Background: Protein misfolding, a universal threat to cells, is dealt with by the ubiquitin-proteasome system.
Results: Ynl155w is a zinc-dependent ubiquitin-binding protein, interacts with proteasome and Cdc48, and is essential for surviving metalloids.
Conclusion: Ynl155w may protect cells from metalloid-induced proteotoxicity by delivering ubiquitinated proteins to Cdc48 and proteasome.
Significance: Ynl155w represents a novel stress response factor for misfolded proteins.
Keywords: Proteasome, Protein Degradation, Protein Misfolding, Stress Response, Ubiquitin
Abstract
Protein misfolding is a universal threat to cells. The ubiquitin-proteasome system mediates a cellular stress response capable of eliminating misfolded proteins. Here we identify Cuz1/Ynl155w as a component of the ubiquitin system, capable of interacting with both the proteasome and Cdc48. Cuz1/Ynl155w is regulated by the transcription factor Rpn4, and is required for cells to survive exposure to the trivalent metalloids arsenic and antimony. A related protein, Yor052c, shows similar phenotypes, suggesting a multicomponent stress response pathway. Cuz1/Ynl155w functions as a zinc-dependent ubiquitin-binding protein. Thus, Cuz1/Ynl155w is proposed to protect cells from metalloid-induced proteotoxicity by delivering ubiquitinated substrates to Cdc48 and the proteasome for destruction.
Introduction
Protein misfolding is toxic to cells and has numerous causes including heat, errors in translation, oxidation, genetic mutation, and aging. Heavy metals and metalloids (e.g. arsenic) are ubiquitous environmental toxins that cause protein misfolding by directly binding proteins and altering their structures. The recognition and elimination of these aberrant proteins is necessary for cell survival.
The ubiquitin-proteasome system is the major pathway for selective protein degradation in eukaryotic cells (1, 2). Substrates requiring destruction by this system are covalently modified by the small protein ubiquitin, which serves as a recognition motif for the proteasome (3). By harboring multiple enzymatic and non-enzymatic functionalities, the proteasome is able to recognize ubiquitinated substrates, and unfold, deubiquitinate, and proteolyze them into small peptides, while largely releasing ubiquitin for reuse.
Prior to destruction by the proteasome, many substrates require additional processing by the homohexameric ATPase Cdc48 (p97 in higher organisms). Cdc48 has numerous cellular functions, many of which require its central role as a force-generating machine (4). For example, Cdc48 can extract proteins from membranes, chromatin, or larger protein assemblies. Like the proteasome, Cdc48 interacts with numerous co-factors that endow it with multiple activities including ubiquitin binding, deubiquitination, and ubiquitination (5).
We describe here a new component of the ubiquitin system, Ynl155w, a zinc finger protein capable of directly interacting with the proteasome and Cdc48. Ynl155w is co-regulated with other elements of the ubiquitin-proteasome system, and its absence markedly compromises survival upon exposure to metalloid toxicity. A second uncharacterized zinc finger protein, Yor052c, shows similar phenotypic properties, suggesting a multicomponent stress response pathway. Ynl155w functions as a zinc-dependent ubiquitin-binding protein, suggesting the hypothesis that Ynl155w protects cells from metalloid toxicity by delivering ubiquitinated proteins to Cdc48 and the proteasome for destruction.
EXPERIMENTAL PROCEDURES
Strains and Plasmids
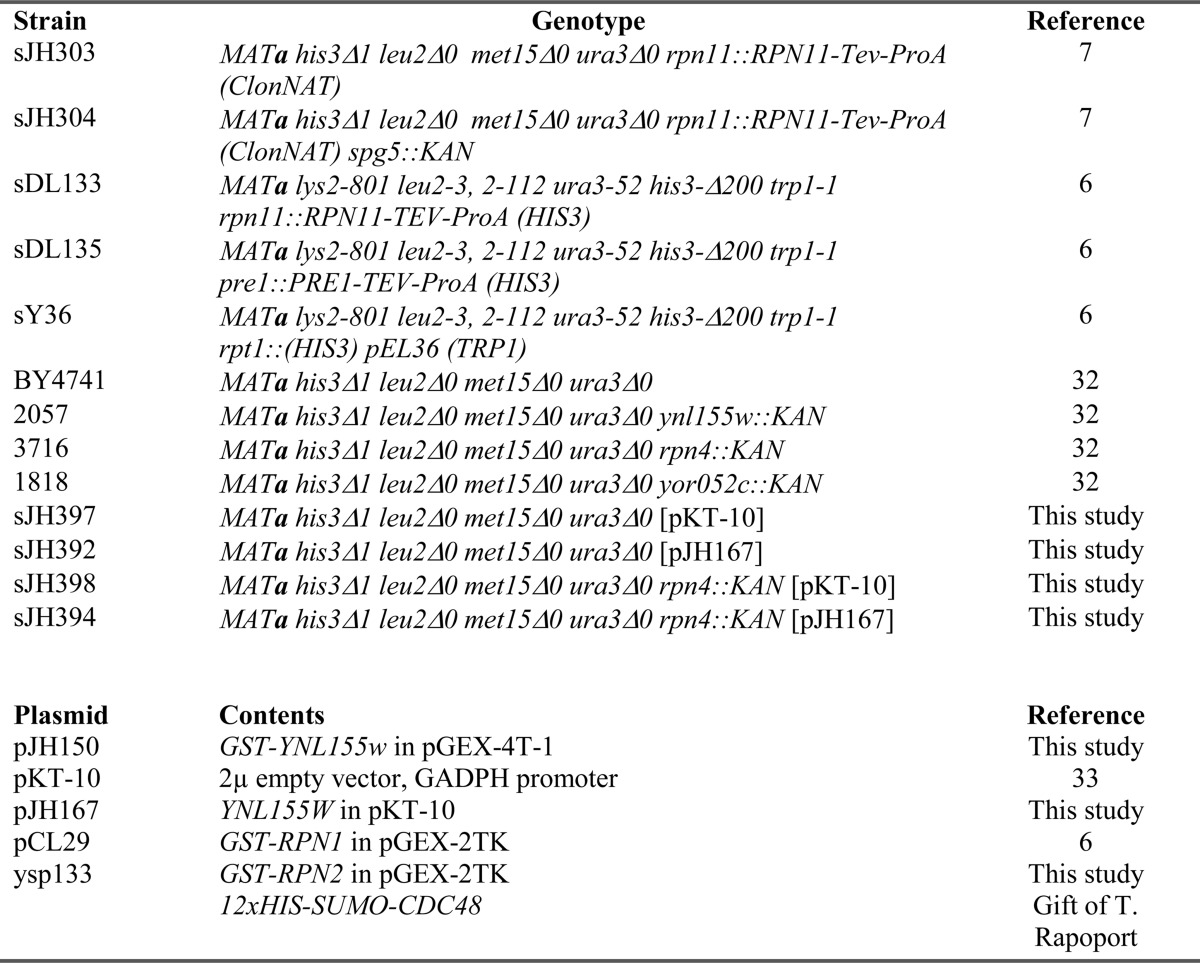
Yeast strains and plasmids are listed in Table 1. Yeast were cultured at 30 °C unless otherwise indicated. YPD medium consisted of 1% yeast extract, 2% Bacto-peptone, and 2% dextrose. Synthetic medium consisted of 0.7% Difco Yeast Nitrogen Base supplemented with amino acids, adenine, and 2% dextrose. Uracil was omitted from this medium for plasmid selection.
