Background: Adora2a encodes A2A adenosine receptor (A2AR) and a new protein (uORF5) translated from an out-of-frame AUG.
Results: uORF5 exists in tissues where the Adora2a transcript is detected and is up-regulated by A2AR activation. uORF5 suppresses AP1-mediated transcription.
Conclusion: Adora2a encodes two distinct proteins (A2AR and uORF5) in an A2AR-dependent manner.
Significance: uORF5 may participate in the functions of A2AR during pathophysiological conditions.
Keywords: Adenosine Receptor, AP1 Transcription Factor, Gene Regulation, Hypoxia, Protein Kinase A (PKA), A2A Adenosine Receptor, Alternative Reading Frame, Dual Coding Gene, Overlapping Reading Frame
Abstract
The A2A adenosine receptor (A2AR) is a G protein-coupled receptor and a major target of caffeine. The A2AR gene encodes alternative transcripts that are initiated from at least two independent promoters. The different transcripts of the A2AR gene contain the same coding region and 3′-untranslated region and different 5′-untranslated regions that are highly conserved among species. We report here that in addition to the production of the A2AR protein, translation from an upstream, out-of-frame AUG of the rat A2AR gene produces a 134-amino acid protein (designated uORF5). An anti-uORF5 antibody recognized a protein of the predicted size of uORF5 in PC12 cells and rat brains. Up-regulation of A2AR transcripts by hypoxia led to increased levels of both the A2AR and uORF5 proteins. Moreover, stimulation of A2AR increased the level of the uORF5 protein via post-transcriptional regulation. Expression of the uORF5 protein suppressed the AP1-mediated transcription promoted by nerve growth factor and modulated the expression of several proteins that were implicated in the MAPK pathway. Taken together, our results show that the rat A2AR gene encodes two distinct proteins (A2AR and uORF5) in an A2AR-dependent manner. Our study reveals a new example of the complexity of the mammalian genome and provides novel insights into the function of A2AR.
Introduction
To optimize the usage of short coding sequences (1), the translation of multiple proteins using different reading frames within the same gene is commonly observed in prokaryotic organisms, including single-stranded DNA phages and Escherichia coli (2–4), and in mitochondria (5, 6). In eukaryotic systems, two genes that produce mRNAs with alternative reading frames (e.g. XLαs/ALEX (7) and prion protein/alternative prion protein (8)) have been reported. The differences in the function and regulation of distinct proteins translated from the same transcript are largely unclear. Recent bioinformatic analyses suggest that the number of dual coding genes in the mammalian genome is probably underestimated (9–11).
Adenosine regulates a variety of physiological functions by activating four different adenosine receptors (A1, A2A, A2B, and A3). The A2A adenosine receptor (A2AR),3 which is encoded by the Adora2a gene, is one of the most well studied G protein-coupled receptors because it is a major target of caffeine and a drug target for several brain disorders (12–15). Previous studies have shown that A2AR is widely expressed throughout the body, with the highest level of expression in the striatum (16–20). The expression of A2AR was shown to be markedly up-regulated during several pathological conditions (e.g. inflammation (21), acute lung injury (16), and hypoxia (22)), suggesting that A2AR plays an important role in stress. Consistent with this notion, agonists of A2AR have been shown to attenuate pathological inflammatory responses (23–27). Stimulation of A2AR triggers multiple signaling pathways, including the cAMP-protein kinase A (PKA)-dependent pathway (28), and regulates a wide variety of downstream targets, such as the cAMP-regulated element-binding protein, nuclear factor-κB, and hypoxia-inducible factor 1, that mediate its effect (29–31).
The expression of the A2AR gene is tightly regulated. We previously demonstrated that the rat A2AR gene contains at least two independent promoters (P1 and P2), which drive the expression of multiple transcripts that contain the same coding region and 3′-untranslated region (UTR) and different 5′-UTRs (U1, 514 bp, initiated from P1; U2, 243 bp, initiated from P2). Both 5′-UTRs negatively suppress the translation of the A2AR protein via an out-of-frame AUG codon (designated uAUG-5), which is located upstream of the start codon of the A2AR protein (20). In the present study, we report that uAUG-5 is a functional start codon of an open reading frame (ORF) that overlaps with the A2AR ORF in the rat Adora2a gene. This upstream ORF encodes a novel 134-amino acid (aa) protein (designated uORF5). The expression of uORF5 was found to moderately suppress the activity of the transcription factor activator protein 1 (AP1) and to regulate expression of several proteins that have been implicated in the MAPK pathway. Because the stimulation of A2AR significantly enhanced the expression of uORF5 in a PKA-dependent manner, uORF5 might contribute to the pathophysiological function of A2AR.
MATERIALS AND METHODS
Reagents
All reagents were purchased from Sigma except where otherwise specified. Forskolin (FK), CGS21680 (CGS), and KT5720 were from Tocris Biosciences (Bristol, UK). SCH58261 was obtained from Sigma/RBI (Natick, MA). Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), and horse serum were purchased from Invitrogen. H89 was from BIOMOL Research Laboratories (Plymouth Meeting, PA). Nerve growth factor (NGF) was obtained from Alomone Labs (Jerusalem, Israel).
Animals and Cell Culture
Rat brain tissues were collected from 12-week-old Sprague-Dawley rats. The experimental procedures were approved by the Institutional Animal Care and Utilization Committee of Academia Sinica (Taiwan).
Rat pheochromocytoma cells (PC12) and human embryonic kidney cells (HEK293T) were obtained from the American Type Culture Collection (Manassas, VA). The PC12 cells were grown in DMEM supplemented with 5% FBS plus 10% horse serum in an incubator at 37 °C and 10% CO2. The A123 cell line, a PKA-deficient variant of PC12 cells (32), was a gift from Dr. J. A. Wagner (Cornell University Medical College). The A123 cells were grown in DMEM supplemented with 5% FBS plus 10% horse serum in an incubator at 37 °C and 10% CO2. The HEK293T cells were grown in DMEM supplemented with 100 units/ml penicillin, 0.1 mg/ml streptomycin, and 10% FBS in an incubator at 37 °C and 5% CO2. For the hypoxia treatment, the cells were placed in a hypoxic incubator (Coy Laboratory Products Inc., Grass Lake, MI) and maintained in a constant environment of 5% O2 and 5% CO2 (balanced with N2) for the specified exposure periods. The oxygen concentration in the chamber was monitored with an oxygen meter and electrode (Coy Laboratory). The medium was preincubated in the hypoxia chamber for 12 h before being used in the hypoxia experiments. For the sorting of transfected PC12 cells, the cells were transfected with pIRES-hrGFP-1a or pIRES-uORF:hrGFP-1a for 48 h. The cells were then suspended in DMEM at a density of 5 × 106 cells/ml and sorted for hrGFP-positive cells using a FACSVantage flow cytometer (BD Biosciences) equipped with an argon ion laser (488 nm, 100 milliwatts). The striatal progenitor ST14A and STHdhQ7 cell lines were kindly provided by Dr. Elena Cattaneo (University of Milan) (33). These cells were grown in DMEM supplemented with 100 units/ml penicillin, 0.1 mg/ml streptomycin, 1 mm sodium pyruvate, 2 mm l-glutamine, and 10% FBS in incubator at 33 °C and 5% CO2 (66).
Plasmids and Cell Transfection
To create the uORF5-V5 and A2AR-V5 expression constructs, DNA fragments containing the Kozak sequence and the coding sequence of A2AR (−65 to +1233, with +1 denoting the translational start site of A2AR; Fig. 1A) or uORF5 (−65 to +340) were amplified from cDNA prepared from the rat striatum using PCR and subcloned into the pcDNA3.1/V5-His-TOPO vector (Invitrogen) following the manufacturer's instructions. Mutation of the translational start codon of uORF5 (uAUG-5, -65m) (20) was created using a two-step PCR approach (34). To prepare the uORF5:hrGFP expression construct, the DNA fragment encoding rat uORF5 was amplified from rat brain cDNA and subcloned into the SacI/XhoI sites of pIRES-hrGFP-1a (Agilent Technologies). The A2AR knockdown constructs were produced by subcloning the 19-nucleotide shRNA sequence of the rat A2AR gene (Adora2a) (Adora2a shRNA-1, 5′-TTACATGGTTTACTACAAC-3′; Adora2a shRNA-2, 5′-GATCATCCGAACCCACGTC-3′) into the pSUPER.neo+gfp vector. The pFA-CMV-GAL4-uORF5 construct was created by subcloning the coding region of uORF5 into the HindIII/KpnI sites of the pFA-CMV vector (Stratagene). 5′-UTR1- and 5′-UTR2-uORF5(HA)-A2AR(V5) were generated from rat striatal cDNA using a two-step RT-PCR approach and subcloned into the pcDNA3.1/V5-His-TOPO vector. The pFA-CMV vector and GAL4-f-luciferase reporter were obtained from Stratagene. The AP1-Luc reporter was obtained from Clontech. The CRE-Luc reporter (35) and the p21-Luc reporter (36) were generous gifts from Dr. Jeffrey J. Y. Yen (Institute of Biomedical Sciences, Academia Sinica) and Dr. Sheau-Yann Shieh (Institute of Biomedical Sciences, Academia Sinica), respectively. The sequences of primers utilized in the generation of the above constructs are listed in Table 1.
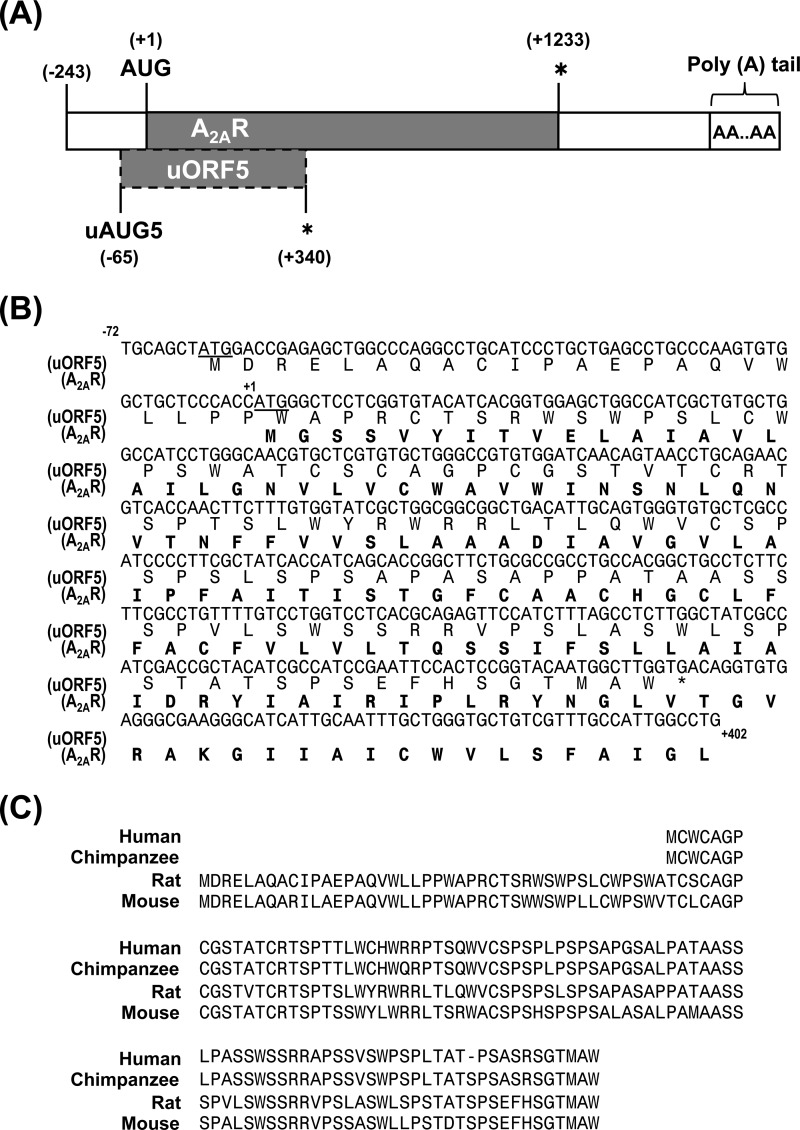
FIGURE 1.
