Background: Proteins of the Sac10b family are widespread in Archaea.
Results: Ssh10b, a member of the Sac10b family from Sulfolobus shibatae, disrupts base pairing in structured RNA through interactions between adjacently bound protein dimers.
Conclusion: Binding by Ssh10b destabilizes RNA secondary structure.
Significance: Proteins of the Sac10b family may play a role in RNA transactions requiring destabilization of RNA secondary structure.
Keywords: Archaea, Crystal Structure, RNA-binding Protein, RNA Structure, RNA-Protein Interaction, Sac10b Family, Ssh10b, Sulfolobus shibatae
Abstract
Proteins of the Sac10b family are highly conserved in Archaea. Ssh10b, a member of the Sac10b family from the hyperthermophilic crenarchaeon Sulfolobus shibatae, binds to RNA in vivo. Here we show that binding by Ssh10b destabilizes RNA secondary structure. Structural analysis of Ssh10b in complex with a 25-bp RNA duplex containing local distortions reveals that Ssh10b binds the two RNA strands symmetrically as a tetramer with each dimer bound asymmetrically to a single RNA strand. Amino acid residues involved in double-stranded RNA binding are similar, but non-identical, to those in dsDNA binding. The dimer-dimer interaction mediated by the intermolecular β-sheet appears to facilitate the destabilization of base pairing in the secondary structure of RNA. Our results suggest that proteins of the Sac10b family may play important roles in RNA transactions requiring destabilization of RNA secondary structure in Sulfolobus.
Introduction
RNA is more susceptible to hydrolysis at higher temperature in vitro (1, 2). However, mRNAs show longer than expected half-lives in hyperthermophilic Archaea (3). The propensity to form higher order structures through base pairing appears to contribute to the thermal stability of RNA in thermophiles (1, 2). On the other hand, destabilization of structured RNA is required to facilitate fundamental processes such as translation and exosome-mediated degradation (4, 5). No proteins capable of destabilizing higher order RNA structures have been found in Archaea.
Members of the Sac10b family are highly conserved among Archaea. All genome-sequenced archaea except for mesophilic euryarchaea of the Methanosarcinales and Halobacteriales encode one to two copies of a Sac10b homolog. However, despite their widespread presence, these proteins vary greatly among different Archaea in gene expression level, nucleic acid binding properties, and presumably physiological function (6–8). Sequences bearing similarity to those of the Sac10b genes have also been detected in some Eukarya such as yeast and humans (9, 10).
Sac10b homologs were initially identified and characterized in Sulfolobus species as small, basic, and abundant DNA-binding proteins capable of binding cooperatively to DNA without apparent sequence specificity and constraining DNA into negative supercoils in a temperature-dependent manner (11–14). It was shown that native Sso10b, a Sac10b homolog from Sulfolobus solfataricus, was acetylated on Lys-16, and acetylation reduced the affinity of the protein for DNA (10). Thus, the Sac10b protein family is also referred to as “Alba,” which stands for “acetylation lowers binding affinity” (10). Given their biochemical properties and cellular abundance in hyperthermophiles, Sac10b homologs have been proposed to play a role in DNA protection and stabilization at high temperature (10, 14, 15). Recently, it was shown that Sso10b was capable of bridging and rigidifying DNA in vitro (16). However, it was also found that Sac10b homologs bound RNA. Ssh10b from Sulfolobus shibatae bound both DNA and RNA, and binding of the protein to DNA was more sensitive to salts than that to RNA in vitro (17). More importantly, RNAs were bound by Ssh10b in vivo, and a significant portion of the protein was associated with ribosomes in a salt-sensitive fashion. In a separate study, both DNase and RNase released Sso10b from cellular nucleic acid-protein complexes, suggesting that this protein was associated with both DNA and RNA in vivo (18). These findings are consistent with a phylogenetic analysis showing that the Sac10b family is related to two eukaryotic protein families involved in RNA binding (9) and with recent reports that Sac10b homologs from Trypanosoma brucei and Toxoplasma gondii bound RNA and were likely involved in translational regulation (19, 20).
Structural studies on several hyperthermophilic members of the Sac10b family have shown that this protein family shares a conserved fold comprising two α-helices and four β-strands that is structurally similar to the C-terminal RNA binding domain of IF3 (15, 21, 22). Recently, the crystal structure of Ape10b2, a Sac10b homolog from Aeropyrum pernix, in complex with a duplex DNA fragment has been determined (23). It reveals the structural determinants for cooperative binding by an Ape10b2 dimer to a duplex DNA fragment in successive minor grooves. However, the structural basis for RNA binding by a Sac10b homolog remains to be understood.
The physiological roles of the Sac10b family are unclear. Increasing evidence suggests that the function of Sac10b homologs may have diverged during evolution. Although these proteins are abundant and bind both DNA and RNA sequences nonspecifically in hyperthermophiles, they are drastically less abundant and either bind specific DNA sequences or bind neither DNA nor RNA in mesophiles or thermophiles (7, 8, 17). Deletion of genes encoding Sac10b homologs in two mesophilic methanogens, Methanococcus voltae and Methanococcus maripaludis, resulted in changes in the expression levels of only a few genes (6, 7). These observations indicate that the Sac10b protein family may have evolved from a more crucial role in hyperthermophiles to a nonessential role in mesophiles.
In this study, we show that binding by Ssh10b destabilized the secondary structure of RNA. We determined the crystal structure of Ssh10b in complex with a 25-bp structured RNA duplex with local mismatches. Ssh10b binds the RNA as a tetramer. The dimer-dimer interaction plays an important role in the destabilization of base pairing in the bound RNA. Our results suggest that proteins of the Sac10b family may play important roles in RNA transactions requiring destabilization of RNA secondary structure in the hyperthermophilic Archaea.
EXPERIMENTAL PROCEDURES
Recombinant Wild-type and Mutant Ssh10b
Recombinant Ssh10b was prepared as described previously with the following modifications (11). The Resource S column fractions containing Ssh10b were pooled and loaded onto a Superdex G75 column (24 ml; GE Healthcare) equilibrated in 30 mm potassium phosphate, pH 6.6 and 150 mm KCl. Fractions containing Ssh10b were pooled and dialyzed against 10 mm Tris-Cl, pH 7.5, 1 mm EDTA, and 1 mm DTT.
Constructs for Ssh10b mutants were prepared by overlap extension PCR (24) using pET30a-Ssh10b as the template (for primer sequences, see Table 1). Sequences of the inserts in all constructs were verified by DNA sequencing. The mutant proteins were overproduced and purified as described previously (11). The proteins eluted from the HiTrap SP column (5 ml; GE healthcare) were dialyzed against 10 mm Tris-Cl, pH 7.5, 1 mm EDTA, and 1 mm DTT. The purified proteins were stored at −80 °C. Protein concentrations were determined by the Lowry method using bovine serum albumin (BSA) as the standard.
TABLE 1.
Oligonucleotides used in the construction of Ssh10b mutants and the preparation of DNA templates for in vitro transcription reactions
Forward and reverse primers are indicated by f and r, respectively.
| Designation | Sequence (5′–3′) |
|---|---|
| K16A(f) | TCTTAATAGGAGCGAAACCAGTAATGAACTATGTTT |
| K16A(r) | ACTGGTTTCGCTCCTATTAAGACTACATTACTTGGAG |
| K17A(f) | TTAATAGGAAAGGCACCAGTAATGAACTA |
| K17A(r) | TGCCTTTCCTATTAAGACTACATTACTTG |
| Y22A(f) | CAGTAATGAACGCTGTTTTAGCAGCATTAACTCTATT |
| Y22A(r) | GCTGCTAAAACAGCGTTCATTACTGGTTTCTTTCCTA |
| R42A(f) | TAATCAAAGCTGCAGGAAGAGCTATTAGCAAAGCCGTAG |
| R42A(r) | ATAGCTCTTCCTGCAGCTTTGATTACTATTTCGCTTACT |
| R44A(f) | AAAGCTAGAGGAGCAGCTATTAGCAAAGCCGTAGATACTG |
| R44A(r) | CTTTGCTAATAGCTGCTCCTCTAGCTTTGATTACTATTTC |
| K16A/Y22A(f) | GGAGCGAAACCAGTAATGAACGCTGTTTTAGCAGCATTAAC |
| K16A/Y22A(r) | AAAACAGCGTTCATTACTGGTTTCGCTCCTATTAAG |
| F60E(f) | AGTAAGGAACAGAGAATTACCAGACAAGATAGAGATTAAA |
| F60E(r) | CTTGTCTGGTAATTCTCTGTTCCTTACTATTTCTACAGTA |
| M20E/L24E/L27E(f) | ACTATGTTGAAGCAGCAGAAACTCTATTAAACCAAGGAGTA |
| M20E/L24E/L27E(r) | TGCTGCTTCAACATAGTTCTCTACTGGTTTCTTTCCTATTAAG |
| ΔN12(f) | GGAGATATACATATGTTAATAGGAAAGAAACCAGTAATGAAC |
| ΔN12(r) | TTTCTTTCCTATTAACATATGTATATCTCCTTCTTAAAGTTAAA |
| For preparation of the DNA template for rGrG(rArCrUrG)8 | CAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCCTATAGTGAGTCGTATTAGAATTCCACAGT/ACTGTGGAATTCTAATACGACTCACTATAG |
RNA Preparation
Partial B2 RNA and rGrG(rArCrUrG)8 were synthesized using a T7 in vitro transcription kit (Promega) according to the manufacturer's procedure. Plasmid 443 containing the partial B2 gene sequence, a generous gift from Jennifer Kugel (25), was linearized with DraI. The linearized fragment was purified by gel electrophoresis for use as the template for the synthesis of the partial B2 RNA. A DNA template for the preparation of rGrG(rArCrUrG)8 was made by annealing a 63-nt4 oligonucleotide with a 30-nt oligonucleotide (Table 1) and subsequent extension of the shorter oligonucleotide by Klenow fragment (Takara). The RNA fragments were dephosphorylated with alkaline phosphatase (New England Biolabs), extracted with phenol/chloroform, precipitated with ethanol, and radiolabeled at the 5′-end with [γ-32P]ATP using T4 polynucleotide kinase.