TABLE 1.
Strains and plasmids
Recombinant Proteins
GST fusions proteins were expressed in Escherichia coli and purified by glutathione-Sepharose affinity chromatography (6). Intact GST fusions were eluted with free glutathione; native proteins were generated by thrombin cleavage. For the experiments shown in Fig. 4, purifications were carried out in the presence of 1 mm zinc sulfate. His12-Sumo-Cdc48 (a gift of T. Rapoport) was expressed in E. coli, purified by nickel-nitrilotriacetic acid affinity chromatography, and eluted with imidazole.
FIGURE 4.
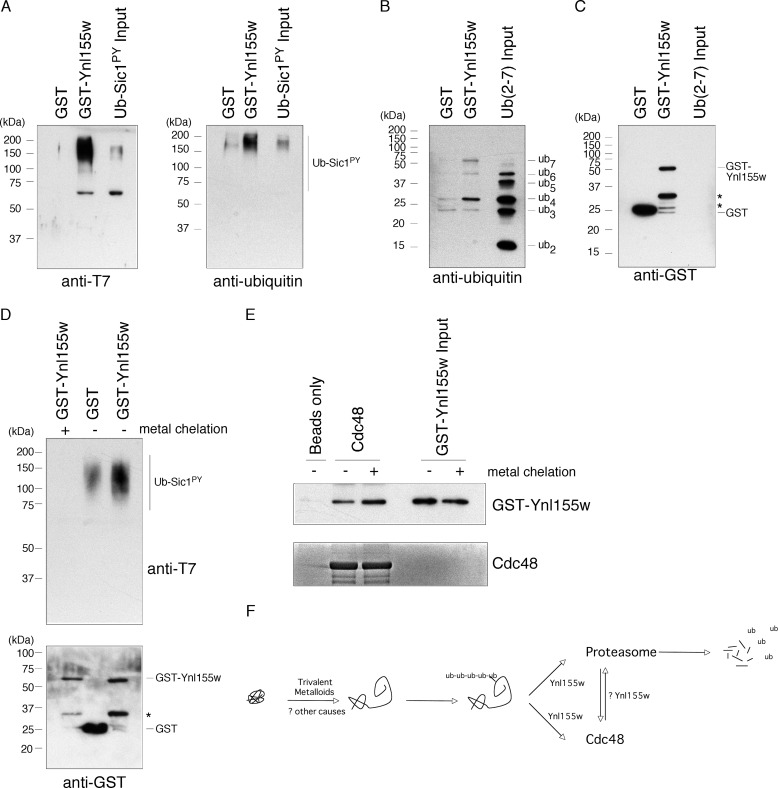
Ynl155w is a zinc-dependent ubiquitin-binding protein. A, binding of T7-tagged ubiquitinated Sic1PY to GST-Ynl155w, as visualized by anti-T7 (left panel) or anti-ubiquitin (right panel) SDS-PAGE immunoblots. Free GST serves as a negative control. B and C, binding of Lys-48-linked free ubiquitin chains (ub2-ub7, as indicated) to GST-Ynl155, as visualized by anti-ubiquitin immunoblot (B). Free GST serves as a negative control. Levels of immobilized GST and GST-Ynl155w were comparable, as visualized by anti-GST antibody (C). Asterisks, GST-Ynl155w breakdown products. D, metal chelation abrogates binding of GST-Ynl155w to ub-Sic1PY, as visualized by anti-T7 immunoblot (upper panel). Anti-GST-immunoblot (lower panel) confirms comparable amounts of GST fusion proteins. Asterisk, GST-Ynl155w breakdown product. E, metal chelation does not affect binding of GST-Ynl155w to Cdc48, as visualized by anti-Ynl155w immunoblot (upper panel). Coomassie staining of the eluates confirms comparable amounts of His12-Sumo-Cdc48 (lower panel). F, schematic illustration of Ynl155w function. Protein misfolding induced by trivalent metalloids, and perhaps other stimuli, results in an increase in ubiquitinated proteins (18). These ubiquitinated substrates are recognized by Ynl155w and delivered to the proteasome and Cdc48, ultimately resulting in their destruction. It remains unclear whether Ynl155w might shuttle between proteasome and Cdc48, or integrate their functions in another manner.
Proteasome and Cdc48 Purification
The proteasome and its subcomplexes were purified from yeast as described (6). Endogenous, genomically encoded TAP-tagged Cdc48 was isolated by IgG affinity chromatography (buffer 50 mm Tris-HCl, pH 7.5, 1 mm EDTA), washed with buffer supplemented with 0.5 m NaCl, and eluted by cleavage with TEV protease. Native gel analysis was carried out as described (7).
Mass Spectrometry
Gel bands were excised, digested with trypsin, and analyzed using a Q Exactive mass spectrometer. Spectra were processed using a Sequest-based in-house software pipeline. Database searching included all entries from the yeast Uniprot database.
Ynl155w Binding Assays
Proteasome, proteasome subcomplexes, and TAP-tagged endogenous Cdc48 were immobilized on IgG resin, challenged with purified Ynl155w or GST-Ynl155w, as indicated, washed with the appropriate purification buffer (see above) supplemented with 50 mm NaCl, and eluted by cleavage with TEV protease. GST fusion proteins were immobilized on glutathione-Sepharose resin, challenged with Ynl155w, washed in buffer supplemented with 50 mm NaCl, and eluted with free glutathione (30 mm). His12-Sumo-Cdc48 was immobilized on nickel-nitrilotriacetic acid resin, challenged with Ynl155w species, washed with buffer containing 50 mm NaCl, and eluted with imidazole.
