Background: Mechanical forces regulate gene expression. The mechanisms are not well understood.
Results: A HES-1 site in the promoter of h2-calponin gene is a tension-regulated repressor responsive to Notch signaling.
Conclusion: Notch regulation plays a role in the mechanoregulation of h2-calponin.
Significance: The findings demonstrated a novel mechanism in the mechanoregulation of h2-calponin gene expression.
Keywords: Actin, Cell Biology, Gene Transcription, Mechanotransduction, Transcription Promoter
Abstract
The essential role of mechanical signals in regulating the function of living cells is universally observed. However, how mechanical signals are transduced in cells to regulate gene expression is largely unknown. We previously demonstrated that the gene encoding h2-calponin (Cnn2) is sensitively regulated by mechanical tension. In the present study, mouse genomic DNA containing the Cnn2 promoter was cloned, and a nested set of 5′ truncations was studied. Transcriptional activity of the Cnn2 promoter-reporter constructs was examined in transfected NIH/3T3, HEK293, and C2C12 cells for their responses to the stiffness of culture substrate. The results showed significant transcriptional activities of the −1.00- and −1.24-kb promoter constructs, whereas the −0.61-kb construct was inactive. The −1.38-, −1.57-, and −2.12-kb constructs showed higher transcriptional activity, whereas only the −1.57- and −2.12-kb constructs exhibited repression of expression when the host cells were cultured on low stiffness substrate. Internal deletion of the segment between −1.57 and −1.38 kb in the −2.12-kb promoter construct abolished the low substrate stiffness-induced repression. Site-specific deletion or mutation of an HES-1 transcription factor binding site in this region also abolished this repression effect. The level of HES-1 increased in cells cultured under a low tension condition, corresponding to the down-regulation of h2-calponin. h2-Calponin gene expression is further affected by the treatment of cells with Notch inhibitor and activator, suggesting an upstream signaling mechanism.
Introduction
Mechanical forces have significant effects on various biochemical and genetic processes in living organisms and contribute to multiple physiological regulations and pathological conditions (1). It is well established that chemical energy can be converted into mechanical forces in living cells by the activity of motor proteins, such as myosin and kinesin/dynein ATPases (2, 3). However, it is much less well understood how mechanical force signals are transduced and converted into chemical signals in cells to regulate biochemical processes and gene expression.
The actin cytoskeleton is a dynamic network in eukaryotic cells capable of bearing forces and undergoing rearrangements in adaptation to mechanical changes in the environment, playing essential functions in cellular mechanical properties and responses to mechanical signals (4). For the dual function of actin cytoskeleton in generating as well as sensing mechanical forces, regulation of actin cytoskeleton is essential for mechanoregulation in eukaryotic cells.
Calponin is an actin filament-associated regulatory protein (5). First found in smooth muscle (6), three isoforms of calponin (h1, h2, and h3) encoded by homologous genes (CNN1, CNN2, and CNN3) have been identified in vertebrates (7–11). h1-Calponin is expressed specifically in differentiated smooth muscle cells (12). h3-Calponin, also called acidic calponin, is found in smooth muscle cells (10) and neuronal tissues (11). In contrast, h2-calponin is present in multiple tissue and cell types, including smooth muscle, lung alveolar cells, endothelial cells, epidermal keratinocytes, fibroblasts, and myeloid leukocytes (9, 13–16).
Calponin binds F-actin with high affinity and inhibits the actin-activated myosin ATPase (17–20) and motor (12, 21, 22) activities. Extensive biochemical and biophysical studies have demonstrated that calponin regulates the function of actin filaments to modify smooth muscle contractility and non-muscle cell motility. For its expression in multiple tissue and cell types, the function and regulation of h2-calponin is of broad biological and medical significances. h2-Calponin stabilizes actin filaments and inhibits actin cytoskeleton-related cellular functions, such as cytokinesis, migration, and phagocytosis (13, 14, 16).
The expression of h2-calponin gene and the degradation of h2-calponin protein are both regulated by mechanical tension in the cytoskeleton (13, 14). In response to tension changes, proteolysis of h2-calponin provides rapid structural and functional modifications in the actin cytoskeleton, whereas its gene regulation conveys chronic and sustained alterations. The regulation of h2-calponin gene expression may be studied as a representative for understanding the mechanisms of mechanoregulation in living cells.
In a previous study, we demonstrated that h2-calponin protein and mRNA was significantly decreased in NIH/3T3 cells when cytoskeleton tension was reduced after blebbstatin inhibition of myosin II motor function (14). In contrast to the endogenous h2-calponin gene, transfective expression of h2-calponin cDNA under the control of CMV promoter was not regulated by mechanical tension (13). This observation suggests that transcriptional control is a primary regulation for the mechanoregulation of h2-calponin gene expression. The promoter of CNN2 gene therefore provides a novel experimental system to investigate how mechanical force signal is transduced in the regulation of gene transcription (23).
In the present study, we characterized the transcriptional activity of h2-calponin gene promoter for its mechanoregulation. We cloned mouse genomic DNA containing the Cnn2 promoter, constructed reporter genes, and analyzed transcriptional activity in several mammalian cell types that express endogenous h2-calponin under mechanoregulation. Using cell culture substrate of different stiffness, transcriptional activities of truncation, deletion, and site-specific mutation promoter constructs were studied to identify an HES-1 (hairy and enhancer of split 1) cis-regulatory element downstream of the Notch signaling pathway.
MATERIALS AND METHODS
Adherent and Floating Cell Cultures
As described previously, NIH/3T3 cells (ATCC CRL1658) were cultured in plastic dishes statically in an attached monolayer or under continuous vibration to prevent attachment to the dishes (26). After cultured at 37 °C in 5% CO2 for 3 days, the cells were washed with PBS, and total cellular protein was extracted for SDS-PAGE and Western blotting analysis.
Cell Culture on Gel Substrates of Different Stiffness
Human embryonic kidney cell line HEK293 (ATCC, CRL1573) and mouse fibroblast line NIH/3T3 were cultured in Dulbecco's modified Eagle's medium containing fetal bovine serum (10%), penicillin (100 IU/ml) and streptomycin (50 IU/ml) at 37 °C in 5% CO2. Monolayer cultures of cells at ∼70% confluence were trypsinized and passed at 1:10 ratio for use in experiments. Mouse skeletal myoblast line C2C12 (ATCC, CRL1772) was cultured with the same media conditions and trypsinized at ∼50% confluence to pass at 1:5 ratio for use in experiments.
To examine the effect of substrate stiffness-dependent cytoskeleton tension on h2-calponin gene expression, HEK293, NIH/3T3, and C2C12 cells were seeded on a thin layer (45 × 50 mm, ∼100-μm thickness) of polyacrylamide gel of different stiffness. Hard (16% gel with an acrylamide:bisacrylamide ratio of 19:1, Ef = ∼75 kilopascal) and soft (5% gel with an acrylamide:bisacrylamide ratio of 500:3, Ef = ∼1 kilopascal) gel substrates were prepared and covalently coated with 0.2 mg/ml type I collagen as described previously (13, 24, 25). After 3 days of culture, the cells were harvested using a cell scraper, washed three times with PBS, and processed for analyses.
SDS-PAGE and Western Blotting
SDS-PAGE and Western blotting were carried out as previously described (13) to examine the expression of endogenous h2-calponin in the cultured cells. The monolayer cells were directly lysed in SDS-PAGE sample buffer containing 2% SDS and analyzed using 12% gel in Laemmli buffer system with an acrylamide:bisacrylamide ratio of 29:1. After electrophoresis, the gels were fixed and stained with Coomassie Blue R-250 to verify sample integrity and normalize protein concentration. Protein bands in unfixed duplicate gels were electrically transferred to nitrocellulose membrane for Western blotting with a rabbit antiserum, RAH2, raised against mouse h2-calponin (26) or a monoclonal antibody against HES-1 (7H11, from Abnova). The calponin or HES-1 band recognized by the first antibody was revealed using alkaline phosphatase-labeled anti-rabbit IgG or anti-mouse IgG second antibody (Santa Cruz Biotechnology) and 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium chromogenic substrate reaction.
Cloning of Mouse h2-Calponin Promoter and Construction of Reporter Genes
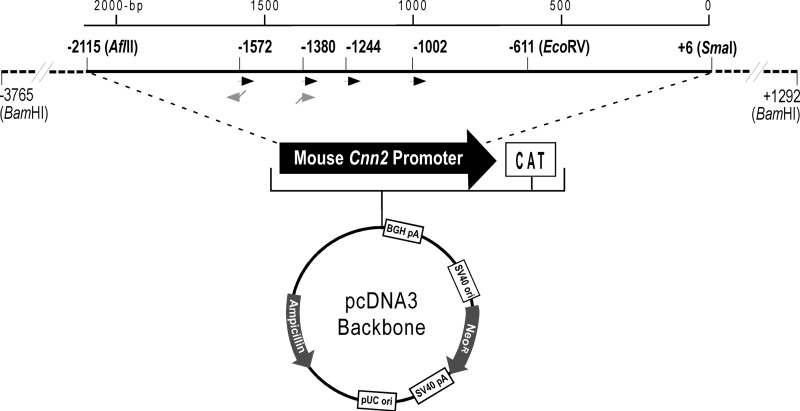
From the screening of a mouse genomic library with a mouse h2-calponin cDNA probe as described previously (16), lambda phage clones containing mouse Cnn2 gene (NC_000076.5) were isolated. A 5,058-bp BamHI restriction fragment containing the promoter of Cnn2 gene (−3,765 to +1,292) was subcloned into plasmid vectors. The region of −3,765 to +6 was further subcloned as a BamHI-SmaI fragment into the pcDNA3.1(+)/chloramphenical acetyltransferase (CAT)3 plasmid vector (Invitrogen) to replace the CMV promoter in front of the CAT coding sequence.