All other oligoribonucleotides used in this study (Table 2) were synthesized at Takara Biotech. A 25-nt RNA for the crystallization trials was heated for 2 min at 70 °C and cooled slowly to room temperature to allow the formation of a dsRNA fragment containing mismatched base pairs. A 30-nt oligo(rA) was labeled at the 5′-end with [γ-32P]ATP using T4 polynucleotide kinase. To prepare a radiolabeled 25-bp dsRNA fragment for electrophoretic mobility shift assay (EMSA), one of the two strands was labeled at the 5′-end with [γ-32P]ATP and subsequently annealed to the complementary strand.
TABLE 2.
RNA fragments used in the present study
| Designation | Sequence (5′–3′) |
|---|---|
| Partial B2 RNA | GGGCUGGUGAGAUGGCUCAGUGGGUAAGAGCACCCGACUGCUCUUCCGAAGGUCAGGAGUUCAAAUCCCAGCA |
| rGrG(rArCrUrG)8 | GGACUGACUGACUGACUGACUGACUGACUGACUG |
| 30-nt oligo(rA) | AAAAAAAAAAAAAAAAAAAAAAAAAAAAAA |
| RNA for crystallization | GGUAAGAGCACCCGACUGCUCUUCC |
| 25-bp dsRNA for EMSA | GGUAAGAGCACCCGACUGCUCUUCC/GGAAGAGCAGUCGGGUGCUCUUACC |
EMSAs
The 32P-labeled partial B2 RNA fragment was heated for 1 min at 70 °C in 10 mm Tris-HCl, pH 7.5 and 10 mm MgCl2 and snap cooled on ice to allow the formation of secondary structure. The 32P-labeled partial B2 RNA, rGrG(rArCrUrG)8, 30-nt oligo(rA), or 25-bp dsRNA (∼10 nm) was incubated for 10 min at 22 °C with indicated amounts of wild-type or mutant Ssh10b in 20 mm HEPES-NaOH, pH 6.9, 1 mm DTT, 100 μg/ml BSA, and 8% glycerol. After incubation, the reaction mixtures were subjected to electrophoresis in a 12 or 15% polyacrylamide gel. Electrophoresis was performed in 0.1× Tris borate-EDTA. The gel was exposed to x-ray film and analyzed by ImageQuant and a Storm phosphorimaging system (GE Healthcare). The apparent dissociation constant (KD) was measured as the concentration of the protein at which half of the input RNA was retarded (26).
RNase T1 Footprinting
32P-Labeled partial B2 RNA was heated for 1 min at 70 °C in 10 mm Tris-HCl, pH 7.5 and 10 mm MgCl2 and snap cooled on ice to allow the formation of secondary structure. The labeled partial B2 RNA fragment (∼10 pmol) was then incubated with different amounts of Ssh10b at 22 °C for 10 min in 20 mm sodium acetate, pH 5.0, 1 mm EDTA, and 0.05 μg/μl yeast tRNA. RNase T1 (1 unit; Roche Applied Science) was added. After 15 min at 22 °C, the mixture was extracted with phenol/chloroform, and RNA was precipitated with ethanol. As a nonstructured RNA control, the radiolabeled partial B2 RNA was digested with RNase T1 (1 unit) for 15 min at 50 °C in 20 mm sodium acetate, pH 5.0, 1 mm EDTA, 7 m urea, and 0.05 μg/μl yeast tRNA as described previously (27). RNA ladders were generated by incubating the radiolabeled RNA for 5 min at 90 °C in 50 mm sodium carbonate, pH 9.0, 1 mm EDTA, and 0.05 μg/μl yeast tRNA. The samples were resolved by electrophoresis in a 15% polyacrylamide gel containing 8 m urea in 1× Tris borate-EDTA. The gel was exposed to x-ray film or analyzed by phosphorimaging (GE Healthcare).
Dimethyl Sulfate (DMS) Footprinting
DMS footprinting was performed as described previously with the following modifications (28). Partial B2 RNA was heated for 1 min at 70 °C in 20 mm HEPES-NaOH, pH 7.5 and 10 mm MgCl2 and snap cooled on ice. The RNA fragment (∼40 pmol) was then incubated with increasing amounts of Ssh10b at 22 °C for 10 min in 50 mm sodium cacodylate, pH 7.5 and 1 mm EDTA. DMS (0.5%) was added, and the mixture was incubated at 22 °C for 5 min. Reactions were quenched and extracted, and then the RNAs were processed for primer extension with Moloney murine leukemia virus reverse transcriptase (New England Biolabs) using a 5′-32P-labeled primer complementary to a stretch of nucleotides (positions 49–73) in the RNA. The samples were resolved by electrophoresis in a 15% polyacrylamide gel containing 8 m urea in 1× Tris borate-EDTA. The gel was exposed to x-ray film.
Blue Native Polyacrylamide Gel Electrophoresis
A sample (∼8 μg) of wild-type or mutant Ssh10b was mixed with 4× loading buffer (200 mm imidazole, 200 mm NaCl, 20% glycerol, and 0.02% Ponceau S) and electrophoresed through a 6–20% gradient blue native polyacrylamide gel (29). The gel was stained with Coomassie Brilliant Blue G-250.
Chemical Cross-linking
Ssh10b (∼ 8 μg) was cross-linked with 3 mm DSP (Dithiobis[succinimidyl propionate], Pierce) in the presence or absence of the 25-bp dsRNA (∼ 5 μg) for 1 h at 22 °C in 20 mm potassium phosphate buffer, pH 7.6, and 50 mm KCl. Cross-linking reactions were stopped by the addition of the sample loading buffer for SDS-PAGE (mercaptoethanol was omitted). The samples were subjected to 15% SDS-PAGE, and the gel was stained with Coomassie Brilliant Blue G-250.
Sequence Alignment
The amino acid sequences of Sac10b homologs were aligned using ClustalX (30), and the alignment was edited with Genedoc.
Crystallization, Data Collection, and Structure Determination
Freshly purified Ssh10b (∼20 mg/ml) was mixed with the 25-bp dsRNA at a molar ratio of 1:1.5. Crystallization experiments were performed using the hanging drop vapor diffusion method at 293 K with 2-μl drops containing a 1 μl of protein solution and 1 μl of reservoir solution equilibrated over 0.5 ml of reservoir solution. Initial crystallization hits were found using an Index kit (Hampton Research, Aliso Viejo, CA). The Ssh10b-RNA complex crystals were grown in 18% (v/v) PEG 5000 monomethyl ether, 100 mm Bis-Tris, pH 7.0, and 200 mm sodium malonate. The Ssh10b-RNA complex data set were collected at beamline 17A of the Photon Factory (High Energy Accelerator Research Organization, Tsukuba, Japan). Prior to data collection, the crystal was transferred to a well solution supplied with 15% (v/v) ethylene glycol as a cryoprotectant.