Phenotypic Analysis of Yeast Mutants
Yeast spot assays were performed by serial dilution of overnight yeast cultures onto the indicated media, and culturing for the indicated periods of time. Sodium arsenite was dissolved in water; antimony trioxide was dissolved in hydrochloric acid.
Ubiquitin Binding Assays
His- and T7-epitope-tagged ubiquitinated Sic1PY (ub-Sic1PY) was prepared as described (8). Lys-48-linked free ubiquitin chains (ub2-ub7) were purchased from Boston Biochem. Equivalent amounts of GST or GST-Ynl155w (50 μg) were immobilized on glutathione-Sepharose resin in buffer (50 mm Tris-HCl, pH 7.5, 1 mm zinc sulfate), challenged with equivalent amounts of ub-Sic1PY (8 nm) or ubiquitin chains (5 μg), washed with buffer supplemented with 50 mm NaCl, and eluted with free glutathione. Metal chelation was carried out for 30 min at 4 °C with 1,10-phenanthroline (5 mm) and EDTA (5 mm).
Antibodies
A polyclonal Ynl155w antibody was raised in rabbits against recombinant GST-Ynl155w (ProteinTech Group). Rpn8 antibodies have been previously described (9). Ubiquitin (Santa Cruz), T7 (Novagen), GST (Santa Cruz), and Pgk1 (Invitrogen) antibodies were commercially available.
RESULTS
Ynl155w Interacts with the Proteasome
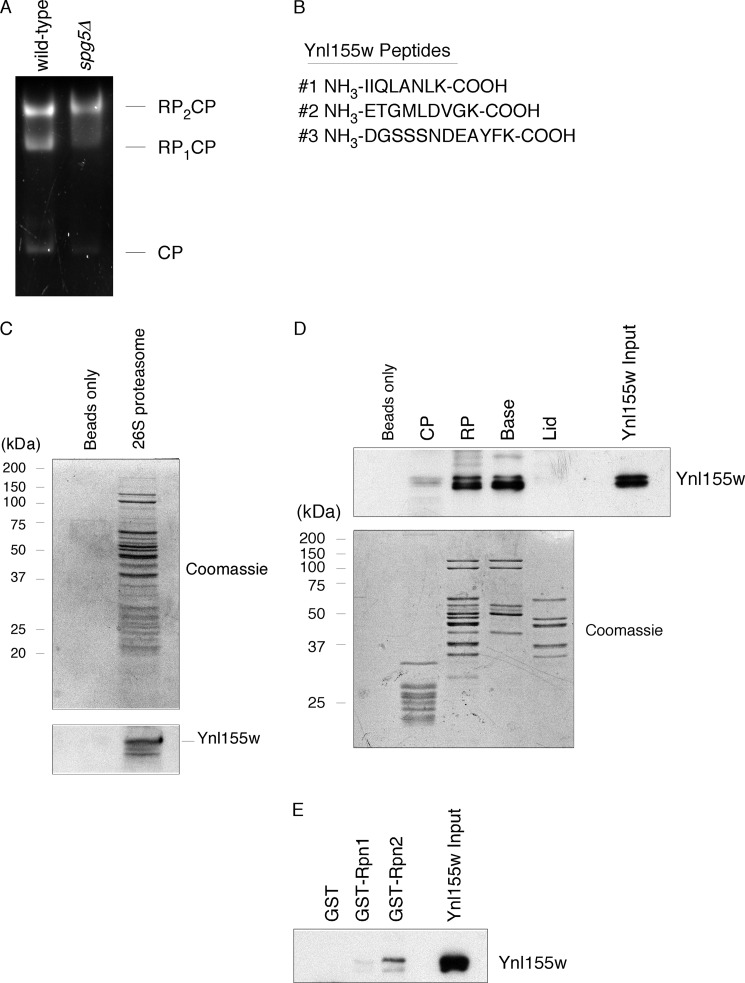
We previously identified Spg5 as a novel regulator of the proteasome under conditions of nutrient starvation (7). During that work, we isolated proteasomes from quiescent wild-type and spg5Δ cells using an affinity purification technique (6), and subjected them to additional purification by native gel electrophoresis. We visualized proteolytically active species using the fluorogenic substrate suc-LLVY-AMC (Fig. 1A), and proteolytically inactive species by immunoblot (7). To identify components of each proteasome subcomplex, we excised individual native gel bands and analyzed them by mass spectrometry. Within spg5Δ proteasomes, but not those of wild-type, we obtained three unique peptides (four in total) from the protein Ynl155w, now known as Cuz1 (Fig. 1B). Two of the peptides were derived from the RP2CP species (Fig. 1A), and two from the RP species (data not shown).
FIGURE 1.
Interaction of Ynl155w with the proteasome. A, native gel electrophoresis of purified wild-type and spg5Δ proteasomes (3 μg), followed by in-gel activity assay with the fluorogenic proteasome substrate, suc-LLVY-AMC. B, proteasomes from panel A were excised from the gel and analyzed by mass spectrometry, yielding three peptides from Ynl155w. C, interaction of purified Ynl155w (2.3 μg) with purified 26 S proteasome (5 μg). Upper panel, Coomassie-stained SDS-PAGE gel. Lower panel, anti-Ynl155w SDS-PAGE immunoblot. D, interaction of purified Ynl155w (70 μg) with RP and base subcomplexes of the proteasome (3 nmol for each subcomplex). Upper panel, anti-Ynl155w immunoblot. Lower panel, Coomassie-stained gel. Input, 0.14%; eluates, 2.2% of total. E, interaction of purified Ynl155w (40 μg) with proteasome subunit Rpn2 (5.3 μg), as visualized by anti-Ynl155w immunoblot. Coomassie staining of eluates confirmed comparable amounts of GST, GST-Rpn1, and GST-Rpn2 (data not shown). Input, 1%; eluates, 2.2% of total.
Ynl155w is a 32-kDa protein that contains an AN1-type zinc finger domain at its N terminus. This protein shows homology to the mammalian proteins AIRAP3 (arsenite inducible RNA-associated protein) and AIRAP-like protein, as well as Aip-1, which is required for longevity in Caenorhabditis elegans (10, 11). Of note, AIRAP has been shown to bind proteasomes (10), although its precise molecular function remains unclear.
We began by confirming the association of Ynl155w with the proteasome. We isolated proteasomes from wild-type cells growing logarithmically, and challenged them with recombinant, purified Ynl155w. We were able to detect binding of Ynl155w to proteasomes, whereas there was no binding to beads alone (Fig. 1C). Thus, Ynl155w can interact, at least in vitro, with proteasomes from growing as well as quiescent cells.
