Background: Molecular features underlying enzyme function in extreme environments are poorly understood.
Results: Identification of the basis for thermostability, halophilicity, and detoxification activity in a mercuric reductase from hot deep-sea brine.
Conclusion: A small number of structural modifications accounts for the enzyme's robustness.
Significance: This work defines novel adaptations that enable enzymes to cope with multiple abiotic stressors simultaneously.
Keywords: Enzyme Kinetics, Enzyme Mechanisms, Enzyme Structure, Metagenomics, Mutagenesis Site-specific, Atlantis II Brine Pool, Extreme Halophilic, Mercuric Reductase, Red Sea Atlantis II, Thermophilic
Abstract
A unique combination of physicochemical conditions prevails in the lower convective layer (LCL) of the brine pool at Atlantis II (ATII) Deep in the Red Sea. With a maximum depth of over 2000 m, the pool is characterized by acidic pH (5.3), high temperature (68 °C), salinity (26%), low light levels, anoxia, and high concentrations of heavy metals. We have established a metagenomic dataset derived from the microbial community in the LCL, and here we describe a gene for a novel mercuric reductase, a key component of the bacterial detoxification system for mercuric and organomercurial species. The metagenome-derived gene and an ortholog from an uncultured soil bacterium were synthesized and expressed in Escherichia coli. The properties of their products show that, in contrast to the soil enzyme, the ATII-LCL mercuric reductase is functional in high salt, stable at high temperatures, resistant to high concentrations of Hg2+, and efficiently detoxifies Hg2+ in vivo. Interestingly, despite the marked functional differences between the orthologs, their amino acid sequences differ by less than 10%. Site-directed mutagenesis and kinetic analysis of the mutant enzymes, in conjunction with three-dimensional modeling, have identified distinct structural features that contribute to extreme halophilicity, thermostability, and high detoxification capacity, suggesting that these were acquired independently during the evolution of this enzyme. Thus, our work provides fundamental structural insights into a novel protein that has undergone multiple biochemical and biophysical adaptations to promote the survival of microorganisms that reside in the extremely demanding environment of the ATII-LCL.
Introduction
Natural environments that encompass multiple abiotic stressors, such as extreme salinity, high temperatures, and high levels of toxic heavy metals, are rare on our planet and are usually difficult to access. In the Red Sea, the brine pool at Atlantis II (ATII)2 Deep is a good example of such an environment. Covering an area of about 60 km2, it is the largest brine pool in the central Red Sea (21° 20.72′ north and 38° 04.59′ east) and is located at a depth of 2000–2200 m. The 200-m-thick pool is stratified into different layers. The bottom layer, referred to as the lower convective layer (LCL), presents an exceptional combination of environmental conditions, characterized by extreme salinity (26%), high temperature (68 °C), hydrostatic pressure, acidic pH (5.3), extremely low levels of light and oxygen, and high concentrations of heavy metals (1–4). The microbial community that resides in the LCL of the ATII (ATII-LCL) therefore offers a unique opportunity to study the biochemical adaptations that enable its members to survive and thrive in such an exacting setting. In an attempt to elucidate how enzymes have evolved strategies for coping simultaneously with extreme salinity, high temperature, and heavy metal toxicity, we sampled the microbial community from ATII-LCL, established a metagenomic dataset based on 454 pyrosequencing, and mined it for sequences encoding enzymes involved in the detoxification of mercury. An operon containing several genes required for mercury detoxification was identified in our dataset, and in this report we describe the product of one of these. MerA ATII-LCL is a mercuric reductase, a key component of an organomercurial detoxification system found in many bacteria that grow in mercury-contaminated environments (5).
Mercuric ion reductase catalyzes the reduction of Hg2+ to Hg0, which is volatile and can be disposed of nonenzymatically (6–8). The enzymes contain flavin adenine dinucleotide (FAD), utilize NADPH as an electron donor, and require an excess of exogenous thiols for activity (9). The presence of thiols such as glutathione ensures that the Hg2+ exists in the cell as the dimercaptide, RS-Hg-SR. Studies of the MerA protein encoded by the mer operon on transposon Tn501 have shown that the enzyme is a homodimer. Each monomer contributes one active site, made up of a pair of redox-active cysteines, to a catalytic core located at the dimer interface (10, 11). A pair of cysteine residues located near the C terminus of one subunit serves to transfer the substrate Hg2+ to the active site in the other subunit (10, 11), which catalyzes the reduction of the metal ion. In addition to the catalytic core, MerA proteins typically have an extended N-terminal domain (NmerA) of around 70 amino acids that contains a pair of cysteines within a highly conserved metal-binding motif (GMTCXXC) (11). Initially, the NmerA cysteine pair appeared to be dispensable for catalysis in vivo, because a double mutant in which both residues were replaced by alanines was found to be fully functional (11). However, it was later shown that, under physiological conditions in which intracellular thiols are depleted, the NmerA domain binds Hg2+ and transfers it from ligands in cytoplasm to the catalytic core for reduction (12, 13). Furthermore, it has been shown that Hg2+ increases H2O2 formation in mitochondria, leading to increased consumption of reduced glutathione (14). Taken together, these results strongly indicate that the pair of cysteines in the NmerA domain of mercuric reductases does have a functional role in vivo, enhancing mercuric ion detoxification by acting as an accessory pathway for delivery of the substrate to the active site (12).
Despite the availability of detailed information concerning the structure-function relationships of MerA, and compelling phylogenetic evidence indicating that MerA originated and evolved in thermophilic microorganisms residing in geothermal environments (15, 16), to our knowledge, no thermophilic and/or halophilic MerA homolog derived from such an environment has yet been characterized in detail.
Thermophilic and halophilic adaptations of proteins remain the subject of intensive study, but it is clear that their hosts have developed diverse strategies to keep them folded in an active state under conditions of elevated temperature and salinity (17). Depending on their optimal growth temperatures, bacteria are generally classified into four groups as follows: psychrophiles grow best at 5–20 °C, mesophiles at 15–45 °C, thermophiles at 45–80 °C, and hyperthermophiles at above 80 °C (18). Two types of physical mechanisms that contribute to protein thermophily are distinguished as “structure-based” and “sequence-based” (19). The features associated with enhanced thermal stability include ionic interactions (20), increase in hydrophobicity and packing density (21, 22), augmentation of hydrogen bonding and van der Waals interactions (23, 24), and specific amino acid substitutions that stabilize protein structure at particularly critical locations (25).
Halophilic microorganisms are broadly classified into slightly, moderately, and extremely halophilic, defined as showing optimal growth in 0.5, 0.5–2.5, and around 4 m NaCl2, respectively (26). Halophilic microorganisms have evolved two strategies to maintain the osmotic pressure of the cytoplasm within its physiological range. The first is based on the biosynthesis and accumulation in the cytoplasm of organic solutes, osmolytes, such as glycine, betaine, and ectoine (27). The second strategy, the salt-in approach, involves the accumulation of molar concentrations of potassium chloride in the cytoplasm (28–30). This, in turn, necessitates adaptation of intracellular proteins to ensure that they remain soluble and active in the presence of such high concentrations of salt. Proteins adapted to high salt often show a predominance of acidic residues on their surfaces (31–33), which interact with water molecules and salt ions, and a corresponding decrease in hydrophobic amino acids and the extent of hydrophobic contact surfaces (31, 34).
The metagenome-derived MerA enzyme from the ATII-LCL described here reveals simple and limited alterations in the primary structure of the protein relative to that of an ortholog from a terrestrial environment (35). These are reflected in critical shifts in the catalytic properties of the enzyme that allow for efficient function under the extreme conditions of its marine habitat. The sequences of the soil and ATII-LCL enzymes are >91% identical, and 67% of the substitutions in the ATII-LCL enzyme are acidic residues. In addition, two short segments near the C-terminal cysteine pair, each containing two basic amino acids and a proline residue, are unique to the ATII-LCL enzyme. These structural features largely account for the ability of MerA ATII-LCL to function efficiently in high salt at high temperature, as site-directed mutagenesis of selected acidic residues and replacement of the two boxes in the ATII-LCL enzyme by the residues found in the soil ortholog reduced the degree of halophilicity and thermostability, respectively, of the former. In addition, the two acidic residues immediately adjacent to the NmerA metal-binding motif in the ATII-LCL protein have a direct effect on both the halophilicity and catalytic efficiency of the enzyme. Presumably, by increasing the efficiency of delivery of Hg2+ ions to the catalytic core for reduction, they also help the host to cope with the high concentrations of mercury present in its hypersaline environment.
EXPERIMENTAL PROCEDURES
Identification of the Coding Sequence of the ATII-LCL MerA
Water samples were collected from the LCL of the Atlantis II brine (2200 m below the surface) in the Red Sea at 21° 20.72′ north and 38° 04.59′ east during the King Abdullah University for Sciences and Technology (KAUST) (Thuwal, Kingdom of Saudi Arabia); Woods Hole Oceanographic Institute (WHOI) (Woods Hole, MA); and Hellenic Center for Marine Research (HCMR) (Anavisso, Greece) oceanographic cruise of the research vessel Aegaeo in March/April 2010. Samples were immediately processed by serial filtration through three 293-mm stainless steel sanitary filter holders (Millipore) containing mixed cellulose-ester filters (nitrocellulose/cellulose acetate) with pore sizes of 3, 0.8, and 0.1 μm (Millipore). Filters were then stored in sucrose buffer (36). Isolation of DNA from microbes trapped on the 0.1-μm filter was performed using a Marine DNA isolation kit (Epicenter). The concentration of the DNA recovered was measured using a NanoDrop 3300 fluorospectrometer (Thermo Scientific) and a Quant-iTTM PicoGreen® dsDNA kit (Invitrogen).