After demonstrating transcriptional activity of the −3.77-kb mouse Cnn2 promoter-CAT reporter construct, we constructed six 5′ serial truncation clones to map the cis-regulatory elements. As illustrated in Fig. 1, −2.12-kb (−2,115 to +6) and −0.61-kb (−611 to +6) clones were constructed from the −3.77-kb clone by restriction enzyme digestion-generated deletions. Four other 5′ truncation Cnn2 promoter constructs, −1.57 kb (−1,572 to +6), −1.38 kb (−1,380 to +6), −1.24 kb (−1,244 to +6), and −1.00 kb (−1,002 to +6), were constructed using PCR-generated DNA fragments from the −3.77-kb template. A common 3′ primer located in the CAT coding sequence was used to pair with different 5′ primers to generate these constructions. The primer pairs introduced a BamHI site at the 5′-end and a NotI restriction site at the 3′-end for unidirectional cloning of the PCR products into pcDNA3.1(+)/CAT reporter plasmid.
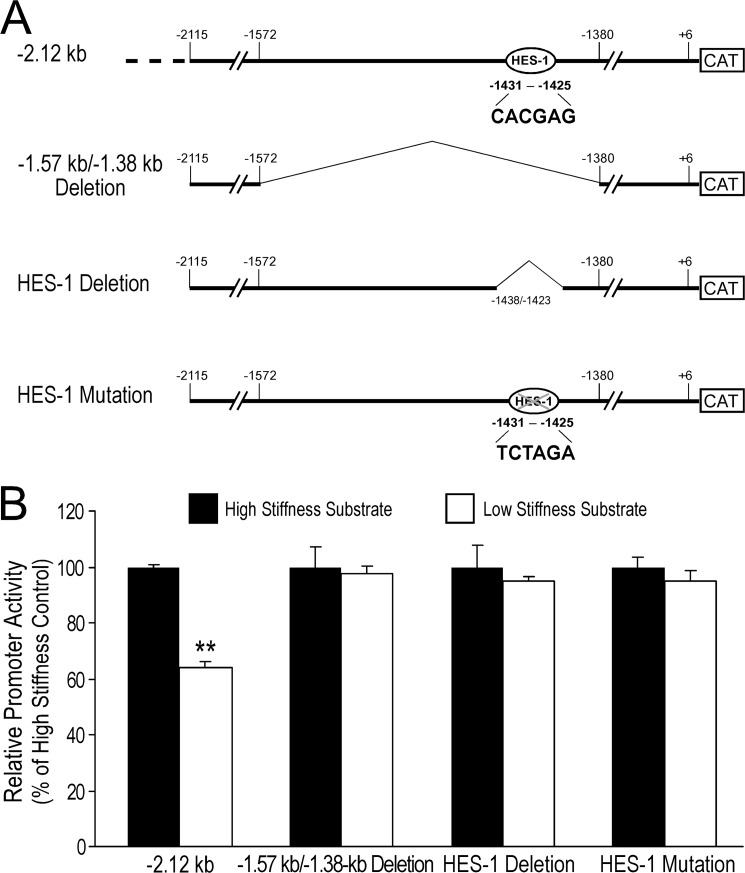
FIGURE 1.
5′ serial truncation and internal deletion constructs of mouse Cnn2 gene promoter. A pcDNA3.1(+)/CAT plasmid was modified by replacing the CMV promoter with a 3.77-kb promoter segment of mouse h2-calponin gene. Six 5′-truncation constructs and an internal deletion construct were derived from this recombinant plasmid. The −2.12- (−2,115 to +6) and −0.61-kb (−611 to +6) constructs were generated using restriction enzyme digestion. The −1.57- (−1,572 to +6), −1.38- (−1,380 to +6), −1.24- (−1,244 to +6), and −1.00-kb (−1,002 to +6) constructs were generated with PCR using customer-designed primers (black arrowheads). The −1,572 to −1,380 internal deletion was made in the −2.12-kb construct with recombinant PCR using deletion primers containing complementary 5′ sequences (gray arrowheads).
A −1,572 to −1,380 internal deletion (−1.57-kb/−1.38-kb deletion) was constructed in the −2.12-kb Cnn2 promoter-reporter gene using recombinant PCR. Two oligonucleotide primers with complementary 5′ sequences were designed for use with flanking primers in the first PCR to amplify the −2,115 to −1,572 and −1,380 to +6 segments of the Cnn2 promoter (Fig. 1). A recombinant DNA fragment was generated by joining the two first PCR products in the second PCR. The internal deletion promoter construct was unidirectionally cloned into pcDNA3.1(+)/CAT reporter plasmid as a BamHI-NotI fragment. The 5′ truncation and internal deletion constructs were verified by restriction enzyme mapping and DNA sequencing.
Transient and Stable Transfections
HEK293 cells were seeded in 24-well plates at 8 × 104 cells/well and cultured for 16 h. For each transfection, 0.04 pmol of the promoter-reporter plasmid was used with liposome reagent (Turbofect, Fermentas) at a 1:4 DNA/liposome (μg/μl) ratio. pEGFP-N1 plasmid encoding GFP under CMV promoter was co-transfected at a molar ratio of 1:20 versus the Cnn2 promoter construct to indicate the efficiency of transfection. DNA and liposome reagent were mixed in 100 μl of DMEM, incubated at room temperature for 15 min, and added to the cell culture medium. After 6 h, the transfection medium was replaced with fresh medium for continuing culture of 24–30 h.
NIH/3T3 and C2C12 cells were transfected similarly. 1 × 105 or 5 × 104 cells/well, respectively, were seeded in 12-well plates. 0.10 pmol of DNA was used for both NIH/3T3 and C2C12 cells at DNA/liposome ratios of 1:2 and 1:4, respectively (μg/μl).
The cultures were examined using an inverted epifluorescence microscope for GFP-positive cells to evaluate the efficiency of transfection. The cells were then washed three times with PBS and lysed for use either directly or after short frozen storage at −80 °C in the quantification of reporter gene expression. For every promoter construct, three or more independent transfection experiments were performed to verify the results.
To establish stable transfections, Cnn2 promoter-reporter plasmid DNA was linearized with restriction enzyme cleavage at the 5′ flanking BamHI (for the −1.57-, −1.38-, −1.24-, and −1.00-kb constructs) or AflII (for the −2.12-kb construct) site. 1 × 106 HEK293 cells were plated on a 100-mm dish for 16 h before the incubation with 0.2 μg of recombinant plasmid DNA mixed with 0.8 μl of liposome reagent for 6 h. After 24 h, the medium was replaced with fresh medium containing G418 at a concentration predetermined to effectively kill untransfected cells in 2 weeks. Cultured in the selection medium for 10–14 days, single colonies of surviving and proliferating cells were picked manually as described previously (26), expanded, verified using PCR for the presence of the CAT reporter gene, examined using Western blot for the normal expression of endogenous h2-calponin, and studied for transcriptional activity of the Cnn2 promoter constructs. At least three original stable transfection clones expressing endogenous h2-calponin at a level similar to that of nontransfected control cells were studied for each promoter construct.
Promoter Analysis
To map the 5′-upstream region of Cnn2 promoter for cis-regulatory elements responsive to mechanoregulation, the stable transfected cell clones were cultured on polyacrylamide gel substrates of high or low stiffness as described above. After 72 h of culture, samples were collected to examine the expression of endogenous h2-calponin using Western blot and transcriptional activity of the Cnn2 promoter-reporter constructs. The results were summarized from at least three original stable transfection clones that have similar normal level expression of endogenous h2-calponin.
CAT-ELISA
CAT-ELISA was performed as previously described (27). Briefly, the concentration of cellular protein extracts was determined using Bradford assay and further normalized by SDS-PAGE gel densitometry. CAT expression of the Cnn2 promoter-reporter constructs was examined with a sandwich ELISA method using the reagent kit from Roche Applied Science. 100 μg of total protein extract of the transfected cells was dissolved in 100 μl of reaction buffer and loaded to each assay well of a 96-well plate. The samples were incubated at 37 °C for 2 h followed by standard ELISA steps, including washes, incubations with anti-CAT antibody and horseradish peroxidase-labeled second antibody, and substrate-enhancer reaction. A415 nm of each assay well was monitored at a series of time points with reference wavelength of 655 nm using an automated microplate reader (Bio-Rad Benchmark). Triplicate wells were examined for each sample. The transcriptional activity of the promoter-reporter constructs was normalized for transfection efficiency determined as the percentage of cells positive for the co-transfective expression of GFP. Absorbance data of soft gel cultures were compared with that of corresponding hard gel cultures to determine the percentage of down-regulation of each promoter construct.