The proteasome is a 2.5-MDa complex composed of 33 distinct subunits, and can be divided into two subcomplexes: the 28-subunit core particle (CP), which houses the proteolytic active sites, and the 19-subunit regulatory particle (RP), which is thought to recognize, unfold, and deubiquitinate substrates (3). The RP can be subdivided into two complexes: base and lid. Using affinity tags on CP, base, and lid subunits, we purified all four proteasome subcomplexes, and again challenged them with Ynl155w. Binding was observed with the RP but not the CP (Fig. 1D). Within the RP, Ynl155w bound to the base but not the lid. The base includes six ATPases (Rpt1–6) as well as two larger related proteins Rpn1 and Rpn2, which show ∼40% sequence identity (12). To test whether Rpn1 or Rpn2 could serve as a receptor for Ynl155w, we isolated recombinant full-length GST-tagged Rpn1 and Rpn2 on resin and challenged these resins with Ynl155w. Binding was detected with GST-Rpn2, but not with GST-Rpn1 or GST alone (Fig. 1E). We were unable to exclude a secondary binding site within one of the six ATPases because of an inability to purify these proteins from bacteria. These results indicate that Ynl155w is a proteasome-associated protein, and specifically recognizes the Rpn2 subunit present within the RP and the base subcomplexes.
Ynl155w Is a Cdc48 Interacting Protein
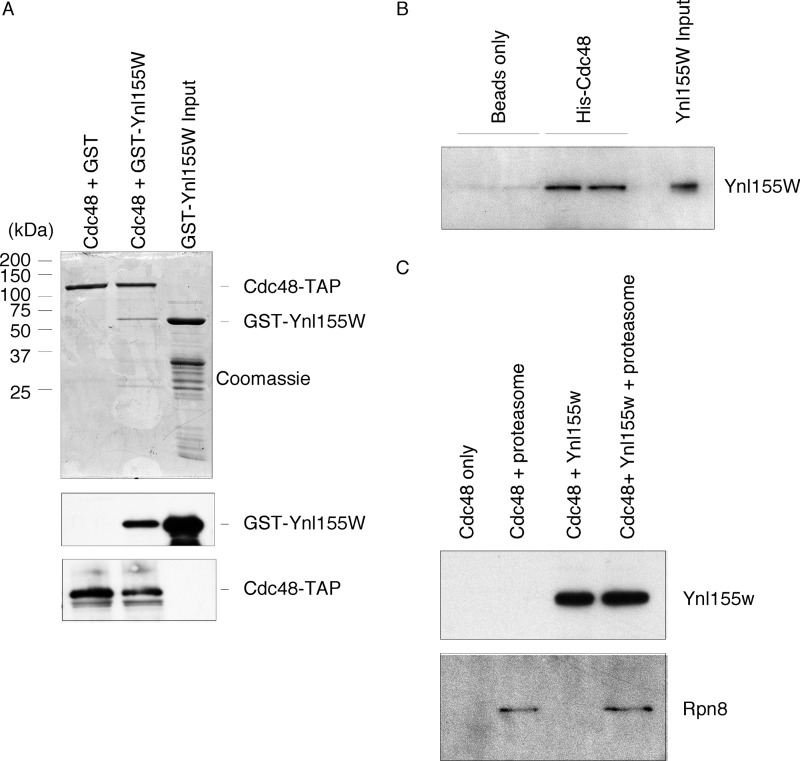
Proteome-wide protein interaction studies suggested that Ynl155w might also interact with the homohexameric ATPase Cdc48 (13). This was of interest as Cdc48, among other roles, is known to function within the ubiquitin-proteasome pathway. To test this possible association, we isolated Cdc48 from yeast cells using a genomically encoded, TAP-tagged Cdc48. Because Cdc48 can associate with a host of factors, we used stringent wash conditions to strip away as many interacting proteins as possible. We then challenged this resin with GST or GST-Ynl155w. We were able to detect binding to GST-Ynl155w, but not GST (Fig. 2A). Indeed, evaluation of the eluates indicated a Coomassie-stainable band for GST-Ynl155w, indicative of strong binding. To confirm that this binding was direct, we utilized purified recombinant His12- and Sumo-tagged Cdc48 that had been expressed in E. coli. Ynl155w retained the capacity to bind recombinant Cdc48 (Fig. 2B). Thus, Ynl155w is both a proteasome- and a Cdc48-interacting protein.
FIGURE 2.
Interaction of Ynl155w with Cdc48. A, GST-Ynl155w (58 μg) interacts with purified endogenous TAP-tagged Cdc48. Upper panel, Coomassie-stained SDS-PAGE gel. Middle panel, anti-GST SDS-PAGE immunoblot. Lower panel, anti-TAP tag immunoblot. B, direct interaction of recombinant purified Ynl155w (60 μg) and His12-Sumo-Cdc48 proteins (30 μg) (in duplicate), as visualized by anti-Ynl155w immunoblot. C, failure to detect a ternary complex between Cdc48 (60 μg), Ynl155w (180 μg), and proteasomes (30 μg). Upper panel, anti-Ynl155w immunoblot. Lower panel, anti-Rpn8 immunoblot.
The ability of Ynl155w to interact with both the proteasome and Cdc48 suggested three models for its function: 1) Ynl155w might bring proteasome and Cdc48 together in a stable ternary complex; 2) Ynl155w might have completely separate functions within the two complexes; and 3) Ynl155w might integrate proteasome and Cdc48 function without formation of a ternary complex, for example, by interacting with them sequentially. To begin to explore these options, we sought to determine whether a proteasome-Ynl155w-Cdc48 ternary complex could be visualized. To do this, we isolated recombinant purified Cdc48 on resin, and added Ynl155w or proteasome holoenzyme (RP2CP) or both. Again, we were able to detect significant binding of Ynl155w to Cdc48 (Fig. 2C). However, addition of proteasome and Ynl155w together did not increase recruitment of proteasome to Cdc48 (Fig. 2C). We also note that, at least under these relatively stringent conditions, there was only minimal interaction between the highly purified proteasome holoenzyme and Cdc48. We next performed the inverse of this experiment, isolating purified proteasomes on resin, and adding back Ynl155w, Cdc48, or both. Again, we were unable to detect recruitment of Cdc48 to proteasome holoenzymes (data not shown). These results disfavor model one, but do not distinguish between models two or three. Accordingly, we consider it unlikely that Ynl155w serves to recruit the proteasome and Cdc48 into a stable complex.