DNA pyrosequencing was carried out with the GS FLX titanium pyrosequencing kit (454 Life Sciences). An ATII-LCL metagenomic dataset was then established from around 4 million sequence reads. The reads were assembled, and ORFs were identified and annotated. An operon containing a MerA ORF was identified by BLAST search against the nr database. The genomic sequence of this ATII-LCL merA gene has been deposited in GenBankTM under accession number KF572479. The ORF was found to show 91% amino acid sequence identity to a putative mercuric reductase from an uncultured bacterium isolated from agricultural soil (35) (GenBankTM under accession number AEV57255.1).
Primary Structure Analysis
Sequence alignment of MerA ATII-LCL with the soil ortholog was performed using the STRAP editor for structural alignment of proteins (37). Identification of the pyridine nucleotide-disulfide oxidoreductase dimerization domain (P-code PF02852.17) of MerA ATII-LCL was achieved by performing a HMMScan search of its amino acid sequence against the Pfam-A database (version 27.0) (38) on the HMMER webserver (39). The statistical significance of the amino acid substitutions observed between MerA ATII-LCL and its ortholog from soil was assessed using Fisher's Exact Independence Test (FET) in R version 2.15.3.
Expression and Purification of Recombinant MerA Enzymes
A single pair of perfectly matched oligonucleotides deduced from the aligned soil and ATII-LCL MerAs was used to amplify the coding sequence of the MerA from the LCL metagenomic DNA. The amplified DNA fragments were cloned in the TOPO TA cloning vector (Invitrogen) and sequenced by the Sanger dideoxy method using the 96-capillary ABI 3730XI DNA sequencer. A clone with a clear halophilic signature was identified and found to show more than 90% identity with the MerA enzyme from soil. Both sequences were synthesized (GenScript) after optimizing codon usage to increase their expression levels in Escherichia coli. Mutations in selected codons (Table 1) were generated using the QuikChange II site-directed mutagenesis kit according to the manufacturer's instructions (Agilent Technologies). Note that all mutants were generated in the ATII-LCL MerA sequence that had been optimized for expression in E. coli.
TABLE 1.
Mutations to substitute residues from the ATII-LCL MerA to their corresponding amino acids in the soil enzyme
| Mutation | From | To |
|---|---|---|
| M1 | E15 and E16 (E15/E16) | A15 and A16 (A15/A16) |
| M4 | E133 and E134 (E133/E134) | G133 and G34 (G133/G134) |
| M15 (box1) | KPAR432–435 (432KPAR435) | LDLT432–435 (432LDLT435) |
| M16 (box2) | KVGKFP465–470 (465KVGKFP470) | DSRTLT465–470 (465DSRTLT470) |
| M15/M16 box1/box2 | 432KPAR435/465KVGKFP470 | 432LDLT435/465DSRTLT470 |
| M17 | E515 and E516 (E515/E516) | A515 and A516 (A515/A516) |
| M18 | E545 and E546 (E545/E546) | A545 and A546 (A545/A546) |
All synthesized genes were cloned into the expression vector ChampionTM pET SUMO (Invitrogen). An overnight culture of transformed E. coli BL21(DE3) cells was diluted 50-fold in fresh LB medium supplemented with 20 μm FAD. Expression of recombinant enzymes was induced at an A600 of 0.5 by the addition of isopropyl β-d-thiogalactoside (final concentration 1 mm) for 3 h at 37 °C. Cultures were centrifuged, lysed by multiple freeze-thaw cycles, and resuspended in TE buffer (10 mm Tris-Cl, pH 7.5, containing 1 mm EDTA) supplemented with 20 μm FAD. The sample was sonicated and cellular debris removed by centrifugation at 14,000 × g for 20 min at 4 °C. The supernatant was filtered and applied to a pre-equilibrated His-Trap column (Amersham Biosciences). The protein was then eluted from the column using increasing concentrations of imidazole. Protein concentration was determined using the BCA protein assay kit (Thermo Scientific). The enzyme concentration was also determined on the basis of flavin absorbance, using an extinction coefficient of 11.3 mm−1 cm−1 for the free FAD at 450 nm. The data obtained with the two assays are entirely compatible with the assumption that ATII-LCL, the soil ortholog, and the ATII-LCL mutants contain 1 FAD eq/monomer. Protein purity was verified by electrophoresis on 12% SDS-polyacrylamide gels. The purified proteins were dialyzed against phosphate-buffered saline and stored at −20 °C until further use.
Mercuric Reductase Assay
Routine enzyme assays were carried out at 37 °C in 80 mm sodium phosphate buffer, pH 7.4, containing 200 μm NADPH, 100 μm HgCl2, and 1 mm β-mercaptoethanol. The enzyme activity was monitored by observing the initial rate of NADPH consumption at 340 nm using a Shimadzu UV-1800 spectrophotometer. The unit of activity is defined as the amount of enzyme that catalyzes Hg2+-dependent oxidation of 1 μmol of NADPH/min (9). In the case of NaCl-dependent activation, the enzyme activity was measured in the presence of the indicated concentrations of NaCl.
To ensure that reducing equivalents from NADPH are being passed to Hg2+ and not to O2 to form H2O2, dissolved O2 was monitored during the course of the assay using the YSI Galvanic DO Sensor (Model 2002). The results revealed that only 0.24% of the total electron flow passes to O2.
Heat Stability Analysis
Replicate samples of each enzyme were incubated at the designated temperatures for 10 min, and the residual activity was then measured. Results are expressed relative to the activity observed at 25 °C.
Inhibition by HgCl2
The enzyme was incubated in the reaction mixture at 37 °C for 10 min in the absence of NADPH, the presence of 500 mm NaCl, and the indicated concentrations of HgCl2. The reaction was then started by the addition of NADPH.
Determination of the Minimum Inhibitory Concentration (MIC) of Hg2+
The nontransformed BL21(DE3) strain and BL21(DE3) cells transformed with the indicated plasmid were plated on LB agar with 1 mm isopropyl β-d-thiogalactoside and 50 μg/ml kanamycin (Sigma). Different concentrations of HgCl2 were added to wells punched in the agar layer, and the plates were incubated for 24 h at 37 °C. The MIC was then determined by measuring the size of the zone of inhibition around each well.
Modeling of the Three-dimensional Structure of ATII-LCL MerA
The three-dimensional structure of the ATII-LCL MerA was built by homology modeling against structures of the N-terminal (Protein Data Bank code 2kt2) (43) and C-terminal (Protein Data Bank code 1zk7) (12) domains of the Tn501 mercuric reductase, using SWISS-MODEL (40–42). Identification of the position of Glu-516 on the surface of the active-site cavity was achieved by manual inspection of the HOLLOW (44) output in PyMOL (PyMOL Molecular Graphics System, Version 1.5.0.1 Schrödinger, LLC). Labeling of specific atoms or residues, superposition of the N- and C-terminal models of ATII-LCL MerA and rendering of the final three-dimensional models were performed using PyMOL.
RESULTS
Mining and Identification of Metagenome-derived MerA ATII-LCL Sequences
We established a metagenomic dataset for the microbial community that resides in the ATII-LCL using 454 pyrosequencing. An ORF that showed >90% homology to a mercuric reductase from a soil bacterium (35) was identified in the assembled reads. To rule out the possibility that the ATII-LCL MerA ORF sequence obtained from assembled metagenomic reads might be the product of chimeric sequences, we decided to amplify the sequence of the identified ORF using ATII-LCL environmental DNA and two perfectly matched oligonucleotides deduced from the sequence of the soil and ATII-LCL orthologs. The amplified products were cloned in the TOPO TA cloning vector (Invitrogen), and clones were sequenced using the dideoxyribonucleotide chain-termination method. A MerA sequence that was rich in acidic amino acids (a typical halophilic signature) was identified and used for all further work (for details see under “Experimental Procedures”).
Structural Differences between ATII-LCL and Soil MerA
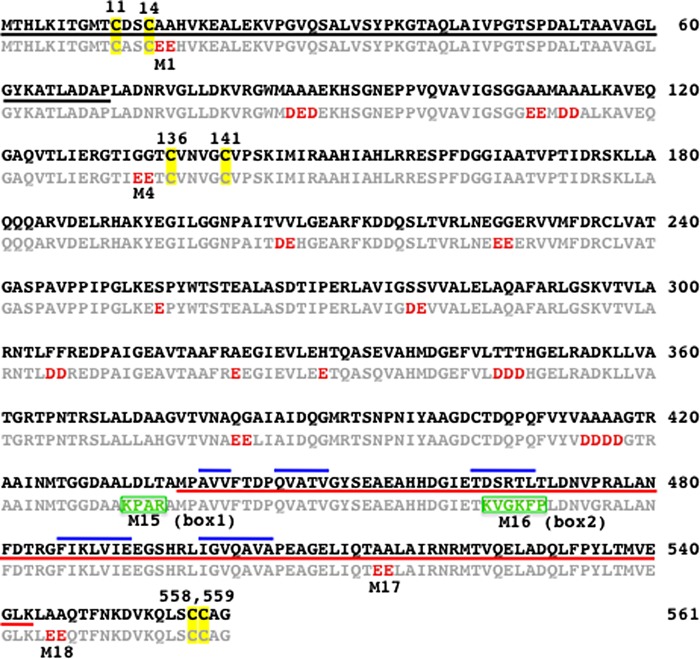
A sequence alignment of MerA ATII-LCL with its soil ortholog is shown in Fig. 1. As expected, both enzymes have the catalytic redox-active cysteine disulfide pair (Cys-136/Cys-141) in the active site (10, 11), and the Hg2+ binding Cys-558/Cys-559 pair located near the C terminus of the polypeptide chain (45, 46). In addition, both molecules possess the NmerA domain that contains the pair of cysteine thiols (Cys-11/Cys-14) in the conserved GMTCXXC sequence motif that has been shown to be involved in Hg2+ binding under glutathione-depleted conditions (12, 13).
