Background: The internal repeats and NTD of yeIF4B stimulate translation initiation.
Results: The minimal number of repeats and conserved motifs in the repeat and NTD necessary for yeIF4B function was determined.
Conclusion: Two repeats provide appreciable function, except when the NTD is missing or eIF4F function is limiting or compromised.
Significance: The results provide a comprehensive description of functionally critical sequence elements in yeIF4B.
Keywords: mRNA, Ribosomes, Translation Initiation Factors, Translation Regulation, Yeast, eIF4A, eIF4B, Translation Initiation, Yeast Translation
Abstract
eIF4B has been implicated in attachment of the 43 S preinitiation complex (PIC) to mRNAs and scanning to the start codon. We recently determined that the internal seven repeats (of ∼26 amino acids each) of Saccharomyces cerevisiae eIF4B (yeIF4B) compose the region most critically required to enhance mRNA recruitment by 43 S PICs in vitro and stimulate general translation initiation in yeast. Moreover, although the N-terminal domain (NTD) of yeIF4B contributes to these activities, the RNA recognition motif is dispensable. We have now determined that only two of the seven internal repeats are sufficient for wild-type (WT) yeIF4B function in vivo when all other domains are intact. However, three or more repeats are needed in the absence of the NTD or when the functions of eIF4F components are compromised. We corroborated these observations in the reconstituted system by demonstrating that yeIF4B variants with only one or two repeats display substantial activity in promoting mRNA recruitment by the PIC, whereas additional repeats are required at lower levels of eIF4A or when the NTD is missing. These findings indicate functional overlap among the 7-repeats and NTD domains of yeIF4B and eIF4A in mRNA recruitment. Interestingly, only three highly conserved positions in the 26-amino acid repeat are essential for function in vitro and in vivo. Finally, we identified conserved motifs in the NTD and demonstrate functional overlap of two such motifs. These results provide a comprehensive description of the critical sequence elements in yeIF4B that support eIF4F function in mRNA recruitment by the PIC.
Introduction
The correct translation initiation codon in most eukaryotic mRNAs is thought to be identified by the scanning mechanism. This process commences with binding of initiator Met-tRNAi to the small (40 S) ribosomal subunit in a ternary complex with eIF2-GTP, in a manner stimulated by eIF1, -1A, -3, and -5, to form the 43 S preinitiation complex (PIC).5 The 43 S PIC then attaches to the mRNA 5′ end following its activation by binding of eIF4F to the m7G cap structure. The eIF4F trimeric complex is composed of cap-binding protein eIF4E, the large scaffolding protein eIF4G, and DEAD-box RNA helicase eIF4A. The RNA helicase activity of eIF4A is thought to facilitate 43 S PIC attachment by resolving secondary structure in cap-proximal mRNA nucleotides. In mammalian cells, interaction between eIF4G and eIF3 is believed to stabilize PIC association with the eIF4F·mRNP, whereas in budding yeast, eIF5-eIF4G interaction might serve the same purpose. In addition to eIF4F and eIF3, eIF4B also plays a key role in stimulating 43 S PIC attachment to mRNA (1, 2).
The best characterized biochemical activity of mammalian eIF4B (meIF4B) is stimulation of the RNA helicase activity of eIF4A (3), which depends on an arginine-rich domain in its C-terminal region and an RNA recognition motif (RRM) in the N-terminal half of the protein that both display RNA binding activity (4–6). However, the molecular mechanism of this stimulation is poorly understood and might involve stabilization of an active conformation of eIF4A, sequestration of ssRNA products of the helicase reaction, or increasing the efficiency of coupling ATP hydrolysis to RNA duplex unwinding (2, 7). meIF4B has also been proposed to stimulate mRNA-43 S PIC association more directly by simultaneous binding to 18 S rRNA, through its RRM domain, and to mRNA via the C-terminal RNA binding domain (8). meIF4B harbors a DRYG-rich repeat that mediates dimerization and interaction with eIF3, which could also stabilize 43 S PIC association with eIF4F·mRNP complexes (9).
Mammalian and yeast eIF4B (yeIF4B) share only ∼20% sequence identity (10, 11), and the only obviously conserved domain is the RRM (Fig. 1A). Until recently, yeIF4B had not been found to promote eIF4A helicase activity in vitro (12, 13), even though yeast eIF4A could be activated by meIF4B (14) and meIF4B could partially replace yeIF4B in a cell-free translation system (10). However, the gene encoding yeIF4B (TIF3) was identified as a dosage suppressor of a temperature sensitive (Ts−) mutation in the eIF4A gene (TIF1) (11), and tif3 mutations interact genetically with mutations in various eIF4 components (15). Moreover, although yeIF4B has not been reported to interact directly with eIF4A, it was found associated with native eIF4G and promoted complex formation between eIF4G and eIF4A in yeast cells (16). Recently, it was reported that yeIF4B can stimulate the RNA unwinding activity of yeast eIF4A and, as observed for meIF4B (7), appears to increase the coupling of ATP hydrolysis to RNA duplex separation by eIF4A (17).
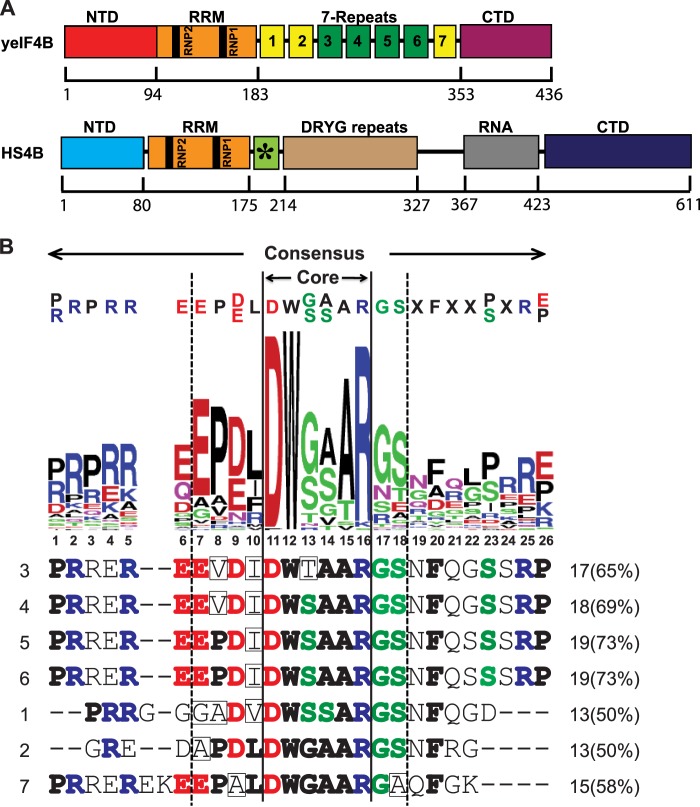
FIGURE 1.
Functional domains of yeast and human eIF4B proteins. A, schematic of yeast and human eIF4B protein functional domains. The schematic of Homo sapiens eIF4B (HS4B) was redrawn with slight modifications from Ref. 31, showing the C-terminal RNA binding domain (RNA) and a region related to the internal repeats in yeIF4B (green box with asterisk), as described under “Results.” B, conserved residues among internal repeats of fungal eIF4B homologs. Upper portion, an alignment of 60 repeats from 10 fungal yeIF4B homologs (5–9 repeats each) was generated using EBI MUSCLE and submitted to WebLogo Berkeley to generate a sequence logo, with the most conserved one or two residues at each position listed above as the consensus sequence. The stretch between Glu-7 and Ser-18 (demarcated by dotted lines), which contains the nearly invariant residues Asp-11, Trp-12, and Arg-16 that, together with residues 13–15, form a highly conserved “Core,” is the region of highest conservation within the logo. Lower portion, alignment of the seven S. cerevisiae yeIF4B repeats generated by EBI MUSCLE, with residues matching or deviating from the consensus sequence shown in boldface or boxed, respectively. Flanking each sequence is its position (1–7) in the repeat array (left) and the number and percentage of residues matching the consensus (right). The logo was constructed from the sequences of yeIF4B homologs from Vanderwaltozyma polyspora, Ashbya gossypii, S. cerevisiae, Kluyveromyces lactis, Lodderomyces elongisporus, Candida dubliniensis, Candida albicans, Meyerozyma guilliermondii, Scheffersomyces stipitis, and Debaryomyces hansenii.
Consistent with these functional links to eIF4F, yeIF4B was found to be required in addition to eIF4F and eIF3 for rapid and stable attachment of reconstituted 43 S PICs to capped, native yeast mRNAs in vitro (18) and for promoting a high apparent affinity of eIF4A for the initiation machinery to stimulate this process (19). Interestingly, yeIF4B can bind directly to eIF3 (18) and 40 S subunits (19), in addition to ssRNA (12, 19), which might enable it to bridge mRNA and the 43 S PIC in addition to promoting eIF4F function.
yeIF4B has an array of seven repeats of a 20–26-amino acid sequence located immediately following the RRM domain (Fig. 1, A and B), which is considered to be unique among the yeast orthologs of this factor (10, 11) and unrelated to the meIF4B DRYG or C-terminal RNA binding domains. The extreme N-terminal (NTD) and C-terminal domains (CTD) of yeIF4B also display no obvious sequence similarity to regions of meIF4B. Functional analysis of yeIF4B variants truncated from the N or C terminus led to the conclusion that the RRM and a subset of internal repeats are required for its ssRNA binding activity, RNA annealing activity, and stimulation of translation in vitro and in vivo, leading to the proposal that RNA annealing is a key function of yeIF4B (20). Surprisingly, however, we showed recently that the yeIF4B RRM and its associated ssRNA binding activity are fully dispensable for wild-type (WT) levels of translation initiation in vivo and WT rates of 43 S PIC attachment to native mRNAs in the reconstituted yeast system (19). By contrast, the 7-repeats domain is crucial for yeIF4B functions both in vivo and in vitro, and interestingly, the NTD also contributes to these activities in a manner partially overlapping with the contribution of the 7-repeats domain. In fact, the NTD is the region most critically required for yeIF4B binding to 40 S subunits in vitro. Interestingly, yeIF4B binds to ribosomal protein Rps20e (homolog of bacterial S10), a portion of which is exposed on the solvent-accessible side of the “head” of the 40 S subunit, and we provided evidence that yeIF4B binding induces structural changes in the ribosome's mRNA entry channel, which might facilitate mRNA loading into the 43 S PIC mRNA-binding cleft (19).
In this study, we sought to determine the minimum number of repeats and the key residues within each repeat that are required for the stimulatory functions of yeIF4B in vivo and in vitro. Interestingly, we determined that just two repeats is sufficient for substantive yeIF4B function, except when the NTD is missing or eIF4F function is limiting, and that the invariant amino acids of the ∼26-aa repeat, Asp, Trp, and Arg of the DWXXXR conserved core, are the only residues essential for repeat function. We also conducted a thorough functional dissection of the NTD and identified two conserved motifs critical for NTD function. Our results provide a comprehensive description of the critical sequence motifs in yeIF4B that support the function of eIF4F in promoting mRNA recruitment by the PIC.
EXPERIMENTAL PROCEDURES
Plasmid Constructions and Yeast Strains
All plasmids employed are listed in Table 1. The TIF3-HIS6 allele resides in the high copy (hc) LEU2 vector YEplac181, hc URA3 vector YEplac195, or single copy (sc) LEU2 vector YCplac111, carrying its native upstream- (747 bp) and downstream (340 bp)-flanking sequences and coding sequences for the His6 epitope inserted immediately before the stop codon (19). Mutant tif3-HIS6 alleles encoding the desired deletions, insertions, or substitutions (as described in Table 2) were amplified by PCR fusion from the template plasmids listed in Table 1, and the resulting PCR products were cloned into YCplac111, YEplac181, YEplac195, or low copy (lc) LEU2 vector pRS315 (21). DNA sequences of the entire PCR-amplified regions were verified in all novel plasmids. Plasmids for expression of yeIF4B proteins in Escherichia coli from the T7 promoter were generated as described previously (19), and PCR products of the appropriate fragments from the relevant yeast shuttle vectors were inserted between the NdeI and XmaI sites of pTYB2 (New England Biolabs) (Table 1). The tif3Δ strain FJZ052 was described previously (19). The amino acids removed, inserted, or substituted for these different yeIF4B variants are listed in Table 3. All yeast strains employed are shown in Table 4.
TABLE 1.
Plasmids used in this study
NA means not applicable, as plasmid was not constructed in this study. Sequences of primers will be provided upon request.