Two overlapping open reading frames coexist in the A2AR gene. A, the major mRNA of the Adora2a gene (GenBankTM accession number BC081727) that contains 5′-UTR (−243/−1) and coding regions for both A2AR (solid box, +1 reading frame) and uORF5 (dashed line box, +2 reading frame). The translation start site of A2AR is located at +1 (AUG), and the translation start site of uORF5 is located at −65 (uAUG5). The asterisks mark the stop codons of A2AR (+1233) and uORF5 (+340). B, translation start sites are underlined. The asterisk marks the stop codon of uORF5. The overlapping region of uORF5 and A2AR is 340 nucleotides. The entire aa sequence of uORF5 and the first 134 aa of A2AR are shown below the DNA sequence. C, alignment of the predicted aa sequences of the human (CA307692), chimpanzee (XM_003953847), rat (BC081727), and mouse (AC137067) uORF5 proteins. Amino acid identity was as follows: rat versus human, 75%; rat versus chimpanzee, 75%; rat versus mouse, 84%.
TABLE 1.
Primer list for plasmid construction
| Constructs | Primers (5′ → 3′) |
|---|---|
| (−65/+340) | CTGCAGCTAT GGACCGAG |
| TCACCAAGCC ATTGTACC | |
| (−65/+1233) | CTGCAGCTAT GGACCGAG |
| TCAGGAAGGG GCAAACT | |
| ((−65/+1233)m) | CTGCAGCTTT GGACCGAG |
| TCAGGAAGGG GCAAACT | |
| 5′-UTR1-uORF5(HA)-A2AR(V5) 5′ | CTAAGCTTACAGGTCAGTGACAAATC |
| AGCGTAATCTGGAACATCGTATGGGTACCAAGCCATTGTACC | |
| 5′-UTR2-uORF5(HA)-A2AR(V5) 5′ | CTAAGCTTGTATCTCAGAACCCTGAA |
| AGCGTAATCTGGAACATCGTATGGGTACCAAGCCATTGTACC | |
| 5′-UTR1 & 5′UTR2-uORF5(HA)-A2AR(V5) 3′ | TACCCATACGATGTTCCAGATTACGCTTGACAGGTGTGAGGG |
| GGAAGGGGCAAACTCTG | |
| A2AR-V5 | CTCCCACCATGGGCTCCT |
| GGAAGGGGCAAACTCTGA | |
| DBD-uORF5 | TGAGCTCATGGACCGAGAGCTG |
| CAGGTACCTCACCAAGCCATTGT | |
| uORF5-V5 | CTGCAGCTATGGACCGAG |
| CCAAGCCATTGTACCGG |
For transient transfection, cells were seeded onto poly-l-lysine (Sigma)-coated plates or coverslips 12–18 h before transfection. Cells at 50–70% confluence were transfected using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. Protein and RNA were extracted from the transfected cells at 48 h post-transfection. The results were obtained from experiments using at least two different plasmid preparations.
Preparation of the Anti-uORF5 Antibody
The DNA sequence encoding uORF5 was amplified from cDNA prepared from the rat striatum using PCR and subcloned into the pET11d vector using standard molecular biology techniques. The resultant plasmid (pET11d-uORF5) was transformed into E. coli BL21-DE3 for the expression of the recombinant uORF5 protein and its subsequent purification using nickel-coupled beads (Novagen, GE Healthcare). The purified recombinant uORF5-His was used to raise a polyclonal antibody in rabbits using standard procedures (37). The resulting antiserum was designated as anti-uORF5. Affinity purification of anti-uORF5 was conducted using a standard procedure in which recombinant uORF5 was bound to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA). In brief, purified uORF5-His was resolved by 15% SDS-PAGE and transferred to PVDF membranes using standard Western blotting procedures. uORF5-His was visualized on the membranes by light staining with Ponceau S. The membranes were cut into small pieces after extensive washing with TBST (500 mm NaCl, 20 mm Tris (pH 7.4), 0.1% Tween 20), blocked in TBST containing 3% BSA for 1 h at room temperature, and incubated with the anti-uORF5 serum with gentle shaking at room temperature overnight. The bound anti-uORF5 antibody was eluted using an acidic glycine buffer (50 mm glycine (pH 2.3), 150 mm NaCl) and neutralized with 1× PBS containing 0.2 m Tris (pH 8). After absorption with the PVDF membranes containing recombinant uORF5, the unbound serum was saved and used as a control (designated as anti-uORF5-absorp).
SDS-PAGE and Western Blotting
Cells were harvested by gentle scraping and then washed with ice-cold PBS. Cell pellets were resuspended in ice-cold radioimmune precipitation assay buffer (10 mm sodium phosphate (pH 7.2), 1% Triton X-114, 0.5% sodium deoxycholate, 150 mm NaCl, and 200× protease inhibitor mixture). Cell fractionation was conducted using a ProteoJet cytoplasmic and nuclear protein extraction kit (Fermentas Inc., Glen Burnie, MD). Briefly, the cells were rinsed twice with PBS and scraped off of the culture dishes in PBS, followed by a centrifugation step at 500 × g for 5 min. Cell pellets were lysed in ice-cold cell lysis buffer containing 1 μm DTT and 1× protease inhibitor mixture (Cell Signaling Technology Inc., Danvers, MA) on ice for 10 min. The lysate was subjected to centrifugation at 500 × g for 7 min, followed by an additional centrifugation step at 20,000 × g for 15 min. The supernatant was collected as the cytoplasmic fraction. The pellet, which contained the nuclei, was washed once with the same buffer, set on ice for 2 min, and then centrifuged at 500 × g for 7 min. The supernatant was removed, and the pellet was suspended in nuclei storage buffer containing 1 μm DTT and 1× protease inhibitor mixture, lysed with nuclei lysis reagent at 4 °C for 15 min, and centrifuged at 20,000 × g for 5 min. The resulting supernatant was collected as the nuclear fraction. Both the cytoplasmic and nuclear fractions were stored at −80 °C until further analysis.
To prepare the brain lysates, brain tissues were homogenized with a glass homogenizer in ice-cold RIPA buffer. The protein concentration was determined by a Bradford assay (Bio-Rad) using bovine serum albumin as the standard. The proteins were resolved using 12 or 15% SDS-PAGE and electrotransferred onto PVDF membranes. The membranes were blocked with 5% nonfat dry milk in TBS containing 0.1% Tween 20 (TBST) for 1 h at room temperature and incubated with the desired primary antibody (anti-A2AR, 1:1000, Santa Cruz Biotechnology, Inc.; anti-uORF5, 1:100; anti-actin, 1:1000, Sigma-Aldrich; anti-tubulin, 1:5000, Sigma-Aldrich; anti-V5, 1:5000, Invitrogen; anti-lamin A/C, 1:1000, GeneTex, Hsinchu City, Taiwan; and anti-HA, 1:1000, GeneTex). Detection of the protein signal was carried out using enhanced chemiluminescence (PerkinElmer Life Sciences).
RNA Purification and Reverse Transcription Quantitative PCR (RT-qPCR)
RNA purification and complementary cDNA synthesis were performed using TRIzol (Invitrogen) and Superscript III (Invitrogen), respectively, following the manufacturer's protocols. RT-qPCR was performed using SYBR Green PCR Master Mix (Applied Biosystems, Invitrogen) on an ABI 7900HT fast real-time PCR system in 384-well plates following the manufacturer's instructions. For each experiment, the cDNA preparation and RT-qPCR analysis were performed at least in triplicate. The expression level of each transcript was normalized to that of the reference gene (glyceraldehyde-3-phosphate dehydrogenase, Gapdh). The sequences of the primers used in this analysis are listed in Table 2.
TABLE 2.