Ynl155w is Co-regulated with Ubiquitin-Proteasome Pathway Components
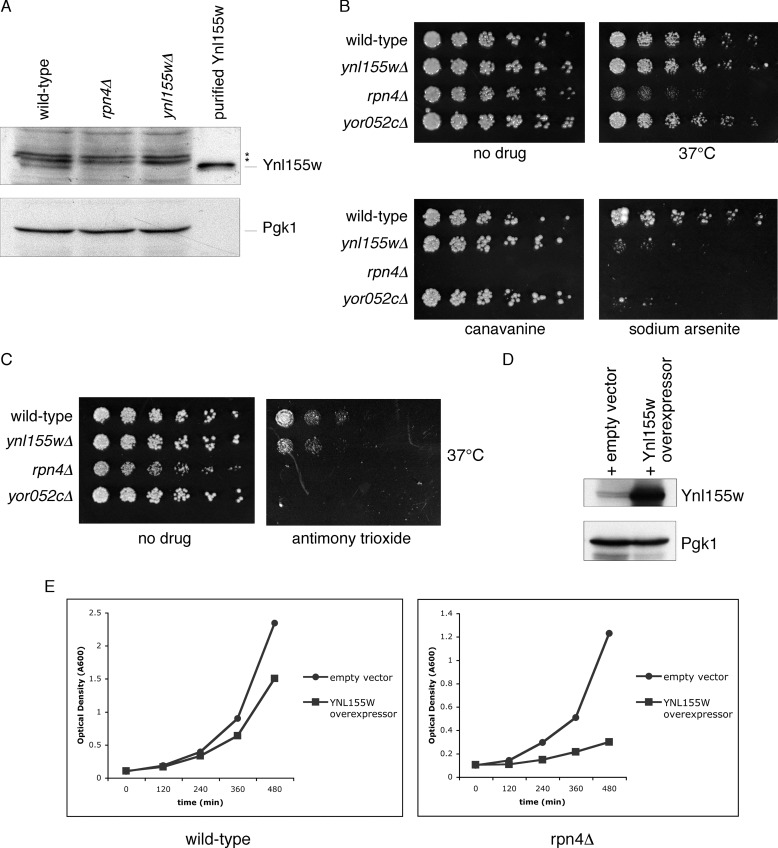
The transcription factor Rpn4 is a master regulator of the ubiquitin-proteasome pathway. It induces the genes encoding all known proteasome subunits as well as several other ubiquitin system components, including Cdc48. Rpn4 recognizes a nine-base sequence in the promoters of these genes, termed the PACE motif (proteasome-associated control element) (14). Moreover, Rpn4 is itself a highly unstable proteasome substrate, creating a homeostatic feedback loop whereby defects in proteasome-mediated degradation result in stabilization of Rpn4, and increased synthesis of ubiquitin-proteasome components until Rpn4 levels are normalized (15–16). A canonical PACE sequence within the 5′-untranslated region of YNL155W is located between residues −191 and −183. To determine whether Rpn4 controls Ynl155w protein levels, we prepared whole cell extracts from wild-type, rpn4Δ, and ynl155Δ cells, and evaluated them by immunoblotting. As shown in Fig. 3A, Ynl155w levels were markedly reduced in the absence of Rpn4, indicating that, at least under normal growth conditions, Rpn4 is the major regulator of Ynl155w levels.
FIGURE 3.
A physiologic role for Ynl155w in the ubiquitin-proteasome system. A, Ynl155w protein levels are dependent on the transcription factor Rpn4. Whole cell extracts from wild-type, rpn4Δ, and ynl155wΔ cells were evaluated by SDS-PAGE immunoblot with antibodies against Ynl155w (upper panel) and Pgk1 loading control (lower panel). Recombinant purified Ynl155w is shown for reference. Asterisks, nonspecific bands. B, ynl155wΔ and yor052cΔ mutants are highly sensitive to arsenic. Three-fold serial dilutions of wild-type, ynl155wΔ, rpn4Δ, and yor052cΔ were spotted onto YPD plates containing no drug, the amino acid canavanine (1.5 μg/ml), and sodium arsenite (1.5 mm), and cultured at 30 or 37 °C for 2–6 days, as indicated. C, ynl155wΔ and yor052cΔ mutants are sensitive to antimony. Three-fold serial dilutions of wild-type, ynl155wΔ, rpn4Δ, and yor052cΔ were spotted onto YPD plates containing no drug or antimony trioxide (0.5 mm) and cultured at 37 °C for 2 days. D, a YNL155W overexpression plasmid increases Ynl155w protein levels. Upper panel, anti-Ynl155w immunoblot. Lower panel, anti-Pgk1 immunoblot (loading control). E, growth inhibition caused by YNL155W overexpression is enhanced in rpn4Δ cells. Wild-type and rpn4Δ cells expressing either an empty vector or a YNL155W overexpression plasmid were cultured in logarithmic phase growth at 30 °C, and culture optical density was assayed at the indicated time points.
Ynl155w Protects Cells from Metalloid-induced Proteotoxicity
The preceding data suggested a role for Ynl155w in protein degradation. We therefore sought in vivo phenotypes for the ynl155Δ mutant that could confirm a physiologic role for Ynl155w in the ubiquitin-proteasome system. Protein misfolding and its associated toxicity, sometimes termed proteotoxicity, can be induced by a variety of stimuli, including heat, abnormal amino acids, translation errors, oxidation, and genetic mutation. We started with two well studied causes of protein misfolding, the abnormal amino acid canavanine, which causes misfolding upon incorporation into nascent proteins, and elevated temperature, which globally affects protein stability. Although it is difficult to definitively exclude a phenotype for any mutant, we were unable to identify a phenotype for the ynl155Δ mutant under these conditions (Fig. 3B). By contrast, the rpn4Δ mutant showed hypersensitivity to both treatments (Fig. 3B). Similarly, we were unable to detect a phenotype for ynl155Δ mutants when exposed to the translation inhibitor cycloheximide or the DNA damaging agents 4-nitroquinolone 1-oxide and hydroxyurea, all conditions under which the rpn4Δ mutant was highly compromised for growth (data not shown).
Because Ynl155w shows sequence homology to the mammalian protein AIRAP, which was initially identified as an arsenite-inducible protein (10), we examined the effect of sodium arsenite on the ynl155Δ mutant. Indeed, this compound proved to be profoundly inhibitory to growth of the ynl155Δ mutant (Fig. 3B). The rpn4Δ mutant showed even greater sensitivity to arsenite (Fig. 3B and data not shown). Importantly, the toxicity of arsenic, particularly in its trivalent form as in sodium arsenite, is thought to reflect the ability of arsenite to cause protein misfolding. Trivalent arsenic causes protein misfolding in vitro and in vivo, induces molecular chaperone function, increases levels of ubiquitinated proteins, and can cause protein aggregation (17, 18). Arsenite is thought to effect protein misfolding by directly binding proteins, and altering their structures (19).