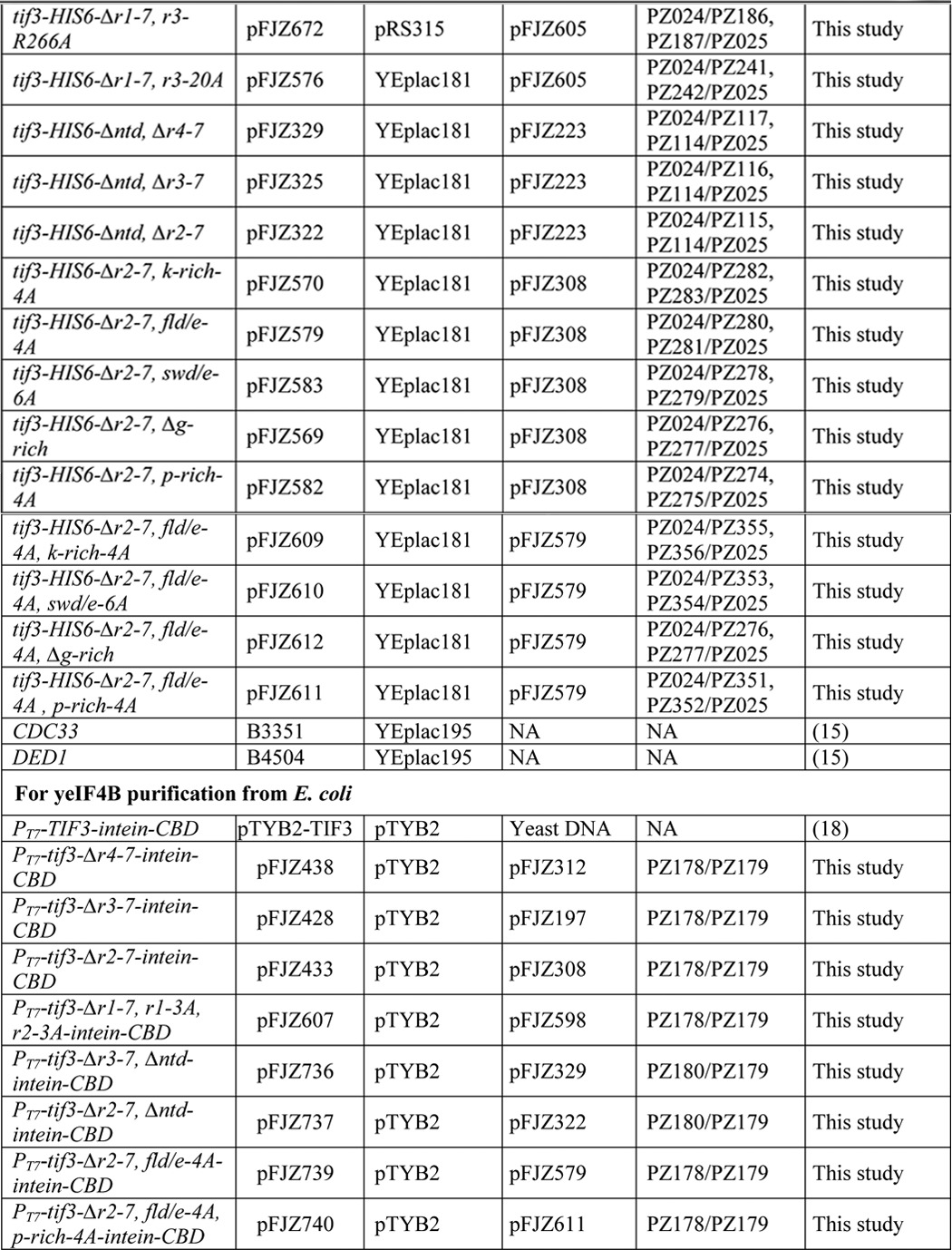
TABLE 2.
Amino acids removed, inserted, or substituted in yeIF4B variants expressed in yeast
| TIF3 allele | Amino acids removed | Amino acids inserted | Amino acids substituted |
|---|---|---|---|
| TIF3-HIS6 | None | None | None |
| tif3-HIS6-Δr1-7 | 183–352 | None | None |
| tif3-HIS6-Δntd | 2–86 | None | None |
| tif3-HIS6-Δctd | 353–436 | None | None |
| tif3-HIS6-Δrrm | 94–182 | None | None |
| tif3-HIS6-Δntd, Δr1-7 | 2–86, 183–352 | None | None |
| tif3-HIS6-Δr4-7 | 251–352 | None | None |
| tif3-HIS6-Δr3-7 | 225–352 | None | None |
| tif3-HIS6-Δr2-7 | 205–352 | None | None |
| tif3-HIS6-Δr1-7, R3, R3 | 183–352 | 225–250, 225–250 | None |
| tif3-HIS6-Δr1-7, R3 | 183–352 | 225–250 | None |
| tif3-HIS6-Δr3-7, r1-3A, r2-3A | 183–352 | 183–224 | D192A, W193A, R197A, D213A, W214A, R218A |
| tif3-HIS6-Δr1-7, r1-3A | 183–352 | 183–204 | D192A, W193A, R197A |
| tif3-HIS6-Δr1-7, r3-3A, r3-3A | 183–352 | 225–250, 225–250 | D235A, W236A, R240A, D235A, W236A, R240A |
| tif3-HIS6-Δr1-7, r3-3A | 183–352 | 225–250 | D235A, W236A, R240A |
| tif3-HIS6-Δr1-7, r3-D235A | 183–352 | 225–250 | D235A |
| tif3-HIS6-Δr1-7, r3-W236A | 183–352 | 225–250 | W236A |
| tif3-HIS6-Δr1-7, r3-R240A | 183–352 | 225–250 | R240A |
| tif3-HIS6-Δr1-7, r3-20A | 183–352 | 225–250 | (Pro-225–Ile-234)A10, (Gly-241–Pro-250)A10 |
| tif3-HIS6-Δntd, Δr4-7 | 2–86, 251–352 | None | None |
| tif3-HIS6-Δntd, Δr3-7 | 2–86, 225–352 | None | None |
| tif3-HIS6-Δntd, Δr2-7 | 2–86, 205–352 | None | None |
| tif3-HIS6-Δr2-7, k-rich-4A | 205–352 | None | KK5-6AA, KK9-10AA |
| tif3-HIS6-Δr2-7, fld/e-4A | 205–352 | None | F16A, L17A, DD19--0AA |
| tif3-HIS6-Δr2-7, swd/e-6A | 205–352 | None | S25A,W26A,EE28-9AA, DD30,32AA |
| tif3-HIS6-Δr2-7, Δg-rich | 205–352, 66–85 | None | None |
| tif3-HIS6-Δr2-7, p-rich-A | 205–352 | None | PPPP94,96,99-100AAAA |
| tif3-HIS6-Δr2-7, fld/e-4A, k-rich-4A | 205–352 | None | KK5-6AA, KK9-10AA, F16A, L17A, DD19-20AA |
| tif3-HIS6-Δr2-7, fld/e-4A, swd/e-6A | 205–352 | None | F16A, L17A, DD19-20AA, S25A,W26A, EE28-9AA, DD30,32AA |
| tif3-HIS6-Δr2-7, fld/e-4A, Δg-rich | 205–352, 66–85 | None | F16A, L17A, DD19-20AA |
| tif3-HIS6-Δr2-7, fld/e-4A, p-rich-4A | 205–352 | None | F16A, L17A, DD19-20AA, PPPP94,96,99-100AAAA |
TABLE 3.
Amino acids removed, inserted, or substituted in purified yeIF4B proteins
| yeIF4B proteins | Amino acids removed | Amino acids inserted | Amino acids substituted |
|---|---|---|---|
| WT | None | None | None |
| Δr4-7 | 251–352 | None | None |
| Δr3-7 | 225–352 | None | None |
| Δr2-7 | 205–352 | None | None |
| Δr1-7, r1-3A, r2-3A | 183–352 | 183–224 | D192A, W193A, R197A, D213A, W214A, R218A |
| Δr3-7, Δntd | 2–86, 225–352 | None | None |
| Δr2-7, Δntd | 2–86, 205–352 | None | None |
| Δr2-7, fld/e-4A | 205–352 | None | F16A, L17A, DD19-20AA |
| Δr2-7, fld/e-4A, p-rich-4A | 205–352 | None | F16A, L17A, DD19-20AA, PPPP94,96,99-100AAAA |
TABLE 4.
S. cerevisiae strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| FJZ052 | MATα his3-Δ1 leu2-Δ0 met15-Δ0 lys2-Δ0 ura3-Δ0 tif3Δ::hisG | 19 |
| F1199 (pSSC120) | MATa his3 ade2 leu2 trp1 ura3 tif1::HIS3 tif2::ADE2 [ptif1-A79V, CEN LEU2] | 35 |
| F1196 (CW04) | MATa his3 ade2 leu2 trp1 ura3 | 11 |
| H4823 (EPY41) | MATa ade2 his3 leu2 trp1 ura3 pep4::HIS3 tif4631::leu2hisG tif4632::ura3 pEP41 [TIF4632-HA, TRP1, CEN4] | 16 |
| H4824 (EPY81) | MATa ade2 his3 leu2 trp1 ura3 pep4::HIS3 tif4631::leu2hisG tif4632::ura3 pEP81 [tif4632-HA-L574F, TRP1, CEN4] | 16 |
| F324 (CB101) | MATα cdc33 leu1 ura3 trp1 ade8 | 36 |
| F2030 (Y10029)a | MATa his3-Δ1 leu2-Δ0 met15-Δ0 ura3-Δ0 ded1-199::KanR | 37 |
| YAS2488 | MATa leu2-3,112 his4-539 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG | 24 |
a The DED1 allele in this mutant was found to encode substitutions W253R and T408I rather than those reported by Li et al. (37) (N. Sen and A. G. Hinnebusch, unpublished observations).
In Vivo Complementation Tests, Western Analysis of yeIF4B Expression, and Polysome Analysis
The hc, lc, or sc LEU2 plasmids containing the appropriate TIF3-HIS6 alleles, or empty vector, were transformed into tif3Δ strain FJZ052, and rates of colony formation from single cells were measured by spotting assays at 14, 18, 30, and 36 °C as described previously (19). Western blot analysis of His6-tagged Tif3 variants was conducted using WCEs prepared by extraction with trichloroacetic acid (TCA) (22) and antibodies against the His6 epitope (Invitrogen) and Gcd6 (23) as described previously (19). Polysome profiles were determined as described previously (19).
Reagents for Biochemical Assays
40 S ribosomal subunits were purified from YAS2488 yeast lysate as described (24), but with the following modifications. Cells were lysed in a liquid nitrogen mill, and lysate was resuspended in 15 ml/liter of culture of 1× Ribo A buffer (20 mm HEPES-KOH (pH 7.4), 100 mm KOAc (pH 7.6), 2.5 mm Mg(OAc)2, 2 mm DTT) plus protease inhibitors (Complete EDTA-free, 1 tablet/50 ml (Roche Applied Science)) and treated with 2.5 units of Turbo DNase (Ambion) for each 10-ml lysate by incubating for 30 min on ice prior to clarifying the lysate for 30 min at 20,000 × g. KCl was added to the supernatant at a final concentration of 400 mm, and the lysate was filtered through glass fiber and 0.8-μm filters before loading onto an 8-ml CIM-QA-8f monolithic anion exchange column (BIA Separations) pre-equilibrated in 1× Ribo A + 400 mm KCl, which allowed large quantities of ribosomes to be rapidly separated from the bulk lysate using high flow rates (10 ml/min). After washing with 25 column volumes of loading buffer, the ribosome fraction was eluted with 10 column volumes of 50% buffer B (1× Ribo A + 900 mm KCl). The eluted peak containing ribosomes was concentrated, puromycin-treated, and loaded onto gradients for separation of subunits as described previously (24). Ribosomal subunits behaved identically to those purified using sucrose cushions in mRNA recruitment and a variety of other assays.6 Initiation factors and tRNA were prepared as described (18, 24), and initiator tRNAMet was methionylated using purified N-terminal His6-tagged E. coli methionyl-tRNA synthetase as described (25). Capped RNA was prepared as described (18) with the following modifications. RNA (5 μm RPL41A transcript) was incubated for 5–10 min at 65 °C and placed on ice before incubating for 1 h at 37 °C in 10–30-μl reactions containing 1× capping buffer (50 mm Tris-HCl (pH 8.0), 6 mm KCl, 1.25 mm MgCl2), 50 μm unlabeled GTP, 0.67 μm [α-32P]GTP (3000 Ci/mmol (PerkinElmer Life Sciences)), 100 μm S-adenosylmethionine, 2 units/μl Ribolock RNase inhibitor (Fermentas), and 150 nm purified vaccinia capping enzyme. Capped RNA was further purified using the RNeasy mini kit (Qiagen).
Analysis of mRNA Recruitment by Reconstituted PICs
Apparent rate constants for mRNA recruitment by reconstituted 43 S PICs were measured as a function of concentration of the appropriate yeIF4B variants or WT yeIF4B, as described previously (19), except that all reactions contained 1× Recon buffer (30 mm HEPES-KOH (pH 7.4), 100 mm KOAc (pH 7.6), 3 mm Mg(OAc)2, 2 mm DTT), 200 nm eIF2·GDPNP·Met-tRNAi ternary complex (formed by preincubating excess GDPNP (for 500 μm final in total reaction volume) with eIF2 (200 nm final) in Recon buffer for 10 min at 26 °C, followed by addition of Met-tRNAi (200 nm final) and incubation for another 5 min), 1 μm eIF1, 1 μm eIF1A, 200 nm eIF5, 30 nm 40 S subunits, 200 nm eIF3, 50 nm eIF4E·eIF4G, 2 μm eIF4A, and 15 nm radiolabeled capped RPL41A mRNA. The amount of each preparation of eIF3, eIF4A, and eIF4E·eIF4G used was determined in titration experiments to give the maximal fraction of mRNA bound to the PIC following 30-min incubations of all components with 300 nm WT yeIF4B. For measurements of eIF4A K½, apparent rate constants were measured as for yeIF4B at 0.1–10 μm eIF4A as indicated (Fig. 6), and yeIF4B was kept constant at the following concentrations, which were determined to be saturating for mRNA recruitment at 2 μm eIF4A (Fig. 5A): WT, 0.3 μm; Δr4-7, 0.3 μm; Δr3-7, 2 μm; Δr2-7, 2 μm.