Search for Transcription Factor Binding Sites
Bioinformatic analysis of transcription factor binding sites was performed using the Genomatix suite software (Genomatix Software GmbH, Munich, Germany). The sequence of the −2.12-kb promoter segment of mouse Cnn2 gene was used as the target sequence for identification of putative transcription factor recognition sites using the MatInspector Version 8.0.5 program (28). The parameters used were the standard (0.75) core similarity and the optimized matrix similarity. Transcription factor binding sites found in the −1,572 to −1,380 and −1,380 to −1,244 regions were considered for potential correlation to mechanoregulation.
HES-1 Deletion and Mutagenesis
A deletion from −1,438 to −1,423 was made in the −2.12-kb construct using recombinant PCR as described above for the −1.57 kb/−1.38 kb deletion. An HES-1 N-box mutation was made in the −2.12-kb construct using recombinant PCR, converting the −1,431 to −1,425 region of the Cnn2 promoter from CACGAG to TCTAGA (a new XbaI site was introduced for easy identification).
Notch Inhibition and Activation Studies
K562 human myelogenous leukemia cells (ATCC, CCL-243) that were growing in culture nonadherently were employed to study the role of Notch signaling in regulating h2-calponin expression. The effects of a Notch inhibitor, N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT), and a Notch activator, phenethyl isothiocyanate (PEITC), were examined. Cells were seeded at 2 × 105/well in a 12-well culture plate in 1 ml of Iscove's medium containing 10% fetal bovine serum and various concentrations of the drug. The solvent control for DAPT was DMF at a concentration of 0.04%, and for PEITC was Me2SO at a concentration of 0.05%. Cells were cultured for 72 h, collected, and centrifuged at 500 × g for 5 min to remove the media. The cell pellets were washed with PBS for three times and lysed in SDS-PAGE sample buffer for Western blotting as described above.
Data Analysis
Densitometry analysis of SDS gels and Western blots were performed using National Institutes of Health Image software (version 1.61) on digital images scanned at 600 dpi. The quantitative data are presented as means ± S.D. unless noted in the figure legend. Statistical analysis was done using the Microsoft Excel or Origin software. The significance of difference was established as p < 0.05.
RESULTS
Substrate Stiffness-regulated Expression of h2-Calponin in Multiple Cell Types
We previously reported that h2-calponin is expressed in smooth muscle and multiple non-muscle tissue and cell types including epithelial, endothelial, fibroblast, and myeloid blood cells (13–16, 26). In the present study, we further demonstrated significant levels of h2-calponin expression in human embryonic kidney cell line HEK293 and mouse skeletal myoblast line C2C12 (Fig. 2).
FIGURE 2.
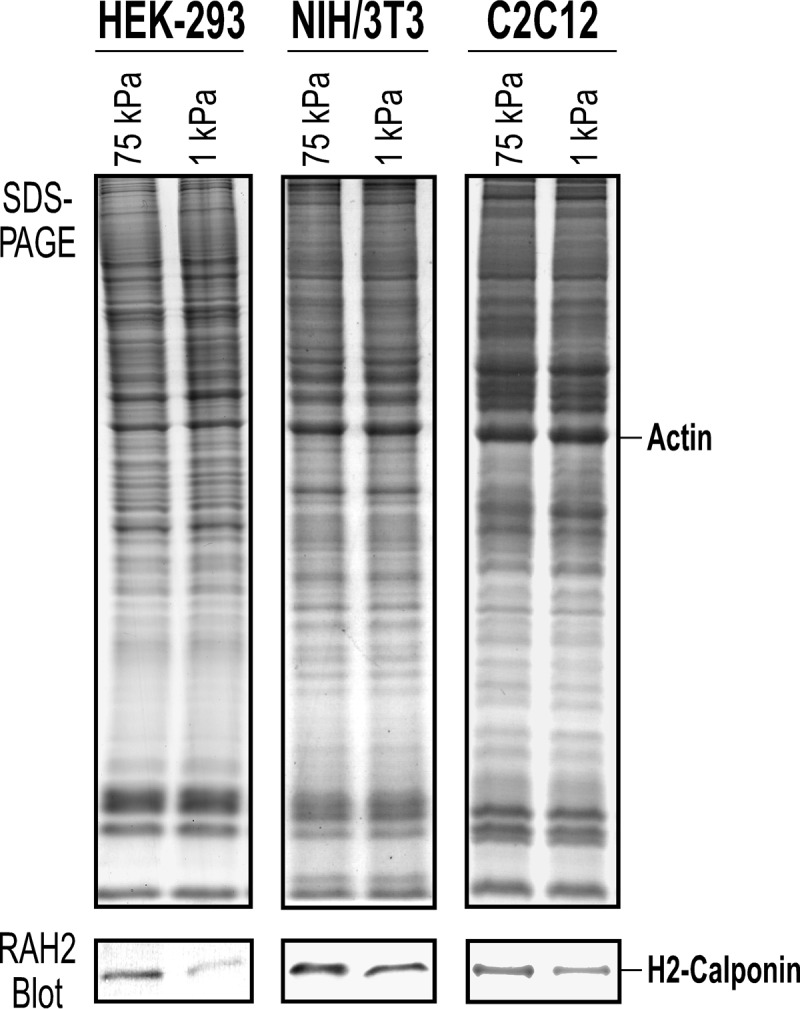
Substrate stiffness-regulated expression of endogenous h2-calponin in HEK293, NIH/3T3, and C2C12 cells. The SDS-PAGE gels and RAH2 Western blots detected significant levels of endogenous h2-calponin in HEK293 epithelial cells, NIH/3T3 fibroblasts, and C2C12 myoblasts. Normalized to the level of actin, the levels of h2-calponin in the three cell lines were significantly decreased when cultured on soft gel substrate (5%, acrylamide:bisacrylamide ratio = 500:3, Ef = ∼1 kilopascal) versus that in cells cultured on hard gel substrate (16%, acrylamide:bisacrylamide ratio = 19:1, Ef = ∼75 kilopascal).
The expression of h2-calponin in cells is dependent on the stiffness of culture substrate. Consistent with our previous observations from studies of epidermal keratinocytes (13) and lung alveolar cells (14), the Western blots in Fig. 2 show that in all of the three cell types examined in the present study, cultures on low stiffness soft polyacrylamide gels corresponding to low tension in the cytoskeleton had a significantly lower level of h2-calponin expression than that in cells cultured on high stiffness hard gel substrate, corresponding to high cytoskeleton tension. Without exception, the regulation of h2-calponin gene expression by mechanical tension built in the cytoskeleton as traction force against the culture substrate (14, 29) has been seen in all cell types studied, indicating a highly conserved mechanism of mechanoregulation.
cis-Regulatory Activities in the 5′ Upstream Region of Mouse Cnn2 Promoter
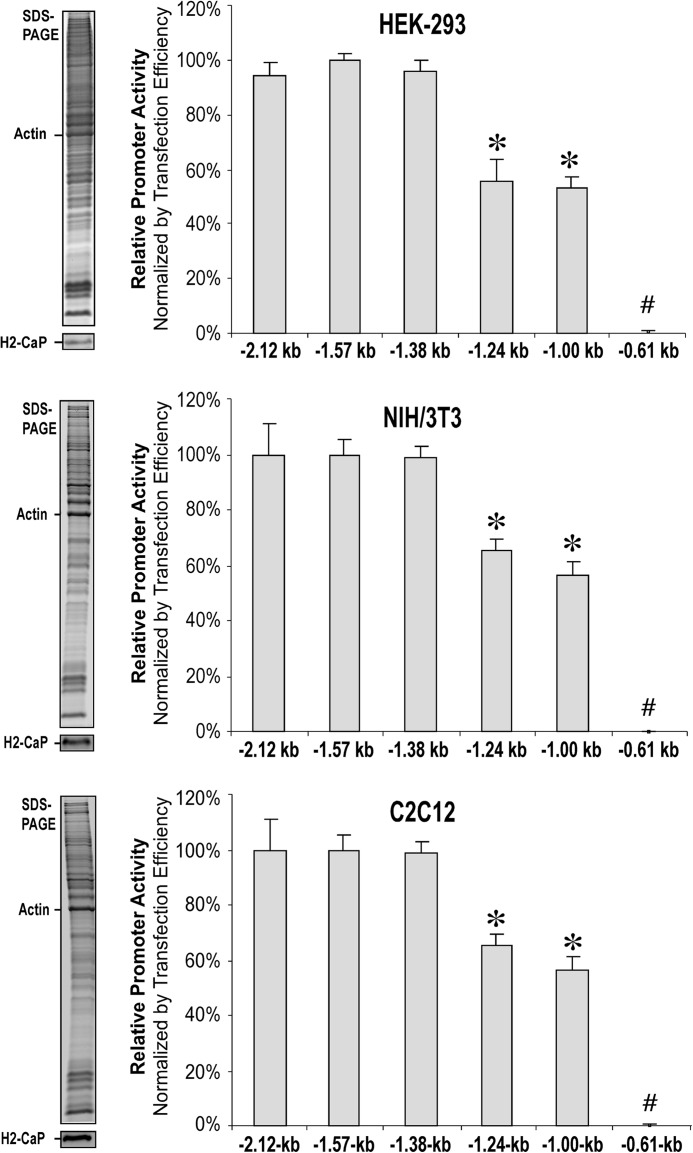
Following the evidence that mechanical tension regulates the level of h2-calponin mRNA (14), the present study mapped the transcriptional regulatory elements in the 5′-upstream region of mouse Cnn2 gene promoter. The results of transient transfection promoter assays in Fig. 3 demonstrated that the −2.12-, −1.57-, and −1.38-kb promoter constructs exhibited high levels of transcriptional activity in HEK293, NIH/3T3, and C2C12 cells cultured on a plastic dish, similar to that of the −3.77-kb construct (data not shown). In contrast, further deletion of a DNA segment of 136 bp (−1,380 to −1,244) resulted in a sharp decrease (∼40%) in transcriptional activity in the −1.24-kb promoter construct. Deletion of the next 242 bp did not cause further change in transcriptional activity in the −1.00-kb construct, whereas removing the downstream 391 bp (−1,002 to −611) completely abolished expression of the CAT reporter gene.