Primer list for RT-qPCR
| Gene name | Primer sequence (5′ → 3′) |
|---|---|
| Mouse B2m | TGTCTCACTGACCGGCCTGTA |
| CAGTTCAGTATGTTCGGCTTC | |
| Mouse Dck | CGAGCCCAGCTAGCCTCTCT |
| TGGTCCACTCTGTTTCATTCA | |
| Mouse Gapdh | TGACATCAAGAAGGTGGTGAA |
| AGAGTGGGAGTTGCTGTTGAAG | |
| Mouse Gas1 | CAGTCCGCAACTTCTTCACC |
| GCTTCTGCTCTCGCATCTCA | |
| Mouse Kif2a | CCAATAATGCACCATCCACC |
| ACCTGCTCTTCCATCTCTACC | |
| Mouse Lrrn3 | AGCAGTGAGGGTGAGCACA |
| AGAGGTTGATTAGAGGAGGGTA | |
| Mouse Npm1 | ATTACACCACCTGTGGTCTTAC |
| CGTCCTCCTCATCTTCATCTTC | |
| Mouse Pdzd2 | ATTGTTTGCCAGTCTTTCCTC |
| TCTGTGCCACTTGGTGCTCT | |
| Mouse Rsal2 | TCATCACCAGCAACTACACCA |
| CATCACCAAGTCAGTCAGAAA | |
| Mouse Rshl1 | TGCCCATTGTTGGAGAGGT |
| TCATACAGGCTGAGGTTGC | |
| Mouse Spag9 | TAAAGGTTCAAGCACTCCCAC |
| TCATCCACATCTCCAATAAGG | |
| Mouse Thbs1 | ACAGCCTCAACAACAGATGC |
| TCACCACAGGTCACAGAACAG | |
| Mouse Tnrc6b | TTGAGAGGCAGAAGCAAGTG |
| GATGTTGTGGGACGTTTAGAG | |
| Rat A2AR | GTCCTGGTCCTCACGCAGAGTTCCATC |
| AGCCATTGTACCGGAGTGGA | |
| Rat Gapdh | TGACATCAAGAAGGTGGTGAA |
| AGAGTGGGAGTTGCTGTTGAAG | |
| Rat Lrrn3 | AGTATTTGTGGCCTGCGTTG |
| GTTGGCTTGTGACAGTGGTT | |
| Rat Pdzd2 | ATGCTCAGATGGTCAGGAGTG |
| ATGTTTGTCAGGATGGATGGA | |
| Rat Rasal2 | TGTTCCTTCAGAGGGTCAGT |
| GAGAGGTATTCGCAGTGTCC | |
| Rat Rshl1 | TGGATGATAGCGGGATGTTG |
| TGTGGCCTGGCTGAATGTGT | |
| uORF5(HA)-A2AR(V5) | GTCCTGGTCCTCACGCAGAGTTCCATC |
| AGCGTAATCTGGAACATCGT |
Immunocytochemistry
Cells were fixed with 4% paraformaldehyde in PBS at room temperature for 30 min, washed three times with PBS, and permeabilized with 0.05% Nonidet P-40/PBS for 10 min. After extensive washing with PBS, the permeabilized cells were blocked in PBS containing 2% BSA and 2% goat serum and stained with the desired primary antibodies (anti-V5, 1:200, Invitrogen; anti-V5, 1:200, Genetex; anti-FLAG 1:160, Sigma; anti-sodium potassium ATPase (Na+/K+-ATPase), 1:200, Abcam Inc., Cambridge, MA), which were reconstituted in PBS containing 1% goat serum and 0.05% Nonidet P-40 at 4 °C for 14–16 h. After extensive washing with PBS, the cells were incubated with the corresponding secondary antibodies conjugated to Alexa Fluor 488 or Alexa Fluor 568 at room temperature for 2 h. The cell nuclei were stained with DAPI. The slides were mounted using Vectashield (Vector Laboratories Ltd., Cambridgeshire, UK). Antibody staining was analyzed using laser confocal microscopy (MRC-1000; Bio-Rad; LSM510, Carl Zeiss, Oberkochen, Germany).
Luciferase Assays
Cells were lysed in the 1× lysis buffer of the Dual-Luciferase Reporter Assay System (Promega, Madison, WI) and centrifuged at 15,000 rpm for 5 min to remove insoluble material. The firefly luciferase activity was determined using a TD-20/20 luminometer (Promega) following the manufacturer's protocol and normalized to the Renilla luciferase activity in the lysate, whose expression was driven by the thymidine kinase promoter. At least three independent replicates were performed for each experiment.
Microarray
RNA was harvested from STHdhQ7 cells transfected with the uORF5:hrGPF expression construct or the empty vector pIRES-hrGFP-1a for 48 h. The presence of DNA contamination was assessed by agarose gel electrophoresis. The RNA quality was evaluated using the Agilent RNA 6000 nanoassay kit (Agilent Technologies, Mississauga, Canada). Only the RNA preparations with an RNA integrity number of >7.0 and an A260/280 absorbance ratio of >1.8 were used in further experiments. The RNA was converted to double-stranded cDNA and amplified using in vitro transcription with the MessageAmp aminoallyl-RNA (aRNA) kit (Ambion, Austin, TX). aRNA probes were labeled with NHS-Cy5 (GE Healthcare) and fragmented using the aRNA fragmentation reagent (Ambion). The labeled probes were hybridized at 50 °C for 16 h to the Mouse Whole Genome OneArray version 4.3 (Phalanx Biotech Group, Hsinchu, Taiwan), which contains 30,968 well characterized genes, in duplicate. Three independent experiments were conducted. The array was then subjected to stepwise washing (wash I, 5 min at 42 °C; wash II, 5 min at 42 °C; wash III, 5 min at 25 °C), exposed to a phosphorimaging cassette, scanned using an Axon 4000B Scanner (Molecular Devices), and analyzed with Genepix software (Molecular Devices, Sunnyvale, CA). The data were processed by global scale normalization.
Statistical Analysis
The results are expressed as the means ± S.E. Each experiment was repeated at least three times. Unless stated otherwise, the statistical analyses were conducted using Student's unpaired t test for comparisons between two groups and one-way analysis of variance followed by a post hoc Holm-Sidak test for comparisons among multiple groups.
RESULTS
The Adora2a Gene Contains Overlapping Alternative ORFs
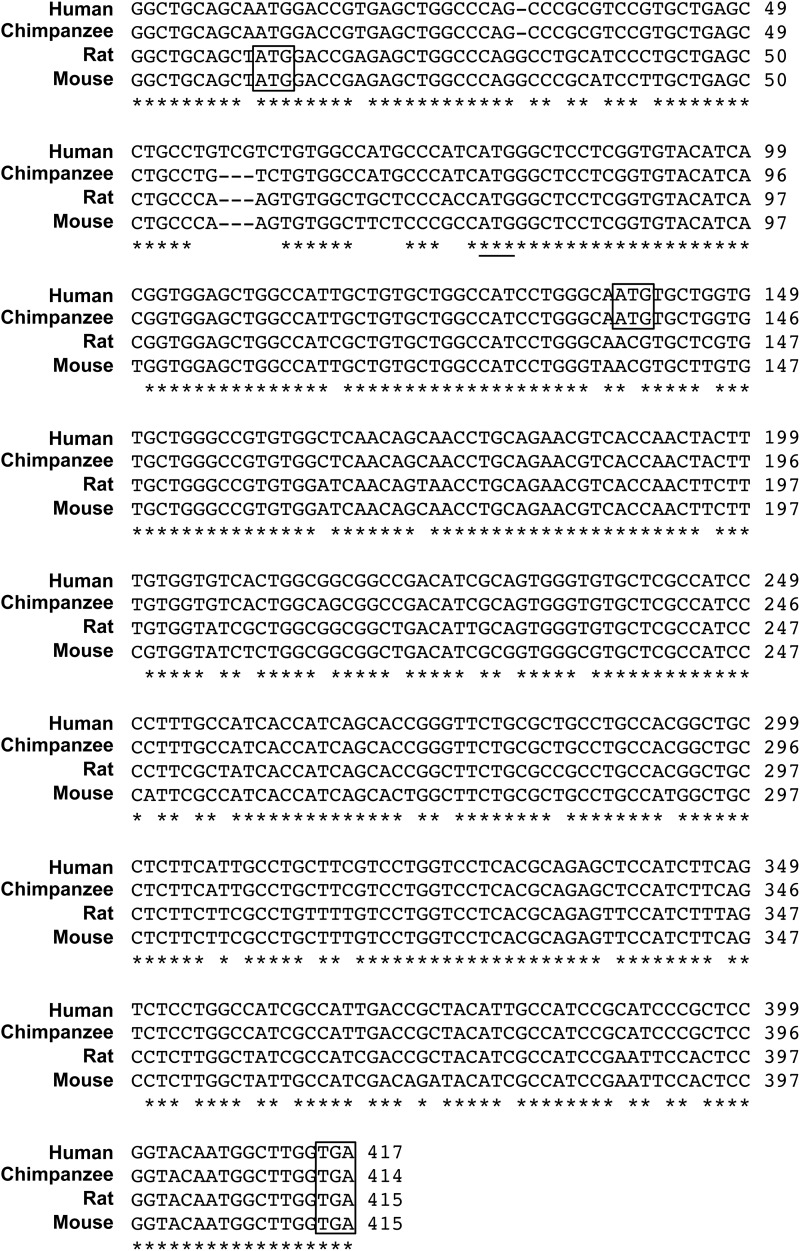
We previously demonstrated that the rat A2AR gene has an upstream, out-of-frame translation start site (uAUG5) that is conserved among species (20). As shown in Fig. 1A, uAUG5 is located 65 nucleotides upstream of the start codon of A2AR. Because uAUG5 is flanked by a nearly perfect Kozak consensus sequence, it is likely to be a functional start codon for an ORF that encodes a novel 134-aa protein (uORF5; Fig. 1B). Because the 5′-UTR of the A2AR gene is conserved across species (Fig. 2) (20), this additional ORF may be present in the Adora2a genes of other species. The mouse uORF5 protein, which is encoded by the new ORF found in the mouse Adora2a gene, shares 84% aa identity with rat ORF5 (Fig. 1C). A shorter uORF5-like protein, which might be encoded by an alternative ORF found in the human and chimpanzee Adora2a genes, shares 75% aa identity with rat uORF5 (Fig. 1C).
FIGURE 2.
Cross-species sequence comparisons of the Adora2a gene. A partial sequence alignment of the Adora2a mRNAs from four different species is presented. The predicted start site and stop site of the uORF5 proteins are boxed. The translational start site of the A2AR protein is underlined. The accession numbers of the human, chimpanzee, rat, and mouse Adora2a genes are CA307692, XM_003953847, BC081727, and AC137067, respectively. Asterisks mark the nucleotides that are identical across these four species.
uORF5 Exists in Vivo as a Product of the Adora2a Gene
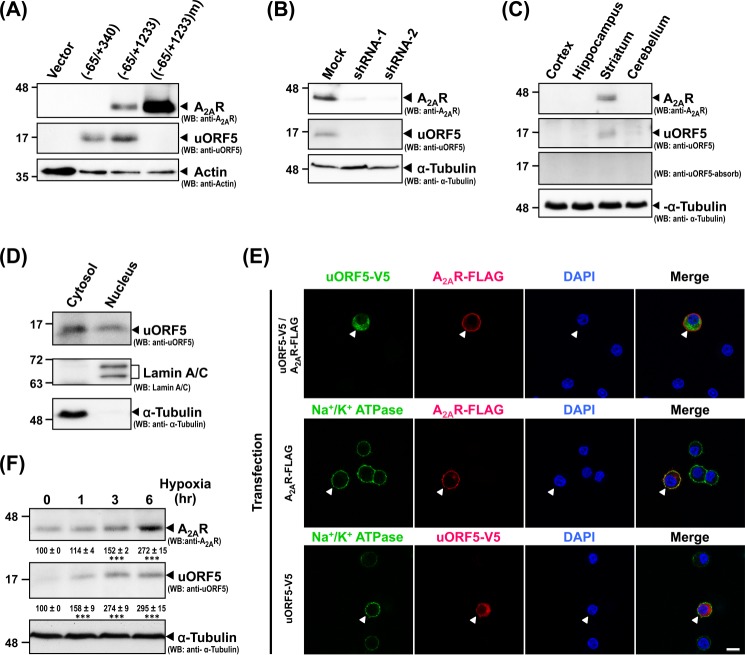
To investigate whether rat uORF5 exists in vivo, we generated a rabbit polyclonal antibody against recombinant uORF5 prepared in E. coli. This anti-uORF5 antibody detected a protein of the predicted size (∼16 kDa) of uORF5 in HEK293 cells transfected with a vector containing the coding sequence of uORF5 (−65/+340) or both ORFs of uORF5 and A2AR (−65/+1233) (Fig. 3A). When the translational start site of uORF5 (uAUG-5) was mutated ((−65/+1233)m), only A2AR (and not uORF5) was detected (Fig. 3A). In the PC12 cell line, which expresses endogenous Adora2a transcripts (28), the anti-uORF5 antibody recognized a band at ∼16 kDa (Fig. 3B). A significant amount of the A2AR protein was also detected in the PC12 cells (Fig. 3B). The transient transfection of two independent shRNAs against the Adora2a transcript effectively eliminated the expression of both the A2AR and uORF5 proteins (Fig. 3B), suggesting that the A2AR and uORF5 proteins are translated from the Adora2a transcripts. Similarly, a 16-kDa protein was detected in the rat striatum, where Adora2a was highly expressed (Fig. 3C) (20). The absorption of anti-uORF5 using the antigen (uORF5-His) bound on PVDF membranes completely eliminated the immunoreactivity of this 16-kDa protein. Taken together, these results suggest that the anti-uORF5 antibody specifically detects uORF5 in a Western blot analysis. In addition, consistent with the expression level of the Adora2a gene, uORF5 is expressed in PC12 cells and the striatum.