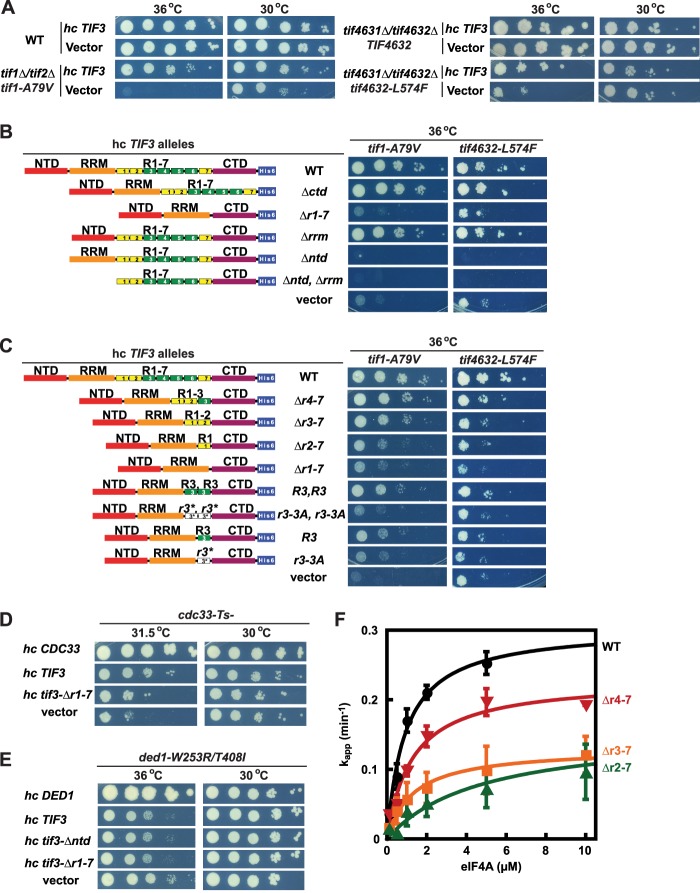
FIGURE 6.
Domains required in overexpressed yeIF4B for suppressing the growth defects conferred by mutations in eIF4F subunits. A and B, seven repeats and NTD are required for suppressing the growth defects of tif1-A79V and tif4632-L574F Ts− mutants. A, serial dilutions of transformants of strains F1196 and F1199 (left) and H4823 and H4824 (right) of the indicated relevant genotypes harboring hc TIF3 or empty vector were spotted on SC-Leu, −Ura (for F1196 and F1199 transformants), or SC-trp, −Leu (for H4823 and H4824 transformants) medium and incubated at 30 °C for 2 days or 36 °C for 3 days. B, serial dilutions of transformants of strains F1199 (left) and H4824 (right) harboring the indicated hc TIF3 alleles were spotted on SC-Leu, −Ura (for F1199 transformants) or SC-Trp, −Leu (for H4824 transformants) medium and incubated at 36 °C for 3 days. C, more than three repeats and the DWXXXR motif are required to suppress the growth defects of tif1-A79V and tif46312-L574F Ts− mutants. Conducted as in B using the indicated hc TIF3 constructs. D, 7-repeats domain is required for suppression of the growth defect of a cdc33-1 Ts− mutant by hc TIF3. Conducted as in A using strain F324 and the indicated hc plasmids but incubating on SC-Ura medium. E, hc TIF3 alleles exacerbate the growth defect of a ded1-W253R/T408I mutant. Conducted as in A using strain F2030 and the indicated hc plasmids but incubating on SC-Ura medium for 2 days at 30 °C or 5 days at 36 °C. F, yeIF4B internal repeats promote functional interaction of eIF4A with the PIC. The fraction of RPL41A mRNA bound to the PIC over time was measured by native gel electrophoresis at the indicated concentrations of eIF4A in the presence of yeIF4B containing 1 (Δr2-7, green triangles, 2 μm), 2 (Δr3-7, orange squares, 2 μm), 3 (Δr4-7, red inverted triangles, 0.3 μm), or 7 (WT, black circles, 0.3 μm) internal repeats. Apparent rate constants, derived from single exponential fits of fraction bound versus time plots, were plotted versus eIF4A concentration and fit with a hyperbolic equation to determine kmax, the maximal rate achieved at this concentration of complexes (30 nm PIC), and K½, the concentration of eIF4A required to achieve the half-maximal rate. Yeast strains in A–E are listed in Table 4.
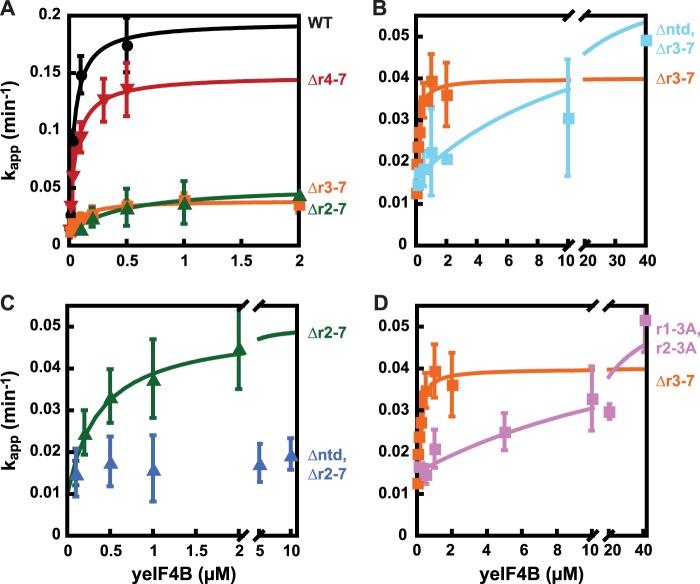
FIGURE 5.
NTD and conserved motifs within the seven repeats have overlapping functions in promoting mRNA recruitment to the preinitiation complex in vitro. The fraction of mRNA bound to the PIC was determined following incubation of 43 S PICs with eIF3, eIF4F, and varying concentrations of WT yeIF4B or yeIF4B variants as indicated and radiolabeled, capped mRNA (yeast RPL41A transcript) to determine the apparent rate of mRNA recruitment. These apparent rates were plotted versus concentration of the yeIF4B variant and fit with a hyperbolic equation to derive kmax, the maximal rate of mRNA recruitment at this concentration of PIC (30 nm), and K½, the concentration of yeIF4B variant required for half-maximal rate of mRNA recruitment. A, just one or two repeats is sufficient for appreciable function, whereas three repeats confer nearly WT yeIF4B activity in vitro. Apparent rates were compared for the indicated concentrations of WT (black circles) or yeIF4B containing three (red inverted triangles), two (orange squares), or one (green triangles) repeats. B and C, NTD overlaps functionally with the 7-repeats domain. Apparent rates were compared at different concentrations of yeIF4B lacking the NTD and containing two repeats (B, cyan squares) or 1 repeat (C, blue triangles). D, conserved Asp, Trp, and Arg residues promote yeIF4B interaction with the PIC. The apparent rates of mRNA recruitment were compared for yeIF4B containing repeats R1–R2 or repeats R1–R2 with the conserved Asp, Trp, and Arg residues in each remaining repeat mutated to Ala (r1–3A, r2–3A, pink squares). Residues deleted, inserted, or substituted in purified yeIF4B proteins are listed in Table 3.
RESULTS
Analysis of Sequence Conservation among the Internal Seven Repeats in eIF4B from Diverse Yeast Species
As described above, yeIF4B can be divided into four distinct domains (Fig. 1A), an NTD, RRM, a segment of seven imperfect repeats (7-repeats domain), and a CTD (20). The third to sixth repeats from the N terminus (repeats 3–6 (R3–R6)) are 26 aa in length and nearly identical in sequence, whereas R1, R2, and R7 are 22, 20, and 24 aa long, respectively, and lack residues found at the N or C terminus of repeats R3–R6 (Fig. 1B). To identify the positions in the repeat most critical for yeIF4B function, we constructed a multiple sequence alignment of the repeats from yeIF4B and its orthologs in 10 fungal species closely related to yeast and derived a graphical “logo” depicting sequence conservation at each of the 26 residues in the set of 60 aligned repeats (Fig. 1B). The central 12 aa at positions 7–18 are the most highly conserved, with Asp at position 11, Trp at position 12, and Arg at position 16 being nearly invariant. Positions 1–5 also tend to be Arg or Pro. Comparing each of the repeats in Saccharomyces cerevisiae eIF4B to the logo reveals that they all display comparable similarity to the consensus sequence within the central 12-aa segment and that (except for R3) they contain a perfect match to the conserved core sequence of DW(G/S)(A/S)AR. Thus, repeats R1, R2, and R7 diverge from the consensus sequence primarily in lacking residues on the N- or C-terminal sides of the central 12-aa segment (Fig. 1B). Based on this analysis, it seemed possible that each of the seven repeats is functional and that repeats R3–R6 are more important than repeats R1, R2, and R7 for yeIF4B activity.
Single Repeat Is Sufficient for Appreciable yeIF4B Function in Vivo
To determine the minimum number of internal repeats required for WT yeIF4B function in the presence of all other domains (NTD, RRM, and CTD), we constructed a set of nested internal deletions of the repeat coding sequences and tested the resulting mutant TIF3 alleles for complementation of the slow growth (Slg−) phenotype of a tif3Δ mutant. The tif3 alleles were expressed from the native promoter on sc or hc plasmids and encode a C-terminal His6 epitope to enable quantification of protein expression by Western analysis of WCEs. Cell suspensions of transformants harboring the different plasmids were spotted on solid medium, and growth was scored semi-quantitatively (from 0 to 10) by determining the highest dilutions at which colony formation occurred and the colony size at that dilution. tif3Δ cells exhibit a Slg− phenotype at all temperatures, but the defect is greatest at lower temperatures (10, 11). As similar results were obtained at 14, 18, 30, and 37 °C (data not shown), only the results of 14 °C complementation tests are presented below.
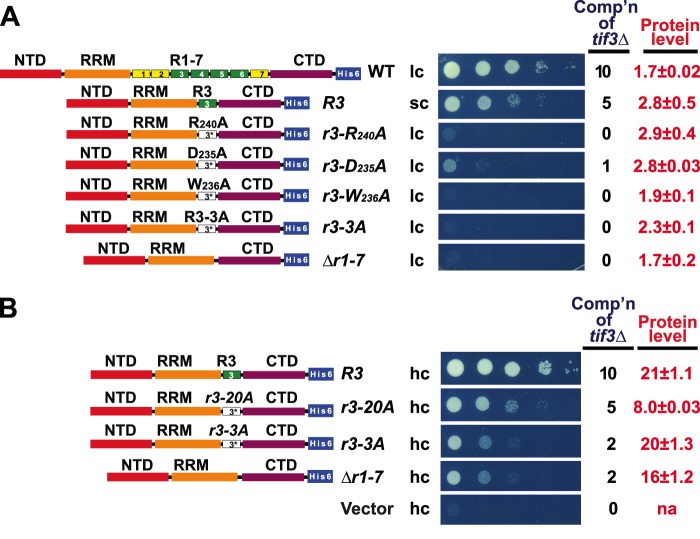
In agreement with our previous findings (19), a precise internal deletion of all seven repeats destroys complementation of the tif3Δ growth defect by the sc Δr1-7 allele, yielding an impairment of colony formation indistinguishable from that displayed by transformants harboring the empty vector (Fig. 2A, sc Δr1-7 versus sc WT and vector). Western blot analysis (Fig. 2B) showed that the tif3-Δr1-7 product is expressed at only ∼60% of WT yeIF4B-His6 and that overexpression of this mutant from a hc plasmid restored low level complementation at a level of protein expression >10-fold higher than WT (Fig. 2A, Δr1-7, hc versus sc) (19). Surprisingly, eliminating the C-terminal four or five repeats in the sc constructs Δr4-7 or Δr3-7, leaving intact only three (R1–R3) or two repeats (R1–R2), respectively, had no effect on complementation compared with that given by the WT allele (Fig. 2A, sc constructs Δr4-7, Δr3-7 versus WT). However, eliminating repeats 2–7 to leave only R1 intact produced an obvious decrease in complementation for the sc allele (Fig. 2A, sc Δr2-7 versus WT), despite the mutant protein being expressed at essentially the WT level (Fig. 2B, protein levels summarized in Fig. 2A). That solitary R1 in the sc Δr2-7 construct is sufficient for appreciable function is evident by comparing it with sc Δr1-7 lacking all repeats, which is completely inactive (Fig. 2A). In fact, overexpressing the Δr2-7 product from an hc plasmid, to levels ∼20-fold above WT, restored full complementation of the tif3Δ growth defect (Fig. 2A, hc and sc Δr2-7 versus WT; Fig. 2B). Thus repeats R1–R2 are sufficient, whereas R1 alone is inadequate, to supply WT complementation of tif3Δ when the mutant proteins are expressed at WT levels, but R1 alone is sufficient for WT function when the mutant protein is highly overexpressed.