FIGURE 3.
Effects of 5′ truncations on the transcriptional activity of mouse Cnn2 promoter. Relative transcriptional activities of the 5′ truncation constructs of mouse Cnn2 promoter were determined in transient transfection experiments. Normalized with the efficiency of transfection, the results showed similar patterns in the three cell types cultured on plastic substrate. The −2.12-, −1.57-, and −1.38-kb promoter constructs had similarly high transcriptional activities. In comparison, transcriptional activities of the −1.24- and −1.00-kb promoter constructs were significantly lower (55.6 ± 7.8 to 68.4 ± 5.2% and 52.9 ± 4.6 to 60.9 ± 5.6%, respectively, of the level of the three longer constructs). The −0.61-kb construct had no detectable transcriptional activity. The expression of endogenous h2-calponin in the transfected HEK293, NIH/3T3, and C2C12 cells is shown in the SDS-PAGE gels and RAH2 Western blots on the left of each panel to demonstrate that a permissive cellular environment was preserved. *, p < 0.05 versus the level of −2.12-, −1.57-, and −1.38-kb groups; #, p < 0.05 versus all other groups. The data were summarized from at least three independent experiments.
The promoter mapping data from transiently transfected cells cultured on plastic substrate of very high stiffness revealed two fundamental regulatory activities in the mouse Cnn2 promoter: the essential promoter activity located in the −1,002 to −611 region is critical to turning on the h2-calponin gene in all of the three cell types tested, and the −1,380 to −1,244 region is necessary for high level transcription of the Cnn2 gene.
A Regulatory Region in Mouse Cnn2 Promoter Responsive to Substrate Stiffness
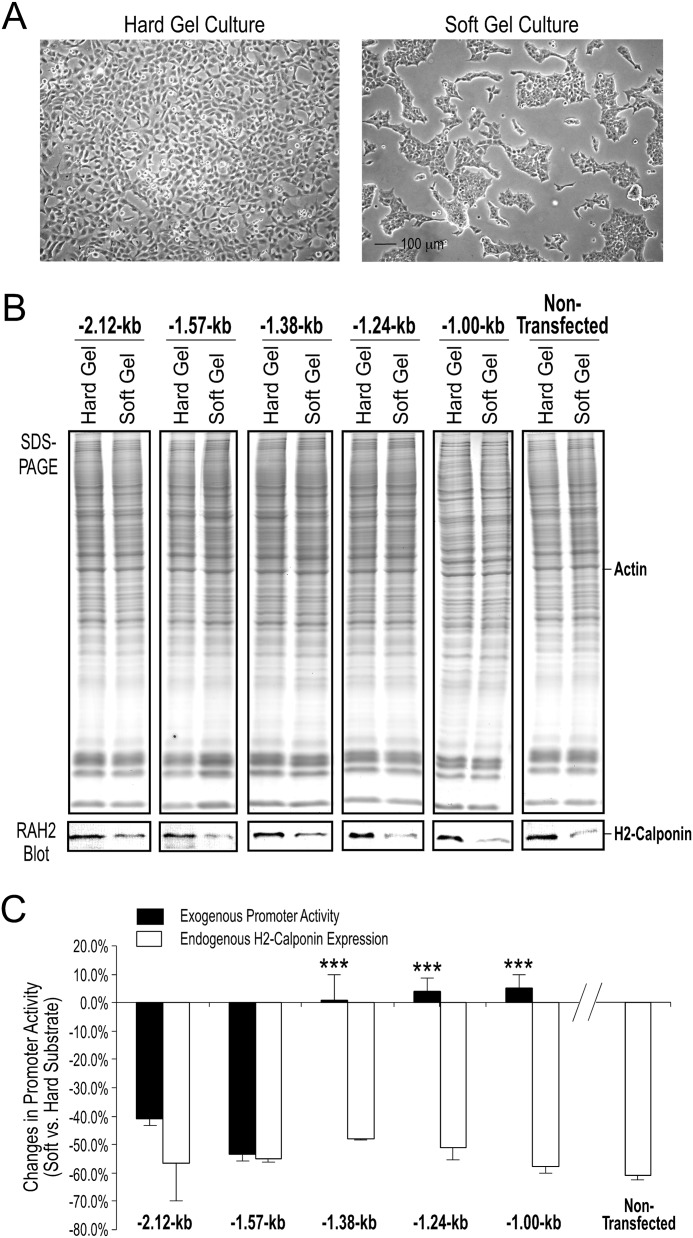
To localize mechanical tension responsive elements in the 5′-upstream region of mouse Cnn2 promoter, stable transfection studies in HEK293 cells showed that the −2.12- and −1.57-kb constructs, but not the −1.38-, −1.24-, and −1.00-kb constructs, exhibited significant decreases in transcriptional activity in cells cultured on low stiffness gel substrate as compared with that in cells cultured on high stiffness substrate (Fig. 4). Although the expression of endogenous h2-calponin in the host cells was still being regulated by mechanical tension (Fig. 4B), the −1.38-, −1.24-, and −1.00-kb Cnn2 promoter-reporter constructs lost the low substrate stiffness-induced repression (Fig. 4C). The data demonstrate the presence of mechanoregulatory cis-elements in the 192 bp of genomic DNA between −1,572 and −1,380 in the 5′-upstream region of the mouse Cnn2 gene.
FIGURE 4.
Responses of 5′ truncated mouse Cnn2 promoters to the stiffness of culture substrate. HEK293 cells were stably transfected with the −2.12-, −1.57-, −1.38-, −1.24-, and −1.00-kb promoter-reporter constructs to map the cis-regulatory elements involved in mechanoregulation. A, anticipated morphological difference of cells cultured for 3 days on polyacrylamide gels with high versus low stiffness. B, SDS-PAGE gels and RAH2 Western blots showed preserved mechanoregulation of the endogenous h2-calponin gene expression in the stable transfected cells as demonstrated by the higher expression in cells cultured on substrate of higher stiffness. Nontransfected HEK293 cells were examined as control. C, CAT-ELISA results determined repressions of transcriptional activity of the −2.12- (down by 40.9 ± 4.1%, n = 3) and −1.57-kb (down by 53.5 ± 5.8%, n = 6) Cnn2 promoter constructs in cells cultured on soft gel as compared with that in cells cultured on hard gels. No soft substrate-induced repression was found with the −1.38- (up by 0.8 ± 9.1%, n = 8), −1.24- (up by 3.9 ± 4.9%, n = 8), and −1.00-kb (up by 5.2 ± 4.5%, n = 11) promoter constructs. The repression of endogenous h2-calponin gene expression in the stable transfected cell lines was quantified using densitometry of the Western blots as control for the preserved cellular regulation (open bars, down by 48.0 ± 1.0 to 60.8 ± 1.4%, similar to that in nontransfected HEK293 cells, n = 3 each). ***, p < 0.001 versus the −2.12- and −1.57-kb groups.