FIGURE 1.
Alignment of the amino acid sequences of MerA ATII-LCL and the soil ortholog. The sequence of the soil enzyme is shown in bold and that of the ATII-LCL in light gray; the NmerA domain is underlined in black; the dimerization domain that is conserved among the homodimeric pyridine nucleotide-disulfide oxidoreductases is underlined in red; and the β-strand structures present in the dimerization domain are overlined in blue. The cysteine pairs 11/14 and 558/559 involved in binding of Hg2+, and cysteines 136/141, which form the disulfide bridge involved in Hg2+ reduction, are highlighted in yellow; negatively charged substitutions in MerA ATII-LCL are shown in red. The two sequences that contribute to thermostability are boxed in green (box1 and box2). The acidic residues marked M in the ATII-LCL sequence were replaced by the corresponding amino acids in the soil enzyme in the indicated ATII-LCL mutants.
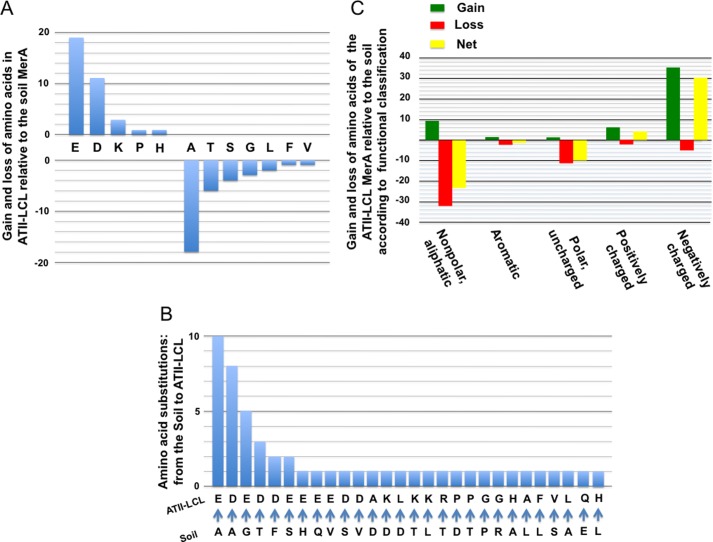
MerA ATII-LCL and its ortholog from soil are both 561 amino acids long and display 91% sequence identity. The 52 differences that distinguish them are accounted for as follows. Most strikingly, the ATII-LCL enzyme shows an overall loss of 18 alanines (2 gains and 20 losses) and an overall gain of 11 aspartic and 19 glutamic acid residues (Asp, 15 gains and 4 losses; Glu, 20 gains and 1 loss) relative to soil enzyme (Fig. 2A). Interestingly, 18 of the 35 acidic residues gained (51%) are found at positions occupied by alanines in the soil homolog and result from mutations in the second position in the Ala codon (Fig. 2B). Moreover, the remaining 17 acidic residues gained in the ATII-LCL MerA correspond to five glycines, three threonines, three serines, two phenylalanines, two valines, one histidine, and one glutamine in the soil enzyme (Fig. 2B). The statistical significance of the amino acid substitutions observed between the ATII-LCL enzyme and its soil ortholog was assessed by FET, and the result highlights the tendency for uncharged side chains to be replaced by charged groups (FET p value = 1.44 × 10−11) and more specifically acidic groups (FET p value = 1.2 × 10−9) in MerA ATII-LCL (Fig. 2C).
FIGURE 2.
Patterns of amino acid substitutions present in the ATII-LCL MerA relative to the soil ortholog. A, amino acid compositions of the ATII-LCL and soil enzymes were compared, and the plot shows the net change in the number of each of the listed amino acids in the ATII-LCL enzyme. B, frequency of substitutions in ATII-LCL plotted against corresponding residue in the soil enzyme. C, patterns of gain, loss, and net change of amino acids in ATII-LCL relative to the soil enzyme based on their functional classifications.
In addition to the acidic amino acid substitutions in the ATII-LCL MerA reported above, we noticed that two short stretches of sequence, one consisting of four (box1) and the other of six (box2) residues, differentiate the two enzymes. The two boxes are located not far from the C-terminal ends of the proteins and, in the ATII-LCL protein, each includes two basic amino acids and a single proline (Fig. 1).
These limited, but chemically distinctive, amino acid divergences between the two orthologs presumably account for the differences in the catalytic properties of the two enzymes, which enable them to function efficiently in their very distinct habitats, the soil and the ATII-LCL. Note that the MerA sequence from the uncultured soil bacterium used in this work as a basis for comparison with the ATII-LCL differs at just three positions from the structurally and functionally well characterized Tn501 MerA enzyme (47) (GenBankTM under accession number CAA77323.1) as follows: Leu-208 (soil) is His in Tn501, Glu-335 is Gln, and Ala-386 is Val.
Phylogenetic Analysis of the ATII-LCL merA Sequence
A maximum likelihood phylogenetic tree (supplemental Fig. S1), including sequences most similar (retrieved from GenBankTM using BLAST) (supplemental Table S1) to the ATII-LCL merA sequence, shows that the ATII-LCL sequence groups with that from the uncultured soil bacterium already mentioned. Nearly all the sequences assigned to an identified species in this tree are from γ-proteobacteria, with only a few from β-proteobacteria. This result suggests that the ATII-LCL merA sequence, as well as the soil bacterium sequence, is derived from members of the γ-proteobacteria.
Cloning, Expression, and Purification of ATII-LCL and Soil MerAs
To study the role of these substitutions on the catalytic properties of the two orthologs, the genes for the 561-amino acid ATII-LCL and soil enzymes were synthesized, and their codon usage was modified to allow optimal expression in E. coli. The genes were cloned into pET SUMO expression vector system (Invitrogen) as described under “Experimental Procedures.” The supplemental Fig. S2 shows SDS-PAGE of the purified proteins. Both proteins were expressed at comparable levels and were found to be soluble.
Thermostability of ATII-LCL and Soil MerAs
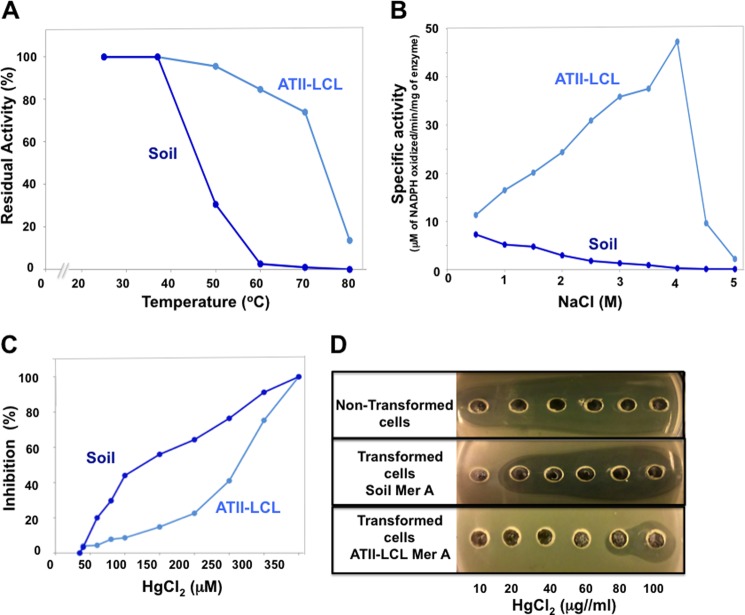
To determine the effect of temperature on enzyme stability, the ATII-LCL and soil enzymes were separately incubated at different temperatures for 10 min, and the residual activity was measured as described under “Experimental Procedures.” As shown in Fig. 3A, ATII-LCL MerA was highly resistant to inactivation by high temperatures in comparison with the soil enzyme. The ATII-LCL MerA retained more than 80% of its activity after a 10-min incubation at 60 °C, although the soil enzyme displayed little to no detectable activity after the same treatment. The temperature required for 50% enzyme inactivation was 75 °C for MerA ATII-LCL and 45 °C for the soil enzyme. This result confirms that ATII-LCL MerA is a thermophilic enzyme, as expected for a gene product found in the ATII-LCL.
FIGURE 3.