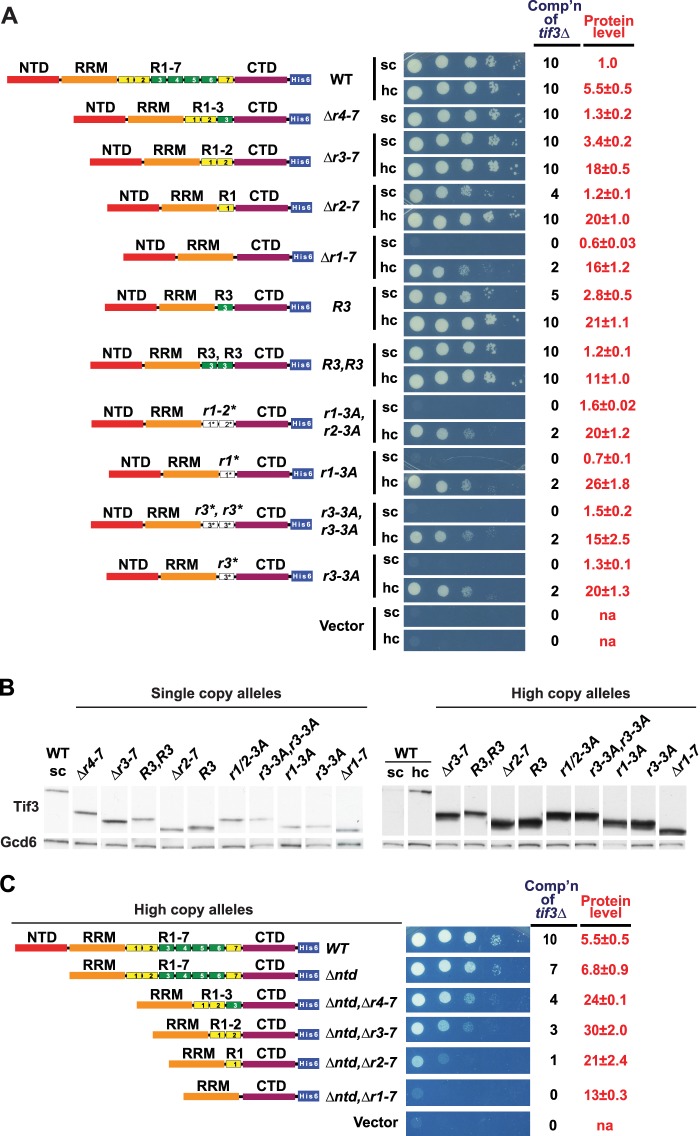
FIGURE 2.
One repeat in overexpressed yeIF4B is sufficient for WT cell growth, dependent on the nearly invariant residues of the core motif DWXXXR. A, requirement of yeIF4B repeats for supporting cell growth. Constructs Δr4-7, Δr3-7, Δr2-7, and Δr1-7 differ from the WT construct by precise deletions of the indicated repeat numbers; constructs R3 and R3,R3 harbor 1 or 2 copies, respectively, of repeat 3 in place of all 7 native repeats; and constructs with “−3A” in the allele designation harbor Ala substitutions of the nearly invariant residues of the DWXXXR motif (D192A, W193A, and R197A in r1-3A; D213A, W214A, and R218A in r2-3A; and D235A, W236A, and R240A in r3-3A). 10-Fold serial dilutions of transformants of tif3Δ strain FJZ052 bearing the indicated TIF3 alleles in hc, sc, or lc plasmids were spotted on SC-Leu medium and incubated at 14 °C for 7 days, and cell growth was scored from 0 (empty vector) to 10 (TIF3+). B, strains described in A were cultured in SC-Leu medium at 14 °C to A600 of ∼1, and WCEs prepared under denaturing conditions were subjected to Western analysis using His6 and Gcd6 antibodies to visualize His6-tagged yeIF4B and Gcd6 (as loading control), respectively. Intensities of yeIF4B-His6 signals relative to Gcd6 were normalized to those for sc WT transformants, and mean and S.E. calculated from replicate determinations are listed under Protein level in A. na, not applicable. C, removal of NTD increases the number of repeats required for yeIF4B function in vivo. Analyses of complementation of tif3Δ (cell growth) and Western analysis of yeIF4B protein levels (data not shown) were conducted as in A and B. Residues deleted, inserted, or substituted in the depicted TIF3 variants are listed in Table 2. Comp'n, complementation ability.
As shown in Fig. 1B, repeats R3–R6 are more closely related than R1, R2, and R7 to the consensus repeat logo. Hence, we asked whether one copy of R3 would be superior to solitary R1 and confer WT function at native levels of yeIF4B expression. However, complementation by the construct with R3 alone was almost indistinguishable from that containing R1 alone, in sc or hc, even though the sc R3 construct was expressed at somewhat higher levels than the corresponding construct containing solitary R1 (Fig. 2, A and B, constructs Δr2-7 and R3). Likewise, an sc construct with two tandem copies of R3 conferred a higher (essentially WT) level of complementation than that given by the solitary R3 construct (Fig. 2, A and B, construct R3,R3 versus R3 and WT). These results confirm our conclusion that a single repeat is insufficient for WT yeIF4B function in vivo unless the mutant protein is overexpressed, whereas two repeats provide WT function at native levels of expression. They also suggest that the residues at the ends of the 26-aa repeat that are present in R3 but lacking in R1 contribute little to repeat function in vivo. Finally, the fact that both 2-repeats constructs, harboring either R1–R2 or two copies of R3, confer WT yeIF4B function suggests that the various repeats perform highly overlapping, if not identical, roles in vivo.
More Repeats Are Required for High Level yeIF4B Function in Vivo When the NTD Is Lacking
We showed previously (19) that eliminating the entire 7-repeats domain had a more dramatic effect on yeIF4B function when the mutant proteins also lacked the NTD, abolishing the residual function of the overexpressed yeIF4B variant, thus indicating functional overlap of the NTD and 7-repeats domain (cf. hc Δr2-7 in Fig. 2A and hc Δntd,Δr2-7 in Fig. 2C). Given our conclusion above that different repeats are functionally redundant, we reasoned that a greater number of repeats might be required for WT function in the absence of the NTD. We tested this prediction by combining the nested repeat deletions described above with a deletion of the NTD and analyzed complementation of the tif3Δ growth defect. As shown previously (19), eliminating the NTD alone reduces complementation by the sc Δntd allele, and overexpressing the mutant protein from an hc plasmid does not improve complementation activity (Fig. 2C, hc Δntd; and data not shown). In contrast to our finding above that one repeat is sufficient for WT growth in the presence of all other domains when the mutant construct is overexpressed, the absence of only four repeats, R4–R7, from the hc construct also lacking the NTD (Δntd,Δr4-7) evokes a marked reduction in complementation despite a higher than WT level of protein expression (Fig. 2C, cf. Δntd and Δntd,Δr4-7). Similar results were obtained for the hc NTD-less construct containing only R1–R2 (Fig. 2C, Δntd,Δr3-7), and the corresponding construct with solitary R1 is almost completely defective (Fig. 2C, Δntd,Δr2-7). Thus, whereas a single repeat (R1 or R3) is sufficient for WT complementation of tif3Δ when the NTD is present in overexpressed yeIF4B (Fig. 2A, Δr2-7 and R3), more than three repeats are required when the NTD is absent (Fig. 2C). These results are fully consistent with functional overlap between the NTD and internal repeats, as well as functional redundancy among the various repeats themselves.
Most Highly Conserved Residues in the Internal Repeats Are Critical for yeIF4B Function in Vivo
The results of our sequence analysis in Fig. 1B identified positions 11, 12, and 16 as the most highly conserved positions in the 26-aa repeats of eIF4B from different yeast species, and all seven repeats of S. cerevisiae eIF4B contain the conserved Asp, Trp, and Arg residues at these positions. To determine whether these residues are important for yeIF4B function in vivo, we introduced triple alanine substitutions of all three conserved residues in various constructs containing only one or two repeats and tested the mutant alleles for complementation activity in vivo. The triple substitutions in the sc constructs harboring R1–R2 or solitary R1 destroyed repeat function, producing levels of complementation identical to that seen in the Δr1-7 construct lacking all seven repeats (Fig. 2A, Δr3-7 versus r1-3A,r2-3A and Δr1-7; Δr2-7 versus r1-3A). The same results were observed when the triple substitutions were made in the constructs harboring solitary R3 or two tandem copies of R3 (Fig. 2A, R3 versus r3-3A and Δr1-7; R3,R3 versus r3-3A,r3-3A). These results indicate that Asp-11, Trp-12, or Arg-16 are critically required for the functions of repeats R1, R2, and R3, and probably the other four repeats as well.
To determine whether all three highly conserved residues are important for repeat function in vivo, we also made single alanine substitutions at positions 11, 12, or 16 in the sc construct containing solitary repeat R3. All of these mutations appeared to completely eliminate R3 function; however, they also lowered the protein level significantly below that of the starting sc R3 construct (data not shown). Accordingly, we generated the same mutations in an R3 allele carried on lc plasmid, from which yeIF4B expression is elevated severalfold compared with that given by the corresponding sc construct (data not shown), and we compared the complementation activities of the resulting lc mutant alleles to that of the sc R3 construct. As shown in Fig. 3A, each of the single substitutions generated in the lc R3 construct essentially eliminated complementation, mimicking the sc Δr1-7 construct lacking all repeats, despite expression levels comparable with that given by the sc R3. Thus, all three highly conserved residues in the DWXXXR motif are critically required for the in vivo function of the internal repeat of yeIF4B.
FIGURE 3.
All three nearly invariant residues of the DWXXXR motif are crucial for yeIF4B repeat function in promoting cell growth. A, repeats labeled with asterisks harbor the indicated single Ala substitutions of conserved residues of the DWXXXR motif of R3, D235A, W236A, and R240A or all three substitutions for r3-3A. B, DWXXXR motif functions when flanking residues are substituted with Ala. The sole repeat in construct r3-20A has the following sequence: (A)10DWTAAR(A)10. A and B, analyses of complementation of tif3Δ (cell growth) and Western analysis of yeIF4B protein levels (data not shown) were conducted as in Fig. 2, A and B. Residues deleted, inserted, or substituted in the depicted TIF3 variants are listed in Table 2. na, not applicable; Comp'n, complementation ability.
We also examined whether the DWXXXR motif might be sufficient for at least partial function of the repeat by altering to Ala all 20 residues flanking the DWTAAR version of the motif found in repeat R3 of the hc solitary R3 construct. Interestingly, appreciable complementation activity was retained for this construct (hc r3-20A) in comparison with both the construct bearing Ala substitutions of only the three nearly invariant residues (hc r3-3A) and that lacking all seven repeats (hc Δr1-7) (Fig. 3B). Thus, although flanking residues contribute to repeat R3 function, all of them taken together are less important than each of the highly conserved residues at positions 11, 12, and 16 for yeIF4B repeat function in vivo.
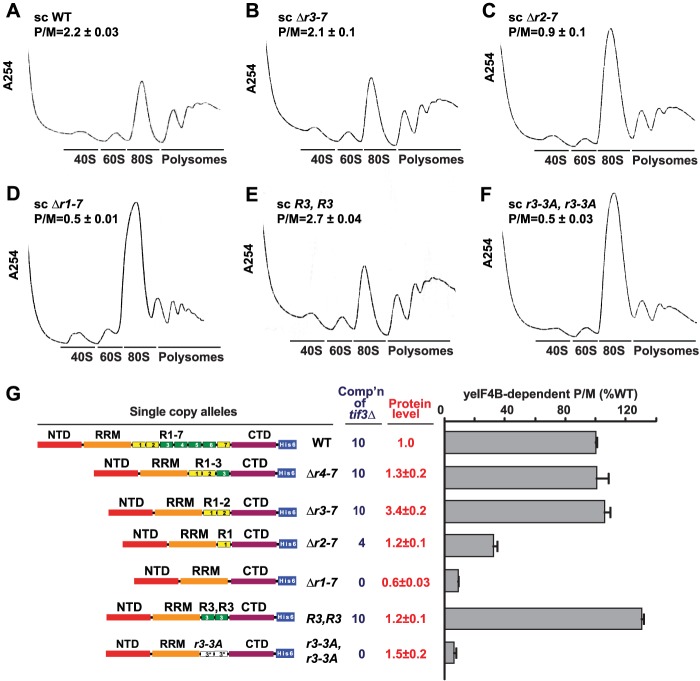
Two yeIF4B Internal Repeats Are Sufficient for Wild-type Translation Initiation in Vivo
Results above indicated that repeats R1–R2 are sufficient, whereas solitary R1 is inadequate, in sc constructs containing all other yeIF4B domains to provide WT complementation of the growth defect of the tif3Δ mutant (Fig. 2A). We wished to demonstrate that complementation of the growth defect by these constructs was mirrored by their ability to stimulate the rate of translation initiation and promote polyribosome assembly in vivo. As shown previously (19), and in agreement with the complementation data, elimination of all seven repeats in the sc Δr1-7 construct provokes a strong reduction in polysome abundance and a commensurate increase in the level of 80 S monosomes (Fig. 4, A versus D and summarized in G, Δr1-7 versus WT). In agreement with the complementation data, removing four or five repeats to leave only R1–R3 or R1–R2 intact, respectively, had no effect on the P/M ratio (Fig. 4, A, B, and G, WT versus Δr3-7 and Δr4-7), whereas removing six repeats and leaving only R1 intact in the sc construct produced a marked reduction in the P/M ratio (Fig. 4, A, C and G; WT versus Δr2-7). Similarly, the sc R3,R3 construct, containing tandem copies of R3, conferred a WT contribution of yeIF4B to the P/M ratio, consistent with its strong complementation activity, whereas the derivative of this construct with −3A mutations in both repeats (r3-3A,r3-3A) supported P/M ratios nearly indistinguishable from that given by deleting all seven repeats in the sc Δr1-7 construct (Fig. 4G). These findings indicate that just two internal repeats, and the highly conserved residues at positions 11, 12, and 16 in each repeat, are required for a WT contribution of yeIF4B to bulk translation initiation in vivo.
FIGURE 4.