FIGURE 3.
uORF5 is endogenously expressed in PC12 cells, which endogenously express A2AR, and the rat striatum. A, HEK293 cells were transfected with the indicated constructs for 48 h. The total lysates were assessed by Western blot analysis (WB) using the indicated antibodies. B, PC12 cells were transfected with the indicated constructs for 48 h. Total lysates were harvested and analyzed by Western blot analysis. C, tissues were collected from 12-week-old SD rats. Total lysates of the cortex, hippocampus, striatum, and cerebellum were harvested to analyze the expression levels of A2AR, uORF5, and tubulin (loading control) by Western blot analysis. D, the cytosolic and nuclear fractions of the PC12 cells were prepared and analyzed by Western blot analysis. E, PC12 cells were transfected with the indicated construct(s) for 48 h, fixed, and subjected to immunofluorescent staining using the indicated antibodies. Arrows mark the transfected cells. The nuclei were labeled with DAPI (blue). F, PC12 cells were subjected to normoxia or hypoxia (5%) for the indicated periods of time. Total cell lysates were collected at the indicated time points and analyzed by Western blot analysis. The data are the means ± S.E. of at least three independent experiments. ***, p < 0.001, compared with the control cells (normoxia).
We next tested the subcellular localization of the uORF5 protein by cell fractionation and immunoblotting. Endogenous uORF5 was detected in both the cytosolic and nuclear PC12 cell fractions (Fig. 3D); lamin A/C and α-tubulin served as markers for the nuclear and cytosolic fractions, respectively. Immunostaining of PC12 cells also confirmed that uORF5 was expressed in the cytoplasm and nuclei but not in the plasma membrane that was labeled by the expression of Na+/K+-ATPase (a plasma membrane marker (38)). Conversely, A2AR was located in the Na+/K+-ATPase-positive plasma membrane (Fig. 3E). Consistent with our hypothesis that the expression of the uORF5 protein is driven by the transcript of the Adora2a gene, the level of the uORF5 protein was markedly elevated in PC12 cells, similar to that of the A2AR protein, upon hypoxia (5% O2, 5% CO2, and balance N2) (Fig. 3F), which was previously shown to up-regulate the Adora2a transcript (22).
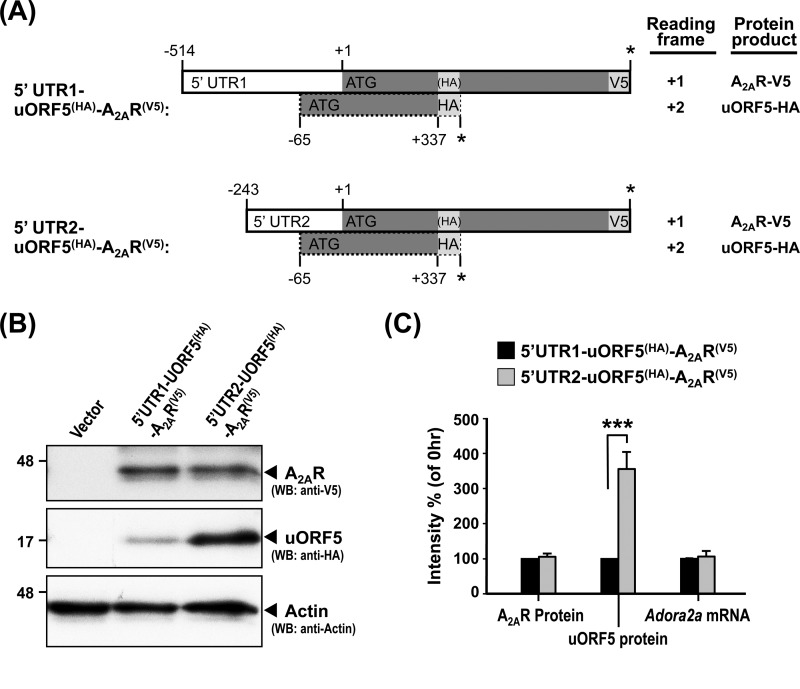
The Different 5′-UTRs of Adora2a mRNA Influence the Expression of the uORF5 Protein
We previously showed that the rat Adora2a gene contains at least two independent promoters, which drive the expression of two transcripts that contain the same coding regions for A2AR and uORF5 and 5′-UTRs of different lengths (5′-UTR1 and 5′-UTR2). To test whether different 5′-UTRs influence the translation of the uORF5 protein, we created a set of constructs with an in-frame HA tag inserted at the 3′-end of the uORF5 ORF (+2 reading frame) (Fig. 4A) and an in-frame V5 tag at the 3′-end of the A2AR ORF (+1 reading frame) (Fig. 4A). These two constructs are designated as 5′-UTR1- and 5′-UTR2-uORF5(HA)-A2AR(V5), respectively. The expression of uORF5-HA and A2AR-V5 in PC12 cells transfected with the indicated construct were detected by Western blot analysis using anti-HA and V5 antibodies (Fig. 4B). RT-qPCR analyses revealed that the levels of the mRNAs derived from 5′-UTR1- and 5′-UTR2-uORF5(HA)-A2AR(V5) were similar (Fig. 4C). As reported previously, no detectable difference in the effects of 5′-UTR1 and 5′-UTR2 on the protein level of A2AR-V5 was found (Fig. 4B). Surprisingly, compared with 5′-UTR2, 5′-UTR1 appeared to exert a negative effect on the expression of the uORF5 protein in PC12 cells (Fig. 4B). This inhibitory effect might be due to the additional uAUG (−335/−186) of a small ORF (150 bp) that is located in the 5′-UTR1-specific region of the Adora2a gene (AF107208) (20). During translational initiation, the small subunit of the ribosome binds to the 5′-cap of the mRNA and recognizes the first AUG, which promotes the assembly of the mature ribosome and initiates translation (39). Thus, the upstream uORF that is located in the 5′-UTR1 can be inhibitory for the translation of the downstream uORF5 cistron (40, 41), as has been observed for the 5′-UTRs of many genes (42–44). Interestingly, this inhibitory effect of the upstream uORF in 5′-UTR1 affected only the expression of the uORF5 protein but not that of the A2AR protein (Fig. 4B). This finding suggests that the expressions of the uORF5 and A2AR proteins are regulated independently.
FIGURE 4.
The expression of uORF5 is repressed by 5′-UTR1. A, schematic of the constructs harboring 5′-UTR1 (−514/−1) or 5′-UTR2 (−243/−1) fused to a dual expression fragment (uORF5(HA)-A2AR(V5)) that encodes both the uORF5 (uORF5(HA)) and A2AR (A2AR(V5)) proteins. The HA sequence (YPYDVPDYA) was inserted in-frame with the C terminus of uORF5 (+2 reading frame) but not with A2AR (+1 reading frame). The V5 sequence (GKPIPNPLLGLDST) was cloned in-frame with the C terminus of A2AR. PC12 cells were transiently transfected with the indicated constructs for 48 h. B, cells were harvested to analyze the protein levels of A2AR and uORF5 by Western blot analysis (WB). The A2AR and uORF5 protein levels were normalized to that of actin (loading control). C, cells were collected for RNA preparation. The level of the Adora2a transcript was determined by RT-qPCR and normalized to gapdh (reference gene). ***, p < 0.001, cells transfected with 5′-UTR1-uORF5(HA)-A2AR(V5) versus 5′-UTR2- uORF5(HA)-A2AR(V5). The values represent the means ± S.E. (error bars) of at least three independent experiments.
uORF5 Suppresses AP1-mediated Promoter Activity
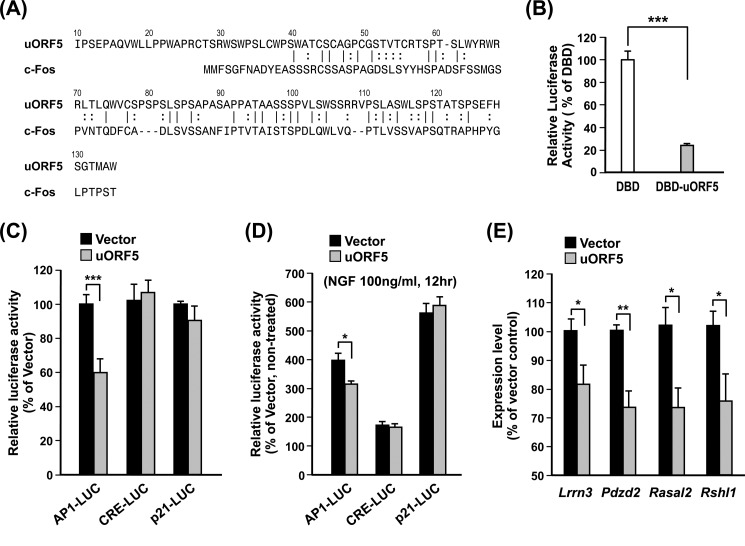
We next characterized the function of uORF5. Functional annotation of the uORF5 protein using LALIGN suggests that uORF5 shares 31.5% aa identity with the N terminus (aa 1–89) of c-Fos, where an activation domain (HOB1-N) is known to reside (Fig. 5A) (45). To examine whether uORF5 modulates transcription, we subcloned the coding sequence of uORF5 (−62 to +340) into the pFA-CMV vector so that uORF5 was fused to the C terminus of the GAL4 DNA-binding domain (DBD) (23). Cotransfection of the resultant construct (pFA-CMV-DBD:uORF5) with a GAL4-responsive luciferase reporter in the striatal progenitor cell line ST14A showed that DBD:uOF5, when compared with DBD alone, markedly suppressed the GAL4 promoter activity (Fig. 5B). Thus, uORF5 might possess the ability to regulate the transcription machinery. Given its similarity to the N terminus of c-Fos, we next assessed whether uORF5 modulates the activity of AP1 (i.e. the c-Fos·c-Jun complex) (46). Consistent with the above finding, the expression of uORF5 suppressed the activity of the AP1, but not the CRE or p21, luciferase reporter in ST14A cells (Fig. 5C). To confirm whether uORF5 modulates AP1 activity, PC12 cells were treated with NGF to activate AP1 (47). Promoter analyses showed that NGF enhanced the promoter activities mediated by AP1, CRE, and p21 in PC12 cells (data not shown). The elevated expression of uORF5 selectively reduced the NGF-induced AP1-mediated, but not the CRE- or p21-mediated, promoter activity (Fig. 5D). Collectively, these data suggest that uORF5 might, at least partially, function as a modulator of AP1-dependent transcription.