Arsenic shares properties with heavy metals. We therefore examined cadmium chloride, nickel sulfate, and cobalt chloride. The rpn4Δ mutant showed marked sensitivity to all three compounds, but ynl155Δ did not (data not shown). Because all three metals were present as divalent cations, we wondered if the trivalence of arsenite was critical to its toxicity to the ynl155Δ mutant. To test this, we utilized the metalloid antimony in its trivalent state, and detected a growth defect in the ynl155Δ mutant (Fig. 3C). The antimony phenotype was less pronounced than with arsenic, so we repeated the assay in liquid culture form. Mock treated wild-type and ynl155Δ cells showed similar doubling times (1.5 h for both); in the presence of antimony trioxide, ynl155Δ cells showed a longer doubling time (2.1 versus 2.4 h; data not shown). Thus, Ynl155w shows remarkable specificity in protecting cells from trivalent metalloids.
Yor052c, a Ynl155w-related Protein, Also Protects Cells from Metalloid Toxicity
Yor052c is a 17-kDa uncharacterized zinc finger protein that shows sequence homology to Ynl155w, particularly within the zinc finger domain. In addition, the YOR052C promoter contains a PACE motif for Rpn4 recognition, as noted above for YNL155W. We therefore examined the same phenotypes in the yor052cΔ mutant. We observed that the yor052cΔ mutant behaved similarly to ynl155wΔ, showing the same pattern of growth inhibition with arsenic and antimony, but not with other stressors of the ubiquitin system (Fig. 3, B and C). Importantly, despite the similarity of the two proteins, the strong phenotype of each mutant alone implies a high degree of non-redundancy in their cellular functions.
YNL155W Overexpression Is Toxic to Cells
We next examined the consequences of overexpressing YNL155W by utilizing a YNL155W plasmid driven by the GADPH promoter. Evaluation of cell lysates confirmed a strong increase in Ynl155w protein levels (Fig. 3D). We evaluated the effect of this plasmid on the growth rates of wild-type and rpn4Δ strains. As shown in Fig. 3E, YNL155W overexpression slowed growth in the wild-type strain under normal culture conditions. The magnitude of growth inhibition was greatly increased in the rpn4Δ mutant: doubling time increased by 20% in the wild-type (1.7 versus 2.1 h), but by 136% in the rpn4Δ mutant (2.2 versus 5.2 h). This toxic effect of YNL155W overexpression indicates that tight control of Ynl155w levels is important for cellular function. Phenotypic exacerbation in the rpn4Δ background implies that proper function of Ynl155w occurs within the context of the concerted cellular stress response mediated by Rpn4.
Ynl155w Is a Zinc-dependent Ubiquitin-binding Protein
Ynl155w contains an AN1-type zinc finger domain at its N terminus. This domain, consisting of six cysteines and two histidines, has potential to coordinate two zinc ions. Although zinc finger proteins were originally described as nucleic acid-binding proteins, it is now clear that they can perform a variety of cellular functions. Of note, the zinc finger domain is currently thought to represent the second most common motif found in ubiquitin-binding proteins (20). We therefore sought to test whether Ynl155w might be a novel ubiquitin-binding protein. We began by generating polyubiquitinated Sic1PY (ub-Sic1PY) in vitro. This substrate is competent for recognition, deubiquitination, and degradation by purified proteasomes (21, 8). We immobilized purified GST-Ynl155w on resin, and challenged it with ub-Sic1PY. We observed strong binding of this substrate to GST-Ynl155w; by contrast, free GST showed little or no binding (Fig. 4A). To confirm this result, we chose an unrelated substrate: free ubiquitin chains ranging in size from ub2 to ub7 and linked through lysine 48. There was little or no binding of diubiquitin or triubiquitin to GST-Ynl155w (Fig. 4B). However, tetraubiquitin and longer chains showed significant binding to GST-Ynl155w. Note that ub7, which is not an abundant component of the input material, is highly concentrated in the GST-Ynl155w-bound material (Fig. 4B). That GST-Ynl155w prefers ubiquitinated species with four or more ubiquitin molecules is interesting given that a four-ubiquitin chain has been proposed as the minimum chain length necessary for recognition by the proteasome (22). Levels of GST and GST-Ynl155w were comparable in these experiments (Fig. 4C). Finally, we tested a third substrate consisting of a single ubiquitin fused to green fluorescent protein (ub-m-GFP) (23). GST-Ynl155w appeared to show no specific binding to this substrate, consistent with the apparent preference of Ynl155w for polyubiquitinated substrates (data not shown).
Although many zinc finger proteins coordinate zinc ions, some coordinate other metals, and some do not bind metal at all. To evaluate the importance of zinc binding for Ynl155w function, we immobilized purified GST-Ynl155w on resin, and then split the resin sample, maintaining one-half in zinc-containing buffer and briefly treating the other half with the metal chelators 1,10-phenanthroline and EDTA. Mock-treated GST-Ynl155w again bound ub-Sic1PY (Fig. 4D). By contrast, the metal-chelated GST-Ynl155w was essentially devoid of the ubiquitin binding function. To test if this effect was specific, we applied control or metal-chelated GST-Ynl155w to immobilized His12-Sumo-Cdc48. Metal-chelated GST-Ynl155w was fully competent to bind Cdc48 (Fig. 4E). Taken together, these results indicate that Ynl155w is a novel zinc-dependent ubiquitin-binding protein.
DISCUSSION
Ynl155w Mediates a Novel Stress Response
All cells are exposed to environmental stresses, and the ability to respond to these insults is critical for survival. The ubiquitin-proteasome system, by virtue of its ability to destroy proteins, plays a central role in responding to stress-induced protein misfolding. In yeast, the transcription factor Rpn4 coordinates a broad stress response pathway to deal with misfolded proteins. Because Rpn4 is itself a substrate of the proteasome, conditions that compromise proteasome-mediated degradation stimulate coordinated synthesis of ubiquitin pathway components until the protein degrading capacity of the cell is restored (15). Furthermore, the RPN4 gene is itself subject to complex transcriptional regulation, suggesting that Rpn4 may respond to numerous cellular inputs (24–25). Feedback control of proteasome function is seen in mammals as well as yeast, although the principal mediator of the response in higher organisms, Nrf1, is not related to Rpn4 by sequence (26, 27).