Two repeats harboring the DWXXXR motif are essential for WT translation initiation in vivo. A–F, polysome profiles of tif3Δ transformants harboring selected sc TIF3 alleles. The indicated strains from Fig. 2A were cultured in SC-Leu at 30 °C to A600 of ∼1, with cycloheximide added prior to harvesting. WCEs were separated by sucrose density gradient centrifugation and scanned at 254 nm. Mean P/M ratios and S.E. from multiple replicates are given. G, P/M ratios for each strain were determined as in A–F. The mean P/M ratio for vector transformants was subtracted from the mean P/M ratios for mutant or WT transformants, and the resulting values were normalized to that obtained for WT to yield yeIF4B-dependent P/M ratios. Protein levels and complementation ability (Comp'n) from Fig. 2A are included for comparison. Residues deleted, inserted, or substituted in the depicted TIF3 variants are listed in Table 2.
One or Two Internal Repeats Support Appreciable but Not Wild-type yeIF4B Function in Promoting mRNA Recruitment to the PIC in Vitro
We previously demonstrated that yeIF4B accelerates and stabilizes mRNA binding to reconstituted PICs in vitro. This reaction is monitored using radiolabeled, capped RPL41A mRNA, which migrates more slowly in native gel electrophoresis when bound to PICs than does free mRNA or mRNPs, enabling measurement of both the kinetics and end points of mRNA recruitment (18). Plotting the apparent rate constants of mRNA recruitment as a function of yeIF4B concentration and fitting the data with a hyperbolic binding equation gives two parameters that can be compared as follows: kmax, the maximal apparent rate constant of mRNA recruitment for each variant at the concentration of preinitiation complexes used (30 nm), and K½, the concentration of each variant that produces a rate constant that is one-half the kmax (19). The K½ value is a measure of the apparent affinity of productive interaction of the varied factor with its other key binding partners in the system.
We recently showed that deletion of internal repeats 1–7 from yeIF4B dramatically increased the K½ value for mRNA recruitment to the PIC relative to that observed with the WT factor, indicating that the internal repeats domain promotes productive interaction of yeIF4B with the translation initiation machinery. To determine the number of repeats required for a WT level of yeIF4B function, we measured kmax and K½ values for mRNA recruitment to the PIC in the presence of WT yeIF4B or yeIF4B containing 1–3 internal repeats (Fig. 5A). In agreement with our in vivo results above (Fig. 2A), yeIF4B harboring only the three repeats, R1–R3, provides a nearly WT level of function in this assay (Fig. 5A; Δr4-7 versus WT), with kinetic parameters (kmax = 0.16 min−1 and K½ = 35 nm) only marginally different from those observed for WT yeIF4B (kmax = 0.20 min−1, K½ = 39 nm; Table 5). However, whereas yeIF4B containing only repeats R1–R2 (Δr3-7) conferred essentially WT function in vivo, in vitro this 2-repeats variant (Fig. 5A) showed an ∼5-fold reduction in kmax (0.04 min−1) and an ∼4-fold increase in K½ (160 nm) compared with the WT protein (Table 5). Likewise, the 1-repeat variant containing only R1 (Δr2-7) showed the same marked defect in rate of mRNA recruitment (kmax = 0.04 min−1), and an even more pronounced ∼12-fold defect in K½ (450 nm) compared with WT (Fig. 5A and Table 5, Δr2-7 versus WT). These data indicate that three or more repeats are needed for robust interaction of yeIF4B with the PIC to promote rapid rates of mRNA recruitment at relatively low yeIF4B concentrations. However, the 1- and 2-repeats variants still confer substantial activity, promoting a rate of recruitment 4-fold above the value seen in the absence of yeIF4B (0.01 min−1) when they are present at elevated concentrations (Fig. 5A).
TABLE 5.
In vitro parameters for yeIF4B stimulation of mRNA recruitment to the translation preinitiation complex
NS means no stimulation; these proteins did not promote the rate or extent of mRNA recruitment beyond that observed in the absence of yeIF4B. Values are means of at least three experiments ± average deviation. The concentration of eIF4A used in these experiments was 2 μm.
| yeIF4B variant | eIF4B titration |
|
|---|---|---|
| K½ | kmax | |
| nm | min−1 | |
| WT | 39 ± 5.2 | 0.20 ± 0.02 |
| Δr4-7 | 35 ± 8.2 | 0.16 ± 0.02 |
| Δr3-7 | 160 ± 16 | 0.04 ± 0.01 |
| r1-3A,r2-3A | >5,000 | 0.05 ± 0.02 |
| Δntd,Δr3-7 | >10,000 | 0.04 ± 0.02 |
| Δr2-7 | 450 ± 140 | 0.04 ± 0.01 |
| fld/e-4A,Δr2-7 | >10,000 | 0.16 ± 0.09 |
| fld/e-4A,p-rich-4A,Δr2-7 | NS | NS |
| Δntd,Δr2-7 | NS | NS |
NTD and Core DWR Sequence of the Repeats Have Overlapping Functions in Promoting yeIF4B Interaction with the PIC for Efficient mRNA Recruitment in Vitro
We next determined the effect of deleting the NTD from these constructs. As noted above, elimination of all but two repeats allowed considerable activity by otherwise intact yeIF4B (Table 5; Δr3-7, K½ = 160 nm, kmax = 0.04 min−1); however, deletion of the NTD from this 2-repeats variant evoked a dramatic increase in K½ for mRNA recruitment to a value of >10,000 nm with essentially no change in maximal rate (kmax = 0.05 min−1) (Fig. 5B and Table 5, Δntd,Δr3-7 versus Δr3-7). Moreover, whereas the construct containing only R1 and an intact NTD (Δr2-7) displays a kmax (0.04 min−1), which is ∼5-fold higher than that observed with no yeIF4B (kapp = 0.01 min−1; Fig. 5A), and only an ∼10-fold defect in K½ relative to WT yeIF4B (Table 5), removing the NTD from this construct abolished the ability to enhance mRNA recruitment even at very high concentrations of the mutant protein (up to 10 μm) (Fig. 5C and Table 5, Δntd,Δr2-7 versus Δr2-7). These data indicate that this mutant cannot interact productively with the PIC. These last findings demonstrate that the NTD is essential for the residual function displayed by the 1-repeat yeIF4B variant and is also required for high apparent affinity interaction of the 2-repeats variant with the PIC. By contrast, we showed previously that the NTD could be removed from otherwise intact yeIF4B with only a modest effect on the rate (reduction in kmax of ∼60%) and no increase in the K½ values for mRNA recruitment (19). These findings demonstrate that the NTD shares overlapping function with the first two repeats of yeIF4B in stimulating mRNA recruitment to the PIC in vitro.
Next, we wished to evaluate the importance of the conserved Asp, Trp, and Arg residues of the core repeat sequence in promoting mRNA recruitment. To do so, we substituted all three residues with Ala in each of the two remaining repeats (R1–R2) of the Δr3-7 construct (Fig. 5D). Substitution of this DWR motif in the 2-repeats construct dramatically increased the K½ from 160 to >5000 nm but did not affect kmax (Fig. 5D and Table 5, Δr3-7 versus r1–3A,r2–3A), indicating that these residues are critical for the ability of repeats R1–R2 to promote productive interaction of yeIF4B with the PIC. Together, these in vitro data indicate that the core DWR sequence of the repeats and the NTD of yeIF4B share overlapping functions in promoting interaction of yeIF4B with the PIC to allow rapid and efficient mRNA recruitment.
It is interesting that the Δr1-7 construct lacking all seven repeats produced a nearly WT maximal rate in this assay (kmax = 0.28 min−1; Ref. 19 and data not shown) that is considerably higher than the maximal rates determined here (kmax = 0.04–0.05 min−1, see Table 5) for the 1- and 2-repeats constructs (Δr2-7 and Δr3-7) and also the 2-repeats construct harboring substituted DWR motifs (r1–3A,r2–3A). However, the no-repeat construct and also the r1–3A,r2–3A construct containing two repeats with substituted DWR motifs both exhibit K½ values in the micromolar range, of 1200 and >5000 nm, respectively (Table 5) (19). Considering that the NTD is essential for the residual function of these variants (Table 5) (19), a potential explanation for these findings is that the presence of one or two repeats (even nonfunctional repeats with substituted DWR motifs) inhibits the overlapping NTD function, so deleting all of the repeats allows for a higher rate of mRNA recruitment, but only at very high yeIF4B concentrations.
NTD and 7-Repeats Domain Are Required for the Ability of Overexpressed yeIF4B to Suppress Growth Defects Conferred by Mutations in eIF4F Components
It was shown previously that overexpressing WT yeIF4B mitigates the temperature-sensitive (Ts−) growth defect conferred by the tif1-A79V allele, encoding the eIF4A-A79V variant, in a strain lacking the functionally redundant isoform of eIF4A encoded by TIF2 (11). More recently, we showed that yeIF4B overexpression similarly suppresses mutations in eIF4G1 and eIF4G2 that impair their physical interactions with eIF4A (in cells containing only that eIF4G isoform), restoring eIF4G·eIF4A complex formation in vivo (16). These and other genetic findings (10) indicate that yeIF4B promotes eIF4F function in yeast cells.
In addition to the in vivo evidence for interaction between yeIF4B and eIF4F, we previously reported a functional interaction between yeIF4B and eIF4A in vitro (19). We measured the apparent rates of mRNA recruitment to the PIC in our reconstituted yeast system at various concentrations of eIF4A in the presence or absence of WT yeIF4B or variants lacking the NTD, RRM, or 7-repeats domain to determine how the apparent affinity of eIF4A for the initiation machinery is affected by yeIF4B. Our results indicated that yeIF4B is critical for eIF4A interaction with the initiation machinery, as yeIF4B omission resulted in a >40-fold increase in the K½ value of eIF4A for mRNA recruitment to the PIC. Both the NTD and 7-repeats domains were found to be important for this functional interaction of yeIF4B and eIF4A that promotes rapid and stable mRNA recruitment to the PIC, whereas the RRM was found to be dispensable (19).
To determine which domains of yeIF4B are required for its ability to rescue eIF4A-A79V function in vivo, we tested various hc TIF3 alleles for the ability to suppress the temperature-sensitive growth (Ts−) phenotype conferred by the tif1-A79V allele. As expected from previous findings (11, 16), overexpressing full-length yeIF4B from an hc TIF3 plasmid mitigates the Slg− phenotype and restores growth at 36 °C of a tif1Δ tif2Δ strain containing episomal tif1-A79V as the only source of eIF4A (Fig. 6A, left). Examining other hc TIF3 alleles revealed that this suppression requires the seven repeats and NTD, whereas the RRM and CTD are dispensable for this activity of overexpressed yeIF4B (Fig. 6B). These findings are in agreement with our previous observations summarized above that the NTD and seven repeats are critical for promoting eIF4A activity in vitro (19). Interestingly, the hc TIF3 alleles lacking the NTD or both NTD and RRM conferred a dominant-negative effect, strongly impairing growth of the tif1-A79V strain even at a temperature (30 °C) where it normally displays only a moderate Slg− phenotype (Fig. 6B, cf. last three constructs for 36 °C; data not shown for 30 °C).
We obtained highly similar results for a tif4631Δ tif4632Δ strain harboring episomal tif4632-L574F as the only source of eIF4G (Fig. 6, A and B). The eIF4G2 mutant encoded by tif4632-L574F is defective for association with eIF4A in a manner rescued by overexpressing WT yeIF4B (16). Overexpressing the Δrrm variant, but not Δr1-7 or Δntd, rescued growth of the tif4632-L574F mutant at 36 °C, whereas overexpressing the Δntd and Δntd,Δrrm variants exacerbated the growth defect. Furthermore, overexpressing WT yeIF4B, but not the Δr1-7 mutant, improved the growth of a cdc33-1 mutant, expressing a thermolabile form of eIF4E (26) at the semi-permissive temperature of 31.5 °C where it confers a strong Slg− phenotype (Fig. 6D). The dominant-negative effect of the hc tif3-Δntd allele was again observed, as viable transformants of the cdc33-1 strain were not recovered using this TIF3 construct. In contrast to these findings, overexpressing either WT yeIF4B or the Δntd or Δr1-7 variants exacerbated, rather than suppressed, the Slg− phenotype conferred by the ded1-W253R,T408I allele at 36 °C (Fig. 6E). Ded1 is another DEAD-box helicase implicated in promoting translation initiation in yeast cells (27).
Interestingly, we found that the hc tif3-Δr4-7 construct, encoding yeIF4B-harboring repeats R1–R3, conferred less complete suppression of the Ts− phenotype of tif1-A79V than did hc WT TIF3 (Fig. 6C). As might be expected, overexpressing yeIF4B variants containing only one or two repeats, including those harboring solitary R1, R1–R2, or one or two copies of R3, were even less effective than the R1–R3 construct in suppressing tif1-A79V (Fig. 6C). Thus, to compensate for a defect in eIF4A function by overexpressing yeIF4B, a greater number of internal repeats (more than three) is required than is needed to provide WT yeIF4B function in otherwise WT cells, where only one repeat is required in overexpressed yeIF4B (Fig. 2A). The DWXXXR motif is essential for the ability of overexpressed yeIF4B to compensate for the eIF4A-A79V defect as alanine substitutions of the conserved residues in constructs encoding one or two copies of repeat R3 abolished suppression of the tif1-A79V Ts− phenotype (Fig. 6C).