The data set was processed with MOSFLM and scaled with SCALA from the CCP4 program suite (31). The structure of the Ssh10b-RNA complex was solved by molecular replacement with PHASER of the CCP4 program suite (31) using the apo-Ssh10b structure as a search model (Protein Data Bank code 1H0X). RNA was readily traceable and manually built with Coot (32). Structural refinement was performed with PHENIX (33). The statistics of data collection and refinement are summarized in Table 3. The quality of the final model was checked by MolProbity (34). All structural figures were rendered in PyMOL.
TABLE 3.
Data collection and refinement statistics
Values in parentheses are for highest resolution shell. r.m.s., root mean square.
| Sso10b-RNA complex | |
|---|---|
| Data collection | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 169.89, 69.76, 48.13 |
| α, β, γ (°) | 90, 96.34, 90 |
| Wavelength (Å) | 0.9642 |
| Resolution (Å) | 50-2.00 (2.11-2.00) |
| Rmerge (%) | 7.4 (40.7) |
| Average I/σ(I) | 11.0 (3.1) |
| Completeness (%) | 99.2 (98.7) |
| Redundancy | 3.6 (3.6) |
| Refinement | |
| Resolution (Å) | 50-2.00 |
| No. reflections | 37,589 |
| Rwork/Rfree (%) | 21.20/24.86 |
| No. atoms | |
| Protein | 2,664 |
| RNA | 1,050 |
| Water | 310 |
| B-factors (Å2) | |
| Protein | 38.6 |
| RNA | 44.9 |
| Water | 42.0 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.7 |
RESULTS
Binding by Ssh10b Destabilizes the Secondary Structure of RNA
We have previously found that Ssh10b binds to RNA in S. shibatae (17). It is known that native RNA tends to fold or misfold into secondary and tertiary structures in vivo, and structured RNAs often contain stretches of short A-form helices (typically <10 base pairs) (35). To determine whether RNA binding by Ssh10b was sensitive to the structure of the bound RNA, we compared the binding of Ssh10b to a partial short interspersed element-encoded mouse B2 RNA molecule containing short A-form helices (Fig. 1B) (25) with that to an unstructured single-stranded RNA (rGrG(rArCrUrG)8 or 30-nt oligo(rA)) by using electrophoretic mobility shift assays. The mouse B2 RNA was used because it was structurally well studied, and the availability of the patterns of cleavage of the RNA with different ribonucleases would be useful to the analysis of the effect of Ssh10b on RNA secondary structure (see below). Judging from the amount of Ssh10b required for retarding half of the input RNA, Ssh10b bound slightly more tightly to the structured partial B2 RNA than to unstructured single-stranded RNAs (with a ∼4-fold difference in apparent KD) (Fig. 1A). Furthermore, binding of Ssh10b to both structured and unstructured RNAs was not drastically affected by the presence of KCl at concentrations up to 0.4 m (data not shown). Therefore, it appears that structured RNA is preferentially bound by Ssh10b over unstructured RNA.
FIGURE 1.
Effect of Ssh10b binding on RNA structure. A, binding of Ssh10b to partial B2 RNA, rGrG(rArCrUrG)8, and 30-nt oligo(rA). Ssh10b was incubated with a 5′-32P-labeled RNA fragment for 10 min at 22 °C. Samples were subjected to electrophoresis in a 12 or 15% polyacrylamide gel. The gel was exposed to x-ray film. Protein concentrations in lanes 1–7 were 0, 0.01, 0.04, 0.16, 0.63, 2.5, and 10 μm, respectively. Apparent KD values represent an average of at least three independent measurements. B, a diagram showing the predicted secondary structure of the partial B2 RNA. Sites cleaved by RNase T1 or modified by DMS in the absence of Ssh10b are marked by ▿ or ○, respectively, whereas sites that became sensitive to RNase T1 cleavage or DMS modification in the presence of Ssh10b are indicated by ▾ or ●, respectively. The sequence of the RNA used for crystallization is indicated by the gray rectangle. C, footprinting analysis of RNase T1 cleavage of partial B2 RNA in the presence of Ssh10b. 5′-32P-Labeled B2 RNA was incubated with Ssh10b at 22 °C for 10 min. RNase T1 (1 unit) was added, and the mixture was incubated at 22 °C for 15 min. RNase T1 ladders were prepared by carrying out the cleavage reaction under denaturing conditions. RNA ladders were generated by incubating the radiolabeled RNA for 5 min at 90 °C under alkaline conditions. Reaction products were subjected to electrophoresis in 15% polyacrylamide containing 8 m urea. The gel was exposed to x-ray film. D, footprinting analysis of DMS modification of partial B2 RNA in the presence of Ssh10b. B2 RNA was incubated with Ssh10b at 22 °C for 10 min. DMS (0.5%) was added, and the samples were incubated at 22 °C for 5 min. Modifications were mapped by primer extension with reverse transcriptase. Reaction products were subjected to electrophoresis in 15% polyacrylamide containing 8 m urea. The gel was exposed to x-ray film.
To learn whether Ssh10b binding would influence RNA secondary structure, we carried out an RNase T1 footprinting analysis of the interaction of the protein with partial B2 RNA containing secondary structures. RNase T1 cleaves unpaired rGs and, thus, is used to probe the secondary structure of RNA. In our control experiment, we found that an unstructured single-stranded RNA fragment was equally sensitive to cleavage by RNase T1 both in the presence and in the absence of Ssh10b, and cleavage was not affected by the protein at concentrations used in this study (data not shown). In our footprinting assays on the partial B2 RNA, G6, G7, G9, G14, G15, G23, G24, and G28 were weakly cleaved in the absence of Ssh10b, apparently because they are located in the stem region of the RNA and are base-paired (Fig. 1, B and C). However, they were significantly cleaved when Ssh10b was added at protein/RNA molar ratios of ≥4. Notably, cleavage of these sites began to increase at protein concentrations below that required to saturate the input RNA (i.e. an Ssh10b/RNA molar ratio of ∼10). It appears that, upon binding, Ssh10b destabilized base pairing of the secondary structure in partial B2 RNA and rendered the Gs in the stem region of the RNA sensitive to cleavage by RNase T1. It was noticed that G11, G20, and G22 in a bulged region of the stem and G36 in a loop at the top were only slightly cleaved in the absence of Ssh10b, but the cleavage was significantly enhanced in the presence of Ssh10b. The reduced sensitivity of these Gs to RNase T1 in the absence of Ssh10b may suggest that they were sterically constrained in an unknown fashion. We also tested RNase I, which cleaves single-stranded RNA, in the footprinting experiments. Ssh10b did not alter the cleavage pattern at lower protein/RNA molar ratios but uniformly protected the B2 RNA from cleavage when the RNA was maximally bound by the protein (data not shown). Presumably, Ssh10b-bound sites on the RNA were readily accessible to RNase T1 but not to RNase I.
The influence of Ssh10b binding on the structure of partial B2 RNA was further examined by DMS footprinting, which permits sensitive detection of single-stranded adenosines and cytidines in RNA. As shown in Fig. 1D, several As and Cs were methylated to various extents in the absence of Ssh10b. Among them, unpaired bases in the top loop and the bulged regions (e.g. A12, A19, and A37) were most strongly methylated. Some base-paired As and Cs, located next to the unpaired regions (e.g. A10 and C18), were less strongly modified. However, in the presence of Ssh10b at protein/RNA molar rations of ≥1.25, several As and Cs located in the base-paired region either became significantly sensitive to DMS methylation (e.g. C16, A32, C46, and C47) or modified to a greater extent than in the absence of the protein (e.g. A10 and C18). Enhanced methylation at C33, C34, C35, and C38 in the loop region is again suggestive of the presence of local constraints for these Cs. In both RNase T1 and DMS footprinting experiments, nucleotides in the middle of the upper stem region (e.g. A29 and G30) appeared more resistant to destabilization by Ssh10b than those near the unpaired regions. It was also found that DMS was able to detect the structural changes in partial B2 RNA at slightly lower Ssh10b/RNA ratios than RNase T1. This discrepancy may result from the difference in sensitivity of the two methods to the structural changes in RNA. Taken together, our data indicate that binding by Ssh10b destabilizes the secondary structure of RNA.
Structure of the Ssh10b-RNA Complex
To gain insight into the structural basis of the interaction of Ssh10b with structured RNA, we sought to determine the crystal structure of a complex between Ssh10b and a 25-nt RNA fragment derived from partial B2 RNA (as shown in Fig. 1B, gray rectangle). We chose to use this RNA fragment in crystallization trials primarily because of our interest in gaining a structural understanding of the footprinting experiments on the interaction between Ssh10b and partial B2 RNA and the ability of the RNA to self-fold into a hairpin structure. In the crystal structure, the high quality electron density unambiguously identified the RNA in the complex as a 25-bp double-stranded self-annealing product of the input RNA molecule (Fig. 2, A and B). This was not entirely surprising because the intermolecular annealing was favored over the intramolecular annealing when the RNA existed at a very high concentration (i.e. ∼1.65 mm) during the crystallization of the complex (36). The 25-bp dsRNA duplex in the crystal contains two mispaired regions, each with two consecutive mismatches, in the middle and a single base insertion near each end (Fig. 2C).