Effects of temperature, salt, and mercury concentration on the catalytic activities of ATII-LCL and soil MerAs. A, thermostability. The ATII-LCL and soil MerAs were incubated at the indicated temperature for 10 min, and the residual enzymatic activities were assayed under standard conditions. B, effect of NaCl concentration on MerA activity. The enzymatic activities were measured in reaction mixtures containing the indicated concentrations of NaCl. C, sensitivity to HgCl2. The enzymes were incubated in the assay mixture for 10 min, in the absence of NADPH and presence of the indicated concentrations of HgCl2. The enzyme activities were then measured by addition of NADPH to the assay mixture. D, determination of the MIC for HgCl2. Solutions containing increasing concentrations of HgCl2 were placed in wells punched in LB-agar plates (supplemented with 50 μg/ml kanamycin and 1 mm isopropyl β-d-thiogalactoside) inoculated with E. coli transformants. The plates were incubated for 24 h at 37 °C. The radii of the clear zones are a measure of the toxic effect of HgCl2 on bacterial growth.
Effects of High Salt on ATII-LCL and Soil MerAs
To examine if the ATII-LCL MerA requires salt for its enzymatic activity, and if the enzyme is active in high salt, we measured the activities of both enzymes in the presence of various salt concentrations, from 0 to 5 m NaCl. MerA ATII-LCL is highly salt-dependent, and its enzymatic activity increases with the increasing NaCl concentration, displaying maximum activity at 4 m NaCl. The soil enzyme however is not activated by NaCl and was found to have no detectable activity at 4 m NaCl (Fig. 3B).
Inhibition of ATII-LCL and Soil MerAs by HgCl2 and Growth Resistance of E. coli Transformants in the Presence of Mercury
We measured the effect of HgCl2 on the enzymatic activities of the purified proteins, and we also examined the degree of resistance of E. coli transformants expressing the ATII-LCL and the soil enzymes to HgCl2.
To assess the inhibitory effect of HgCl2 on the enzymatic activity, ATII-LCL and soil MerAs were incubated in reaction mixture in the absence of NADPH and in the presence of the indicated concentrations of HgCl2 for 10 min at 37 °C. The reactions were then started by the addition of NADPH, and the initial rates of NADPH oxidation were measured (for details see under “Experimental Procedures”). Fig. 3C shows that ATII-LCL MerA was highly resistant to inhibition by HgCl2 relative to the soil enzyme as judged by the IC50 values of 100 and 270 μm for the soil and ATII-LCL enzymes, respectively. It should be noted here that the β-mercaptoethanol concentration used in the assay mixture (1 mm) was sufficient to fully complex Hg2+ as RS-Hg-SR, because increasing the concentration to 2 mm had no effect on either the Hg2+ inhibition profiles or the IC50 values obtained for any of the enzymes tested.
To estimate the impact of ATII-LCL MerA on the level of HgCl2 resistance shown by E. coli cells, we measured the MICs of the salt for nontransformed cells, and we transformed cells expressing the soil or the ATII-LCL enzyme by exposing growing cells to concentrations of HgCl2 that varied from 10 to 100 μg/ml. Untransformed E. coli cells were sensitive to all concentrations of HgCl2, although the soil transformant cells displayed visible growth inhibition at concentrations of 20 μg/ml HgCl2. The MIC for ATII-LCL transformants, however, was about 80 μg/ml, indicating that the ATII-LCL MerA enhances mercury detoxification and confers increased resistance to Hg2+ on cells that express it (Fig. 3D).
Taken together, these results indicate that the ATII-LCL MerA is a thermophilic and extremely halophilic enzyme that tolerates elevated concentrations of Hg2+, and it renders E. coli cells resistant to high levels of mercury.
Dissection of Structure-Function Relationships in MerA ATII-LCL by Targeted Mutagenesis
It is intriguing that the amino acid sequence of the metagenomically derived MerA ATII-LCL, with its thermophilicity, extreme halophilicity, and high HgCl2 detoxification activity differs by only 9% from that of a soil ortholog that lacks these properties. This provides a unique opportunity to analyze the functional effects of the limited number of alterations observed. To this end, we replaced selected residues in the ATII-LCL protein by those found in the corresponding positions in the soil enzyme by site-specific mutagenesis. Seven mutations were generated near critical sites along the ATII-LCL polypeptide chain. All mutations involved the replacement of either acidic residues or the two basic/proline boxes by the corresponding residues in the soil enzyme (see Table 1 and Fig. 1). The effects of these replacements on the response of the mutants to temperature, salt, and HgCl2 are reported below.
All mutants were cloned in the pET SUMO expression vector, expressed, and purified as described under “Experimental Procedures.” The supplemental Fig. S2 shows SDS-PAGE of the purified mutant proteins.
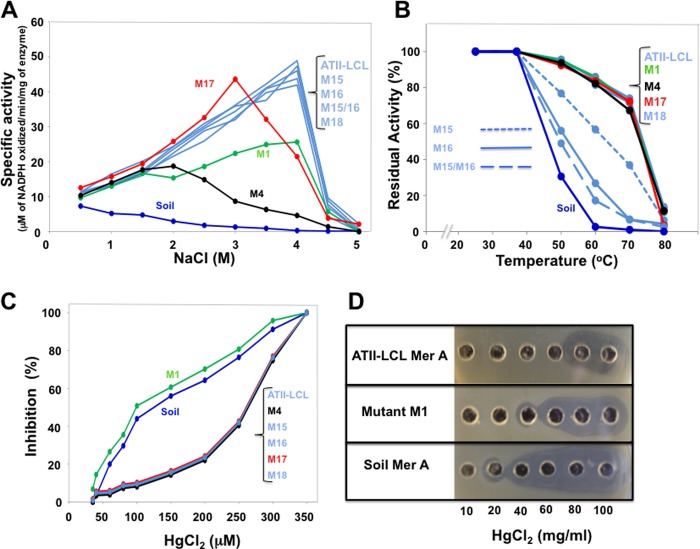
Three of the mutations that replaced acidic amino acids, M1, M4, and M17, by the uncharged residues found in the soil enzyme (Table 1) showed clear and distinctive effects on the activation of the ATII-LCL enzyme by NaCl. Although mutant M1 still displayed maximum activity at 4 m NaCl, its peak activity was less than 50% that of the wild type. Mutants M4 and M17, however, showed altered responses to NaCl insofar as their activities peaked at 2 and 3 m NaCl, respectively (Fig. 4A). Although mutation M17 had a relatively minor effect on the level of peak activity, mutant M4 reached only 40% that of the wild-type enzyme (Fig. 4A). Despite the fact that the two glutamic acids (Glu-545/Glu-546) mutated in M18 (Fig. 1) are only 12 residues from the cysteine pair involved in Hg2+ binding process (Cys-598/Cys-559), their replacement by alanines (M18) failed to alter the enzyme's response to NaCl (Fig. 3A).
FIGURE 4.
Properties of the ATII-LCL MerA mutants. Mutants generated by site-directed mutagenesis (M1, M4, M15, M16, the double mutant M15/M16, M17, and M18) were examined as described in Fig. 4. Refer to Table 1 for details regarding each mutant.
All of the mutations that replaced acidic amino acids and affected the response of the mutants (M1, M4, and M17) to NaCl had no measurable effect on thermostability of the mutated enzymes (Fig. 4B). Interestingly, however, the two basic/proline boxes (Fig. 1 and Table 1) were found to be involved in the thermostability of the enzyme.
Replacement of the four residues of box1 or the six residues of box2 by the amino acids found in the mesophilic soil enzyme produced a drastic effect on the thermostability of ATII-LCL enzyme. Incubation of the mutant enzymes M15 (box1) and M16 (box2) for 10 min at 60 °C led to loss of 40 and 70% of activity, respectively, although the double mutant M15/M16 showed 80% inactivation (Fig. 4B). All mutants in which acidic residues were replaced (M1, M4, M17, and M18), like the wild-type ATII-LCL, retained more than 80% of their activity at this temperature (Fig. 4B).
The only mutation that affected the resistance of the ATII-LCL MerA to inhibition by Hg2+ was that in mutant M1, which is located within the NmerA domain (Figs. 1 and 4C). In fact, mutant M1 has an inhibitory profile comparable with that of the soil enzyme. Interestingly, this mutation lies right next to the conserved metal-binding domain containing the pair of cysteines (Cys-11/Cys-14) that is involved in the binding of Hg2+ under glutathione-depleted conditions (12).
Comparison of the Kinetic Parameters of MerA ATII-LCL, Its Mutants, and the Soil Enzyme
To analyze the effects of these mutations on the enzymatic properties of the enzyme, we determined the Km value for Hg2+ and the kcat value for the soil enzyme and MerA ATII-LCL and its mutant derivatives. Although mutations of the basic/proline boxes 1 and 2 (M15, M16, and M15/M16) substantially reduced the thermostability of the proteins (see above), they had no apparent effect on the Km or the kcat values of the mutant enzymes (Table 2). This result indicates that loss of thermostability does not affect the kinetic parameters of catalysis, but rather it is due to structural alterations that make it more sensitive to heat.
TABLE 2.