The foregoing genetic results indicate that overexpressing yeIF4B specifically rescues a function carried out in vivo by each of the components of eIF4F, but not by Ded1, in a manner requiring the NTD and three or more internal repeats of yeIF4B. The exacerbating effects of hc tif3-Δntd on the growth of eIF4A, eIF4G, and eIF4E mutants might be explained by proposing that the yeIF4B variant lacking only the NTD competes for WT yeIF4B in associating with eIF4F or the PIC and that the absence of the NTD exacerbates defects in other eIF4F components, whereas the variant lacking the seven repeats cannot compete with WT yeIF4B. In this view, the NTD is an “effector” domain that enhances eIF4F function, whereas the seven repeats are crucial for interaction of yeIF4B with eIF4F, the PIC, or mRNA. The notion that the NTD promotes eIF4F function is consistent with our finding that more than three repeats are required both for complementation of tif3Δ by an sc TIF3 allele lacking the NTD (Fig. 2C) and for suppression of tif1-A79V by hc TIF3 (Fig. 6C), because both mutations compromise eIF4A activity and impose a requirement for a greater number of repeats.
Three Internal Repeats in yeIF4B Are Required to Promote Nearly Wild-type eIF4A Activity in Vitro
The above genetic results suggested that more than three internal repeats in yeIF4B are required for its ability to effectively promote eIF4A/eIF4F function. To test the validity of this interpretation, we compared the effects of having different numbers of internal repeats in yeIF4B on the eIF4A concentration dependence of mRNA recruitment to the PIC (Fig. 6F and Table 6). We found that in the presence of the Δr4-7 variant containing three repeats (R1–R3), a nearly WT rate of mRNA recruitment could be achieved at a saturating concentration of eIF4A (Δr4-7, kmax = 0.26 min−1; WT, kmax = 0.35 min−1) with a modest change in the concentration dependence of eIF4A (K½ = 1.8 μm) compared with WT yeIF4B (K½ = 1.0 μm). In the presence of the yeIF4B variants with two repeats (R1–R2) or solitary R1, there were somewhat larger defects in both the K½ of eIF4A (Δr3-7, K½ = 2.4 μm; Δr2-7, K½ = 2.8 μm) and kmax for mRNA recruitment (Δr3-7, kmax = 0.13 min−1; Δr2-7, kmax = 0.11 min−1; Fig. 6F and Table 6). These data indicate that more than three repeats are needed to achieve a fully WT enhancement of eIF4A function and that three repeats promote eIF4A activity more effectively than do one or two repeats. The effects of the repeat deletions on the K½ for eIF4A indicate that the first three repeats promote the ability of eIF4B to productively interact with eIF4A. Moreover, the diminished kmax values, which are measured at saturating concentrations of eIF4A, indicate that the first three repeats also affect the intrinsic ability of eIF4B to promote mRNA recruitment to the PIC in addition to enhancing productive interaction with eIF4A.
TABLE 6.
In vitro parameters for eIF4A stimulation of mRNA recruitment to the translation preinitiation complex
Values are reported ± average deviation of three or more experiments. The concentration of each eIF4B variant used (WT, 0.3 μm; Δr4-7, 0.3 μm; Δr3-7, 2 μm; Δr2-7, 2 μm) was saturating with respect to the rate of mRNA recruitment at 2 μm eIF4A.
| yeIF4B variant | eIF4A titration |
|
|---|---|---|
| K½ | kmax | |
| nm | min−1 | |
| WT | 1.0 ± 0.23 | 0.35 ± 0.08 |
| Δr4-7 | 1.8 ± 0.17 | 0.26 ± 0.03 |
| Δr3-7 | 2.4 ± 0.74 | 0.13 ± 0.03 |
| Δr2-7 | 2.8 ± 0.90 | 0.11 ± 0.05 |
It should be noted that the concentration of eIF4A required for the maximal rate is somewhat higher for the 1-, 2-, and 3-repeats yeIF4B variants than the concentration of eIF4A (2 μm) that was included for the yeIF4B K½ measurements described above in Fig. 5 and Table 5. Accordingly, the decreases in rates measured for these yeIF4B variants that are defective in promoting eIF4A activity are a combination of an ∼3-fold defect in ability to promote mRNA recruitment at saturating concentrations of eIF4A (kmax values in Table 6) and 2–3-fold increases in the K½ for eIF4A (Table 6). As the K½ for eIF4A in the absence of yeIF4B is >15 μm (19), the variant containing only a single repeat clearly confers substantial stimulation of eIF4A function in this assay. As discussed further below, combining these results with the fact that the eIF4A concentration in yeast cells (∼50 μm (28)) is much higher than the concentration used in our in vitro experiments helps to explain why variants containing only one or two repeats provided strong complementation of the tif3Δ mutation in vivo, while exhibiting substantial kinetic defects in vitro (Table 5).
Identification of Critical Sequence Elements in the yeIF4B NTD
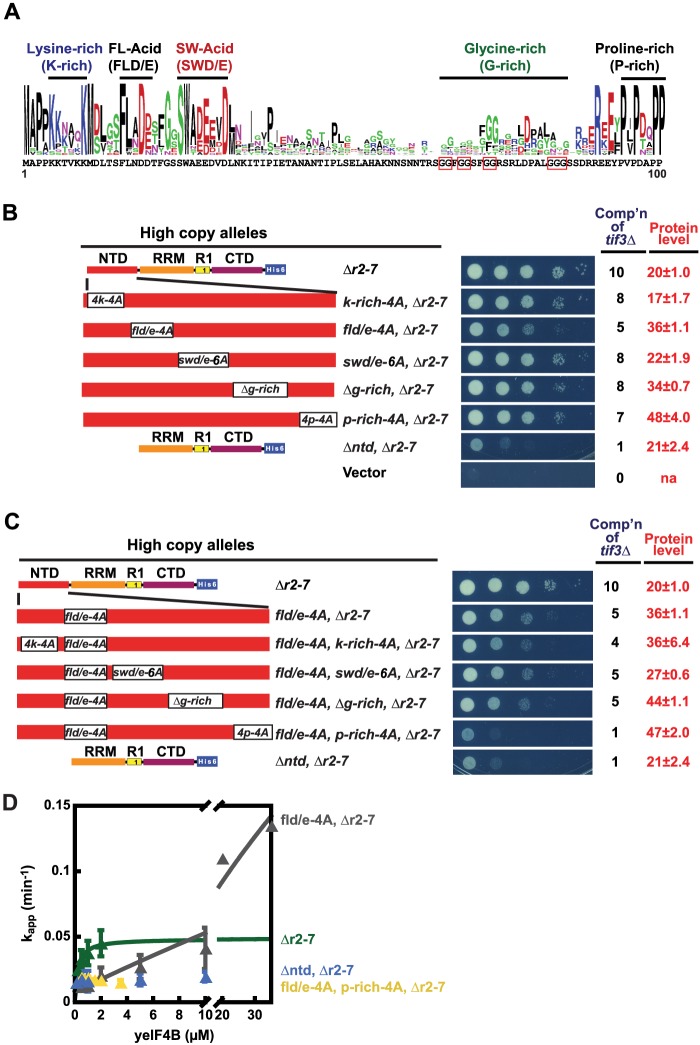
In an effort to uncover the critical sequence elements in the yeIF4B NTD, we constructed a sequence logo for this domain from the multiple alignments described above of yeIF4B sequences from different yeast species. The resulting logo revealed four highly conserved motifs, designated on the basis of their most prominent conserved residues as K-rich (Lys-rich), FL(D/E) (Phe, Leu, acidic), SW(D/E) (Ser, Trp, acidic), and P-rich (Pro-rich), plus a less conserved, Gly-rich stretch designated G-rich (Fig. 7A). To examine the importance of these motifs for NTD function, we substituted the prominent conserved amino acids in the K-rich, FL(D/E), SW(D/E), or P-rich segments or deleted the entire G-rich element in the hc Δr2-7 construct. (The exact residues substituted in each case are listed in Table 2.) The NTD mutations were generated in the hc Δr2-7 construct based on the results above (Fig. 2C) indicating that eliminating the NTD produces the greatest reduction in yeIF4B function in the context of solitary R1, because of functional overlap between the NTD and the 7-repeats domain.
FIGURE 7.
Identification of functional motifs in the yeIF4B NTD. A, conserved motifs in the NTDs of fungal eIF4Bs. Sequence logos were generated for the NTDs of fungal eIF4B as described in Fig. 1B using NTD sequences from 14 fungal eIF4B proteins. Five conserved motifs were identified and designated by the labels shown above. B, FLD/E is the most critical motif identified in the yeIF4B NTD. Constructs k-rich-4A, fld/e-4A, swd/e-6A, and p-rich-4A are derivatives of hc Δr2-7 harboring Ala substitutions of residues indicated in Table 3, whereas Δg-rich harbors a deletion of residues 66–85. C, FL(D/E) and P-rich NTD motifs have partially overlapping functions in promoting cell growth. Analyses of complementation of tif3Δ (cell growth) and Western analysis of yeIF4B protein levels (data not shown) were conducted for B and C as described in Fig. 2, A and B. Residues deleted or substituted in the depicted TIF3 variants are listed in Table 2. D, conserved motifs in the NTD promote yeIF4B interaction with the PIC. Apparent rate constants for mRNA recruitment to the PIC were measured and plotted as described in Fig. 5 at the indicated concentrations of R1-containing yeIF4B variants as follows: Δr2-7, green triangles; fl(d/e)-4A,Δr2-7, gray triangles; ΔntdΔr2-7, blue triangles; fl(d/e)-4A,p-rich-4A,Δr2-7, yellow triangles. Residues deleted or substituted in purified yeIF4B proteins are listed in Table 3.
As shown in Fig. 7B, only the Ala substitutions of the FL(D/E) element (fld/e-4A) produced a marked reduction in complementation of the tif3Δ growth defect, with relatively smaller decreases for the other four mutations, compared with the parental construct hc Δr2-7 (Fig. 7B). Because the hc fld/e-4A Δr2-7 allele confers substantially greater complementation compared with the hc Δntd, Δr2-7 construct lacking the entire NTD, we then combined the fld/e-4A mutation with each of the other four NTD mutations and repeated the complementation analysis. Interestingly, only the substitution of the Pro-rich segment reduced the complementation activity when combined with substitution of the FL(D/E) element, and this combination of substitutions was indistinguishable from deletion of the entire NTD (Fig. 7C). These findings identify the FL(D/E) and P-rich segments as the most critical sequence elements in the NTD in vivo and show that each element can provide partial NTD function in the absence of the other one.
To gain further understanding of the function of these elements in the NTD, we purified derivatives of the single-repeat protein (Δr2-7) harboring the two NTD substitutions that were most detrimental to function in vivo, and we tested the effects of these mutations in our in vitro reconstituted system for mRNA recruitment, as described above. Introducing the fld/e-4A substitution evoked a >10-fold increase in K½ relative to the analogous protein with an intact NTD (Fig. 7D and Table 5; fl(d/e)-4A,Δr2-7, K½ >10,000 nm versus Δr2-7, K½ = 450 nm), indicating that mutation of the FL(D/E) motif impairs yeIF4B interaction with components of the PIC. Surprisingly, introduction of the fld/e-4A substitution also increased the extrapolated kmax by ∼3-fold (Fig. 7D and Table 5; fl(d/e)-4A,Δr2-7, kmax = 0.16 versus Δr2-7, kmax = 0.04). Similar to our suggestion above that the presence of only one or two repeats can inhibit the overlapping function of the NTD, perhaps the NTD can also restrict the overlapping function of solitary repeat R1, and mutating the FL(D/E) motif of the NTD relieves this inhibitory effect.
The fl(d/e)-4A,Δr2-7 variant just described, although strongly impaired, displays appreciable yeIF4B function compared with the Δr2-7 variant lacking the NTD entirely, which shows no detectable stimulation of mRNA recruitment (Fig. 7D and Table 5; fl(d/e)-4A,Δr2-7 versus Δntd,Δr2-7). This implies that the NTD lacking the FL(D/E) motif retains residual function, consistent with the ability of this mutant to partially suppress the growth defect of the tif3Δ strain. By contrast, the combination of fld/e-4A and p-rich-4A substitutions made in the Δr2-7 variant abolished the stimulation of mRNA recruitment at all concentrations of the mutant protein tested (Fig. 7D and Table 5; fl(d/e)-4A,p-rich-4A,Δr2-7), thus indicating complete inactivation of NTD function. These findings are in accordance with the additive effects of the FL(D/E) and P-rich motif substitutions on NTD function in vivo (Fig. 7C).
DISCUSSION
In this study, we have determined the minimum number of internal repeats needed in yeIF4B to provide a WT level of translation initiation in vivo at native or elevated levels of yeIF4B expression, and to compensate for mutations in eIF4A, eIF4G, or eIF4E when yeIF4B is being overexpressed in yeast cells. We found that only two repeats, either the R1–R2 combination or two copies of R3, are sufficient for full complementation of the cell growth and polysome assembly defects in tif3Δ mutant cells when the yeIF4B variants are expressed from sc alleles at roughly WT levels of expression. Surprisingly, a single repeat (R1 or R3) was sufficient to rescue WT cell growth when the yeIF4B proteins were overexpressed by ∼20-fold. It is noteworthy that R1–R2 are somewhat degenerate in lacking certain highly conserved residues flanking the nearly invariant core region, whereas R3 is a “perfect” repeat (Fig. 1B). Thus, despite their sequence differences, the perfect and imperfect repeats appear to be functionally interchangeable in vivo rather than being dedicated to distinct functions. The fact that a single repeat is sufficient at high expression levels of yeIF4B might seem incompatible with a scenario in which different functionally interchangeable repeats cooperate to mediate multiple simultaneous interactions with eIF4F or the PIC. However, given the functional overlap between the NTD and repeats, it is possible that when the 1-repeat variants of yeIF4B are overexpressed, binding to the PIC is mediated by the NTD, and the single repeat executes another function in mRNA recruitment, such as interacting with eIF4F or mRNA.