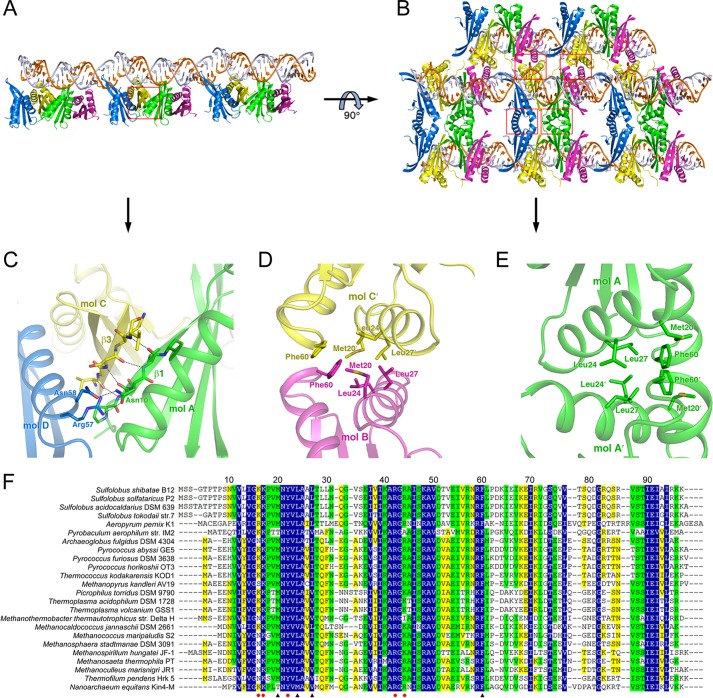
FIGURE 2.
Overall structure of the Ssh10b-RNA complex and interactions between Ssh10b and the RNA. A, ribbon diagrams of the Ssh10b-RNA complex and that rotated by 90° about the vertical axis. The four Ssh10b monomers are colored in green, magenta, yellow, and blue, respectively. The secondary structural elements of monomer A are labeled. Two complementary RNA strands are colored in orange and white, respectively. B, electron density of the RNA in the Ssh10b-RNA complex. C, a schematic diagram summarizing the Ssh10b-RNA interactions. All hydrogen bonds between Ssh10b and the 25-bp dsRNA are shown as dashed lines. D, E, and F, close-up views of the interfaces between the RNA and molecule A, B, or D. Interacting residues and nucleotides are shown as sticks and labeled. Hydrogen bonds are shown as dashed lines. mol, molecule.
The crystal structure of the Ssh10b-RNA complex was determined at 2.0-Å resolution (Fig. 2A). Each asymmetric unit contains four Ssh10b molecules (molecules A, B, C, and D) assembled into two compact dimers (dimers AB and CD) and a copy of 25-bp RNA duplex. The structures of the four Ssh10b molecules and their dimeric assembly closely resemble those of apo-Sso10b (15). Each dimer is shaped like a basket with two handles. The RNA lies on one side of each of the two dimers in an extended cleft formed by two pairs of handles (Fig. 2, A and B). The overall structure of the RNA in the complex is similar to that of a typical A-form dsRNA as characterized by the deep narrow major groove and the shallow wide minor groove. However, slight distortion occurs in the middle of the RNA fragment due to the presence of four mismatched base pairs. A nucleotide insertion at the third position from each end of the RNA duplex also contributes to the structural distortion (Fig. 2A).
In the Ssh10-RNA structure, dimers AB and CD form a pair of clamps diagonally binding to the RNA in the major groove. Monomers A and C interact with the two RNA strands, respectively, in the same manner (Fig. 2C). Taking monomer A as an example, Ssh10b-RNA interactions occur primarily on loops β1-α1 and β2-α2 (Fig. 2D). The side chain of Lys-16 in loop β1-α1 is extended into the deep major groove and interacts with the C13 and C14 phosphate groups of the RNA backbone through hydrogen bonds. A water molecule bridges the side chain of Lys-16 and the base of C14 through a network of hydrogen bonds. The neighboring Lys-17 residue hydrogen bonds to the C11 phosphate group. Tyr-22 at the N terminus of α1 interacts with the backbone of C12 through hydrogen bonding. Its side chain hydroxyl group also interacts with the main chain amino groups of Lys-7 and Gly-15, stabilizing the conformation of loop β1-α1. On the other hand, Arg-42 and Arg-44 of loop β2-α2 form hydrogen bonds with O4 of the C13 ribose group and the C15 phosphate group, respectively (Fig. 2D). The interactions between monomer C and the other RNA strand are schematically shown in Fig. 2C. Very few interactions are observed between monomer B or D and the RNA. The side chain of Lys-16 in the β1-α1 loop of monomer B sticks into the minor groove of the RNA, interacting with the base of U23 directly and with the base of U22 via a water molecule through hydrogen bonds, whereas the main chain carbonyl group of this residue forms hydrogen bonds with the 2′-hydroxyl group of the A5 ribose group and the phosphate group of G6 of the other RNA strand (Fig. 2E). The side chain of Lys-16 in the β1-α1 loop of monomer D extends into the major groove at the terminus of a symmetry-related RNA molecule (Fig. 2F), forming hydrogen bonds with the U22 and U23 phosphate groups of the RNA backbone. It is noteworthy that the β3-β4 loop, or the handle of the basket, in monomer A, B, or C shows no direct interactions with the RNA, providing no interpretable electron density for model building. However, the β3-β4 loop of monomer D can be modeled because it inserts into the vacant space near the distorted insertion at the third position from the 5′-end of one of the symmetry-related RNA strands, interacting with the nearby nucleotides, presumably as a result of crystal packing (Fig. 2F).
Extensive interactions are observed at the interface between dimers AB and CD. Four main chain hydrogen bonds between two parallel β-strands, β1 of monomer A and β3 of monomer C, allow the formation of an extended intermolecular β-sheet. Asn-10 at the N terminus of β1 in molecule A interacts with Arg-57 and Asn-58 at the C terminus of α2 in molecule D through two hydrogen bonds, further strengthening the interactions between the two dimers (Fig. 3, A and C). Moreover, a hydrophobic patch consisting of the side chains of Met-20, Leu-24, Leu-27, and Phe-60 is located on the surface of helices α1 and α2. The hydrophobic patches of monomers A and D are in close contact with those of their 2-fold axis-related monomers, whereas the patches of monomers B and D alternately interact with those of their 2-fold screw axis-related monomers (Fig. 3, B, D, and E). The homotypic interactions between the hydrophobic patches mediate crystal packing in the complex. Sequence alignments reveal that amino acid residues making up the hydrophobic patch are highly conserved among all members of the Sac10b family (Fig. 3F). A close inspection shows that the homotypic interactions identified in this study are also present in the crystal structures of other Sac10b proteins as well as the Ape10b2-dsDNA complex (23).
FIGURE 3.
Binding of Ssh10b as a tetramer to the structured RNA and homotypic interactions between two Ssh10b dimers bound to two separate RNA molecules in the complex. A, a ribbon diagram showing the interaction between two Ssh10b dimers adjacently bound to the RNA along the screw axis of the RNA. B, a ribbon diagram of Ssh10b dimer packing in the complex crystal as obtained by rotating the diagram in A by 90° about the horizontal axis. C, a close-up view of the interfaces between two adjacently bound Ssh10b dimers. The interacting residues are labeled and shown as sticks. Hydrogen bonds are shown as dashed lines. D and E, a close-up view of the homotypic interactions contributed by the hydrophobic patches of the interacting Ssh10b dimers in the complex crystal. Residues comprising the hydrophobic patch are labeled and shown as sticks. F, a sequence alignment of members of the Sac10b family. Residues involved in RNA binding and dimer-dimer interaction are indicated with closed circles (red) and closed triangles (black), respectively. mol, molecule.