FIGURE 5.
uORF5 suppressed AP1-mediated promoter activity. A, alignment of uORF5 and the N terminus (aa 1–105) of rat c-Fos (XP_234422). B, ST14A cells were transfected with vectors encoding the GAL4 DBD or the chimeric DBD:uORF5 protein, a firefly luciferase reporter gene driven by five copies of the GAL4-binding element, and a thymidine kinase promoter-driven Renilla luciferase (as an internal control) for 48 h. The activity of firefly luciferase was measured and normalized to that of Renilla luciferase. ***, p < 0.001, cells with transfected DBD versus DBD-uORF5. C, cells were cotransfected with the uORF5 expression construct or the empty pcDNA3.1 vector and a firefly luciferase reporter driven by six copies of AP1 (AP1-LUC), three copies of CRE (CRE-LUC), or the p21 promoter (p21-LUC) plus an internal control (thymidine kinase promoter-driven Renilla luciferase) at a molar ratio of 5:4:1 for 48 h. The indicated firefly luciferase activity was normalized to the activity of Renilla luciferase. ***, p < 0.001, cells transfected with empty vector versus uORF5. D, PC12 cells were cotransfected with the uORF5 expression construct or the empty pcDNA3.1 vector and AP1-LUC, CRE-LUC, or the p21-LUC plus thymidine kinase promoter-driven Renilla luciferase (as an internal control) for 48 h. The cells were then treated with or without NGF (100 ng/ml) as indicated for 12 h. The AP1-, CRE-, and p21 promoter-mediated expression of firefly luciferase was normalized to the activity of Renilla luciferase. Relative activities of the indicated promoter in the NGF-treated cells to the non-treated cells are shown. *, p < 0.05, cells transfected with empty vector versus uORF5. E, PC12 cells were transiently transfected with hrGFP or uORF5:hrGFP for 48 h. The transfected cells were sorted for the expression of hrGFP and harvested for RNA preparation. The expression of the selected genes was measured by RT-qPCR and normalized to that of gapdh (reference gene). *, p < 0.05; **, p < 0.01, cells transfected with empty vector versus uORF5. The values represent the means ± S.E. (error bars) of at least three independent experiments.
To identify the potential downstream targets of uORF5, microarray gene profiling was performed to identify the genes whose expression was altered in the presence of uORF5. Striatal progenitor cells (STHdhQ7) were transfected with hrGFP or uORF5:hrGFP for 48 h. The transfected cells were sorted by the expression of hrGFP using flow cytometry and then harvested for RNA preparation and gene profiling by microarray. In three independent experiments, uORF5 altered the expression of only a small number of genes (Table 3). Of the 12 genes selected for further analyses, RT-qPCR was used to validate that the expression levels of five genes (Pdzd2, Lrrn3, Spag9, Rshl1, and Rasal2) were decreased by the expression of uORF5 in the STHdhQ7 cells (Table 3). Except for Spag9, which was not detected in the PC12 cells, the overexpression of uORF5:hrGFP in PC12 cells also led to the suppression of the previously mentioned uORF5-responsive genes (Pdzd2, Lrrn3, Rshl1, and Rasal2) when compared with cells expressing hrGFP alone (Fig. 5E). Literature mining suggests that these four uORF5-responsive genes might be involved in the regulation of the MAPK pathway (48–52).
TABLE 3.
Overexpression of uORF5-altered gene expression profiles in STHdhQ7 cells
STHdhQ7 cells were transfected with an uORF5:hrGFP or hrGFP expression construct for 48 h. The transfected cells were sorted by flow cytometry and harvested for RNA preparation. The RNA was analyzed by microarray and RT-qPCR. Twelve genes whose expression was altered at least ±1.5-fold in two of the three independent microarray experiments were selected for further verification using RT-qPCR. The data are the means ± S.E. of three independent experiments.
| Gene | Gene symbol | GenBankTM accession no. | Microarray 1 decrease | Microarray 2 decrease | Real-time PCR |
|---|---|---|---|---|---|
| % | % | ||||
| β2-Microglobulin precursor | B2m | AK019389 | −51.7 | −42.6 | 4.9 ± 10.2 |
| c-Jun-amino-terminal kinase-interacting protein 4 | Spag9 | AK147431 | −59.0 | −36.7 | −18.4 ± 3.2a |
| Deoxycytidine kinase | Dck | BC060062 | −64.0 | −50.7 | −2.6 ± 7.3 |
| Growth arrest-specific protein 1 precursor | Gas1 | AI595188 | −44.2 | −33.2 | −14.3 ± 8.4 |
| Kinesin-like protein KIF2A | Kif2a | AC154257 | −55.3 | −35.5 | −15.1 ± 8.6 |
| Leucine-rich repeat neuronal protein 3 | Lrrn3 | AC167364 | −63.5 | −41.7 | −20.6 ± 7.4b |
| Nucleophosmin 1 | Npm1 | CA540276 | −47.4 | −49.4 | −7.6 ± 0.0 |
| PDZ domain-containing 2 | Pdzd2 | AC133282 | −68.1 | −60.8 | −18.7 ± 4.5b |
| RAS protein activator-like 2 | Rasal2 | AC119220 | −48.6 | −40.9 | −21.9 ± 5.5b |
| Radial spoke head-like protein 1 | Rshl1 | AC170864 | −56.6 | −52.2 | −25.2 ± 11.2b |
| Thrombospondin-1 precursor | Thbs1 | AL845495 | 251.4 | 224.1 | −0.9 ± 9.2 |
| Trinucleotide repeat-containing gene 6B protein | Tnrc6b | AC125540 | −48.9 | −73.1 | −20.1 ± 10.8 |
a p < 0.01, cells transfected with uORF5:hrGFP versus hrGFP.
b p < 0.05, cells transfected with uORF5:hrGFP versus hrGFP.
A2AR Activation Increases the Expression of the uORF5 Protein
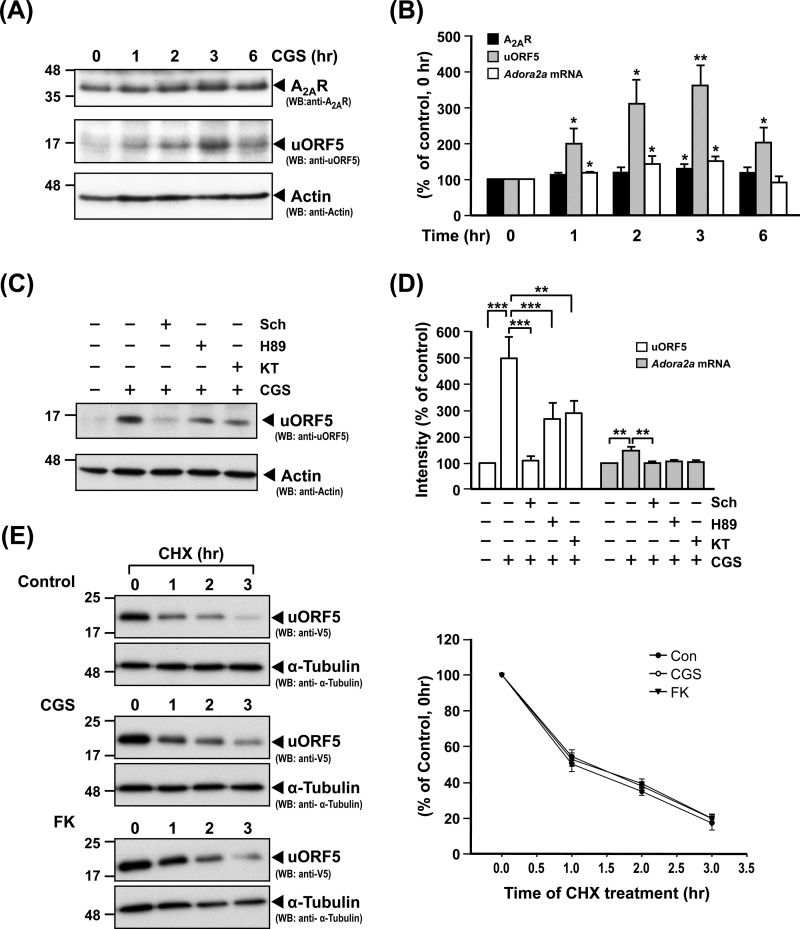
To investigate whether the uORF5 and A2AR proteins, which are translated from the Adora2a transcript, are functionally related, we activated A2AR in PC12 cells using an A2AR agonist (CGS) for 1–6 h and analyzed the protein levels of A2AR and uORF5 using a Western blot analysis. The activation of A2AR did not affect the level of the A2AR protein during the period of time tested, but it greatly elevated the level of the uORF5 protein (Fig. 6A). Although the level of the Adora2a transcript was also slightly enhanced, the increases in the Adora2a transcript and the A2AR protein were much lower than that of the uORF protein (Fig. 6B). For example, when compared with those of the control (non-treated) cells (100%), the levels of the Adora2a transcript, A2AR protein, and uORF5 protein after a 3-h treatment with CGS were 150.1 ± 13.3, 129.2 ± 11.8, and 359.6 ± 56.7% (mean ± S.E., three independent experiments), respectively. This enhancing effect of CGS on uORF5 was mediated by A2AR because it was found to be blocked by Sch58261 (Sch), an A2AR-selective inhibitor (Fig. 6C). Because stimulation of the A2AR is known to activate adenylyl cyclase and the cAMP/PKA pathway, we tested whether inhibition of PKA using H89 or KT5720 affected the enhancing effects of CGS on the uORF5 protein and Adora2a mRNA using an RT-qPCR and Western blot analysis. As shown in Fig. 6, C and D, H89 and KT5720 effectively prevented the enhancing effects of CGS, suggesting that activation of A2AR increased the protein level of uORF5 and the A2AR mRNA level of A2AR through a PKA-dependent pathway.
FIGURE 6.