We demonstrated that a larger number of repeats is required to supply a WT level of yeIF4B function in vivo when the NTD is absent and when overexpression of yeIF4B is employed to compensate for the reduced activity of the eIF4A-A79V mutant, as well as the eIF4G2-L574F variant that impairs eIF4A activity by reducing eIF4G2-eIF4A association (16). We also obtained strong genetic evidence that the yeIF4B NTD functions in parallel with eIF4A (and other components of eIF4F), as overexpressing yeIF4B variants lacking the NTD greatly exacerbate the growth defects conferred by eIF4A-A79V, eIF4G2-L574F, and the cdc33-1 mutation (impairing eIF4E stability), but otherwise it has little effect on the growth of WT cells or of a ded1 Ts− mutant. In fact, overexpression of WT yeIF4B reduces, rather than enhances, the growth of the ded1 mutant at the nonpermissive temperature. Results very similar to those for the ded1 mutant were obtained when WT yeIF4B or the NTD-less variant was overexpressed in an eIF3a mutant with a Slg-phenotype (tif32-box6) (29) (data not shown), indicating that excess yeIF4B specifically rescues defects in components of eIF4F.
The synthetic-sick/lethal interaction observed on overexpressing the NTD-less version of yeIF4B in eIF4F mutants can be explained by proposing that the yeIF4B NTD normally stimulates eIF4F activity or promotes an eIF4F-dependent step of initiation, so replacing WT yeIF4B by the NTD-less variant in initiation complexes reduces eIF4F activity or impairs the same step of initiation stimulated by eIF4F. Considering the absence of analogous genetic interactions between the high copy TIF3 or tif3-Δntd alleles and the ded1 mutation, and also published evidence that Ded1 (but not eIF4A or yeIF4B) is crucial for processive scanning through an exceptionally long 5′UTR of a reporter mRNA (30), a potential explanation for these results is that overexpressed yeIF4B primarily stimulates the function of eIF4F in promoting 43 S attachment to mRNA 5′ ends versus subsequent scanning of the 5′UTR, and this particular activity of eIF4F is impaired by overexpressing the dominant-negative NTD-less yeIF4B variant. Our finding that more than three repeats are needed to provide WT 7-repeats domain function in situations where eIF4F activity is reduced (absence of yeIF4B NTD or in rescuing eIF4A/eIF4G mutants) implies that the repeats function in concert to enhance eIF4F's role in PIC attachment to mRNA.
This conclusion is fully consistent with our analysis of the biochemical activities of different yeIF4B variants in the reconstituted yeast system. The in vitro analysis of yeIF4B mutants lacking different numbers of internal repeats confirmed that appreciable recruitment of RPL41A mRNA to the PIC is observed with only a single repeat (R1) present in the Δr2-7 variant but that very high concentrations of this 1-repeat protein are needed for function. We also found that the 2-repeats (R1–R2) variant Δr2-7 was relatively less defective in its interaction with the initiation machinery in the in vitro assay, as indicated by an ∼3-fold decrease in K½ relative to the 1-repeat variant (Table 5). These findings are consistent with our in vivo observations as follows: (i) greater complementation of the tif3Δ Slg− phenotype was conferred by the repeats R1–R2 versus solitary R1 when the yeIF4B variants were expressed at roughly native levels from sc plasmids, and (ii) the solitary R1 variant must be overexpressed to achieve WT complementation. Presumably overexpression of the solitary R1 variant in vivo compensates for its elevated K½ to achieve the same level of complementation conferred by the 2-repeats variant. However, the fact that the kmax for both the R1 and R1–R2 variants is 5-fold below the level observed with WT yeIF4B (Table 5) makes it unclear how a WT level of complementation can be achieved with these constructs in vivo.
One factor mitigating this conundrum is that the eIF4A concentration employed when measuring the K½ values for yeIF4B variants (2 μm; Table 5) is subsaturating for variants with a defect in recruiting eIF4A to the PIC or mRNA. When eIF4A concentrations were increased to determine the nature of these effects (Fig. 6F and Table 6), the rates of mRNA recruitment for the R1 and R1–R2 variants increased to within a factor of ∼3 of the value measured for WT yeIF4B and were nearly indistinguishable from one another (Table 6). As noted above, this ameliorating effect of elevated eIF4A concentration is expected to operate at the in vivo concentration estimated for eIF4A of ∼50 μm. To account for the complete complementation of tif3Δ afforded by the (overexpressed) R1 and R1–R2 variants, it could be proposed that their predicted ∼3-fold reductions in mRNA recruitment at physiological eIF4A concentrations does not reduce the overall rate of bulk translation because another step in the initiation pathway is rate-limiting or because other factors or conditions prevail in vivo to compensate for these moderate reductions in rate of mRNA recruitment by the PIC.
As noted above, three or more repeats were found to be required in overexpressed yeIF4B to efficiently suppress different mutations affecting eIF4F components. Consistent with this, the presence of three repeats (R1–R3) in the Δr4-7 variant substantially improved yeIF4B function in stimulating 48 S PIC assembly, decreasing K½, and increasing kmax by a factor of ∼4 compared with the corresponding values for the R1–R2 variant (Table 5). Even at high eIF4A concentrations, the kmax value is higher for the R1–R3 variant versus the R1–R2 variant, whereas four or more repeats appear to be required to achieve WT K½ and kmax values in this assay (Fig. 7D and Table 6). These findings are in complete agreement with our conclusion that four or more repeats are required in overexpressed yeIF4B to rescue the functions of defective eIF4F components at high physiological concentrations of eIF4A in vivo.
Another notable accomplishment of this study is the demonstration that each of the most highly conserved residues of the yeIF4B internal repeat, Asp, Trp, and Arg of the core motif DWxxxR, is critically required for repeat function in vivo and in vitro. In addition, we identified evolutionarily conserved motifs in the NTD and demonstrated that the FL(D/E) motif, containing conserved Phe, Leu, and acidic residues, is the most important conserved element in the NTD. Substituting the FL(D/E) motif in a construct containing only one repeat R1 (where the NTD is critical for complementation of tif3Δ) substantially reduces but does not abolish NTD function in vivo, which requires additionally substituting the P-rich motif. Thus, the FL(D/E) and P-rich elements have partially overlapping functions in vivo. Our in vitro analysis confirmed the overlapping contributions of the FL(D/E) and P-rich elements to the ability of the NTD to support the function of yeIF4B in stimulating mRNA recruitment by the PIC.
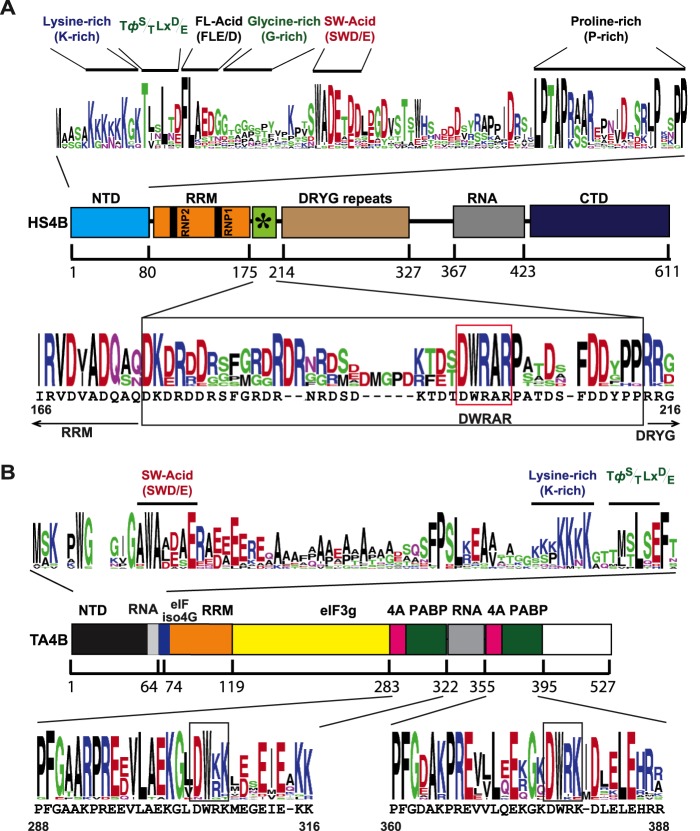
It is interesting that conserved motifs can be identified in the NTD of mammalian eIF4B that resemble the conserved motifs we identified in the yeIF4B NTD, although they occur in a somewhat different order (Fig. 8A). Two conserved motifs in the NTD of plant eIF4B also resemble the yeast SW(D/E) and K-rich motifs (Fig. 8B) but again occur in a different order than that seen in yeast or mammalian eIF4B. Although the K-rich motif is required for the RNA binding activity lodged in the NTD of wheat eIF4B (31), we found little evidence for its function in yeIF4B. The NTDs of plant and mammalian eIF4B appear to share a conserved motif, TΦ(S/T)LX(D/E) (with Φ designating a hydrophobic residue) located just N-terminal to the FL(E/D) motif in mammalian eIF4B and at the C terminus of the plant eIF4B NTD (Fig. 8, A and B), which was shown for the plant factor to bind a shorter isoform of eIF4G called eIFiso4G (31). This motif is not evident in yeIF4B, and the FL(E/D) motif found here to be the most critical element in the yeIF4B NTD is not evident in plant eIF4B (Fig. 8B). Thus, eIF4B from these different groups of eukaryotes seems to differ in both the number and order of short motifs present in the NTD. It is unknown which of the conserved NTD motifs are important for the functions of mammalian or plant eIF4B.
FIGURE 8.
Mammalian and plant eIF4B proteins share conserved yeIF4B NTD motifs, and meIF4B harbors a conserved sequence similar to the yeIF4B internal repeats. Protein sequence logos for mammalian and plant eIF4B NTDs and repeat elements were generated as described in Fig. 1B. A, functional domains and sequence conservation in meIF4B. Upper portion, five conserved motifs in the meIF4B NTD were designated by the labels shown above the logos. The G-rich motif is between the FL(E/D) and SW(D/E) motifs, instead of following the SW(D/E) motif as occurs in the yeIF4B NTD (Fig. 7A). Middle portion, schematic of functional domains in eIF4B from H. sapiens (HS4B), as described in Fig. 1A. Lower portion, conserved residues in the region between the RRM and DRYG domains of meIF4B. The human eIF4B sequence between Ile-166 and Gly-216 is shown below the logo. A DWRAR motif highlighted in the red box resembles the DWXXXR motif in yeIF4B repeats. The position of the sequence element in human eIF4B that is similar to the yeIF4B repeat consensus is indicated by the green box containing an asterisk in the schematic of human eIF4B shown in the middle portion. The upper logo for meIF4B NTDs was constructed from the sequences of animal eIF4B homologs from H. sapiens, Gallus gallus, Lapemis hardwickii, Danio rerio, Salmo salar, Tetraodon nigroviridis, Strongylocentrotus purpuratus, Marinitoga piezophila, Ciona intestinalis, Nematostella vectensis, Trichoplax adherens, Nasonia vitripennis, and Apis mellifera. The lower logo for the HS4B repeat was generated from the sequences of animal eIF4B homologs from H. sapiens, Mus musculus, Dicentrarchus labrax, L. hardwickii, G. gallus, Callithrix jacchus, Oreochromis niloticus, Anolis carolinensis, Sarcophilus harrisii, Takifugu rubripes, Oryzias latipes, and Xenopus tropicalis. B, functional domains and sequence conservation in plant eIF4B. Upper portion, SW(D/E), K-rich, and TΦ(S/T)LX(D/E) motifs identified in the plant eIF4B NTD are designated by labels above the logos. Middle portion, schematic of functional domains in eIF4B from wheat (Triticum aestivum) (TA4B), redrawn with slight modifications from Ref. 31, showing binding sites for eIFiso4G, eIF3g, eIF4A (4A), or RNA. Lower portion, sequence logos for the most highly conserved residues in the ∼40-residue repeats identified in plant eIF4B as eIF4A and poly(A)-binding protein-binding sites (31). The sequences outlined with black boxes superficially resemble the DWXXXR motif in yeIF4B repeats. Both upper and lower logos were generated from the sequences of plant eIF4B homologs from Arabidopsis thaliana (eIF4B1 and eIF4B2), Triticum aestivum, Capsella rubella, Eutrema salsugineum, Prunus persica, Populus trichocarpa, Theobroma cacao, Cucumis sativus, Citrus clementine, Solanum lycopersicum, Ricinus communis, Zea mays, Sorghum bicolor, Genlisea aurea, Setaria italic, Oryza sativa Japonica, Echinochloa phyllopogon.
Interestingly, human eIF4B also contains a sequence related to one of the yeIF4B seven repeats located just C-terminal to the RRM (Fig. 8A), the same location where the seven repeats reside in yeIF4B. We found evidence that this repeat-related segment in human eIF4B can augment the complementation activity of a TIF3 construct lacking all seven repeats, albeit not to the same extent as conferred by yeast single repeats R1 or R3.7 It will be interesting to learn whether these conserved elements in human eIF4B contribute to its known functions in stimulating eIF4F activity and mRNA recruitment by the PIC.