Amino Acid Residues Involved in Ssh10b Binding to RNA
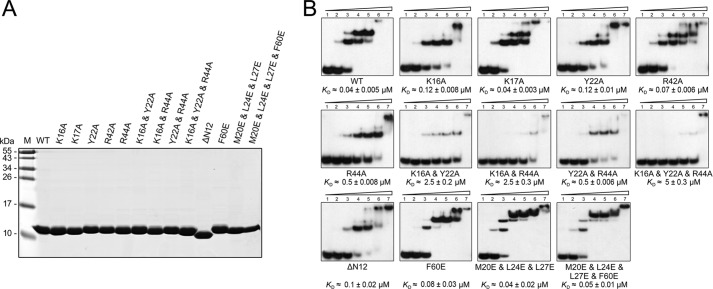
As shown in the Ssh10b-RNA complex structure, five amino acid residues, i.e. Lys-16, Lys-17, Tyr-22, Arg-42, and Arg-44, are involved in RNA binding. To verify the roles of these residues, we constructed Ssh10b mutant proteins containing an alanine substitution for each of the residues (Fig. 4A). The ability of the mutant proteins to bind RNA was examined by EMSA. K17A bound to the RNA as well as the wild-type protein, suggesting a weak interaction between Lys-17 and the RNA. The apparent binding affinities of K16A, Y22A, and R44A for the RNA were 3–12-fold lower than that of wild-type Ssh10b, whereas R42A was less than 2-fold weaker in binding affinity than the wild-type protein (Fig. 4B). We also prepared double and triple alanine substitution mutants at residues Lys-16, Tyr-22, and Arg-44 (Fig. 4A) and tested their RNA binding ability (Fig. 4B). Binding of Y22A/R44A to the RNA decreased by ∼10-fold, and that of K16A/Y22A and K16A/R44A for the RNA was lowered by >60-fold as compared with that of wild-type Ssh10b. The K16A/Y22A/R44A triple mutant barely bound the RNA. Therefore, Lys-16, Tyr-22, and Arg-44 are key residues in the interaction of Ssh10b with RNA.
FIGURE 4.
Preparation of mutant Ssh10b proteins and analysis of their binding to dsDNA. A, SDS-polyacrylamide gel electrophoresis of purified wild-type and mutant Ssh10b. Recombinant wild-type and mutant Ssh10b proteins were purified. A sample (∼5 μg) of each protein was subjected to electrophoresis in a 15% SDS-polyacrylamide gel. The gel was stained with Coomassie Brilliant Blue G-250. Lane M, molecular mass standards. B, binding of mutant Ssh10b to dsRNA. Wild-type or mutant Ssh10b was incubated with a 5′-32P-labeled 25-bp dsRNA fragment for 10 min at 22 °C. Samples were subjected to electrophoresis in a 15% polyacrylamide gel. The gel was exposed to x-ray film. Protein concentrations in lanes 1–7 were 0, 0.01, 0.04, 0.16, 0.63, 2.5, and 10 μm, respectively. Apparent KD values represent an average of at least three independent measurements.
Structural Basis for Destabilization of RNA Secondary Structure by Ssh10b
Sac10b homologs have been suggested to interact with dsDNA as a tetramer (22, 23). As revealed in the Ssh10b-RNA complex structure, two adjacent Ssh10b dimers bound to a distorted dsRNA stretch containing mismatches are in close contact through an extended intermolecular β-sheet. In agreement with this finding, when increasing amounts of Ssh10b were allowed to bind the 25-bp dsRNA, two shifts were generated with the first and second shifts corresponding to one and two Ssh10b dimers in complex with the RNA, respectively (Fig. 4B). In the Ssh10b-RNA structure, two dimers bound to the two RNA strands interact through the N terminus and the adjacent β1 in monomer A and the third β-sheet in monomer C. To understand the role of the dimer-dimer interaction of Ssh10b, we constructed a deletion mutant lacking the N-terminal 12 residues of the protein (ΔN12) (Fig. 4A). Loss of the dimer-dimer interaction lowered the affinity of the protein for dsRNA by 2–3-fold and as expected significantly reduced the amount of the protein tetramer formed on the RNA, suggesting that the intermolecular β-sheet permits cooperative binding of the RNA by the protein (Fig. 4B).
We also examined the ability of ΔN12 in destabilizing the secondary structure of RNA. In the RNase T1 footprinting assays, the sensitivity of the target Gs in B2 RNA to RNase T1 cleavage in the presence of ΔN12 was substantially lower than that in the presence of the wild-type protein at a protein/RNA molar ratio of 10 (Fig. 5). Based on these results, we conclude that the intermolecular N-terminal interactions between the two monomers from two adjacently located Ssh10b dimers increase the affinity of Ssh10b for dsRNA and play an important role in destabilizing the secondary structure of the bound RNA.
FIGURE 5.
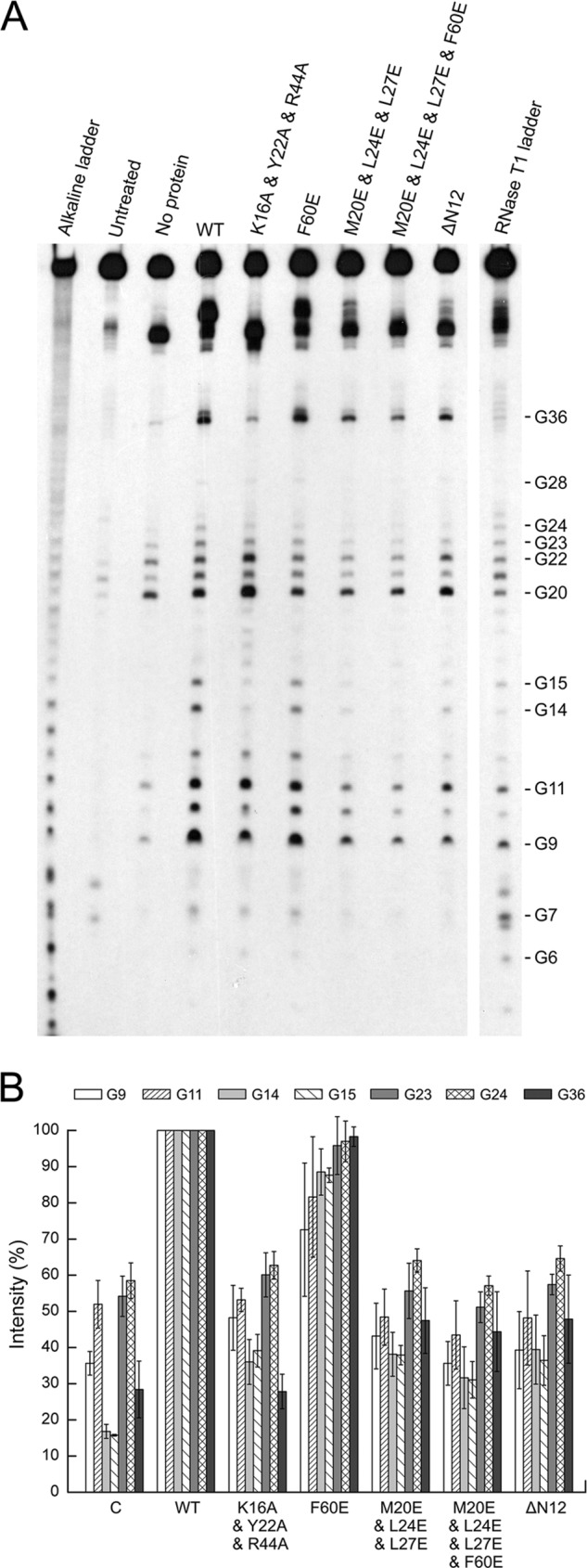
Footprinting analysis of the interaction of Ssh10b mutants with partial B2 RNA. A 5′-32P-labeled partial B2 RNA was incubated with wild-type or mutant Ssh10b at a protein/RNA molar ratio of 10 for 10 min at 22 °C. RNase T1 (1 unit) was added, and the mixture was incubated for 15 min at 22 °C. RNase T1 ladders were produced by performing the above reactions in the absence of Ssh10b under denaturing conditions. Alkaline ladders were generated by incubating the radiolabeled RNA for 5 min at 90 °C under alkaline conditions. Reaction products were subjected to electrophoresis in 15% polyacrylamide containing 8 m urea. The gel was exposed to x-ray film (A), and RNA cleavage by RNase T1 was analyzed by using a phosphorimaging system (B). The data shown represent an average of at least three independent measurements. Error bars represent S.D.