Kinetic parameters of the soil, ATII-LCL, and MerA mutants
Kinetic parameters of the soil, ATII-LCL, and MerA mutants were determined in the presence of 500 mm NaCl.
| Enzyme/mutation | Km | kcat | kcat/Km |
|---|---|---|---|
| μm | s−1 | μm−1 s−1 | |
| ATII-LCL MerA | 8.65 | 22.5 | 2.60 |
| Soil MerA | 14.69 | 12.2 | 0.83 |
| M15 box1 | 9.74 | 22.5 | 2.31 |
| M16 box2 | 9.56 | 21.6 | 2.26 |
| M15/16 box1/box2 | 8.05 | 22.2 | 2.76 |
| M1 | 16.54 | 10.8 | 0.65 |
| M4 | 13.55 | 11.8 | 0.87 |
| M17 | 11.23 | 20 | 1.78 |
| M18 | 10.23 | 21.8 | 2.13 |
In contrast, mutations of the acidic residues that affected the extreme halophilicity of the enzyme were directly correlated with changes in Km and kcat. Compared with the wild-type MerA, the N-terminal M1 mutation results in a 2-fold reduction in kcat and a 1.9-fold increase in Km. The same was observed with M4, which shows a 1.6-fold increase in Km and a 1.9-fold decrease in kcat. Although both mutations increased the Km value, decreased the kcat value, and reduced activation by NaCl, only M4 altered the halophilic character of the enzyme from extremely to slightly halophilic, shifting the position of peak activity from 4 to 2 m NaCl. It is important to note that mutation M1 is adjacent to Cys-11 and Cys-14 within the NmerA metal-binding site, although the M4 mutation is separated by just one residue from the redox-active pair Cys-136 and Cys-141 in the catalytic site. In contrast to mutations M1 and M4, mutation M17 affected the affinity and the turnover number of the mutated enzyme only slightly, increasing the Km value by 1.29-fold and decreasing the kcat value by 1.15-fold. Even though this mutation altered Km and kcat values only subtly, it had a significant effect on the behavior of the enzyme with respect to activation by NaCl; maximal activation is observed at 3 m (halophilic) rather than 4 m NaCl (extremely halophilic). Mutation M18, however, had no noticeable effect on Km, kcat, or the halophilicity of the enzyme.
Finally, as shown in Fig. 4C, the only mutation that affected the resistance of the enzyme to inhibition by Hg2+ was the substitution of the glutamic acid pair in M1. Interestingly, this mutation and mutation M4 were found to substantially decrease the catalytic activity and the efficiency of the enzyme (increase the Km and lower the kcat, see Table 2). However, only M1 was found to alter resistance to Hg2+. To explore if the loss of resistance to Hg2+ in the M1 mutant is reflected in the efficiency of in vivo detoxification and the survival of E. coli cells at high concentrations of Hg2+, we determined the MIC for E. coli transformants expressing the soil enzyme, ATII-LCL, and mutant M1. Fig. 4D shows that indeed mutation M1 substantially affects detoxification efficiency, reducing the MIC value from 80 to 40 μg/ml Hg2+. This result highlights the importance of the Glu-15 and Glu-16 pair in the ATII-LCL enzyme for its detoxification function and its contribution to the survival of its host in the ATII-LCL.
All the kinetic parameters described above were measured in the presence of 500 mm NaCl. To determine whether the salt concentration has an effect on the kinetic parameters or alters the relative values of the Km and the kcat for the soil and ATII-LCL enzymes and its mutants, we also measured these parameters in the presence of the concentration of NaCl that gave maximum activity for the respective enzymes and mutants as determined in Fig. 4A, i.e. ATII-LCL, M4, M15, M16, M15/M16, M18, and M1 at 4 m NaCl; M17, at 3 m NaCl; M4 at 2 m NaCl, and the soil enzyme in the absence of added NaCl. The results are presented in supplemental Table S2. Although the kinetic parameters reported in supplemental Table S2 show slight changes in the Km and kcat values compared with those determined at 500 mm NaCl, the ratios between the Km and kcat values for the soil, ATII-LCL and mutant enzymes did not differ significantly than those at 500 mm NaCl. Interestingly, the Km reported in the literature (9) for the structurally and functionally well characterized Tn501 MerA enzyme (12 μm) is virtually identical to that of the soil enzyme (11.99 μm; supplemental Table S2), although the latter rises to 14.7 μm in the presence of 500 mm NaCl (Table 2).
Because MerA is a two-substrate enzyme, we determined the Km value for NADPH to ensure that the kinetic values reported above were obtained at saturating concentrations of NADPH. We observed little difference in the Km value for NADPH between the soil, ATII-LCL, and its mutant enzymes. The observed Km value for the enzymes were all between 4.4 and 6.3 μm (supplemental Table S2), although the NADPH concentration used to assay the enzymes was 200 μm, equivalent to around 40-fold excess over the Km. The fact that the affinities of NADPH for the soil, ATII-LCL, and mutant enzymes are broadly comparable indicates that the primary property targeted during the evolution of the ATII-LCL enzyme has been its affinity for Hg2+. Obviously, this adaptation process has not required changes in the enzyme's affinity for the electron donor.
Location of the Glutamic Residues and the Two Basic/Proline Boxes in the Three-dimensional Structure of ATII-LCL MerA
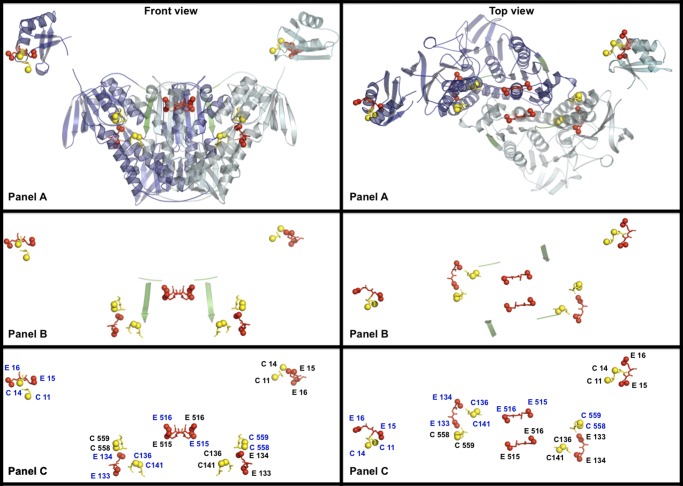
The three-dimensional structure of the ATII-LCL MerA catalytic core determined by homology modeling against Tn501 MerA (12) is presented in Fig. 5 (see also supplemental Fig. S3). The model shows the homodimer structure of the catalytic core of the enzyme, which contains the C-terminal pair of cysteines (Cys-558 and Cys-559) involved in binding of Hg2+ from one subunit and the cysteine redox-active pair of the second subunit (Cys-136 and Cys-141) (12), present in the catalytic pocket of the enzyme (see also supplemental Fig. 3). The pair of glutamic acids (Glu-133 and Glu-134) that contributes to increasing the affinity of the enzyme for Hg2+ and to the high efficiency of Hg2+ reduction is placed in close proximity to the binding and the reducing cysteine pairs in the catalytic pocket of the enzyme. The model also shows that the carboxylic group of Glu-516, one of the Glu-515 and Glu-516 pair, is located on the proposed entry path of Hg(SR)2 (12) leading to the catalytic pocket of the enzyme.
FIGURE 5.
Three-dimensional structure models of the homodimer ATII-LCL MerA. Homology modeling based on the Tn501 Mer reductase (12) was carried out as described under “Experimental Procedures.” Front and top views are presented. Yellow spheres denote the sulfur atoms of the cysteines involved in Hg2+ binding and reduction; red spheres represent the side-chain carboxylic oxygens of glutamic acid residues that were mutated in this work. A shows the C- and N-terminal domains portrayed in schematic form; one subunit is shown in dark blue and the other in cyan. Box1 (random coiled loop) and box2 (β-strand) are highlighted in green. B highlights the disposition of box1 and box2 and the functionally important cysteine pairs and nearby glutamic acid residues, which are labeled in C according to the subunit to which they belong (see supplemental Fig. 3 for the structure of the monomer).
The two basic/proline boxes that contribute to the thermostability of the enzyme are also located in a critical position in the molecule. Box1, an unstructured loop, is located just at the beginning of the dimerization domain (Figs. 1 and 5A). This domain, which is present in ATII-LCL MerA, is known to be conserved among the homodimer pyridine nucleotide-disulfide oxidoreductases (48). The second box (box2), however, is located within the dimerization sequence arranged in a β-strand structure (Figs. 1 and 5A; see also supplemental Fig. 3). It is important to note that replacement of box2 by the motif found in the soil enzyme had a more drastic effect on the thermostability of the enzyme than the alteration of box1 (Fig. 4B).
In addition, Fig. 5 show the three-dimensional structure of the NmerA (43), separately from the catalytic core, containing the pair of glutamic (Glu-15/Glu-16), adjacent to the pair of cysteines (Cys-11/Cys-14) involved in binding of Hg2+ when glutathione levels are depleted (12, 43).
The model indicates that the negatively charged residues that are linked to the halophilicity of the ATII-LCL MerA are sited near residues involved in the binding and reduction of Hg2+, whereas those that affect the thermostability are positioned near/within the functionally essential dimerization domain.
DISCUSSION
The lower convective layer of the Atlantis II brine pool is a unique environment characterized by a combination of abiotic factors, i.e. high temperature, elevated concentrations of heavy metals, and extreme salinity, that place severe demands on the physiology of the organisms found there (3, 4). The majority of the microorganisms that thrive in this environment are difficult to culture, which greatly constrains our ability to understand the biochemical adaptations that allow them to function under such extreme chemical and physical conditions. The emerging fields of metagenomic (49–51) and deep-sea sampling (52) allow one to address these questions by studying the reconstructed genomes of microorganisms that reside in this unique and unexplored environment.