We have identified a novel arm of the Rpn4-mediated stress response that protects cells from protein misfolding induced by metalloid compounds. Under these conditions, Ynl155w is essential for cell survival. Ynl155w is a zinc-dependent ubiquitin-binding protein and interacts with both the proteasome and Cdc48. Thus, Ynl155w appears to protect cells from metalloid-induced protein misfolding by delivering ubiquitinated substrates to Cdc48 and the proteasome, thereby facilitating their destruction (Fig. 4F). Sá-Moura and colleagues (28) have also recently characterized Ynl155w, and their results are concordant with some our findings, specifically that Ynl155w interacts with the proteasome and Cdc48, and they have named the protein Cuz1 (Cdc48-associated ubiquitin-like domain/zinc finger protein-1). The various functions of Ynl155w appear to be distributed across the protein. We find that ubiquitin binding requires the N-terminal zinc finger. By contrast, Sá-Moura et al. (28) show that a C-terminal ubiquitin-like domain is necessary for Cdc48 binding. Zinc chelation did not affect Cdc48 binding (Fig. 4E), which is consistent with this model. Precise localization of the proteasome binding site remains to be determined. Finally, recent reports suggest that Cdc48 may directly interact with the CP of the proteasome (29, 30). However, in our experiments, CP is associated with RP, which occludes the potential binding site for Cdc48, and thus precludes evaluation of CP-Cdc48 interaction.
We also report here the first phenotypic characterization of Yor052c, a zinc finger protein with sequence homology to Ynl155w. Like YNL155W, YOR052C contains a PACE motif within its promoter (data not shown) (28), suggesting that it is also regulated by Rpn4. We find that yor052cΔ mutants show a similar phenotypic profile as ynl155wΔ mutants: both are essential for survival when cells are treated with metalloid compounds, but apparently dispensable for growth under several other conditions that cause protein misfolding. These overlapping phenotypic profiles, in conjunction with their sequence similarity, suggest a common function for the two proteins, but the strong phenotype of each mutant is consistent with significant non-redundant roles. We propose that Yor052c be named Tmc1 for trivalent metalloid sensitive, Cuz1-related protein-1.
Ynl155w/Cuz1 Is a Novel Ubiquitin Receptor
Our data add Ynl155w to a growing list of ubiquitin-binding proteins. The sheer number of ubiquitin receptors indicates a high degree of complexity in recognizing and processing ubiquitinated substrates. Ynl155w is distinguished by its ability to interact with both Cdc48 and the proteasome, its regulation by Rpn4, and its highly specific phenotypic profile. These data suggest that Ynl155w may recognize only a subset of misfolded ubiquitinated proteins. At a minimum, Ynl155w appears essential for surviving metalloid toxicity, suggesting that proteins damaged by metalloids may represent its preferred substrates. Identifying the spectrum of Ynl155w substrates, as well as the physical and chemical determinants for recognition by Ynl155w, represent important goals. It is worth noting that one of the two mammalian proteins related to Ynl155w, AIRAP-L, contains two ubiquitin-interacting motifs (11). Although this has not yet been tested experimentally, these findings suggest that the ubiquitin binding function of Ynl155w may be conserved in higher organisms.
Within the proteasome, Rpn1 and Rpn2 appear to serve as a scaffold for a variety of substrate processing factors (6, 12, 31). Rpn1 binds the ubiquitin receptors Rad23 and Dsk2, and the deubiquitinating enzyme Ubp6. Rpn2 binds the ubiquitin receptor Rpn13 and the ubiquitinating enzyme Hul5. Thus the interaction of Ynl155w with Rpn2 suggests that Ynl155w is well positioned to deliver substrates to the proteasome in a manner that allows for further processing prior to destruction.
Trivalent Metalloids and Human Disease
Ynl155w and Yor052c protect cells from the toxicity of trivalent metalloids. Although the two proteins are likely to function in other cellular contexts, their specificity for trivalent metalloids suggests that they may provide insight into the nature and function of these medically relevant compounds. Acute promyelocytic leukemia is caused by a cytogenetic translocation resulting in the fusion protein PML-RARα. Trivalent arsenic has helped transform treatment of this disease: by directly binding PML-RARα, arsenic induces its misfolding and subsequent proteasomal destruction, eliminating this oncogene (19). Antimony is used to treat cutaneous leishmaniasis, although the molecular basis for its effect is not known. The possibility exists that metalloids and heavy metals, by virtue of their ability to alter protein homeostasis, may have other clinically useful properties. A better understanding of the nature of their toxicity as well as their associated cellular response pathways may allow for the design of novel therapies.
Acknowledgments
We thank Tom Rapoport, G. W. Hwang, and Akira Naganuma for plasmids.
This work was supported by National Institutes of Health Grant GM043601 (to D. F.).
- AIRAP
- arsenite inducible RNA-associated protein
- CP
- core particle
- RP
- regulatory particle
- PACE
- proteasome-associated control element.