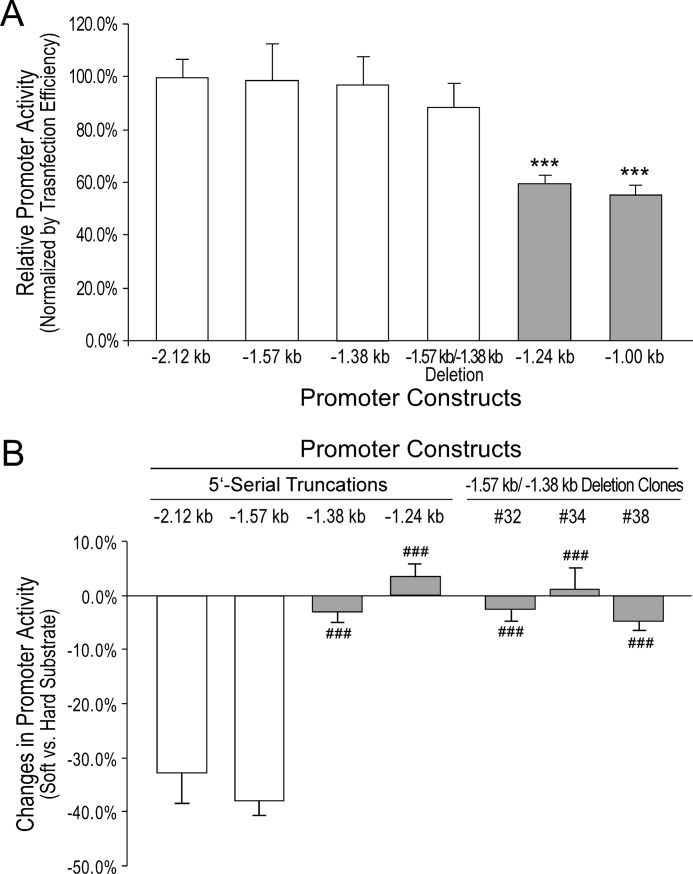
To exclude any additional effect of the −2,115 to −1,572 region on the mechanoregulatory activity of the downstream −1,572 to −1,380 segment, internal deletion of the −1,572 to −1,380 segment reproduced the abolishment of tension regulation, like that in the −1.38-kb truncation construct. Transient transfection experiments showed that in cells cultured on a plastic dish, the 192-bp internal deletion construct had high level transcriptional activity similar to that of the −2.12-, −1.57-, and −1.38-kb 5′-truncation constructs (Fig. 5A). Stable transfection experiments then showed that similar to the −1.38-kb truncated promoter construct, the −1.57- to −1.38-kb internal deletion abolished the low stiffness substrate-induced repression (Fig. 5B). Therefore, the 192-bp genomic DNA segment from −1,572 to −1,380 of mouse Cnn2 promoter functions in the mechanoregulation independent of the upstream regions.
FIGURE 5.
Internal deletion of the −1.57- to −1.38-kb segment diminished the low substrate stiffness-induced repression of mouse Cnn2 promoter. A, CAT-ELISA data from promoter analysis in transiently transfected HEK293 cells cultured on plastic dishes showed that the −1.57- to −1.38-kb internal deletion did not affect the high level transcription in cells on high stiffness substrate. The expression level of the internal deletion construct was similar to that of the −2.12- −1.57-, and −1.38-kb 5′ truncation constructs and significantly higher than the levels of the −1.24- and −1.00-kb truncation constructs. B, stable transfection experiments showed that deletion of the −1.57- to −1.38-kb segment in the −2.12-kb promoter construct abolished the soft substrate-induced suppression of transcription as compared with that seen with the −2.12- and −1.57-kb constructs. This loss of regulation is similar to that in the −1.38- and −1.24-kb truncation constructs. ***, p < 0.001 versus the −2.12-, −1.57-, −1.38-, and 1.57- to −1.38-kb internal deletion groups. ###, p < 0.001 versus the −2.12- and −1.57-kb groups. The results were summarized from experiments using three or more original stable transfected clones in each group.
Specific Transcription Factor Binding Sites in Mouse Cnn2 Promoter
The localization of a mechanoregulatory repressor activity in a genomic DNA segment of less than 200 bp allowed a focused search for relevant transcription factor binding sites. Of the 25 tentative binding sites predicted in the −1,572 to −1,380 segment, there are sites for potential transcriptional repressors, including two CCCTC-binding factor sites (30), one PAX5 site (paired box protein 5) (31), and one HES-1 site (32) (Fig. 6A). Among them, HES-1 has been observed with a function in mechanoregulation downstream of the Notch signaling pathway (33). Notch regulation has been demonstrated for a role in conducting mechanoregulation (34, 35) via HES-1 (36, 37).
FIGURE 6.
Deletion or mutation of an HES-1 site abolished the low tension-induced repression of mouse Cnn2 promoter. A, using the MatInspector software, a binding site for transcriptional repressor HES-1 is found in the −1,572 to −1,380 region of mouse Cnn2 gene. This site is removed in the −1,572 to −1,380 and site-specific deletions of the −2.12-kb promoter construct and mutated by the substitution of 6 base pairs. B, CAT-ELISA data from stable transfection experiments showed that similar to what is seen with the 192-bp (−1,572 to −1,380) deletion, specific HES-1 deletion or mutation abolished the soft substrate-induced repression of Cnn2 promoter. Wild type −2.12-kb promoter construct was used as control. **, p < 0.01 versus the high stiffness group.
To investigate the role of the HES-1 regulatory site in the mechanoregulation of the mouse Cnn2 gene, stable transfection experiments showed that the selective deletion of the 6-bp HES1 N-box located between −1,431 and −1,425 (Fig. 6A) abolished the soft substrate-induced repression of the −2.12-kb Cnn2 promoter (Fig. 6B). A similar effect was seen with the 6-bp mutation in the HES-1 binding site (Fig. 6). The effects of site-specific HES-1 deletion and mutation on the mechanoregulation of Cnn2 promoter are similar to that of the 192-bp internal deletion and the −1.38-kb truncation.
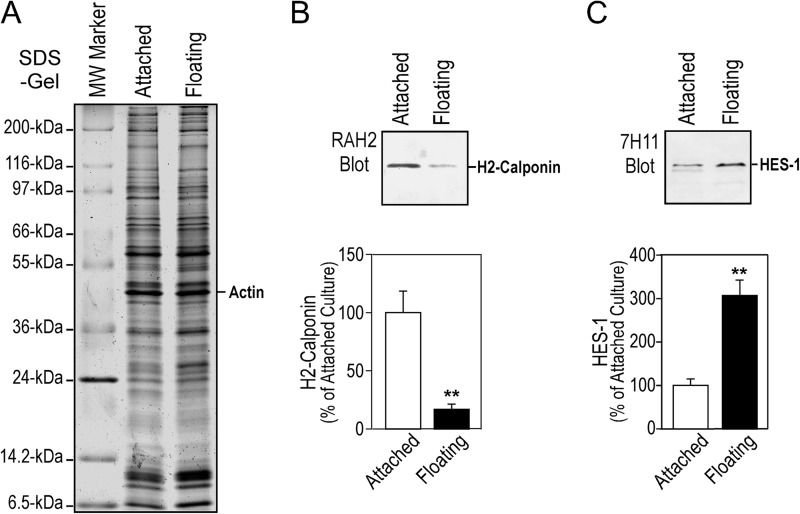
To verify the role of HES-1 in the expression of h2-calponin, we have examined the relative abundance of HES-1 in protein extracts from cells in adherent or floating cultures, in which the higher or lower cytoskeleton tension produces higher or lower level of h2-calponin expression (26). The Western blot results in Fig. 7 show that the level of HES-1 was significantly higher in floating versus adherent cells, counter-correlated with h2-calponin expression. The 5′-regulatory elements identified in mouse Cnn2 promoter and their functional relationships are summarized in Fig. 8.
FIGURE 7.
Substrate anchorage-dependent expression of h2-calponin in NIH/3T3 cells is counter-correlated with the level of HES-1. NIH/3T3 cells were cultured in plastic dishes steadily as an adherent monolayer or under continuous vibration that prevented the cells from anchoring on the dish for 3 days (26). Total cellular protein was examined with SDS-PAGE (14% gel with acrylamide:bisacrylamide ratio of 180:1) (A) and Western blotting using anti-h2-calponin polyclonal antibody RAH2 (B) and anti-HES-1 monoclonal antibody 7H11 (C). Normalized by the level of actin, the results showed that floating cells under low tension expressed lower level of h2-calponin as expected, which corresponded to a higher level of HES-1. **, p < 0.001.
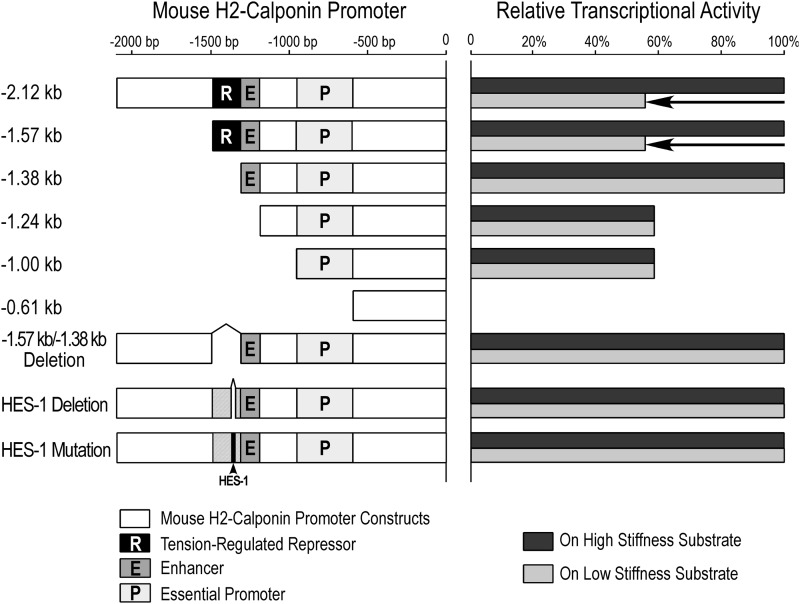
FIGURE 8.