In this report, we analyze and describe the properties of a mercuric reductase named MerA ATII-LCL from this remote deep-sea environment. Having identified the coding sequence in a metagenomic dataset, MerA ATII-LCL was expressed in E. coli in parallel with an ortholog from an uncultured soil bacterium. We then compared the catalytic properties of the enzymes, and we performed comprehensive site-directed mutagenesis to dissect and understand the structural features that contribute to the enzyme's halophilicity, thermostability, and high resistance to Hg2+.
The results show that the amino acid sequence of MerA ATII-LCL differs from that of its mesophilic ortholog from an uncultured soil microorganism (35) by just 9%. Yet, the two enzymes differ strikingly in their properties and responses to three abiotic factors. The major sequence differences in MerA ATII-LCL were found to be located at critical positions in its structure and can be summarized as follows: 1) most of the substitutions replace nonpolar with acidic amino acids and thus reduce the hydrophobicity of the protein; 2) two short sequences, each characterized by a proline and two basic residues, are unique to the ATII-LCL enzyme.
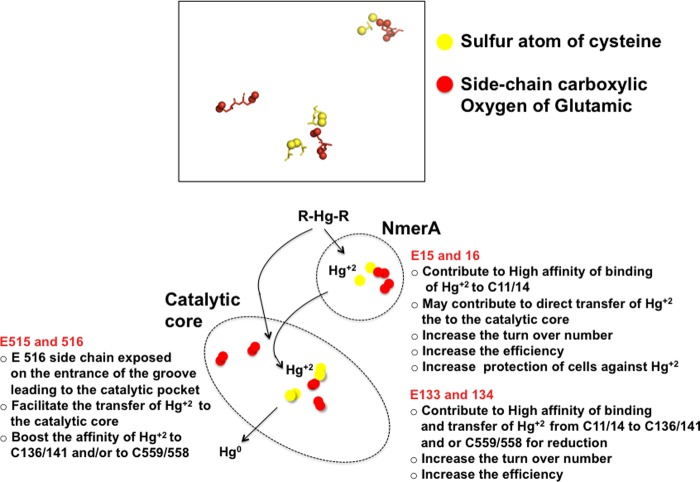
The kinetic characterization of three mutant derivatives of MerA ATII-LCL (M1, M4, and M17), in which negatively charged amino acids were replaced by the residues present at the corresponding position in the soil enzyme, showed that these substitutions produced different effects on the substrate affinity of the enzyme and its turnover number, as indicated by their apparent Km and kcat values. The pair of glutamic acids (Glu-515/Glu-516) replaced in M17, which lie near the C-terminal pair of cysteines (Cys-558/Cys-559) involved in the binding of Hg2+ (45), was shown to be involved in enhancing the enzyme's affinity for Hg2+. The three-dimensional model presented in Fig. 5 (see also supplemental Fig. 3) shows that the carboxylic group of Glu-516 is located at the entrance to the groove leading to the catalytic pocket.
The pair of glutamic acids mutated in strain M4 (Glu-133/Glu-134), located in the catalytic pocket adjacent to the redox-active pair of cysteines (Cys-136/Cys-141) (10, 11) and near the C-terminal cysteine pair (Cys-558/Cys-559), was shown to be required for the substrate affinity for Hg2+ and high catalytic turnover number of the ATII-LCL enzyme. Interestingly, replacement of this pair of glutamic acids resulted in an enzyme with a catalytic efficiency similar to that of the soil ortholog. In summary, both pairs of glutamic acids (Glu-133/ Glu-134 and Glu-515/Glu-516) are required for the high catalytic efficiency of the enzyme; most probably Glu-515/Glu-516 promote the binding of Hg2+ to the C-terminal Cys-558/Cys-559 and Glu-133/Glu-134 facilitate the transfer of Hg2+ to the C-terminal binding cysteine pair (summarized in Fig. 6).
FIGURE 6.
Summary of the proposed functions of glutamic acid residues that affected the catalytic properties and the halophilicity of ATII-LCL MerA.
Finally, the pair of glutamic acids (Glu-15/Glu-16) adjacent to the cysteine pair (Cys-11/Cys-12) in the NmerA domain was shown to be a major contributor to the high catalytic efficiency of MerA ATII-LCL, as its mutation in M1 (Glu-15/Glu-16 to Ala-15/Ala-16) resulted in an enzyme with low affinity for Hg2+ and a low turnover number and a catalytic efficiency similar to that of the soil enzyme and mutant M4. Therefore, it seems that the presence of Glu-15/Glu-16 next to the metal-binding motif in the NmerA domain is required for maximum efficiency of binding and delivery of Hg2+ to the catalytic core for reduction (summarized in Fig. 6 and discussed below).
The Glu-to-Ala mutations that affected the catalytic activities of the ATII-LCL enzyme, as described above, also have direct effects on the enzyme's tolerance toward NaCl. The mutations that reduced the catalytic efficiency of the enzyme (M1 and M4) resulted in enzymes whose activity was less resistant to increases in NaCl concentrations. Interestingly, although mutant M1 still shows maximum activity at 4 m NaCl, in mutant M4 peak activity is shifted to 2 m NaCl. In mutant M17, a distinctive shift in peak activity from 4 to 3 m NaCl is observed, but the level of the peak is similar to that in the wild type.
It is also conceivable that the various glutamic acid pairs located near critical positions involved in the binding and reduction of Hg2+ contribute to electrostatic repulsion of the high concentrations of chloride anions, which might otherwise interfere with efficient detoxification of Hg2+, and they have been selected partly to serve this function.
Based on these results, MerA ATII-LCL appears to derive from a microorganism belonging to a lineage that adopted the salt-in strategy to cope with the hypersaline environment of the ATII-LCL, in which the concentration of NaCl reaches 4 m. At this salt concentration, mutants M1, M4, and M17 were found to display 50, 8, and 44% of the activity observed for MerA ATII-LCL (the soil enzyme shows no detectable activity in 4 m NaCl). Therefore, these three pairs of negatively charged side chains in the ATII-LCL MerA are crucial for the enzyme's ability to function with high efficiency in the presence of salt concentrations typical of those in environments inhabited by other microorganisms that have exploited the salt-in strategy (28–30).
Signatures of halophilicity have been identified mostly by structural comparisons of proteins from halophilic and nonhalophilic microorganisms (31, 34, 53–55), but they have not been experimentally analyzed in detail (55–58). The work described here presents an analysis of selected, evolutionarily adaptive acidic residues that shows them to confer site-specific levels of halophilicity on MerA that varied from 0.5 to 4 m NaCl. In addition, the data indicate that these acidic residues are not just required for the salt tolerance, solubility, and protection of the enzyme from aggregation (31–33, 59, 60) but also to increase the catalytic efficiency of the enzyme in the presence of elevated concentrations of salt.
The second of the adaptations acquired by MerA ATII-LCL, compared with the soil enzyme, is its thermophilic character. In this regard, the presence of the two basic/proline boxes, box1 and box2, is an additional structural feature that distinguishes it from its terrestrial counterpart. Substitution of any of the three glutamic pairs mutated in M1, M4, and M17, respectively, had no effect on the thermostability of the enzyme, but replacement of either box alone, or both together, clearly affected the temperature stability of the mutants. Because the catalytic properties of the enzyme measured at 37 °C were not appreciably changed in any of the three mutants in which the boxes were altered (M15, M16, and M15/M16), we concluded that these two segments are mainly involved in stabilizing the structure of the enzyme. Each box includes a proline residue, which has been shown to be involved in the thermostability of various enzymes (25, 61). Proline is unique in having a pyrrolidine ring, which reduces the structural flexibility of polypeptide regions containing it, and therefore it provides structural stabilization at critical locations in proteins. It is intriguing to note that box2 lies within the dimerization domain of MerA ATII-LCL (Figs. 1 and 5A; see also supplemental Fig. 3). Moreover, the residues in box2 form a β-strand that is oriented in an antiparallel β-sheet structure that falls within the dimerization domain of MerA ATII-LCL (Figs. 1 and 5A). Antiparallel β-sheet structures are known to stabilize proteins, especially when the number of strands present is greater than 2 (62). MerA has five β-strands within its dimerization domain (48). Therefore, the presence of a proline residue at the end of the β-strand of box2 adds stability to the antiparallel β-sheets, which may help to stabilize the MerA ATII-LCL structure at high temperatures. The unstructured loop formed by box1 falls just at the beginning of the dimerization domain, and its proline residue most probably contributes to the stability of the dimer, although its contribution to thermostability of the enzyme is much less than that of box2.
These results allow us to distinguish sequences that contribute to extreme halophilicity from those required for thermostability, because removal of the two boxes resulted in a MerA enzyme that is still halophilic but is almost completely inactivated by incubation for 10 min at 70 °C (the ATII-LCL enzyme and all other mutants retain more than 70% of their activity when treated in the same manner). It is important to note that mutation of box1 and box2 did not have any effect on the enzyme's resistance to Hg2+. A more detailed understanding of the roles of the prolines and the basic amino acids in the two boxes in conferring thermostability on the enzyme will require further analysis by targeted mutagenesis of each residue and determination of the crystal structure of MerA ATII-LCL (work in progress).
The third notably adaptive property of MerA ATII-LCL is its resistance to high levels of Hg2+. The Hg2+ concentration required for 50% inhibition of the enzyme activity is at least 2.7-fold higher than for the soil enzyme. The only mutation that affected this property was the substitution of alanines for Glu-15 and Glu-16 in mutant M1. This mutation, located directly adjacent to the cysteine pair Cys-136/Cys-141 in the NmerA domain, resulted in an enzyme with low catalytic efficiency and abolished the high Hg2+ resistance characteristic of the ATII-LCL enzyme. In addition, it also greatly reduced the in vivo detoxification ability of E. coli cells expressing the mutant enzyme.
