., Tate J., Thimmajanarthanan M., Thomas P. D., Wu C. H., Yeats C., Yong S. Y. (2012) InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 40, D306–D312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Punta M., Coggill P. C., Eberhardt R. Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., Clements J., Heger A., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A., Finn R. D. (2012) The Pfam protein families database. Nucleic Acids Res. 40, D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr., Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. (1977) The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 112, 535–542 [DOI] [PubMed] [Google Scholar]
- 47. Thoden J. B., Wohlers T. M., Fridovich-Keil J. L., Holden H. M. (2001) Human UDP-galactose 4-epimerase. Accommodation of UDP-N-acetylglucosamine within the active site. J. Biol. Chem. 276, 15131–15136 [DOI] [PubMed] [Google Scholar]
- 48. Shaw M. P., Bond C. S., Roper J. R., Gourley D. G., Ferguson M. A., Hunter W. N. (2003) High-resolution crystal structure of Trypanosoma brucei UDP-galactose 4′-epimerase: a potential target for structure-based development of novel trypanocides. Mol. Biochem. Parasitol. 126, 173–180 [DOI] [PubMed] [Google Scholar]
- 49. Ishiyama N., Creuzenet C., Lam J. S., Berghuis A. M. (2004) Crystal structure of WbpP, a genuine UDP-N-acetylglucosamine 4-epimerase from Pseudomonas aeruginosa: substrate specificity in UDP-hexose 4-epimerases. J. Biol. Chem. 279, 22635–22642 [DOI] [PubMed] [Google Scholar]
- 50. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 51. Eswar N., John B., Mirkovic N., Fiser A., Ilyin V. A., Pieper U., Stuart A. C., Marti-Renom M. A., Madhusudhan M. S., Yerkovich B., Sali A. (2003) Tools for comparative protein structure modeling and analysis. Nucleic Acids Res. 31, 3375–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Demendi M., Ishiyama N., Lam J. S., Berghuis A. M., Creuzenet C. (2005) Toward a better understanding of the substrate specificity of the UDP-N-acetylglucosamine C4 epimerase WbpP. Biochem. J. 389, 173–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jörnvall H., Persson B., Krook M., Atrian S., Gonzàlez-Duarte R., Jeffery J., Ghosh D. (1995) Short-chain dehydrogenases/reductases (SDR). Biochemistry 34, 6003–6013 [DOI] [PubMed] [Google Scholar]
- 54. Varki A., Cummings R. D., Esko J. D., Freeze H. H., Stanley P., Marth J. D., Bertozzi C. R., Hart G. W., Etzler M. E. (2009) Symbol nomenclature for glycan representation. Proteomics 9, 5398–5399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Paul B. C., El-Ganiny A. M., Abbas M., Kaminskyj S. G., Dahms T. E. (2011) Quantifying the importance of galactofuranose in Aspergillus nidulans hyphal wall surface organization by atomic force microscopy. Eukaryot. Cell 10, 646–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Henry C., Latgé J.-P., Beauvais A. (2012) β1,3-Glucans are dispensable in Aspergillus fumigatus. Eukaryot. Cell 11, 26–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Verwer P. E., van Duijn M. L., Tavakol M., Bakker-Woudenberg I. A., van de Sande W. W. (2012) Reshuffling of Aspergillus fumigatus cell wall components chitin and β-glucan under the influence of caspofungin or nikkomycin Z alone or in combination. Antimicrob. Agents Chemother. 56, 1595–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fortwendel J. R., Juvvadi P. R., Pinchai N., Perfect B. Z., Alspaugh J. A., Perfect J. R., Steinbach W. J. (2009) Differential effects of inhibiting chitin and 1,3-β-d-glucan synthesis in ras and calcineurin mutants of Aspergillus fumigatus. Antimicrob. Agents Chemother. 53, 476–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Valiante V., Jain R., Heinekamp T., Brakhage A. A. (2009) The MpkA MAP kinase module regulates cell wall integrity signaling and pyomelanin formation in Aspergillus fumigatus. Fungal Genet. Biol. 46, 909–918 [DOI] [PubMed] [Google Scholar]
- 60. Alam M. K., Kaminskyj S. G. W. (2013) Aspergillus galactose metabolism is more complex than that of Saccharomyces: the story of GalDGAL7 and GalEGAL1. Botany 91, 467–477 [Google Scholar]
- 61. Alam M. K., El-Ganiny A. M., Afroz S., Sanders D. A., Liu J., Kaminskyj S. G. (2012) Aspergillus nidulans galactofuranose biosynthesis affects antifungal drug sensitivity. Fungal Genet. Biol. 49, 1033–1043 [DOI] [PubMed] [Google Scholar]
- 62. Timson D. J. (2007) Galactose metabolism in Saccharomyces cerevisiae. Dyn. Biochem. Process Biotech. Mol. Biol. 1, 63–73 [Google Scholar]
- 63. Slepak T., Tang M., Addo F., Lai K. (2005) Intracellular galactose 1-phosphate accumulation leads to environmental stress response in yeast model. Mol. Genet. Metab. 86, 360–371 [DOI] [PubMed] [Google Scholar]
- 64. Mellado E., Dubreucq G., Mol P., Sarfati J., Paris S., Diaquin M., Holden D. W., Rodriguez-Tudela J. L., Latgé J. P. (2003) Cell wall biogenesis in a double chitin synthase mutant (chsG−/chsE−) of Aspergillus fumigatus. Fungal Genet. Biol. 38, 98–109 [DOI] [PubMed] [Google Scholar]
- 65. Gastebois A., Mouyna I., Simenel C., Clavaud C., Coddeville B., Delepierre M., Latgé J.-P., Fontaine T. (2010) Characterization of a new β(1–3)-glucan branching activity of Aspergillus fumigatus. J. Biol. Chem. 285, 2386–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]