Summary of the regulatory elements in the 5′ upstream region of mouse Cnn2 promoter. The Cnn2 promoter constructs studied are aligned in the left panel. The right panel summarizes the transcriptional activities of each construct in cells cultured on high (black bars) versus low (gray bars) stiffness substrates. Several regulatory activities are found in the mouse Cnn2 promoter. The essential promoter activity (P) located in the −1,002 to −611 region is required to turn on the expression of Cnn2 gene. The −1,380 to −1,244 region (E) is required for high level expression. The promoter and enhancer activities are both independent of the stiffness of culture substrate. The low tension-induced repressor activity (R) located in the −1,572 to −1,380 region is based on the function of an HES-1 site.
Notch Inhibitor and Activator Regulates the Expression of h2-Calponin in a Tension-dependent Manner
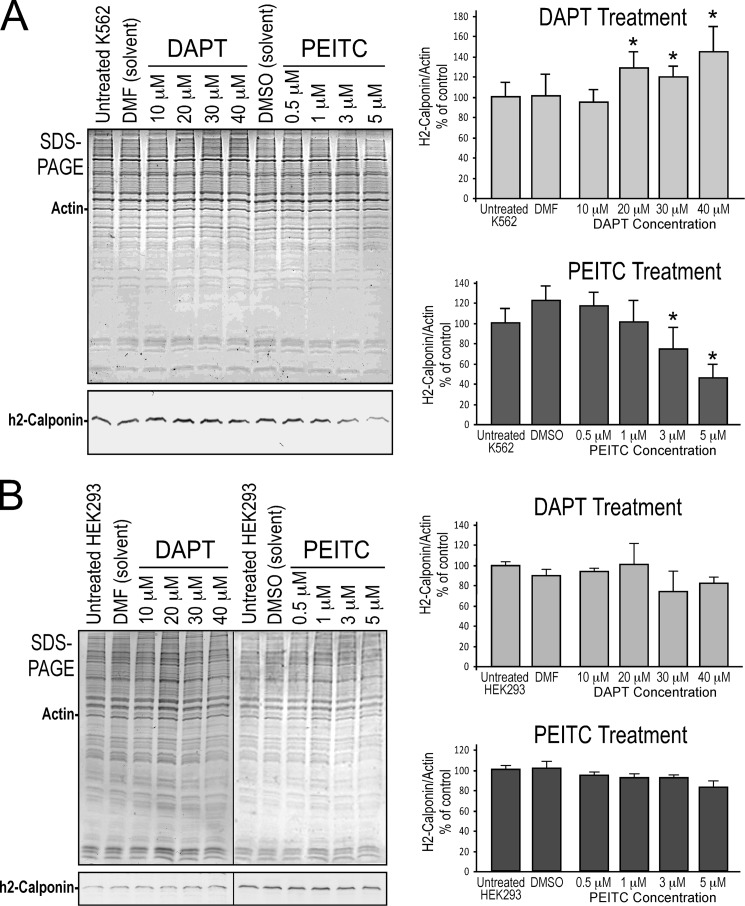
Employing K562 cells, a human myeloid blood cell line that expresses a significant level of h2-calponin in nonadherent cultures in which a low cytoskeleton tension is anticipated, Notch inhibitor DAPT increased the level of h2-calponin expression (Fig. 9A) presumably by diminishing the low tension-induced repression of Cnn2 gene. Consistently, Notch activator PEITC significantly decreased the level of h2-calponin expression (Fig. 9A). In contrast, DAPT and PEITC had minimum effects on the expression of h2-calponin in HEK293 cells growing on plastic substrate as an adherent monolayer (Fig. 9B), suggesting that the higher tension in adherent cells has a dominant effect over the Notch activator. The results suggest that Notch signaling plays a role in the regulation of h2-calponin gene expression, which is dependent on mechanical tension, where the higher tension built in the cytoskeleton of adherent versus floating cells dominantly up-regulates h2-calponin expression (13, 14).
FIGURE 9.
Notch inhibitor and activator regulate h2-calponin expression in a mechanical tension-dependent manner. A, SDS-PAGE and Western blots showed the effects of Notch inhibitor DAPT and activator PEITC on the expression of h2-calponin in nonadherent K562 cells. Normalized to actin and compared with the untreated or solvent-treated controls, the results demonstrate that DAPT increased and PEITC decreased h2-calponin expression. B, similar treatments did not produce significant change in HEK293 cells cultured in plastic as an adherent monolayer. The results suggest a role of Notch signaling in the regulation of h2-calponin gene expression, which was dependent on substrate stiffness-produced mechanical tension in the cell. *, p < 0.05; **, p < 0.01 versus control. The data are presented as means ± S.E. n = 3 in each experimental group.
DISCUSSION
Mechanosignaling of cellular activities includes dynamic and static stimulations (40–42). Examples of dynamic signaling have been reported such as stretch-activated membrane channels and cilia- or hair cell-mediated signaling (43–45). In the present study, we investigated the regulatory effect of static traction force in the cytoskeleton on the expression of h2-calponin gene that encodes a protein regulating the function of actin cytoskeleton in multiple types of eukaryotic cells (5).
Several cell signaling pathways have been suggested for functions in cytoskeleton-based mechanoregulation of eukaryotic cells. For example, the angiotensin II receptor-related pathway (46), the integrin-mediated pathway (47), and the G protein-coupled receptor pathway (48). Despite the direct relevance of h2-calponin to the mechanoregulated cytoskeleton function, the regulation of Cnn2 gene expression remains to be investigated in detail.
Key features of the mechanoregulation of h2-calponin gene expression are its sensitivity and responses to intracellular tension built against the stiffness of extracellular matrix or culture substrate (13, 14). Our Cnn2 promoter mapping data confirmed the mechanical tension regulation of h2-calponin gene expression at the transcriptional level.
An early report suggested that DNA methylation could suppress h2-calponin expression to counter the trisomy 21 gene dosage in Down syndrome (49). However, human genome sequencing data later revealed that the human CNN2 gene is located in chromosome 19 (NC_000019.9) instead of chromosome 21, although the later contains a CNN2-like potentially pseudo gene (50). Therefore, our present study is the first study of the repressor regulation of h2-calponin gene expression.
It is known that mechanical tension regulates h2-calponin at the levels of both gene expression and protein turnover (13, 14). However, the transfective expression of h2-calponin under a viral promoter that is not regulated by the cellular mechanoregulation showed no decrease but an increase in the level of h2-calponin protein when the cells were cultured on soft versus hard gel substrate (14). This observation suggests that transcriptional regulation is a primary determinant for the cellular response to mechanical signals, at least in the case of h2-calponin. Focusing on transcriptional regulation, the present study employed cell culture substrates of different stiffness to generate tension signals and investigated the promoter activity of the mouse Cnn2 gene. The results identified three 5′-upstream regions with distinct regulatory functions (Fig. 8). Because h2-calponin gene is under similar mechanoregulation in multiple cell types, the following findings may indicate common regulatory mechanisms.
Cnn2 Promoter Transfected into Cells Responded to Mechanoregulation Same as That of Endogenous Cnn2 Gene
Our study demonstrated that when transfected into cultured cells, the cloned mouse Cnn2 promoter retained mechanoregulated transcriptional activity. Summarized in Fig. 8, the critical role of the 391-bp segment between −1,002 and −611 indicates an essential promoter activity that merits further investigation to explore how the cell type- and differentiation state-specific expression of calponin isoform genes (26) is stringently controlled. Similarly, the enhancer activity identified between −1,380 and −1,244 is worth further study for its role in determining the level of Cnn2 gene expression in a permissive cellular environment. The promoter analysis in transfected cells further led to the interesting discovery of the cis-regulatory element in the −1,572 and −1,380 region responsible for the low stiffness substrate-induced repression of Cnn2 gene.
It is important to note that the essential promoter element in the −1.00-kb truncation construct, the −1,380 to −1,244 region required for high level expression, and the −1,572 and −1,380 mechanoregulated repressor region of Cnn2 gene all remained functional when transfected into cultured cells. Similar repression regulation of the Cnn2 promoter occurs in both transient transfection without chromatin integration and multiple clones of stable transfected cells with random chromatin integrations. Therefore, chromatin structure and integration sites are unlikely to play major roles in the mechanoregulation of h2-calponin gene expression. This observation supports a primary role for trans-regulatory factors.