Activation of A2AR leads to increased expression of the uORF5 protein. A and B, PC12 cells were pretreated with adenosine deaminase (ADA; 1 unit/ml) for 6 h and then treated with CGS (100 nm) for the indicated periods of time. A, Total cell lysates were harvested for the measurement of A2AR and uORF5 by Western blot analysis. B, The levels of the A2AR and uORF5 proteins were normalized to that of actin (loading control). Total RNA was collected for the determination of the Adora2a mRNA level by RT-qPCR. The level of Adora2a mRNA was normalized with that of gapdh (reference gene). *, p < 0.05; **, p < 0.01, compared with the control (0 h). C and D, PC12 cells were pretreated with adenosine deaminase for 6 h, incubated with or without the indicated inhibitor (H89, 1 μm; KT5720, 1 μm; Sch58261, 1 μm) for 1 h, and then treated for 3 h with CGS (100 nm). C, total cell lysates were harvested for the measurement of uORF5 protein by Western blot analysis. D, the uORF5 protein level was normalized to that of actin (loading control). Total RNA was collected for the determination of the Adora2a mRNA level by RT-qPCR. The level of Adora2a mRNA was normalized with that of gapdh (reference gene). ***, p < 0.001; **, p < 0.01. E, to evaluate the stability of the uORF5 protein, PC12 cells were transfected with uORF5-V5 for 48 h. The transfected cells were pretreated with adenosine deaminase for 6 h and treated with CGS or FK for 1 h. Cycloheximide (100 μg/ml) was then added to the medium to block protein synthesis. Total lysates were collected at the indicated times. The level of uORF5-V5 was measured by Western blot analysis and normalized with that of α-tubulin (internal control). The values represent the means ± S.E. (error bars) of at least three independent experiments.
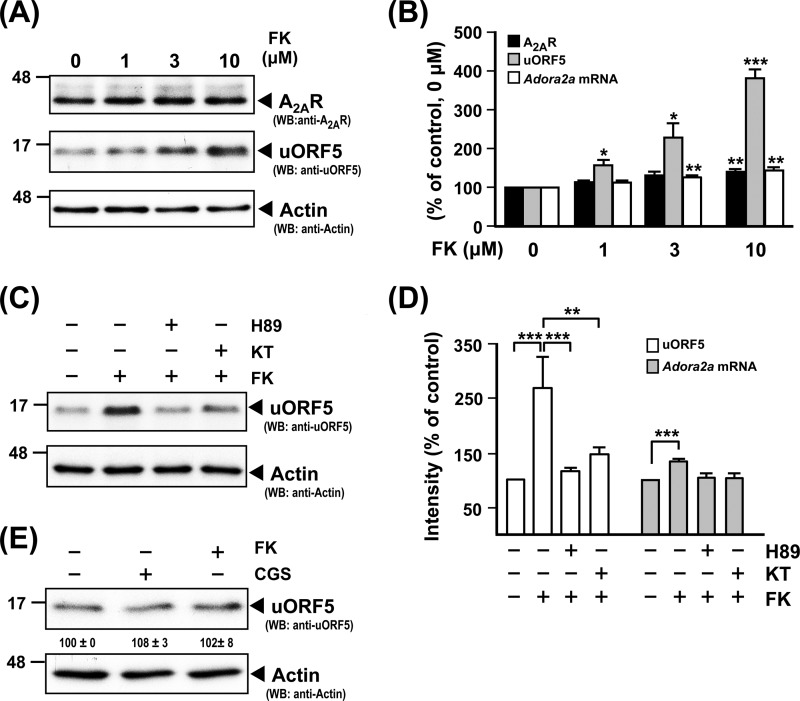
To validate that PKA regulates uORF expression, we next tested whether activation of PKA through the direct stimulation of adenylyl cyclase using FK altered the protein level of uORF5. After a 3-h incubation, FK dose-dependently increased expression levels of the uORF5 protein, A2AR protein, and Adora2a mRNA (Fig. 7, A and B). At a low dosage of FK (1 μm), only the uORF5 protein was enhanced. No effect on levels of the A2AR protein or Adora2a mRNA was found. However, at a high dosage of FK (10 μm), slight increases in levels of Adora2a mRNA and the A2AR protein were detected. When compared with those of control (non-treated) cells (100%), levels of the Adora2a transcripts, A2AR protein, and uORF5 protein after a 3-h treatment with FK at 10 μm were 143.9 ± 8.2, 140.7 ± 9.1, and 382.1 ± 23.2% (mean ± S.E., three independent experiments), respectively. Such enhancing effects of FK at 3 μm were mediated by PKA because they were blocked by both H89 and KT5760 (Fig. 7, C and D). In addition, we showed that neither CGS nor FK enhanced the protein level of uORF5 in a PKA-deficient PC12 mutant (A123 cells; Fig. 7E), further suggesting that PKA mediates expression of the uORF5 protein.
FIGURE 7.
PKA regulates expression of the uORF5 protein. A and B, PC12 cells were pretreated with adenosine deaminase (ADA; 1 unit/ml) for 6 h and then treated with FK of the indicated concentration. A, total cell lysates were harvested for the measurement of A2AR and uORF5 protein by Western blot analysis. B, levels of the A2AR and uORF5 proteins were normalized to that of actin (loading control). Total RNA was collected for the determination of the Adora2a mRNA level by RT-qPCR. The level of Adora2a mRNA was normalized with that of gapdh (reference gene). *, p < 0.05; **, p < 0.01; ***, p < 0.001, compared with the control (0 μm). C and D, PC12 cells were pretreated with adenosine deaminase for 6 h, incubated with or without the indicated inhibitor (H89, 10 μm; KT5720, 1 μm) for 1 h, and then treated for 3 h with FK (3 μm). C, total cell lysates were harvested for the measurement of uORF5 protein by Western blot analysis. D, the uORF5 protein level was normalized to that of actin (loading control). Total RNA was collected for the determination of the Adora2a mRNA level by RT-qPCR. The level of Adora2a mRNA was normalized with that of gapdh (reference gene). ***, p < 0.001; **, p < 0.01. E, A123 cells were pretreated with adenosine deaminase for 6 h and then treated with the indicated drug (CGS, 100 nm; FK, 3 μm). Total cell lysates were harvested for the measurement of uORF5 protein by Western blot analysis. The uORF5 protein level was normalized to that of actin (loading control). The data are the means ± S.E. (error bars) of at least three independent experiments.
To determine whether A2AR increases the level of the uORF5 protein by altering its stability, uORF5-V5 was exogenously expressed in PC12 cells treated with CGS or FK, as indicated. The stability of the uORF5 protein was determined by treating the cells with cycloheximide (100 μg/ml) to block new protein synthesis. The protein level of uORF5 was assessed by Western blot analysis and normalized to that of α-tubulin (internal control). As shown in Fig. 6E, uORF5 has a short half-life of ∼1 h. The protein degradation curves of the uORF5 protein in all of the conditions tested were similar, suggesting that the activation of A2AR or the PKA pathway did not affect the protein stability of uORF5. Therefore, the enhancing effect of A2AR signaling on the uORF protein is likely to occur at the post-translational level.
DISCUSSION
Other than the rat Adora2a gene, only three other dual coding genes containing alternative reading frames have been reported to date (7, 8, 53). For these three genes, a novel protein product was found to be translated from a transcript that was initially identified for the translation of another protein. For example, Klemke et al. (7) demonstrated that the rat XLαs gene encodes the extra large Gα subunit (XLαs) and a structurally irrelevant protein (ALEX), which is translated from the downstream +1 reading frame of the same mRNA. Intriguingly, ALEX can bind to the XL-domain of XLαs and, therefore, might regulate the function of XLαs. Another well characterized example is the PRNP gene. This gene encodes the prion protein (PrP) and the AltPrP protein that is translated from the +3 frame. AltPrP is detected in the mitochondria of human astrocytoma cell line U-118 and is up-regulated by endoplasmic reticulum stress and proteasomal inhibition (8). No functional interaction between the PrP and AltPrP proteins has been reported. During the preparation of this manuscript, a new dual coding gene (ATXN1) containing alternative reading frames was reported (53). The newly identified Alt-ATXN1 protein is translated from a downstream AUG (+3 frame) of the ATXN1 gene and functions as a novel interacting protein of the ATXN1 protein. In the present study, we showed that the rat Adora2a gene encodes a novel protein (uORF5) using a frame that differs from that used for the A2AR protein (Fig. 1). The expression of uORF5 was detected in rat tissues where the Adora2a transcript is markedly expressed (Fig. 3C) and under conditions in which Adora2a is up-regulated (e.g. hypoxia) (Fig. 3F). In addition, an ORF for a similar mouse uORF5 protein, which shares 84% aa identity with rat uORF5, was detected in the mouse Adora2a gene. In the human and chimpanzee genomes, the ORF for a shorter uORF5-like protein, which shares 75% aa identity with rat uORF5, was also detected. These uORF5-like proteins in humans and chimpanzees might use an AUG codon, which is flanked by a weak Kozak sequence (39) and is located downstream of the start codon of the A2AR protein (Figs. 1C and 2). The strength of the Kozak sequence is not a concern because ALEX, one of the two proteins encoded by the dual coding gene XLαs/Gαs, is known to be translated from a downstream AUG codon without an optimal Kozak sequence (7). It was proposed that when the first AUG is surrounded by a weak Kozak motif, the second AUG (with or without an optimal Kozak sequence) can be used to initiate protein translation due to leaky scanning (54). This is important because the AUG of the human A2AR protein is flanked with an imperfect Kozak sequence (Fig. 2). To determine whether the human uORF5-like protein exists in vivo would require additional tools (e.g. a specific antibody) and is certainly worth further investigation. These uORF5-like proteins from other species are likely to possess functions that are similar to that of the rat uORF5 protein due to their high homology with rat uORF5 (75–84% aa identity). These dual coding genes provide a new layer of complexity and plasticity in eukaryotic proteomes (55).
Database annotations of mammalian genomes suggest that the number of dual coding genes is higher than has been reported (9–11). Specifically, Ribrioux et al. (11) identified 217 potential dual coding genes, which have Kozak sequences located at their start codons and are conserved among the human, mouse, and rat genomes. It would be of great interest to further evaluate whether the proteins translated from these predicted dual coding genes exist in vivo. The functional interaction between two structurally irrelevant proteins that are translated from the same transcript, as demonstrated for the XLαs/ALEX gene (7) and the A2AR/uORF5 gene (Fig. 6), suggest that these dual coding genes might serve as tightly orchestrated sets of signals for the regulation of cellular functions. For example, through a PKA-dependent pathway (Figs. 6 and 7), the activation of A2AR increases the expression of the uORF5 protein, which, in turn, regulates the expression of a set of genes (Spag9, Lrrn3, Rasal2, and Rshl1) that are involved in the MAPK pathway (Fig. 5E and Table 3). Specifically, Spag9 is associated with c-Jun NH2-terminal kinases and may participate in the activation of the p38 MAPK pathway (49). The overexpression of Lrrn3 promotes the epidermal growth factor-induced phosphorylation of MAPK (48). Rasal2 is highly homologous to the GTPase-activating protein-related domain of Ras GTPase-activating proteins, which regulate the activation of Ras and are involved in the Ras-MAPK pathway (52, 56). Rshl1 inhibits the activation of ERK1/2 kinase during cell cycle progression (51). These data suggest that uORF5 may serve as a novel signaling molecule that mediates, at least in part, the functions of A2AR. Because the uORF5 and A2AR proteins are translated from the same transcript, we were not able to specifically down-regulate the expression of uORF5 without affecting the level of the A2AR protein using the standard siRNA approach to test the above hypothesis. Given that the uORF5 protein shares more than 30% amino acid identity with the activation domain of c-Fos (Fig. 5A), the potential role of A2AR/uORF5 in the regulation of gene transcription mediated by the c-Fos-containing AP1 complex is of great interest. A number of earlier studies showed that the activation of A2AR alters the level of c-Fos and, therefore, might affect AP1-mediated gene expression in the brain in a spatially dependent manner (57–59). Further investigation is required to determine whether uORF5 contributes to the modulation of AP1 activity by A2AR in vivo.