Higher Order Oligomerization of Ssh10b
There is another intermolecular interface between two symmetry-related Ssh10b dimers that involves a hydrophobic patch (Phe-60, Met-20, Leu-24, and Leu-27). This hydrophobic patch is highly conserved in the Sac10b family. To learn more about the role of this intermolecular interface, we constructed three mutant Ssh10b proteins, i.e. F60E, M20E/L24E/L27E, and M20E/L24E/L27E/F60E (Fig. 4A). Glutamic acid substitution of the hydrophobic amino acid residues would presumably result in the disruption of the hydrophobic interaction between the two patches. We found that the binding affinities of the three mutant proteins were unchanged, but the gel shift patterns of the mutant proteins were altered as compared with that of wild-type Ssh10b (Fig. 4B). F60E appeared to form the second shift on the 25-bp dsRNA fragment at lower protein concentrations than that for wild-type Ssh10b, whereas M20E/L24E/L27E and M20E/L24E/L27E/F60E were able to generate an additional shift on the duplex RNA as compared with the wild-type protein. Although the gel shift patterns of the mutant proteins remain to be understood, they appear to reflect the smaller area of the binding interface in the mutants (data not shown).
The dimer-dimer interaction through the hydrophobic patches is probably involved in the higher order oligomerization of Ssh10b. To test this possibility, we determined the ability of wild-type and mutant Ssh10b proteins to form oligomers by both blue native PAGE and chemical cross-linking with dithiobis(succinimidyl propionate). As revealed by blue native PAGE, wild-type Ssh10b appeared to form a ladder of tetramer, octamer, 12-mer, 16-mer, etc. with decreasing staining intensity (Fig. 6A). The N-terminal deletion mutant generated a ladder similar to that of the wild-type protein, suggesting that the dimer-dimer interaction through the intermolecular β-sheet is not involved in oligomerization of the protein. By contrast, the ability of M20E/L24E/L27E and M20E/L24E/L27E/F60E to form higher order oligomers was significantly reduced. In the dithiobis(succinimidyl propionate) cross-linking experiments, wild-type Ssh10b, ΔN12, F60E, and K16A/Y22A/R44A were all cross-linked into dimers, trimers, and larger oligomers, whereas M20E/L24E/L27E and M20E/L24E/L27E/F60E were cross-linked only into dimers in the absence of the RNA (Fig. 6B). Addition of the RNA to the reactions resulted in an increase in cross-linking efficiency for wild-type Ssh10b, ΔN12, and F60E. Cross-linking of M20E/L24E/L27E and to a lesser extent M20E/L24E/L27E/F60E into larger oligomers was also slightly enhanced in the presence of the RNA. However, a detectable difference existed in mobility between the dimer of M20E/L24E/L27E cross-linked in solution and that cross-linked on the RNA, raising the possibility that the oligomers cross-linked in solution may differ from those on the RNA. K16A/Y22A/R44A, which lost the ability to bind RNA, was cross-linked with similar efficiencies whether the RNA was added or not. Therefore, the intermolecular interactions through the hydrophobic patches appear to contribute to the ability of Ssh10b to form higher order protein complexes in solution.
FIGURE 6.
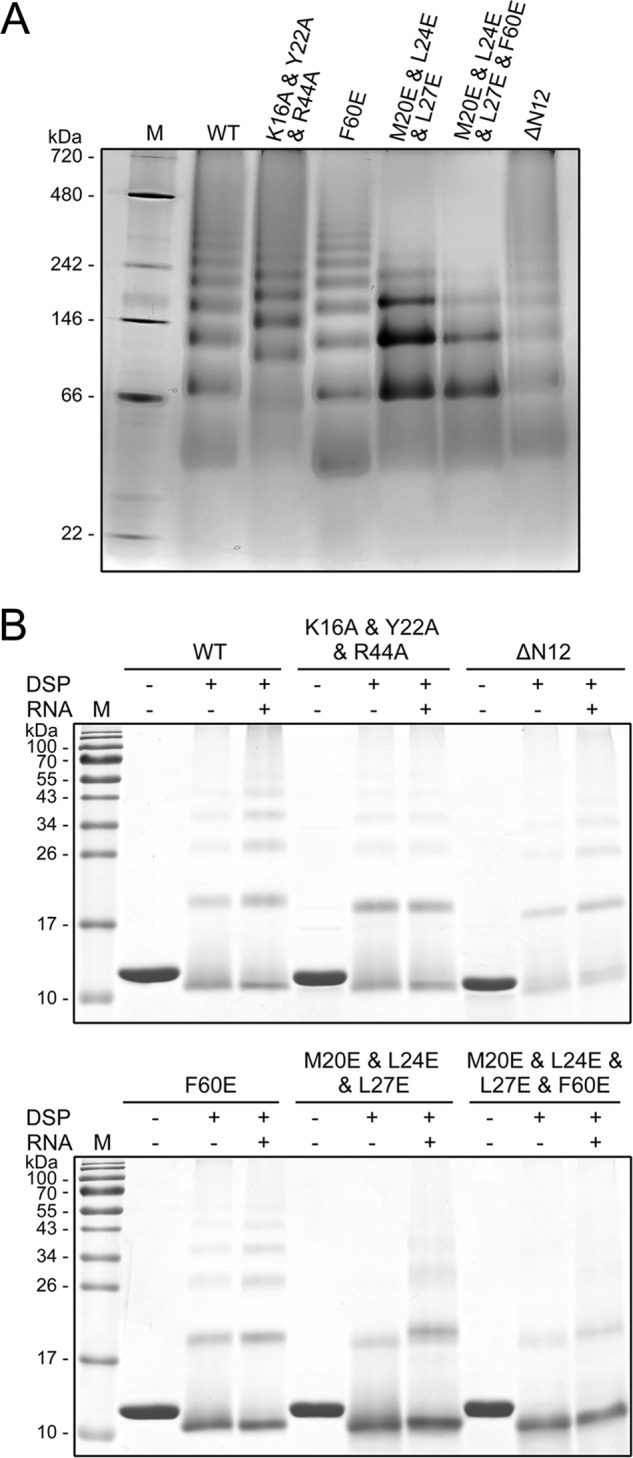
Oligomerization states of mutant Ssh10b. A, blue native polyacrylamide gel electrophoresis of wild-type and mutant Ssh10b. Wild-type or mutant Ssh10b (∼8 μg) was loaded onto a 6–20% gradient blue native polyacrylamide gel. The gel was stained with Coomassie Brilliant Blue. Lane M, molecular mass standards with molecular masses indicated. B, chemical cross-linking of wild-type and mutant Ssh10b. Wild-type and mutant Ssh10b were cross-linked alone or in complex with the 25-bp dsRNA with dithiobis(succinimidyl propionate) (DSP). After 1 h at 22 °C, reactions were terminated and analyzed by 15% SDS-PAGE. Molecular mass standards are indicated.
DISCUSSION
Native RNA molecules are often found to form secondary and tertiary structures, which are important to their function and stability (2, 37–39). Intriguingly, binding by Ssh10b destabilized the secondary structure of RNA presumably by weakening base pairing between the RNA strands. It appears that Ssh10b binding would on one hand serve to protect the bound RNA and on the other hand enhance the availability of structured RNA for RNA transactions such as translation and exosome-mediated RNA degradation that require destabilization of RNA secondary and tertiary structures.
To understand the structural basis of binding and destabilization of structured RNA by Ssh10b, we solved the crystal structure of the protein in complex with a 25-bp dsRNA containing mismatches and insertions. Three residues, Lys-16, Tyr-22, and Arg-44, constitute the dsRNA binding site of Ssh10b and contribute to structure-specific dsRNA binding by interacting with the RNA backbone and inserting into the characteristic deep narrow major groove of dsRNA. By comparison, four residues, Arg-13, Arg-40, Arg-42, and Asn-43, are involved in dsDNA binding in the Ape10b2-DNA complex (23). Arg-40 and Asn-43 in Ape10b2 correspond to Arg-42 and Ala-45 in Ssh10b, respectively. Arg-42 in Ssh10b has a moderate role in RNA binding, whereas Ala-45 is not in contact with the RNA. Notably, whereas Tyr-22 in Ssh10b, which is highly conserved in the Sac10b family, interacts with the RNA, Tyr-19, its counterpart in Ape10b2, is not in contact with the DNA (23). Arg-13 and Arg-42 in Ape10b2 or Lys-16 and Arg-44 in Ssh10b are key residues involved in both DNA and RNA binding (23). It appears that the amino acid residues involved in nucleic acid binding interact primarily with the phosphate backbone through polar interaction. Based on these results, we conclude that proteins of the Sac10b family do not distinguish between dsDNA and dsRNA in binding, but the modes of binding are different.