The role of trans-regulatory factor mechanoregulation of h2-calponin gene expression is consistent with the observation that the mechanoregulation of h2-calponin expression was dependent on the amount of tension over time. Our previous cyclic stretch studies demonstrated a down-regulation of h2-calponin expression quantitatively dependent on the duration of the relaxation phase of the stretching cycle, during which the cells experienced low tension (14). In contrast to an all-or-none switching mechanism, this phenomenon may reflect an accumulative concentration effect of trans-regulatory factors.
Role of Notch/HES-1 Pathway in the Regulation of Cnn2 Gene Expression Dependent on Mechanical Tension
The −1.38-kb truncation and the −1,572 to −1,380 internal deletion constructs both exhibited unregulated high level expressions on soft gel substrate (Figs. 4 and 5). This observation was confirmed in numerous transient and stable transfection experiments. In floating cells that were experiencing lower cytoskeleton tension than that in cells in adherent cultures (26), the decrease in h2-calponin expression was accompanied with a significant increase in HES-1 counter-correlated with h2-calponin expression (Fig. 7). These data suggested that the control of Cnn2 gene expression by cellular tension is not a high tension-stimulated up-regulation but a low tension-induced repression (Fig. 8).
Based on this hypothesis, the search for transcriptional factor binding sites in the −1,572 to −1,380 segment of mouse Cnn2 promoter was focused on transcription repressors and identified a binding site for HES-1, an established transcription repressor (39). Site-specific deletion or mutation experiments confirmed that this HES-1 site is a critical regulatory element in the mechanoregulation of Cnn2 gene (Fig. 6).
HES-1 has been reported to suppress transcription via the retinoid X receptor (RXR), a transcription factor that functions under the influence of transcription repressors (38) such as HES-1 (39). Plausibly, two RXR binding sites are found adjacently in the downstream −1,380 to −1,244 region that is responsible for the high level transcriptional activity of the mouse Cnn2 promoter (Figs. 3 and 6). The functional relationship of this HES-1/RXR pair will be investigated in follow-up studies.
Further supporting the role of HES-1 in mechanoregulation of Cnn2 gene expression, HES-1 is known to function as a downstream signaling molecule in the Notch pathway that has been suggested to mediate mechanoregulation (36, 37). The role of Notch regulation on h2-calponin expression is demonstrated by the effects of Notch inhibitor and activator in a tension-dependent manner (Fig. 9). The data are consistent with a role of Notch-RBP J-HES-1 pathway (51) in the mechanoregulation of h2-calponin gene expression. However, a definitive demonstration awaits further experiments that show a loss of mechanoregulation in cells lacking HES-1 and binding of HES-1 to the endogenous Cnn2 promoter.
Transcriptional Regulation of h2-Calponin Gene and Mechanostability of Cytoskeleton
The mechanical tension-regulated expression of Cnn2 gene and the function of h2-calponin in stabilizing actin cytoskeleton present a novel model to study cellular mechanoregulation. When sensing mechanical stimuli, the cytoskeleton, as a dynamic tensegrity network, rearranges and redistributes to reduce the overall stress (4). At the same time, the force signal is transmitted into the nuclei to modulate adaptive gene expressions (23). In this regulation, the level of h2-calponin gene expression is high at high cytoskeleton tension to make the actin cytoskeleton more stable and vice versa.
Our data showed that the mechanoregulation of h2-calponin gene expression is primarily a low tension-induced repression of transcription. Therefore, a post-transcriptional down-regulation of h2-calponin in the actin cytoskeleton, such as that produced by low tension-induced proteolysis (14), would decrease F-actin stability and in turn decrease the tension signal transduced into and sensed by the cell, resulting in a repression of gene transcription to maintain a low level equilibrium of h2-calponin in the cell.
Transfective expression of h2-calponin under a viral promoter that lacks the mechanoregulation showed no decrease but a significant increase in h2-calponin protein when the cells were cultured on soft versus hard gel substrate (14). Therefore, low cytoskeleton tension may actually promote accumulation of h2-calponin protein in the cell; thus the down-regulation at the transcriptional level would be critical to the effectiveness of cellular response to mechanical signals. Further testing this hypothesis will help to understand the role of h2-calponin in cytoskeleton function and dynamics.
Functional Implications
The mechanoregulation of Cnn2 gene expression and the function of h2-calponin in cytoskeleton stability and dynamics indicate broad physiological and pathological significance. Tissues and cells originating from all three embryonic germ layers have been found to express h2-calponin. Multiple immortalized cell lines including epithelial, fibroblasts, myeloid, and myoblasts express high levels of h2-calponin under the regulation of cytoskeleton tension (5).
Based on anatomical and functional features, these naturally h2-calponin-positive cell types can be placed into three classes: the first is cells bearing constant or dynamic mechanical tension, such as epithelial and endothelial cells and smooth muscle cells in the wall of various internal organs; the second is migrating cells such as fibroblasts and macrophages; the third group is proliferating cells including myoblasts and other stem cells. This phenotypic classification may help to understand the physiological roles of h2-calponin as a mechanoregulatory molecule in different cell types. The present study laid a foundation for future investigations on the regulation and function of calponin, a widely distributed and abundant but yet poorly understood, cytoskeleton regulatory protein.
Acknowledgments
We thank Hui Wang for technical assistance. Dr. Qi-Quan Huang and Dr. Xin Wang participated in the cloning of mouse Cnn2 gene promoter, construction of some of the promoter-reporter genes, and initial characterization when this research group was at Case Western Reserve University.
This work was supported, in whole or in part, by National Institutes of Health Grant HL-086720 (to J. P. J.).
- CAT
- chloramphenical acetyltransferase
- DAPT
- N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester
- Ef
- elastic force
- PEITC
- phenethyl isothiocyanate
- RXR
- vitamin D receptor/retinoic acid X receptor.
REFERENCES
- 1. Wang N., Tytell J. D., Ingber D. E. (2009) Mechanotransduction at a distance. Mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 10, 75–82 [DOI] [PubMed] [Google Scholar]
- 2. Rayment I. (1996) Kinesin and myosin. Molecular motors with similar engines. Structure 4, 501–504 [DOI] [PubMed] [Google Scholar]
- 3. Mikhailenko S. V., Oguchi Y., Ishiwata S. (2010) Insights into the mechanisms of myosin and kinesin molecular motors from the single-molecule unbinding force measurements. J. R. Soc. Interface 7, (Suppl. 3) S295–S306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kang J., Steward R. L., Kim Y., Schwartz R. S., LeDuc P. R., Puskar K. M. (2011) Response of an actin filament network model under cyclic stretching through a coarse grained Monte Carlo approach. J. Theor. Biol. 274, 109–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu K. C., Jin J. P. (2008) Calponin in non-muscle cells. Cell Biochem. Biophys. 52, 139–148 [DOI] [PubMed] [Google Scholar]
- 6. Takahashi K., Hiwada K., Kokubu T. (1986) Isolation and characterization of a 34,000-dalton calmodulin- and F-actin-binding protein from chicken gizzard smooth muscle. Biochem. Biophys. Res. Commun. 141, 20–26 [DOI] [PubMed] [Google Scholar]
- 7. Takahashi K., Nadal-Ginard B. (1991) Molecular cloning and sequence analysis of smooth muscle calponin. J. Biol. Chem. 266, 13284–13288 [PubMed] [Google Scholar]
- 8. Nishida W., Kitami Y., Hiwada K. (1993) cDNA cloning and mRNA expression of calponin and SM22 in rat aorta smooth muscle cells. Gene 130, 297–302 [DOI] [PubMed] [Google Scholar]
- 9. Strasser P., Gimona M., Moessler H., Herzog M., Small J. V. (1993) Mammalian calponin. Identification and expression of genetic variants. FEBS Lett. 330, 13–18 [DOI] [PubMed] [Google Scholar]
- 10. Applegate D., Feng W., Green R. S., Taubman M. B. (1994) Cloning and expression of a novel acidic calponin isoform from rat aortic vascular smooth muscle. J. Biol. Chem. 269, 10683–10690 [PubMed] [Google Scholar]
- 11. Trabelsi-Terzidis H., Fattoum A., Represa A., Dessi F., Ben-Ari Y., der Terrossian E. (1995) Expression of an acidic isoform of calponin in rat brain. Western blots on one- or two-dimensional gels and immunolocalization in cultured cells. Biochem. J. 306, 211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Winder S. J., Sutherland C., Walsh M. P. (1991) Biochemical and functional characterization of smooth muscle calponin. Adv. Exp. Med. Biol. 304, 37–51 [DOI] [PubMed] [Google Scholar]