The mechanism by which A2AR/PKA increases the level of the uORF5 protein (Fig. 6C) is currently unknown. Because the stability of the uORF5 protein was not affected by PKA (Fig. 6E) and because the enhanced level of the Adora2a transcript was not sufficient to account for the marked enhancement of the uORF5 protein during A2AR activation (Fig. 6B), we reasoned that the activation of PKA might increase the translation of uORF5. The alternative usage of start codons present on the same transcript (60, 61) probably does not contribute to the selective enhancement of the uORF protein because the level of the A2AR protein was not significantly affected (Fig. 6B). For the very same reason, the PKA-mediated increase in the length of the poly(A) tail (62) is unlikely to be the cause for the increase in the uORF protein level. One interesting possibility is the alternative usage of the two A2AR promoters (P1 and P2), which produce different 5′-UTRs (5′-UTR1 and 5′-UTR2) in the Adora2a transcript (20). However, the expression level of 5′-UTR1 is much lower than that of 5′-UTR2 in PC12 cells, as demonstrated in our previous study (20). Therefore, a promoter switch from P1 to P2 probably does not contribute to the increase in the uORF5 protein by A2AR activation either. Further experiments are needed to delineate the regulation of uORF5 by A2AR-mediated cAMP signaling.
Our findings suggest that uORF5 plays a fine-tuning role because the elevated expression of uORF5 only moderately reduced the expression of its target genes (Fig. 5E and Table 3). Moreover, uORF5-mediated A2AR signaling is likely to function in tissues where the Adora2a transcript is highly expressed (e.g. the striatum) (Fig. 3C) (63) or under conditions in which the Adora2a transcript is significantly up-regulated (e.g. hypoxia) (Fig. 3F) (22). In addition to hypoxia, the expression of Adora2a mRNA is also highly elevated during inflammation (21). It has been well documented that the activation of A2AR suppresses inflammation in peripheral blood mononuclear cells and neutrophils (18, 64, 65). Because the expression of uORF5 is up-regulated during A2AR activation (Fig. 6), it is very likely that uORF5 is expressed in these inflamed cells and contributes to the anti-inflammatory effect of A2AR. Our findings suggest that the rat Adora2a gene encodes not only A2AR but also a transcriptional regulator, uORF5, in response to A2AR stimulation (Fig. 5); provide novel insights into our current understanding of the function of A2AR; and suggest the utility of future investigations into the potential dual coding genes in the mammalian genome.
This work was supported by National Science Council Grants NSC96-2321-B-001-015, NSC97-2321-B-001-012, and 100-2320-B-001-0110MY3 and the Institute of Biomedical Sciences of Academia Sinica.
- A2AR
- A2A adenosine receptor
- aRNA
- aminoallyl-RNA
- AP1
- activator protein 1
- CGS
- CHX
- cycloheximide
- DBD
- DNA-binding domain
- FK
- forskolin
- PrP
- prion protein
- RT-qPCR
- reverse transcription quantitative PCR
- XLαs
- extra large Gα subunit
- aa
- amino acid(s)
- hrGFP
- humanized Renilla reniformis-derived GFP.
REFERENCES
- 1. Huynen M. A., Konings D. A., Hogeweg P. (1993) Multiple coding and the evolutionary properties of RNA secondary structure. J. Theor. Biol. 165, 251–267 [DOI] [PubMed] [Google Scholar]
- 2. Barrell B. G., Air G. M., Hutchison C. A., 3rd (1976) Overlapping genes in bacteriophage phiX174. Nature 264, 34–41 [DOI] [PubMed] [Google Scholar]
- 3. Shaw D. C., Walker J. E., Northrop F. D., Barrell B. G., Godson G. N., Fiddes J. C. (1978) Gene K, a new overlapping gene in bacteriophage G4. Nature 272, 510–515 [DOI] [PubMed] [Google Scholar]
- 4. Oppenheim D. S., Yanofsky C. (1980) Translational coupling during expression of the tryptophan operon of Escherichia coli. Genetics 95, 785–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F., Schreier P. H., Smith A. J., Staden R., Young I. G. (1981) Sequence and organization of the human mitochondrial genome. Nature 290, 457–465 [DOI] [PubMed] [Google Scholar]
- 6. Bibb M. J., Van Etten R. A., Wright C. T., Walberg M. W., Clayton D. A. (1981) Sequence and gene organization of mouse mitochondrial DNA. Cell 26, 167–180 [DOI] [PubMed] [Google Scholar]
- 7. Klemke M., Kehlenbach R. H., Huttner W. B. (2001) Two overlapping reading frames in a single exon encode interacting proteins. A novel way of gene usage. EMBO J. 20, 3849–3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vanderperre B., Staskevicius A. B., Tremblay G., McCoy M., O'Neill M. A., Cashman N. R., Roucou X. (2011) An overlapping reading frame in the PRNP gene encodes a novel polypeptide distinct from the prion protein. FASEB J. 25, 2373–2386 [DOI] [PubMed] [Google Scholar]
- 9. Liang H., Landweber L. F. (2006) A genome-wide study of dual coding regions in human alternatively spliced genes. Genome Res. 16, 190–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chung W. Y., Wadhawan S., Szklarczyk R., Pond S. K., Nekrutenko A. (2007) A first look at ARFome. Dual-coding genes in mammalian genomes. PLoS Comput. Biol. 3, e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ribrioux S., Brüngger A., Baumgarten B., Seuwen K., John M. R. (2008) Bioinformatics prediction of overlapping frameshifted translation products in mammalian transcripts. BMC Genomics 9, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kulisevsky J., Poyurovsky M. (2012) Adenosine A2A-receptor antagonism and pathophysiology of Parkinson's disease and drug-induced movement disorders. Eur. Neurol. 67, 4–11 [DOI] [PubMed] [Google Scholar]
- 13. Von Lubitz D. K., Lin R. C., Jacobson K. A. (1995) Cerebral ischemia in gerbils. Effects of acute and chronic treatment with adenosine A2A receptor agonist and antagonist. Eur. J. Pharmacol. 287, 295–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chiang M. C., Chen H. M., Lai H. L., Chen H. W., Chou S. Y., Chen C. M., Tsai F. J., Chern Y. (2009) The A2A adenosine receptor rescues the urea cycle deficiency of Huntington's disease by enhancing the activity of the ubiquitin-proteasome system. Hum. Mol. Genet. 18, 2929–2942 [DOI] [PubMed] [Google Scholar]
- 15. Chou S. Y., Lee Y. C., Chen H. M., Chiang M. C., Lai H. L., Chang H. H., Wu Y. C., Sun C. N., Chien C. L., Lin Y. S., Wang S. C., Tung Y. Y., Chang C., Chern Y. (2005) CGS21680 attenuates symptoms of Huntington's disease in a transgenic mouse model. J. Neurochem. 93, 310–320 [DOI] [PubMed] [Google Scholar]
- 16. Eckle T., Koeppen M., Eltzschig H. K. (2009) Role of extracellular adenosine in acute lung injury. Physiology 24, 298–306 [DOI] [PubMed] [Google Scholar]
- 17. Weaver D. R., Reppert S. M. (1992) Adenosine receptor gene expression in rat kidney. Am. J. Physiol. 263, F991–F995 [DOI] [PubMed] [Google Scholar]
- 18. Leibovich S. J., Chen J. F., Pinhal-Enfield G., Belem P. C., Elson G., Rosania A., Ramanathan M., Montesinos C., Jacobson M., Schwarzschild M. A., Fink J. S., Cronstein B. (2002) Synergistic up-regulation of vascular endothelial growth factor expression in murine macrophages by adenosine A(2A) receptor agonists and endotoxin. Am. J. Pathol. 160, 2231–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weaver D. R. (1993) A2a adenosine receptor gene expression in developing rat brain. Brain Res. Mol. Brain Res. 20, 313–327 [DOI] [PubMed] [Google Scholar]
- 20. Lee Y. C., Chang C. W., Su C. W., Lin T. N., Sun S. H., Lai H. L., Chern Y. (1999) The 5′ untranslated regions of the rat A2A adenosine receptor gene function as negative translational regulators. J. Neurochem. 73, 1790–1798 [DOI] [PubMed] [Google Scholar]
- 21. Murphree L. J., Sullivan G. W., Marshall M. A., Linden J. (2005) Lipopolysaccharide rapidly modifies adenosine receptor transcripts in murine and human macrophages. Role of NF-κB in A(2A) adenosine receptor induction. Biochem. J. 391, 575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kobayashi S., Millhorn D. E. (1999) Stimulation of expression for the adenosine A2A receptor gene by hypoxia in PC12 cells. A potential role in cell protection. J. Biol. Chem. 274, 20358–20365 [DOI] [PubMed] [Google Scholar]
- 23. Fields S., Song O. (1989) A novel genetic system to detect protein-protein interactions. Nature 340, 245–246 [DOI] [PubMed] [Google Scholar]
- 24. Impellizzeri D., Di Paola R., Esposito E., Mazzon E., Paterniti I., Melani A., Bramanti P., Pedata F., Cuzzocrea S. (2011) CGS 21680, an agonist of the adenosine (A2A) receptor, decreases acute lung inflammation. Eur. J. Pharmacol. 668, 305–316 [DOI] [PubMed] [Google Scholar]
- 25. Konrad F. M., Neudeck G., Vollmer I., Ngamsri K. C., Thiel M., Reutershan J. (2013) Protective effects of pentoxifylline in pulmonary inflammation are adenosine receptor A2A dependent. FASEB J. 27, 3524–3535 [DOI] [PubMed] [Google Scholar]
- 26. Koizumi S., Odashima M., Otaka M., Jin M., Linden J., Watanabe S., Ohnishi H. (2009) Attenuation of gastric mucosal inflammation induced by indomethacin through activation of the A2A adenosine receptor in rats. J. Gastroenterol. 44, 419–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moore C. C., Martin E. N., Lee G. H., Obrig T., Linden J., Scheld W. M. (2008) An A2A adenosine receptor agonist, ATL313, reduces inflammation and improves survival in murine sepsis models. BMC Infect Dis. 8, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chern Y., Lai H. L., Fong J. C., Liang Y. (1993) Multiple mechanisms for desensitization of A2a adenosine receptor-mediated cAMP elevation in rat pheochromocytoma PC12 cells. Mol. Pharmacol. 44, 950–958 [PubMed] [Google Scholar]