By analogy with other mercuric reductases, the second Hg2+-binding site (Cys-11/Cys-14) in the NmerA domain is assumed to cooperate with the C-terminal pair of cysteine thiols (Cys-558/Cys-559) to protect cells against Hg2+ when levels of low molecular weight thiols are low (12). This should facilitate the efficient transfer of Hg2+ from proteins and other carriers to the active site of the enzyme by a ligand-exchange mechanism (63, 64). The results presented here corroborate this role of NmerA domain. This is underlined by the presence of the adjacent pair of glutamic acids in the ATII-LCL enzyme, which contribute to its capacity to operate in a hypersaline environment. The fact that Glu-15/Glu-16 (M1) added critical catalytic advantages to the ATTII-LCL enzyme toward extreme salinity and high efficiency in detoxification of Hg2+ presents strong evidence of the pivotal importance of the NmerA's Cys-11/Cys-14 for microorganisms to survive in the ATII-LCL environment.
The ATII-LCL MerA molecule therefore represents a novel enzyme, which has undergone multiple evolutionary adaptations that enhance its stability and catalytic efficiency, thus allowing its host to cope with the harsh environmental conditions found in the ATII-LCL unique environment. The relatively simple structural changes that underpin its remarkable functional attributes make ATII-MerA a potential model for further research to address environmental adaptation of enzymes and proteins to environments characterized by multiple abiotic stressors.
Acknowledgments
We thank Drs. Amy S. Bower, Stephen A. Swift, and Abdulaziz Al-Suwailem for technical advice and important information regarding sample collection. We are grateful to Dr. Paul Hardy for critical reading of the manuscript.

This article contains supplemental Figs. 1–3, Tables 1 and 2, and additional references.
This paper is dedicated to the memory of the late Provost of The American University in Cairo, Dr. Medhat Haroun.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) KF572479.
- ATII
- Atlantis II
- LCL
- lower convective layer
- MIC
- minimum inhibitory concentration
- FET
- Fisher's Exact Independence Test.
REFERENCES
- 1. Backer H., Schoell M. (1972) New deeps with brines and metalliferous sediments in the Red Sea. Nat. Phys. Sci. 240, 153–158 [Google Scholar]
- 2. Miller A. R., Densmore C. D., Degens E. T., Hathaway J. C., Manheim F. T., McFarlin P. J., Pocklington R., Jokelin A. (1966) Hot brines and recent iron deposits in deeps of the Red Sea. Geochim. Cosmochim. Acta 30, 341–360 [Google Scholar]
- 3. Swift S. A., Bower A. S., Schmitt R. W. (2012) Vertical, horizontal, and temporal changes in temperature in the Atlantis II and Discovery hot brine pools, Red Sea. Deep-Sea Res. I 64, 118–128 [Google Scholar]
- 4. Antunes A., Ngugi D. K., Stingl U. (2011) Microbiology of the Red Sea (and other) deep-sea anoxic brine lakes. Environ. Microbiol. Rep. 3, 416–433 [DOI] [PubMed] [Google Scholar]
- 5. Barkay T., Wagner-Döbler I. (2005) Microbial transformations of mercury: potentials, challenges, and achievements in controlling mercury toxicity in the environment. Adv. Appl. Microbiol. 57, 1–52 [DOI] [PubMed] [Google Scholar]
- 6. Schottel J. L. (1978) The mercuric and organomercurial detoxifying enzymes from a plasmid-bearing strain of Escherichia coli. J. Biol. Chem. 253, 4341–4349 [PubMed] [Google Scholar]
- 7. Summers A. O. (1986) Organization, expression, and evolution of genes for mercury resistance. Annu. Rev. Microbiol. 40, 607–634 [DOI] [PubMed] [Google Scholar]
- 8. Barkay T., Miller S. M., Summers A. O. (2003) Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol. Rev. 27, 355–384 [DOI] [PubMed] [Google Scholar]
- 9. Fox B., Walsh C. T. (1982) Mercuric reductase. Purification and characterization of a transposon-encoded flavoprotein containing an oxidation-reduction-active disulfide. J. Biol. Chem. 257, 2498–2503 [PubMed] [Google Scholar]
- 10. Miller S. M., Moore M. J., Massey V., Williams C. H., Jr., Distefano M. D., Ballou D. P., Walsh C. T. (1989) Evidence for the participation of Cys558 and Cys559 at the active site of mercuric reductase. Biochemistry 28, 1194–1205 [DOI] [PubMed] [Google Scholar]
- 11. Moore M. J., Walsh C. T. (1989) Mutagenesis of the N- and C-terminal cysteine pairs of Tn501 mercuric ion reductase: consequences for bacterial detoxification of mercurials. Biochemistry 28, 1183–1194 [DOI] [PubMed] [Google Scholar]
- 12. Ledwidge R., Patel B., Dong A., Fiedler D., Falkowski M., Zelikova J., Summers A. O., Pai E. F., Miller S. M. (2005) NmerA, the metal binding domain of mercuric ion reductase, removes Hg2+ from proteins, delivers it to the catalytic core, and protects cells under glutathione-depleted conditions. Biochemistry 44, 11402–11416 [DOI] [PubMed] [Google Scholar]
- 13. Rossy E., Champier L., Bersch B., Brutscher B., Blackledge M., Covès J. (2004) Biophysical characterization of the MerP-like amino-terminal extension of the mercuric reductase from Ralstonia metallidurans CH34. J. Biol. Inorg. Chem. 9, 49–58 [DOI] [PubMed] [Google Scholar]
- 14. Lund B. O., Miller D. M., Woods J. S. (1993) Studies on Hg(II)-induced H2O2 formation and oxidative stress in vivo and in vitro in rat kidney mitochondria. Biochem. Pharmacol. 45, 2017–2024 [DOI] [PubMed] [Google Scholar]
- 15. Barkay T., Kritee K., Boyd E., Geesey G. (2010) A thermophilic bacterial origin and subsequent constraints by redox, light and salinity on the evolution of the microbial mercuric reductase. Environ. Microbiol. 12, 2904–2917 [DOI] [PubMed] [Google Scholar]
- 16. Boyd E. S., Barkay T. (2012) The mercury resistance operon: from an origin in a geothermal environment to an efficient detoxification machine. Front. Microbiol. 3, 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Madigan M. T., Oren A. (1999) Thermophilic and halophilic extremophiles. Curr. Opin. Microbiol. 2, 265–269 [DOI] [PubMed] [Google Scholar]
- 18. Vieille C., Zeikus G. J. (2001) Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 65, 1–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Berezovsky I. N., Shakhnovich E. I. (2005) Physics and evolution of thermophilic adaptation. Proc. Natl. Acad. Sci. U.S.A. 102, 12742–12747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vetriani C., Maeder D. L., Tolliday N., Yip K. S., Stillman T. J., Britton K. L., Rice D. W., Klump H. H., Robb F. T. (1998) Protein thermostability above 100°C: a key role for ionic interactions. Proc. Natl. Acad. Sci. U.S.A. 95, 12300–12305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schumann J., Böhm G., Schumacher G., Rudolph R., Jaenicke R. (1993) Stabilization of creatinase from Pseudomonas putida by random mutagenesis. Protein Sci. 2, 1612–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hennig M., Sterner R., Kirschner K., Jansonius J. N. (1997) Crystal structure at 2.0 Å resolution of phosphoribosyl anthranilate isomerase from the hyperthermophile Thermotoga maritima: possible determinants of protein stability. Biochemistry 36, 6009–6016 [DOI] [PubMed] [Google Scholar]
- 23. Berezovsky I. N., Tumanyan V. G., Esipova N. G. (1997) Representation of amino acid sequences in terms of interaction energy in protein globules. FEBS Lett. 418, 43–46 [DOI] [PubMed] [Google Scholar]
- 24. Jaenicke R., Böhm G. (1998) The stability of proteins in extreme environments. Curr. Opin. Struct. Biol. 8, 738–748 [DOI] [PubMed] [Google Scholar]
- 25. Watanabe K., Masuda T., Ohashi H., Mihara H., Suzuki Y. (1994) Multiple proline substitutions cumulatively thermostabilize Bacillus cereus ATCC7064 oligo-1,6-glucosidase. Irrefragable proof supporting the proline rule. Eur. J. Biochem. 226, 277–283 [DOI] [PubMed] [Google Scholar]
- 26. Kushner D. J. (ed) (1978) Life in High Salt and Solute Concentrations, pp. 317–368, Academic Press, London [Google Scholar]
- 27. Oren A. (2008) Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Systems 4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Christian J. H., Waltho J. A. (1962) Solute concentrations within cells of halophilic and non-halophilic bacteria. Biochim. Biophys. Acta 65, 506–508 [DOI] [PubMed] [Google Scholar]
- 29. Eisenberg H., Mevarech M., Zaccai G. (1992) Biochemical, structural, and molecular genetic aspects of halophilism. Adv. Protein Chem. 43, 1–62 [DOI] [PubMed] [Google Scholar]