- 13. Hossain M. M., Crish J. F., Eckert R. L., Lin J. J., Jin J. P. (2005) h2-Calponin is regulated by mechanical tension and modifies the function of actin cytoskeleton. J. Biol. Chem. 280, 42442–42453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hossain M. M., Smith P. G., Wu K., Jin J. P. (2006) Cytoskeletal tension regulates both expression and degradation of h2-calponin in lung alveolar cells. Biochemistry 45, 15670–15683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tang J., Hu G., Hanai J., Yadlapalli G., Lin Y., Zhang B., Galloway J., Bahary N., Sinha S., Thisse B., Thisse C., Jin J. P., Zon L. I., Sukhatme V. P. (2006) A critical role for calponin 2 in vascular development. J. Biol. Chem. 281, 6664–6672 [DOI] [PubMed] [Google Scholar]
- 16. Huang Q. Q., Hossain M. M., Wu K., Parai K., Pope R. M., Jin J. P. (2008) Role of H2-calponin in regulating macrophage motility and phagocytosis. J. Biol. Chem. 283, 25887–25899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Winder S. J., Walsh M. P. (1990) Smooth muscle calponin. Inhibition of actomyosin MgATPase and regulation by phosphorylation. J. Biol. Chem. 265, 10148–10155 [PubMed] [Google Scholar]
- 18. Abe M., Takahashi K., Hiwada K. (1990) Effect of calponin on actin-activated myosin ATPase activity. J. Biochem. 108, 835–838 [DOI] [PubMed] [Google Scholar]
- 19. Horiuchi K. Y., Chacko S. (1991) The mechanism for the inhibition of actin-activated ATPase of smooth muscle heavy meromyosin by calponin. Biochem. Biophys. Res. Commun. 176, 1487–1493 [DOI] [PubMed] [Google Scholar]
- 20. Winder S. J., Allen B. G., Fraser E. D., Kang H. M., Kargacin G. J., Walsh M. P. (1993) Calponin phosphorylation in vitro and in intact muscle. Biochem. J. 296, 827–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shirinsky V. P., Biryukov K. G., Hettasch J. M., Sellers J. R. (1992) Inhibition of the relative movement of actin and myosin by caldesmon and calponin. J. Biol. Chem. 267, 15886–15892 [PubMed] [Google Scholar]
- 22. Haeberle J. R. (1994) Calponin decreases the rate of cross-bridge cycling and increases maximum force production by smooth muscle myosin in an in vitro motility assay. J. Biol. Chem. 269, 12424–12431 [PubMed] [Google Scholar]
- 23. Shivashankar G. V. (2011) Mechanosignaling to the cell nucleus and gene regulation. Annu. Rev. Biophys. 40, 361–378 [DOI] [PubMed] [Google Scholar]
- 24. Wang Y. L., Pelham R. J., Jr. (1998) Preparation of a flexible, porous polyacrylamide substrate for mechanical studies of cultured cells. Methods Enzymol. 298, 489–496 [DOI] [PubMed] [Google Scholar]
- 25. Engler A., Bacakova L., Newman C., Hategan A., Griffin M., Discher D. (2004) Substrate compliance versus ligand density in cell on gel responses. Biophys. J. 86, 617–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hossain M. M., Hwang D. Y., Huang Q. Q., Sasaki Y., Jin J. P. (2003) Developmentally regulated expression of calponin isoforms and the effect of h2-calponin on cell proliferation. Am. J. Physiol. Cell Physiol 284, C156–C167 [DOI] [PubMed] [Google Scholar]
- 27. Huang Q. Q., Jin J. P. (1999) Preserved close linkage between the genes encoding troponin I and troponin T, reflecting an evolution of adapter proteins coupling the Ca2+ signaling of contractility. J. Mol. Evol. 49, 780–788 [DOI] [PubMed] [Google Scholar]
- 28. Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. (2005) MatInspector and beyond. Promoter analysis based on transcription factor binding sites. Bioinformatics 21, 2933–2942 [DOI] [PubMed] [Google Scholar]
- 29. Yeung T., Georges P. C., Flanagan L. A., Marg B., Ortiz M., Funaki M., Zahir N., Ming W., Weaver V., Janmey P. A. (2005) Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskeleton 60, 24–34 [DOI] [PubMed] [Google Scholar]
- 30. Phillips J. E., Corces V. G. (2009) CTCF. Master weaver of the genome. Cell 137, 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eberhard D., Jiménez G., Heavey B., Busslinger M. (2000) Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. EMBO J. 19, 2292–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kageyama R., Ohtsuka T., Kobayashi T. (2007) The Hes gene family. Repressors and oscillators that orchestrate embryogenesis. Development 134, 1243–1251 [DOI] [PubMed] [Google Scholar]
- 33. Liu Z. J., Shirakawa T., Li Y., Soma A., Oka M., Dotto G. P., Fairman R. M., Velazquez O. C., Herlyn M. (2003) Regulation of Notch1 and Dll4 by vascular endothelial growth factor in arterial endothelial cells. Implications for modulating arteriogenesis and angiogenesis. Mol. Cell. Biol. 23, 14–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Teixeira A. I., Ilkhanizadeh S., Wigenius J. A., Duckworth J. K., Inganäs O., Hermanson O. (2009) The promotion of neuronal maturation on soft substrates. Biomaterials 30, 4567–4572 [DOI] [PubMed] [Google Scholar]
- 35. Chen J., Zolkiewska A. (2011) Force-induced unfolding simulations of the human Notch1 negative regulatory region. Possible roles of the heterodimerization domain in mechanosensing. PLoS One 6, e22837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morrow D., Sweeney C., Birney Y. A., Cummins P. M., Walls D., Redmond E. M., Cahill P. A. (2005) Cyclic strain inhibits Notch receptor signaling in vascular smooth muscle cells in vitro. Circ. Res. 96, 567–575 [DOI] [PubMed] [Google Scholar]
- 37. Morrow D., Sweeney C., Birney Y. A., Guha S., Collins N., Cummins P. M., Murphy R., Walls D., Redmond E. M., Cahill P. A. (2007) Biomechanical regulation of hedgehog signaling in vascular smooth muscle cells in vitro and in vivo. Am. J. Physiol. Cell Physiol. 292, C488–C496 [DOI] [PubMed] [Google Scholar]
- 38. Allenby G., Bocquel M. T., Saunders M., Kazmer S., Speck J., Rosenberger M., Lovey A., Kastner P., Grippo J. F., Chambon P. (1993) Retinoic acid receptors and retinoid X receptors. Interactions with endogenous retinoic acids. Proc. Natl. Acad. Sci. U.S.A. 90, 30–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chow E. K., Castrillo A., Shahangian A., Pei L., O'Connell R. M., Modlin R. L., Tontonoz P., Cheng G. (2006) A role for IRF3-dependent RXRα repression in hepatotoxicity associated with viral infections. J. Exp. Med. 203, 2589–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang N., Naruse K., Stamenović D., Fredberg J. J., Mijailovich S. M., Tolić-Nørrelykke I. M., Polte T., Mannix R., Ingber D. E. (2001) Mechanical behavior in living cells consistent with the tensegrity model. Proc. Natl. Acad. Sci. U.S.A. 98, 7765–7770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Janmey P. A., Winer J. P., Murray M. E., Wen Q. (2009) The hard life of soft cells. Cell Motil. Cytoskeleton 66, 597–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hoffman B. D., Grashoff C., Schwartz M. A. (2011) Dynamic molecular processes mediate cellular mechanotransduction. Nature 475, 316–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sachs F. (2010) Stretch-activated ion channels. What are they? Physiology 25, 50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Corey D. (2003) Sensory transduction in the ear. J. Cell Sci. 116, 1–3 [DOI] [PubMed] [Google Scholar]
- 45. Géléoc G. S., Holt J. R. (2003) Developmental acquisition of sensory transduction in hair cells of the mouse inner ear. Nat. Neurosci. 6, 1019–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. de Cavanagh E. M., Ferder M., Inserra F., Ferder L. (2009) Angiotensin II, mitochondria, cytoskeletal, and extracellular matrix connections. An integrating viewpoint. Am. J. Physiol. Heart Circ. Physiol. 296, H550–H558 [DOI] [PubMed] [Google Scholar]
- 47. Li J., Zhao Z., Wang J., Chen G., Yang J., Luo S. (2008) The role of extracellular matrix, integrins, and cytoskeleton in mechanotransduction of centrifugal loading. Mol. Cell Biochem. 309, 41–48 [DOI] [PubMed] [Google Scholar]
- 48. Tzima E. (2006) Role of small GTPases in endothelial cytoskeletal dynamics and the shear stress response. Circ. Res. 98, 176–185 [DOI] [PubMed] [Google Scholar]
- 49. Kuromitsu J., Yamashita H., Kataoka H., Takahara T., Muramatsu M., Sekine T., Okamoto N., Furuichi Y., Hayashizaki Y. (1997) A unique downregulation of h2-calponin gene expression in Down syndrome. A possible attenuation mechanism for fetal survival by methylation at the CpG island in the trisomic chromosome 21. Mol. Cell. Biol. 17, 707–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Masuda H., Tanaka K., Takagi M., Ohgami K., Sakamaki T., Shibata N., Takahashi K. (1996) Molecular cloning and characterization of human non-smooth muscle calponin. J. Biochem. 120, 415–424 [DOI] [PubMed] [Google Scholar]
- 51. Kageyama R., Ishibashi M., Takebayashi K., Tomita K. (1997) bHLH transcription factors and mammalian neuronal differentiation. Int. J. Biochem. Cell Biol. 29, 1389–1399 [DOI] [PubMed] [Google Scholar]