- 29. De Ponti C., Carini R., Alchera E., Nitti M. P., Locati M., Albano E., Cairo G., Tacchini L. (2007) Adenosine A2a receptor-mediated, normoxic induction of HIF-1 through PKC and PI-3K-dependent pathways in macrophages. J. Leukoc. Biol. 82, 392–402 [DOI] [PubMed] [Google Scholar]
- 30. Lukashev D., Ohta A., Apasov S., Chen J. F., Sitkovsky M. (2004) Cutting edge. Physiologic attenuation of proinflammatory transcription by the Gs protein-coupled A2A adenosine receptor in vivo. J. Immunol. 173, 21–24 [DOI] [PubMed] [Google Scholar]
- 31. Cheng H. C., Shih H. M., Chern Y. (2002) Essential role of cAMP-response element-binding protein activation by A2A adenosine receptors in rescuing the nerve growth factor-induced neurite outgrowth impaired by blockage of the MAPK cascade. J. Biol. Chem. 277, 33930–33942 [DOI] [PubMed] [Google Scholar]
- 32. Ginty D. D., Glowacka D., DeFranco C., Wagner J. A. (1991) Nerve growth factor-induced neuronal differentiation after dominant repression of both type I and type II cAMP-dependent protein kinase activities. J. Biol. Chem. 266, 15325–15333 [PubMed] [Google Scholar]
- 33. Ehrlich M. E., Conti L., Toselli M., Taglietti L., Fiorillo E., Taglietti V., Ivkovic S., Guinea B., Tranberg A., Sipione S., Rigamonti D., Cattaneo E. (2001) ST14A cells have properties of a medium-size spiny neuron. Exp. Neurol. 167, 215–226 [DOI] [PubMed] [Google Scholar]
- 34. Higuchi R., Krummel B., Saiki R. K. (1988) A general method of in vitro preparation and specific mutagenesis of DNA fragments. Study of protein and DNA interactions. Nucleic Acids Res. 16, 7351–7367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen W., Yu Y. L., Lee S. F., Chiang Y. J., Chao J. R., Huang J. H., Chiong J. H., Huang C. J., Lai M. Z., Yang-Yen H. F., Yen J. J. (2001) CREB is one component of the binding complex of the Ces-2/E2A-HLF binding element and is an integral part of the interleukin-3 survival signal. Mol. Cell Biol. 21, 4636–4646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ou Y. H., Chung P. H., Sun T. P., Shieh S. Y. (2005) p53 C-terminal phosphorylation by CHK1 and CHK2 participates in the regulation of DNA-damage-induced C-terminal acetylation. Mol. Biol. Cell 16, 1684–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Harlow E., Lane D. (1988) Antibodies: A Laboratory Manual, 1st Ed., pp. 59–137, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 38. Lambrecht N., Munson K., Vagin O., Sachs G. (2000) Comparison of covalent with reversible inhibitor binding sites of the gastric H,K-ATPase by site-directed mutagenesis. J. Biol. Chem. 275, 4041–4048 [DOI] [PubMed] [Google Scholar]
- 39. Kozak M. (1999) Initiation of translation in prokaryotes and eukaryotes. Gene 234, 187–208 [DOI] [PubMed] [Google Scholar]
- 40. Jackson R. J., Hellen C. U., Pestova T. V. (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hood H. M., Neafsey D. E., Galagan J., Sachs M. S. (2009) Evolutionary roles of upstream open reading frames in mediating gene regulation in fungi. Annu. Rev. Microbiol. 63, 385–409 [DOI] [PubMed] [Google Scholar]
- 42. Jousse C., Bruhat A., Carraro V., Urano F., Ferrara M., Ron D., Fafournoux P. (2001) Inhibition of CHOP translation by a peptide encoded by an open reading frame localized in the chop 5′UTR. Nucleic Acids Res. 29, 4341–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Watatani Y., Ichikawa K., Nakanishi N., Fujimoto M., Takeda H., Kimura N., Hirose H., Takahashi S., Takahashi Y. (2008) Stress-induced translation of ATF5 mRNA is regulated by the 5′-untranslated region. J. Biol. Chem. 283, 2543–2553 [DOI] [PubMed] [Google Scholar]
- 44. Lee J., Park E. H., Couture G., Harvey I., Garneau P., Pelletier J. (2002) An upstream open reading frame impedes translation of the huntingtin gene. Nucleic Acids Res. 30, 5110–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brown H. J., Sutherland J. A., Cook A., Bannister A. J., Kouzarides T. (1995) An inhibitor domain in c-Fos regulates activation domains containing the HOB1 motif. EMBO J. 14, 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Eferl R., Wagner E. F. (2003) AP-1. A double-edged sword in tumorigenesis. Nat Rev. Cancer 3, 859–868 [DOI] [PubMed] [Google Scholar]
- 47. Tong L., Perez-Polo J. R. (1996) Effect of nerve growth factor on AP-1, NF-κB, and Oct DNA binding activity in apoptotic PC12 cells. Extrinsic and intrinsic elements. J. Neurosci. Res. 45, 1–12 [DOI] [PubMed] [Google Scholar]
- 48. Fukamachi K., Matsuoka Y., Ohno H., Hamaguchi T., Tsuda H. (2002) Neuronal leucine-rich repeat protein-3 amplifies MAPK activation by epidermal growth factor through a carboxyl-terminal region containing endocytosis motifs. J. Biol. Chem. 277, 43549–43552 [DOI] [PubMed] [Google Scholar]
- 49. Kelkar N., Standen C. L., Davis R. J. (2005) Role of the JIP4 scaffold protein in the regulation of mitogen-activated protein kinase signaling pathways. Mol. Cell. Biol. 25, 2733–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jagadish N., Rana R., Selvi R., Mishra D., Garg M., Yadav S., Herr J. C., Okumura K., Hasegawa A., Koyama K., Suri A. (2005) Characterization of a novel human sperm-associated antigen 9 (SPAG9) having structural homology with c-Jun N-terminal kinase-interacting protein. Biochem. J. 389, 73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Grummt M. V. K. M. (2006) Development and Application of a High Throughput Cell Based Assay to Identify Novel Modulators of ERK1/2 Activation and Functional Characterisation of the Candidate Radial Spokehead Like (Rshl1). Ph.D. dissertation, University of Heidelberg [Google Scholar]
- 52. Noto S., Maeda T., Hattori S., Inazawa J., Imamura M., Asaka M., Hatakeyama M. (1998) A novel human RasGAP-like gene that maps within the prostate cancer susceptibility locus at chromosome 1q25. FEBS Lett. 441, 127–131 [DOI] [PubMed] [Google Scholar]
- 53. Bergeron D., Lapointe C., Bissonnette C., Tremblay G., Motard J., Roucou X. (2013) An out-of-frame overlapping reading frame in the ataxin-1 coding sequence encodes a novel ataxin-1 interacting protein. J. Biol. Chem. 288, 21824–21835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xu H., Wang P., Fu Y., Zheng Y., Tang Q., Si L., You J., Zhang Z., Zhu Y., Zhou L., Wei Z., Lin B., Hu L., Kong X. (2010) Length of the ORF, position of the first AUG and the Kozak motif are important factors in potential dual-coding transcripts. Cell Res. 20, 445–457 [DOI] [PubMed] [Google Scholar]
- 55. Kochetov A. V. (2008) Alternative translation start sites and hidden coding potential of eukaryotic mRNAs. BioEssays 30, 683–691 [DOI] [PubMed] [Google Scholar]
- 56. Vojtek A. B., Der C. J. (1998) Increasing complexity of the Ras signaling pathway. J. Biol. Chem. 273, 19925–19928 [DOI] [PubMed] [Google Scholar]
- 57. Svenningsson P., Le Moine C., Fisone G., Fredholm B. B. (1999) Distribution, biochemistry, and function of striatal adenosine A2A receptors. Prog. Neurobiol. 59, 355–396 [DOI] [PubMed] [Google Scholar]
- 58. Pinna A., Wardas J., Cristalli G., Morelli M. (1997) Adenosine A2A receptor agonists increase Fos-like immunoreactivity in mesolimbic areas. Brain Res. 759, 41–49 [DOI] [PubMed] [Google Scholar]
- 59. Satoh S., Matsumura H., Koike N., Tokunaga Y., Maeda T., Hayaishi O. (1999) Region-dependent difference in the sleep-promoting potency of an adenosine A2A receptor agonist. Eur. J. Neurosci. 11, 1587–1597 [DOI] [PubMed] [Google Scholar]
- 60. Okazaki S., Ito T., Ui M., Watanabe T., Yoshimatsu K., Iba H. (1998) Two proteins translated by alternative usage of initiation codons in mRNA encoding a JunD transcriptional regulator. Biochem. Biophys. Res. Commun. 250, 347–353 [DOI] [PubMed] [Google Scholar]
- 61. Delmas V., Laoide B. M., Masquilier D., de Groot R. P., Foulkes N. S., Sassone-Corsi P. (1992) Alternative usage of initiation codons in mRNA encoding the cAMP-responsive-element modulator generates regulators with opposite functions. Proc. Natl. Acad. Sci. U.S.A. 89, 4226–4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Emanuel R. L., Iwasaki Y., Arbiser Z. K., Velez E. M., Emerson C. H., Majzoub J. A. (1998) Vasopressin messenger ribonucleic acid regulation via the protein kinase A pathway. Endocrinology 139, 2831–2837 [DOI] [PubMed] [Google Scholar]
- 63. Schiffmann S. N., Fisone G., Moresco R., Cunha R. A., Ferré S. (2007) Adenosine A2A receptors and basal ganglia physiology. Prog. Neurobiol. 83, 277–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. McColl S. R., St-Onge M., Dussault A. A., Laflamme C., Bouchard L., Boulanger J., Pouliot M. (2006) Immunomodulatory impact of the A2A adenosine receptor on the profile of chemokines produced by neutrophils. FASEB J. 20, 187–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sun W. C., Berghaus L. J., Moore J. N., Hurley D. J., Vandenplas M. L., Thompson R., Linden J. (2010) Lipopolysaccharide and TNF-α modify adenosine A(2A) receptor expression and function in equine monocytes. Vet. Immunol. Immunopathol. 135, 289–295 [DOI] [PubMed] [Google Scholar]
- 66. Cattaneo E., Conti L. (1998) Generation and characterization of embryonic striatal conditionally immortalized ST14A cells. J. Neurosci. Res. 53, 223–234 [DOI] [PubMed] [Google Scholar]