REFERENCES
- 1. Finley D., Ulrich H. D., Sommer T., Kaiser P. (2012) The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics 192, 319–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Varshavsky A. (2012) The ubiquitin system, an immense realm. Annu. Rev. Biochem. 81, 167–176 [DOI] [PubMed] [Google Scholar]
- 3. Matyskiela M. E., Martin A. (2013) Design principles of a universal protein degradation machine. J. Mol. Biol. 425, 199–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buchberger A. (2013) Roles of cdc48 in regulated protein degradation in yeast. Subcell Biochem. 66, 195–222 [DOI] [PubMed] [Google Scholar]
- 5. Stolz A., Hilt W., Buchberger A., Wolf D. H. (2011) Cdc48. A power machine in protein degradation. Trends Biochem. Sci. 36, 515–523 [DOI] [PubMed] [Google Scholar]
- 6. Leggett D. S., Hanna J., Borodovsky A., Crosas B., Schmidt M., Baker R. T., Walz T., Ploegh H., Finley D. (2002) Multiple associated proteins regulate proteasome structure and function. Mol. Cell 10, 495–507 [DOI] [PubMed] [Google Scholar]
- 7. Hanna J., Waterman D., Boselli M., Finley D. (2012) Spg5 protein regulates the proteasome in quiescence. J. Biol. Chem. 287, 34400–34409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee B. H., Lee M. J., Park S., Oh D. C., Elsasser S., Chen P. C., Gartner C., Dimova N., Hanna J., Gygi S. P., Wilson S. M., King R. W., Finley D. (2010) Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 467, 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hanna J., Hathaway N. A., Tone Y., Crosas B., Elsasser S., Kirkpatrick D. S., Leggett D. S., Gygi S. P., King R. W., Finley D. (2006) Deubiquitinating enzyme ubp6 functions noncatalytically to delay proteasomal degradation. Cell 127, 99–111 [DOI] [PubMed] [Google Scholar]
- 10. Stanhill A., Haynes C. M., Zhang Y., Min G., Steele M. C., Kalinina J., Martinez E., Pickart C. M., Kong X. P., Ron D. (2006) An arsenite-inducible 19S regulatory particle-associated protein adapts proteasomes to proteotoxicity. Mol. Cell 23, 875–885 [DOI] [PubMed] [Google Scholar]
- 11. Yun C., Stanhill A., Yang Y., Zhang Y., Haynes C. M., Xu C. F., Neubert T. A., Mor A., Philips M. R., Ron D. (2008) Proteasomal adaptation to environmental stress links resistance to proteotoxicity with longevity in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 105, 7094–7099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rosenzweig R., Bronner V., Zhang D., Fushman D., Glickman M. H. (2012) Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome. J. Biol. Chem. 287, 14659–14671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrín-Alvarez J. M., Shales M., Zhang X., Davey M., Robinson M. D., Paccanaro A., Bray J. E., Sheung A., Beattie B., Richards D. P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M. M., Vlasblom J., Wu S., Orsi C., Collins S. R., Chandran S., Haw R., Rilstone J. J., Gandi K., Thompson N. J., Musso G., St Onge P., Ghanny S., Lam M. H., Butland G., Altaf-Ul A. M., Kanaya S., Shilatifard A., O'Shea E., Weissman J. S., Ingles C. J., Hughes T. R., Parkinson J., Gerstein M., Wodak S. J. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643 [DOI] [PubMed] [Google Scholar]
- 14. Mannhaupt G., Schnall R., Karpov V., Vetter I., Feldmann H. (1999) Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 450, 27–34 [DOI] [PubMed] [Google Scholar]
- 15. Xie Y., Varshavsky A. (2001) RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome. A negative feedback circuit. Proc. Natl. Acad. Sci. U.S.A. 98, 3056–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang X., Xu H., Ha S. W., Ju D., Xie Y. (2010) Proteasomal degradation of Rpn4 in Saccharomyces cerevisiae is critical for cell viability under stressed conditions. Genetics 184, 335–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jacobson T., Navarrete C., Sharma S. K., Sideri T. C., Ibstedt S., Priya S., Grant C. M., Christen P., Goloubinoff P., Tamás M. J. (2012) Arsenite interferes with protein folding and triggers formation of protein aggregates in yeast. J. Cell Sci. 125, 5073–5083 [DOI] [PubMed] [Google Scholar]
- 18. Kirkpatrick D. S., Dale K. V., Catania J. M., Gandolfi A. J. (2003) Low-level arsenite causes accumulation of ubiquitinated proteins in rabbit renal cortical slices and HEK293 cells. Toxicol. Appl. Pharmacol. 186, 101–109 [DOI] [PubMed] [Google Scholar]
- 19. Zhang X. W., Yan X. J., Zhou Z. R., Yang F. F., Wu Z. Y., Sun H. B., Liang W. X., Song A. X., Lallemand-Breitenbach V., Jeanne M., Zhang Q. Y., Yang H. Y., Huang Q. H., Zhou G. B., Tong J. H., Zhang Y., Wu J. H., Hu H. Y., de Thé H., Chen S. J., Chen Z. (2010) Arsenic trioxide controls the fate of the PML-RARα oncoprotein by directly binding PML. Science 328, 240–243 [DOI] [PubMed] [Google Scholar]
- 20. Randles L., Walters K. J. (2012) Ubiquitin and its binding domains. Front. Biosci. 17, 2140–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Saeki Y., Isono E., Toh-E A. (2005) Preparation of ubiquitinated substrates by the PY motif-insertion method for monitoring 26S proteasome activity. Methods Enzymol. 399, 215–227 [DOI] [PubMed] [Google Scholar]
- 22. Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J. 19, 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guterman A., Glickman M. H. (2004) Complementary roles for Rpn11 and Ubp6 in deubiquitination and proteolysis by the proteasome. J. Biol. Chem. 279, 1729–1738 [DOI] [PubMed] [Google Scholar]
- 24. Hahn J. S., Neef D. W., Thiele D. J. (2006) A stress regulatory network for co-ordinated activation of proteasome expression mediated by yeast heat shock transcription factor. Mol. Microbiol. 60, 240–251 [DOI] [PubMed] [Google Scholar]
- 25. Owsianik G., Balzil L., Ghislain M. (2002) Control of 26S proteasome expression by transcription factors regulating multidrug resistance in Saccharomyces cerevisiae. Mol. Microbiol. 43, 1295–1308 [DOI] [PubMed] [Google Scholar]
- 26. Radhakrishnan S. K., Lee C. S., Young P., Beskow A., Chan J. Y., Deshaies R. J. (2010) Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell 38, 17–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Steffen J., Seeger M., Koch A., Krüger E. (2010) Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Mol. Cell 40, 147–158 [DOI] [PubMed] [Google Scholar]
- 28. Sá-Moura B., Funakoshi M., Tomko R. J., Jr., Dohmen R. J., Wu Z., Peng J., Hochstrasser M. (2013) A conserved protein with AN1 zinc-finger and ubiquitin-like domains modulates Cdc48 (p97) function in the ubiquitin-proteasome pathway. J. Biol. Chem. 288, 33682–33696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barthelme D., Sauer R. T. (2013) Bipartite determinants mediate an evolutionarily conserved interaction between Cdc48 and the 20S peptidase. Proc. Natl. Acad. Sci. U.S.A. 110, 3327–3332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barthelme D., Sauer R. T. (2012) Identification of the Cdc48·20S proteasome as an ancient AAA+ proteolytic machine. Science 337, 843–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Elsasser S., Gali R. R., Schwickart M., Larsen C. N., Leggett D. S., Müller B., Feng M. T., Tübing F., Dittmar G. A., Finley D. (2002) Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell Biol. 4, 725–730 [DOI] [PubMed] [Google Scholar]
- 32. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 33. Hwang G. W., Ishida Y., Naganuma A. (2006) Identification of F-box proteins that are involved in resistance to methylmercury in Saccharomyces cerevisiae. FEBS Lett. 580, 6813–6818 [DOI] [PubMed] [Google Scholar]