Structural analysis of the Ssh10b-dsRNA complex shows that Ssh10b dimers bind in successive major grooves on the same side of the dsRNA. Each Ssh10b dimer bound asymmetrically to the two RNA strands. By comparison, in the Ape10b2-dsDNA complex, an Ape10b2 dimer binds to the two DNA strands in the minor groove in a similarly asymmetrical fashion (23). However, apparent differences exist between binding of the 25-bp dsRNA by multiple Ssh10b dimers and predicted binding of a long dsDNA fragment by multiple Ape10b2 dimers. In the Ssh10b-dsRNA complex structure, two neighboring Ssh10b dimers bound symmetrically in successive major grooves are related by a pseudo 2-fold axis perpendicular to the screw axis of dsRNA and form a close intermolecular β-sheet. This intermolecular β-sheet allows the two neighboring Ssh10b dimers to assemble into a tetramer and bind to dsRNA cooperatively. In the Ape10b2-dsDNA complex structure, however, Ape10b2 dimers bind in successive minor grooves on both sides of dsDNA in the same fashion (23). Although the intermolecular β-sheet is also formed between two adjacent Ape10b2 dimers on the same side of dsDNA, the interactions between the two Ape10b2 dimers are notably weaker than those between two cooperatively bound Ssh10b dimers in the Ssh10b-dsRNA complex (23). The strong intermolecular interactions allow Ssh10b to bind dsRNA as stable tetramers and, thus, to exhibit greater binding affinity for the RNA than it would when bound as dimers (40). This is consistent with the finding that RNA was more tightly bound than DNA by Ssh10b (7). Notably, a stretch of structured RNA between the two cooperatively bound Ssh10b dimers was not in the standard A-form. The configuration of this RNA stretch may have arisen from the structural distortion of the bound dsRNA containing base mismatches, a structural mimicry of destabilized dsRNA. This observation implies that cooperative binding of Ssh10b facilitates structural destabilization of dsRNA. In agreement with this, mutations that either reduced RNA binding by the protein or affected the formation of the intermolecular β-sheet between the adjacent Ssh10b dimers resulted in a decrease in the ability of Ssh10b to destabilize the secondary structure of RNA.
A hydrophobic patch surrounding Phe-60 mediates another set of intermolecular interactions that are highly conserved among the majority of the Sac10b proteins. The homotypic interactions between Ssh10b dimers through the patches are responsible primarily for protein oligomerization. The patch-mediated dimer-dimer interaction appears to play a negligible role in the binding of RNA by Ssh10b because substitution of either Phe-60 or all four key residues of the patch by Glu resulted in little change in the affinity of the protein for the RNA. This is in contrast with the report that an F60A mutant reduced the affinity of Sso10b for DNA by 10-fold (41). It is speculated that the patch-mediated Ssh10b interaction may help bring RNA strands together.
The physiological significance of the interaction of the Sac10b proteins with RNA in hyperthermophilic archaea is unclear. Given their cellular abundance, sequence-nonspecific interaction with RNA, simultaneous binding of both strands in duplex RNA as tetramers, and ability to destabilize the secondary structure of RNA, the Sac10b proteins may function as an RNA chaperone. Binding by these proteins may facilitate folding of RNA into and help maintain it in a functionally accessible form. In addition, when bound by the protein, RNA may become less susceptible to decay at temperature optimal for the growth of the organism. Therefore, we speculate that the Sac10b proteins serve a role in RNA transactions such as translation in hyperthermophilic archaea.
It is known that leaderless mRNAs are common in Archaea (42, 43). A gene encoded by a monocistronic transcript and that located at the 5′-proximal end of an operon often lack a Shine-Dalgarno sequence and generally have little or no sequence upstream of a translational start point in Sulfolobus species (42, 44, 45). In bacteria, IF3 and ribosomal protein S1 play key roles in the translation of mRNAs lacking the Shine-Dalgarno sequence (46–49). S1 is the only ribosomal protein not stably associated with ribosome (37). This large ribosomal protein disrupts RNA secondary structures in vitro (37, 50). It is involved in the binding of mRNA to the small ribosomal subunit (37, 49) and induces structural changes in the mRNAs during the initiation of translation in vivo (37, 51, 52). Sulfolobus species encode no homologs of S1 or IF3. However, the Sac10b proteins are similar to the C-terminal RNA binding domain of IF3 in structure (15, 21, 22). It has been shown that all of the functions of the bacterial IF3 are displayed by the isolated C-terminal domain of IF3 in vitro (53, 54). This, together with the resemblance of Ssh10b to S1 in destabilizing RNA secondary structure, raises the possibility that the Sac10b proteins are involved in the translation of leaderless mRNAs in Archaea. This possibility has gained support from two recent studies. Mani et al. (20) found that three Alba domain proteins (Alba1, Alba2, and Alba3) from T. brucei were cytoplasmic RNA-binding proteins, and Alba2 and Alba3 were partially co-sedimented with polyribosomes in sucrose gradients. They speculated that the three proteins play regulatory roles in the initiation of translation. Gissot et al. (19) showed that Alba homologs from T. gondii bound RNA and co-purified with proteins in the translation machinery. These findings led the authors to suggest that the Alba homologs are involved in the translational control of gene expression in the parasite.
In conclusion, Ssh10b may serve to facilitate RNA transactions requiring destabilization of RNA secondary structure. Further studies are underway to elucidate the physiological functions of the Sac10b family in general and examine the potential role of the Sac10b proteins in archaeal translation in particular.
Acknowledgment
We thank the staff at beamline 17A of the Photon Factory (Grant 2011G044, High Energy Accelerator Research Organization, Tsukuba, Japan) for technical assistance during data collection.
This work was supported by National Natural Science Foundation of China Grants 31130003 (to L. H.), 31000022 (to L. G.), and 31100535 (to J. D.); Chinese Ministry of Science and Technology 973 Program Grants 2011CB910304 and 2011CB911103 (to D.-C. W.); and Chinese Academy of Sciences Grant KSCX2-EW-J-3 (to D.-C. W.).
The atomic coordinates and structure factors (code 3WBM) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- nt
- nucleotide
- ds
- double-stranded
- DMS
- dimethyl sulfate
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Forterre P. (1995) Thermoreduction, a hypothesis for the origin of prokaryotes. C. R. Acad. Sci. III 318, 415–422 [PubMed] [Google Scholar]
- 2. Poole A. M., Jeffares D. C., Penny D. (1998) The path from the RNA world. J. Mol. Evol. 46, 1–17 [DOI] [PubMed] [Google Scholar]
- 3. Bini E., Dikshit V., Dirksen K., Drozda M., Blum P. (2002) Stability of mRNA in the hyperthermophilic archaeon Sulfolobus solfataricus. RNA 8, 1129–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Green R., Noller H. F. (1997) Ribosomes and translation. Annu. Rev. Biochem. 66, 679–716 [DOI] [PubMed] [Google Scholar]
- 5. Lorentzen E., Conti E. (2005) Structural basis of 3′ end RNA recognition and exoribonucleolytic cleavage by an exosome RNase PH core. Mol. Cell 20, 473–481 [DOI] [PubMed] [Google Scholar]
- 6. Heinicke I., Müller J., Pittelkow M., Klein A. (2004) Mutational analysis of genes encoding chromatin proteins in the archaeon Methanococcus voltae indicates their involvement in the regulation of gene expression. Mol. Genet. Genomics 272, 76–87 [DOI] [PubMed] [Google Scholar]
- 7. Liu Y., Guo L., Guo R., Wong R. L., Hernandez H., Hu J., Chu Y., Amster I. J., Whitman W. B., Huang L. (2009) The Sac10b homolog in Methanococcus maripaludis binds DNA at specific sites. J. Bacteriol. 191, 2315–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu Y. F., Zhang N., Yao H. W., Pan X. M., Ge M. (2011) Mth10b, a unique member of the Sac10b family, does not bind nucleic acid. PLoS One 6, e19977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aravind L., Iyer L. M., Anantharaman V. (2003) The two faces of Alba: the evolutionary connection between proteins participating in chromatin structure and RNA metabolism. Genome Biol. 4, R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bell S. D., Botting C. H., Wardleworth B. N., Jackson S. P., White M. F. (2002) The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 296, 148–151 [DOI] [PubMed] [Google Scholar]