We found no evidence for an element related to the yeast 7-repeats motif in the vicinity of the putative RRM in plant eIF4B (data not shown). A repeat of ∼40 residues has been identified in the C-terminal half of plant eIF4B that was implicated in binding eIF4A and poly(A)-binding protein (Fig. 8B) (31). Each of these repeats contains a conserved DW(R/K)K motif, which resembles the critical DWXXXR motif in the yeast 7-repeats sequence. However, like meIF4B, yeIF4B has not been found to bind directly to eIF4A in the absence of an RNA molecule that could bridge their interaction (6, 32, 33).8 In addition, poly(A)-binding protein was not present in our in vitro experiments that demonstrated the role of the yeIF4B 7-repeats domain in promoting 43 S PIC attachment to mRNA. Hence, we consider it unlikely that the ∼40-residue repeat in plant eIF4B is related in sequence or function to the yeast 7-repeats domain.
The functional importance of domains of yeIF4B that facilitate its binding to the 40 S subunit, and its ability to alter accessibility to hydroxyl radical cleavage of 18 S rRNA residues near the entry channel, previously led us to propose that yeIF4B might stabilize mRNA binding to the ribosome by extending the mRNA-binding surface and contacting both the mRNA and ribosome or that it allosterically stimulates initial insertion of mRNA into the mRNA binding cleft of the 40 S subunit. As yeIF4B also specifically reduces the K½ for eIF4A in 48 S assembly in vitro, it clearly also facilitates the functional interaction of eIF4A with the PIC and/or mRNA through both its NTD and 7-repeats domain (19), and we showed here that four or more repeats are required for a fully WT level of this yeIF4B activity (Table 6). Perhaps yeIF4B binding to the 40 S subunit helps to recruit eIF4A near the mRNA entry channel at a position where it functions most effectively to stimulate mRNA insertion into the mRNA binding cleft. Alternatively, the repeats might interact with eIF4A or eIF4G to promote productive conformations of the latter factors within eIF4F to increase the rate of unwinding duplex structures in the mRNA 5′ end. In either case, it could be imagined that the presence of multiple repeats ensures high level occupancy by yeIF4B of its relevant binding sites on the 40 S subunit, or within eIF4F, and that the conserved Asp, Trp, and Arg residues are essential for these interactions.
Recent evidence indicates that yeIF4B can increase the coupling between ATP hydrolysis and duplex unwinding by eIF4A·eIF4G complexes, presumably by increasing the dwell time of the closed conformation of the RecA-like domains of eIF4A to allow sufficient time for RNA strand displacement prior to ATP hydrolysis (17). It is possible that the ability of yeIF4B to reduce the K½ value of eIF4A in stimulating mRNA recruitment by the PIC (observed here) reflects an increased dwell time of the productive conformation of eIF4A. However, our yeIF4B preparations that are active in promoting mRNA recruitment by the PIC do not stimulate unwinding by eIF4A/eIF4G (13), consistent with other previous findings (12). Thus, either the exact RNA substrate used in unwinding assays is critical or stimulating the unwinding activity of eIF4A is not the key function of yeIF4B in promoting mRNA recruitment by the PIC, which instead might be to stimulate eIF4A/eIF4G interactions with the PIC. In any event, it would be interesting to determine whether the NTD, RRM, or seven repeats are required for the ability of yeIF4B to stimulate the unwinding activity of eIF4A·eIF4G complexes under the assay conditions of Andreou and Klostermeier (17).
Acknowledgments
We thank Tom Dever and the members of our laboratories for useful comments and suggestions, Eun-Hee Park for sharing observations concerning suppression of tif4632-L574F by various TIF3 alleles, and Neelam Sen for sharing observations about the Ded1 substitutions encoded by ded1-199. We also thank Michael Altmann (University of Bern) for generously sharing antibodies against yeIF4B and Patrick Linder (University of Geneva) for gifts of yeast strains.
This work was supported, in whole or in part, by National Institutes of Health Intramural Research Program Grant (to A. G. H. and F. Z.) and Grant GM62128 (to J. R. L.). This work was also supported by an American Heart Association postdoctoral fellowship (to S. E. W.).
S. E. Walker, A. M. Munoz, and J. R. Lorsch, unpublished results.
F. Zhou and A. G. Hinnebusch, unpublished observations.
S. E. Walker, S. F. Mitchell, and J. R. Lorsch, unpublished observations.
- PIC
- preinitiation complex
- NTD
- N-terminal domain
- CTD
- C-terminal domain
- RRM
- RNA recognition motif
- P/M
- polysome/monosome
- aa
- amino acid
- ssRNA
- single strand RNA
- meIF4B
- mammalian eIF4B
- hc
- high copy
- sc
- single copy
- lc
- low copy
- WCE
- whole cell extract
- mRNP
- mRNA ribonucleoprotein complex
- GDPNP
- 5′-guanylyl imidodiphosphate.
REFERENCES
- 1. Pestova T. V., Lorsch J. R., Hellen C. U. (2007) in Translational Control in Biology and Medicine (Mathews M., Sonenberg N., Hershey J. W., eds) pp. 87–128, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 2. Hinnebusch A. G., Lorsch J. R. (2012) The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb. Perspect. Biol. 4, 1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rogers G. W., Jr., Richter N. J., Lima W. F., Merrick W. C. (2001) Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 276, 30914–30922 [DOI] [PubMed] [Google Scholar]
- 4. Méthot N., Pause A., Hershey J. W., Sonenberg N. (1994) The translation initiation factor eIF-4B contains an RNA-binding region that is distinct and independent from its ribonucleoprotein consensus sequence. Mol. Cell. Biol. 14, 2307–2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Naranda T., Strong W. B., Menaya J., Fabbri B. J., Hershey J. W. (1994) Two structural domains of initiation factor eIF-4B are involved in binding to RNA. J. Biol. Chem. 269, 14465–14472 [PubMed] [Google Scholar]
- 6. Rozovsky N., Butterworth A. C., Moore M. J. (2008) Interactions between eIF4AI and its accessory factors eIF4B and eIF4H. RNA 14, 2136–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Özeş A. R., Feoktistova K., Avanzino B. C., Fraser C. S. (2011) Duplex unwinding and ATPase activities of the DEAD-box helicase eIF4A are coupled by eIF4G and eIF4B. J. Mol. Biol. 412, 674–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Methot N., Pickett G., Keene J. D., Sonenberg N. (1996) In vitro RNA selection identifies RNA ligands that specifically bind to eukaryotic translation initiation factor 4B: the role of the RNA remotif. RNA 2, 38–50 [PMC free article] [PubMed] [Google Scholar]
- 9. Méthot N., Song M. S., Sonenberg N. (1996) A region rich in aspartic acid, arginine, tyrosine, and glycine (DRYFG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interaction with eIF3. Mol. Cell. Biol. 16, 5328–5334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Altmann M., Müller P. P., Wittmer B., Ruchti F., Lanker S., Trachsel H. (1993) A Saccharomyces cerevisiae homologue of mammalian translation initiation factor 4B contributes to RNA helicase activity. EMBO J. 12, 3997–4003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Coppolecchia R., Buser P., Stotz A., Linder P. (1993) A new yeast translation initiation factor suppresses a mutation in the eIF-4A RNA helicase. EMBO J. 12, 4005–4011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Altmann M., Wittmer B., Méthot N., Sonenberg N., Trachsel H. (1995) The Saccharomyces cerevisiae translation initiation factor Tif3 and its mammalian homologue, eIF-4B, have RNA annealing activity. EMBO J. 14, 3820–3827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rajagopal V., Park E. H., Hinnebusch A. G., Lorsch J. R. (2012) Specific domains in yeast eIF4G strongly bias the RNA unwinding activity of the eIF4F complex toward duplexes with 5′-overhangs. J. Biol. Chem. 287, 20301–20312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blum S., Schmid S. R., Pause A., Buser P., Linder P., Sonenberg N., Trachsel H. (1992) ATP hydrolysis by initiation factor 4A is required for translation initiation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 89, 7664–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. de la Cruz J., Iost I., Kressler D., Linder P. (1997) The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 94, 5201–5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Park E. H., Walker S. E., Zhou F., Lee J. M., Rajagopal V., Lorsch J. R., Hinnebusch A. G. (2013) Yeast eukaryotic initiation factor (eIF) 4B enhances complex assembly between eIF4A and eIF4G in vivo. J. Biol. Chem. 288, 2340–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Andreou A. Z., Klostermeier D. (2013) eIF4B and eIF4G jointly stimulate eIF4A ATPase and unwinding activities by modulation of the eIF4A conformational cycle. J. Mol. Biol., 10.1016/j.jmb.2013.09.027 [DOI] [PubMed] [Google Scholar]
- 18. Mitchell S. F., Walker S. E., Algire M. A., Park E. H., Hinnebusch A. G., Lorsch J. R. (2010) The 5′-7-methylguanosine cap on eukaryotic mRNAs serves both to stimulate canonical translation initiation and block an alternative pathway. Mol. Cell 39, 950–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Walker S. E., Zhou F., Mitchell S. F., Larson V. S., Valasek L., Hinnebusch A. G., Lorsch J. R. (2013) Yeast eIF4B binds to the head of the 40 S ribosomal subunit and promotes mRNA recruitment through its N-terminal and internal repeat domains. RNA 19, 191–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Niederberger N., Trachsel H., Altmann M. (1998) The RNA recognition motif of yeast translation initiation factor Tif3/eIF4B is required but not sufficient for RNA strand-exchange and translational activity. RNA 4, 1259–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sikorski R. S., Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reid G. A., Schatz G. (1982) Import of proteins into mitochondria. J. Biol. Chem. 257, 13062–13067 [PubMed] [Google Scholar]
- 23. Cigan A. M., Bushman J. L., Boal T. R., Hinnebusch A. G. (1993) A protein complex of translational regulators of GCN4 is the guanine nucleotide exchange factor for eIF-2 in yeast. Proc. Natl. Acad. Sci. U.S.A. 90, 5350–5354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Acker M. G., Kolitz S. E., Mitchell S. F., Nanda J. S., Lorsch J. R. (2007) Reconstitution of yeast translation initiation. Methods Enzymol. 430, 111–145 [DOI] [PubMed] [Google Scholar]
- 25. Walker S. E., Fredrick K. (2008) Preparation and evaluation of acylated tRNAs. Methods 44, 81–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nielsen K. H., Szamecz B., Valásek L., Jivotovskaya A., Shin B. S., Hinnebusch A. G. (2004) Functions of eIF3 downstream of 48 S assembly impact AUG recognition and GCN4 translational control. EMBO J. 23, 1166–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hinnebusch A. G. (2011) Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol. Mol. Biol. Rev. 75, 434–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. von der Haar T., McCarthy J. E. (2002) Intracellular translation initiation factor levels in Saccharomyces cerevisiae and their role in cap-complex function. Mol. Microbiol. 46, 531–544 [DOI] [PubMed] [Google Scholar]
- 29. Chiu W. L., Wagner S., Herrmannová A., Burela L., Zhang F., Saini A. K., Valásek L., Hinnebusch A. G. (2010) The C-terminal region of eukaryotic translation initiation factor 3a (eIF3a) promotes mRNA recruitment, scanning, and, together with eIF3j and the eIF3b RNA recognition motif, selection of AUG start codons. Mol. Cell. Biol. 30, 4415–4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Berthelot K., Muldoon M., Rajkowitsch L., Hughes J., McCarthy J. E. (2004) Dynamics and processivity of 40 S ribosome scanning on mRNA in yeast. Mol. Microbiol. 51, 987–1001 [DOI] [PubMed] [Google Scholar]
- 31. Cheng S., Gallie D. R. (2006) Wheat eukaryotic initiation factor 4B organizes assembly of RNA and eIFiso4G, eIF4A, and poly(A)-binding protein. J. Biol. Chem. 281, 24351–24364 [DOI] [PubMed] [Google Scholar]
- 32. Marintchev A., Edmonds K. A., Marintcheva B., Hendrickson E., Oberer M., Suzuki C., Herdy B., Sonenberg N., Wagner G. (2009) Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell 136, 447–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nielsen K. H., Behrens M. A., He Y., Oliveira C. L., Jensen L. S., Hoffmann S. V., Pedersen J. S., Andersen G. R. (2011) Synergistic activation of eIF4A by eIF4B and eIF4G. Nucleic Acids Res. 39, 2678–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gietz R. D., Sugino A. (1988) New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74, 527–534 [DOI] [PubMed] [Google Scholar]
- 35. Schwer B., Linder P., Shuman S. (1998) Effects of deletion mutations in the yeast Ces1 protein on cell growth and morphology and on high copy suppression of mutations in mRNA capping enzyme and translation initiation factor 4A. Nucleic Acids Res. 26, 803–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brenner C., Nakayama N., Goebl M., Tanaka K., Toh-e A., Matsumoto K. (1988) CDC33 encodes mRNA cap-binding protein eIF-4E of Saccharomyces cerevisiae. Mol. Cell. Biol. 8, 3556–3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Z., Vizeacoumar F. J., Bahr S., Li J., Warringer J., Vizeacoumar F. S., Min R., Vandersluis B., Bellay J., Devit M., Fleming J. A., Stephens A., Haase J., Lin Z. Y., Baryshnikova A., Lu H., Yan Z., Jin K., Barker S., Datti A., Giaever G., Nislow C., Bulawa C., Myers C. L., Costanzo M., Gingras A. C., Zhang Z., Blomberg A., Bloom K., Andrews B., Boone C. (2011) Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat. Biotechnol. 29, 361–367 [DOI] [PMC free article] [PubMed] [Google Scholar]