- 30. Lanyi J. K. (1974) Salt-dependent properties of proteins from extremely halophilic bacteria. Bacteriol. Rev. 38, 272–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Paul S., Bag S. K., Das S., Harvill E. T., Dutta C. (2008) Molecular signature of hypersaline adaptation: insights from genome and proteome composition of halophilic prokaryotes. Genome Biol. 9, R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fukuchi S., Yoshimune K., Wakayama M., Moriguchi M., Nishikawa K. (2003) Unique amino acid composition of proteins in halophilic bacteria. J. Mol. Biol. 327, 347–357 [DOI] [PubMed] [Google Scholar]
- 33. Frolow F., Harel M., Sussman J. L., Mevarech M., Shoham M. (1996) Insights into protein adaptation to a saturated salt environment from the crystal structure of a halophilic 2Fe-2S ferredoxin. Nat. Struct. Biol. 3, 452–458 [DOI] [PubMed] [Google Scholar]
- 34. Siglioccolo A., Paiardini A., Piscitelli M., Pascarella S. (2011) Structural adaptation of extreme halophilic proteins through decrease of conserved hydrophobic contact surface. BMC Struct. Biol. 11, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sen D., Van der Auwera G. A., Rogers L. M., Thomas C. M., Brown C. J., Top E. M. (2011) Broad-host-range plasmids from agricultural soils have IncP-1 backbones with diverse accessory genes. Appl. Environ. Microbiol. 77, 7975–7983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rusch D. B., Halpern A. L., Sutton G., Heidelberg K. B., Williamson S., Yooseph S., Wu D., Eisen J. A., Hoffman J. M., Remington K., Beeson K., Tran B., Smith H., Baden-Tillson H., Stewart C., Thorpe J., Freeman J., Andrews-Pfannkoch C., Venter J. E., Li K., Kravitz S., Heidelberg J. F., Utterback T., Rogers Y. H., Falcón L. I., Souza V., Bonilla-Rosso G., Eguiarte L. E., Karl D. M., Sathyendranath S., Platt T., Bermingham E., Gallardo V., Tamayo-Castillo G., Ferrari M. R., Strausberg R. L., Nealson K., Friedman R., Frazier M., Venter J. C. (2007) The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5, e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gille C., Frömmel C. (2001) STRAP: editor for STRuctural alignments of proteins. Bioinformatics 17, 377–378 [DOI] [PubMed] [Google Scholar]
- 38. Punta M., Coggill P. C., Eberhardt R. Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., Clements J., Heger A., Holm L., Sonnhammer E. L., Eddy S. R., Bateman A., Finn R. D. (2012) The Pfam protein families database. Nucleic Acids Res. 40, D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hunter S., Jones P., Mitchell A., Apweiler R., Attwood T. K., Bateman A., Bernard T., Binns D., Bork P., Burge S., de Castro E., Coggill P., Corbett M., Das U., Daugherty L., Duquenne L., Finn R. D., Fraser M., Gough J., Haft D., Hulo N., Kahn D., Kelly E., Letunic I., Lonsdale D., Lopez R., Madera M., Maslen J., McAnulla C., McDowall J., McMenamin C., Mi H., Mutowo-Muellenet P., Mulder N., Natale D., Orengo C., Pesseat S., Punta M., Quinn A. F., Rivoire C., Sangrador-Vegas A., Selengut J. D., Sigrist C. J., Scheremetjew M., Tate J., Thimmajanarthanan M., Thomas P. D., Wu C. H., Yeats C., Yong S. Y. (2012) InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 40, D306–D312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 41. Guex N., Peitsch M. C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 42. Schwede T., Kopp J., Guex N., Peitsch M. C. (2003) SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 31, 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ledwidge R., Hong B., Dötsch V., Miller S. M. (2010) NmerA of Tn501 mercuric ion reductase: structural modulation of the pKa values of the metal binding cysteine thiols. Biochemistry 49, 8988–8998 [DOI] [PubMed] [Google Scholar]
- 44. Ho B. K., Gruswitz F. (2008) HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct. Biol. 8, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Distefano M. D., Moore M. J., Walsh C. T. (1990) Active site of mercuric reductase resides at the subunit interface and requires Cys135 and Cys140 from one subunit and Cys558 and Cys559 from the adjacent subunit: evidence from in vivo and in vitro heterodimer formation. Biochemistry 29, 2703–2713 [DOI] [PubMed] [Google Scholar]
- 46. Moore M. J., Miller S. M., Walsh C. T. (1992) C-terminal cysteines of Tn501 mercuric ion reductase. Biochemistry 31, 1677–1685 [DOI] [PubMed] [Google Scholar]
- 47. Brown N. L., Ford S. J., Pridmore R. D., Fritzinger D. C. (1983) Nucleotide sequence of a gene from the Pseudomonas transposon Tn501 encoding mercuric reductase. Biochemistry 22, 4089–4095 [DOI] [PubMed] [Google Scholar]
- 48. Zhang Y., Bond C. S., Bailey S., Cunningham M. L., Fairlamb A. H., Hunter W. N. (1996) The crystal structure of trypanothione reductase from the human pathogen Trypanosoma cruzi at 2.3 Å resolution. Protein Sci. 5, 52–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tyson G. W., Chapman J., Hugenholtz P., Allen E. E., Ram R. J., Richardson P. M., Solovyev V. V., Rubin E. M., Rokhsar D. S., Banfield J. F. (2004) Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428, 37–43 [DOI] [PubMed] [Google Scholar]
- 50. Venter J. C., Remington K., Heidelberg J. F., Halpern A. L., Rusch D., Eisen J. A., Wu D., Paulsen I., Nelson K. E., Nelson W., Fouts D. E., Levy S., Knap A. H., Lomas M. W., Nealson K., White O., Peterson J., Hoffman J., Parsons R., Baden-Tillson H., Pfannkoch C., Rogers Y. H., Smith H. O. (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74 [DOI] [PubMed] [Google Scholar]
- 51. DeLong E. F. (2009) The microbial ocean from genomes to biomes. Nature 459, 200–206 [DOI] [PubMed] [Google Scholar]
- 52. Lutz R. A., Falkowski P. G. (2012) Ocean science. A dive to Challenger Deep. Science 336, 301–302 [DOI] [PubMed] [Google Scholar]
- 53. Britton K. L., Baker P. J., Fisher M., Ruzheinikov S., Gilmour D. J., Bonete M. J., Ferrer J., Pire C., Esclapez J., Rice D. W. (2006) Analysis of protein solvent interactions in glucose dehydrogenase from the extreme halophile Haloferax mediterranei. Proc. Natl. Acad. Sci. U.S.A. 103, 4846–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ortega G., Laín A., Tadeo X., López-Méndez B., Castaño D., Millet O. (2011) Halophilic enzyme activation induced by salts. Sci. Rep. 1, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tadeo X., López-Méndez B., Trigueros T., Laín A., Castaño D., Millet O. (2009) Structural basis for the amino acid composition of proteins from halophilic archaea. PLoS Biol. 7, e1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Esclapez J., Pire C., Bautista V., Martínez-Espinosa R. M., Ferrer J., Bonete M. J. (2007) Analysis of acidic surface of Haloferax mediterranei glucose dehydrogenase by site-directed mutagenesis. FEBS Lett. 581, 837–842 [DOI] [PubMed] [Google Scholar]
- 57. Tokunaga H., Arakawa T., Tokunaga M. (2008) Engineering of halophilic enzymes: two acidic amino acid residues at the carboxy-terminal region confer halophilic characteristics to Halomonas and Pseudomonas nucleoside diphosphate kinases. Protein Sci. 17, 1603–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jolley K. A., Russell R. J., Hough D. W., Danson M. J. (1997) Site-directed mutagenesis and halophilicity of dihydrolipoamide dehydrogenase from the halophilic archaeon, Haloferax volcanii. Eur. J. Biochem. 248, 362–368 [DOI] [PubMed] [Google Scholar]
- 59. Dym O., Mevarech M., Sussman J. L. (1995) Structural features that stabilize halophilic malate dehydrogenase from an archaebacterium. Science 267, 1344–1346 [DOI] [PubMed] [Google Scholar]
- 60. Pieper U., Kapadia G., Mevarech M., Herzberg O. (1998) Structural features of halophilicity derived from the crystal structure of dihydrofolate reductase from the Dead Sea halophilic archaeon, Haloferax volcanii. Structure 6, 75–88 [DOI] [PubMed] [Google Scholar]
- 61. Goihberg E., Dym O., Tel-Or S., Levin I., Peretz M., Burstein Y. (2007) A single proline substitution is critical for the thermostabilization of Clostridium beijerinckii alcohol dehydrogenase. Proteins 66, 196–204 [DOI] [PubMed] [Google Scholar]
- 62. Syud F. A., Stanger H. E., Mortell H. S., Espinosa J. F., Fisk J. D., Fry C. G., Gellman S. H. (2003) Influence of strand number on antiparallel β-sheet stability in designed three- and four-stranded β-sheets. J. Mol. Biol. 326, 553–568 [DOI] [PubMed] [Google Scholar]
- 63. Engst S., Miller S. M. (1998) Rapid reduction of Hg(II) by mercuric ion reductase does not require the conserved C-terminal cysteine pair using HgBr2 as the substrate. Biochemistry 37, 11496–11507 [DOI] [PubMed] [Google Scholar]
- 64. Engst S., Miller S. M. (1999) Alternative routes for entry of HgX2 into the active site of mercuric ion reductase depend on the nature of the X ligands. Biochemistry 38, 3519–3529 [DOI] [PubMed] [Google Scholar]