- 11. Cui Q., Tong Y., Xue H., Huang L., Feng Y., Wang J. (2003) Two conformations of archaeal Ssh10b. The origin of its temperature-dependent interaction with DNA. J. Biol. Chem. 278, 51015–51022 [DOI] [PubMed] [Google Scholar]
- 12. Forterre P., Confalonieri F., Knapp S. (1999) Identification of the gene encoding archeal-specific DNA-binding proteins of the Sac10b family. Mol. Microbiol. 32, 669–670 [DOI] [PubMed] [Google Scholar]
- 13. Grote M., Dijk J., Reinhardt R. (1986) Ribosomal and DNA binding proteins of the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. Biochim. Biophys. Acta 873, 405–413 [Google Scholar]
- 14. Xue H., Guo R., Wen Y., Liu D., Huang L. (2000) An abundant DNA binding protein from the hyperthermophilic archaeon Sulfolobus shibatae affects DNA supercoiling in a temperature-dependent fashion. J. Bacteriol. 182, 3929–3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wardleworth B. N., Russell R. J., Bell S. D., Taylor G. L., White M. F. (2002) Structure of Alba: an archaeal chromatin protein modulated by acetylation. EMBO J. 21, 4654–4662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Laurens N., Driessen R. P., Heller I., Vorselen D., Noom M. C., Hol F. J., White M. F., Dame R. T., Wuite G. J. (2012) Alba shapes the archaeal genome using a delicate balance of bridging and stiffening the DNA. Nat. Commun. 3, 1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guo R., Xue H., Huang L. (2003) Ssh10b, a conserved thermophilic archaeal protein, binds RNA in vivo. Mol. Microbiol. 50, 1605–1615 [DOI] [PubMed] [Google Scholar]
- 18. Marsh V. L., Peak-Chew S. Y., Bell S. D. (2005) Sir2 and the acetyltransferase, Pat, regulate the archaeal chromatin protein, Alba. J. Biol. Chem. 280, 21122–21128 [DOI] [PubMed] [Google Scholar]
- 19. Gissot M., Walker R., Delhaye S., Alayi T. D., Huot L., Hot D., Callebaut I., Schaeffer-Reiss C., Dorsselaer A. V., Tomavo S. (2013) Toxoplasma gondii Alba proteins are involved in translational control of gene expression. J. Mol. Biol. 425, 1287–1301 [DOI] [PubMed] [Google Scholar]
- 20. Mani J., Güttinger A., Schimanski B., Heller M., Acosta-Serrano A., Pescher P., Späth G., Roditi I. (2011) Alba-domain proteins of Trypanosoma brucei are cytoplasmic RNA-binding proteins that interact with the translation machinery. PLoS One 6, e22463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang G., Guo R., Bartlam M., Yang H., Xue H., Liu Y., Huang L., Rao Z. (2003) Crystal structure of a DNA binding protein from the hyperthermophilic euryarchaeon Methanococcus jannaschii. Protein Sci. 12, 2815–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao K., Chai X., Marmorstein R. (2003) Structure of a Sir2 substrate, Alba, reveals a mechanism for deacetylation-induced enhancement of DNA binding. J. Biol. Chem. 278, 26071–26077 [DOI] [PubMed] [Google Scholar]
- 23. Tanaka T., Padavattan S., Kumarevel T. (2012) Crystal structure of archaeal chromatin protein Alba2-double-stranded DNA complex from Aeropyrum pernix K1. J. Biol. Chem. 287, 10394–10402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 25. Espinoza C. A., Goodrich J. A., Kugel J. F. (2007) Characterization of the structure, function, and mechanism of B2 RNA, an ncRNA repressor of RNA polymerase II transcription. RNA 13, 583–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lane D., Prentki P., Chandler M. (1992) Use of gel retardation to analyze protein-nucleic acid interactions. Microbiol. Rev. 56, 509–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kuchino Y., Nishimura S. (1989) Enzymatic RNA Sequencing. Methods Enzymol. 180, 154–163 [DOI] [PubMed] [Google Scholar]
- 28. Tijerina P., Mohr S., Russell R. (2007) DMS footprinting of structured RNAs and RNA-protein complexes. Nat. Protoc. 2, 2608–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wittig I., Braun H. P., Schägger H. (2006) Blue native PAGE. Nat. Protoc. 1, 418–428 [DOI] [PubMed] [Google Scholar]
- 30. Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 32. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 33. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 34. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Russell R. (2008) RNA misfolding and the action of chaperones. Front. Biosci. 13, 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hilbers C. W., Haasnoot C. A., de Bruin S. H., Joordens J. J., van der Marel G. A., van Boom J. H. (1985) Hairpin formation in synthetic oligonucleotides. Biochimie 67, 685–695 [DOI] [PubMed] [Google Scholar]
- 37. Rajkowitsch L., Chen D., Stampfl S., Semrad K., Waldsich C., Mayer O., Jantsch M. F., Konrat R., Bläsi U., Schroeder R. (2007) RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 4, 118–130 [DOI] [PubMed] [Google Scholar]
- 38. Schroeder R., Grossberger R., Pichler A., Waldsich C. (2002) RNA folding in vivo. Curr. Opin. Struct. Biol. 12, 296–300 [DOI] [PubMed] [Google Scholar]
- 39. Weeks K. M. (1997) Protein-facilitated RNA folding. Curr. Opin. Struct. Biol. 7, 336–342 [DOI] [PubMed] [Google Scholar]
- 40. Wilce J., Vivian J., Wilce M. (2012) Oligonucleotide binding proteins: the occurrence of dimer and multimer formation. Adv. Exp. Med. Biol. 747, 91–104 [DOI] [PubMed] [Google Scholar]
- 41. Jelinska C., Petrovic-Stojanovska B., Ingledew W. J., White M. F. (2010) Dimer-dimer stacking interactions are important for nucleic acid binding by the archaeal chromatin protein Alba. Biochem. J. 427, 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moll I., Grill S., Gualerzi C. O., Bläsi U. (2002) Leaderless mRNAs in bacteria: surprises in ribosomal recruitment and translational control. Mol. Microbiol. 43, 239–246 [DOI] [PubMed] [Google Scholar]
- 43. Slupska M. M., King A. G., Fitz-Gibbon S., Besemer J., Borodovsky M., Miller J. H. (2001) Leaderless transcripts of the crenarchaeal hyperthermophile Pyrobaculum aerophilum. J. Mol. Biol. 309, 347–360 [DOI] [PubMed] [Google Scholar]
- 44. Benelli D., Maone E., Londei P. (2003) Two different mechanisms for ribosome/mRNA interaction in archaeal translation initiation. Mol. Microbiol. 50, 635–643 [DOI] [PubMed] [Google Scholar]
- 45. Tolstrup N., Sensen C. W., Garrett R. A., Clausen I. G. (2000) Two different and highly organized mechanisms of translation initiation in the archaeon Sulfolobus solfataricus. Extremophiles 4, 175–179 [DOI] [PubMed] [Google Scholar]
- 46. Moll I., Grill S., Gründling A., Bläsi U. (2002) Effects of ribosomal proteins S1, S2 and the DeaD/CsdA DEAD-box helicase on translation of leaderless and canonical mRNAs in Escherichia coli. Mol. Microbiol. 44, 1387–1396 [DOI] [PubMed] [Google Scholar]
- 47. Moll I., Resch A., Bläsi U. (1998) Discrimination of 5′-terminal start codons by translation initiation factor 3 is mediated by ribosomal protein S1. FEBS Lett. 436, 213–217 [DOI] [PubMed] [Google Scholar]
- 48. Tedin K., Moll I., Grill S., Resch A., Graschopf A., Gualerzi C. O., Bläsi U. (1999) Translation initiation factor 3 antagonizes authentic start codon selection on leaderless mRNAs. Mol. Microbiol. 31, 67–77 [DOI] [PubMed] [Google Scholar]
- 49. Tzareva N. V., Makhno V. I., Boni I. V. (1994) Ribosome-messenger recognition in the absence of the Shine-Dalgarno interactions. FEBS Lett. 337, 189–194 [DOI] [PubMed] [Google Scholar]
- 50. Bear D. G., Ng R., Van Derveer D., Johnson N. P., Thomas G., Schleich T., Noller H. F. (1976) Alteration of polynucleotide secondary structure by ribosomal protein S1. Proc. Natl. Acad. Sci. U.S.A. 73, 1824–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kolb A., Hermoso J. M., Thomas J. O., Szer W. (1977) Nucleic acid helix-unwinding properties of ribosomal protein S1 and the role of S1 in mRNA binding to ribosomes. Proc. Natl. Acad. Sci. U.S.A. 74, 2379–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tedin K., Resch A., Bläsi U. (1997) Requirements for ribosomal protein S1 for translation initiation of mRNAs with and without a 5′ leader sequence. Mol. Microbiol. 25, 189–199 [DOI] [PubMed] [Google Scholar]
- 53. Laursen B. S., Sørensen H. P., Mortensen K. K., Sperling-Petersen H. U. (2005) Initiation of protein synthesis in bacteria. Microbiol. Mol. Biol. Rev. 69, 101–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Petrelli D., Garofalo C., Lammi M., Spurio R., Pon C. L., Gualerzi C. O., La Teana A. (2003) Mapping the active sites of bacterial translation initiation factor IF3. J. Mol. Biol. 331, 541–556 [DOI] [PubMed] [Google Scholar]