Abstract
Background: The newly identified metastasis-associated in colon cancer-1 (MACC1) gene is involved in angiogenesis, epithelial-to-mesenchymal transition (EMT), invasiveness, and metastasis in a variety of malignancies. Overexpression of MACC1 gene is a prognostic marker for poor outcome of hepatocellular carcinoma (HCC) patients. However, the association between genetic polymorphisms of MACC1 gene and poor outcome in HCC has been not been performed. We therefore investigated the correlation of MACC1 single nucleotide polymorphisms (SNPs) with tumor recurrence and overall survival in HCC patients undergoing liver transplantation (LT). Methods: The study included 187 HCC patients treated with LT. Five polymorphisms in the MACC1 gene (rs1990172, rs3735615, rs4721888, rs2241056, rs975263) were genotyped in 183 cases of tumorous tissue sample and 117 cases of adjacent normal tissue sample using SNaP-Shot assays. The association of SNPs with tumor recurrence and overall survival was then analyzed by additive, dominant, recessive, and overdominant models in a cohort of 156 HCC patients. Results: In terms of tumor recurrence, heterozygous of SNP rs1990172 and SNP rs975263 showed a significant high risk of relapse using univariate and multivariate analysis (overdominant, HR(95%CI)=2.27 [1.41-3.66], P=0.001; HR(95%CI)=2.16 [1.37-3.39], P=0.001). But the difference between heterozygous of these two SNPs and overall survival did not reach a significance in all models. The other three investigated SNPs were not significantly associated with tumor recurrence and overall survival (P>0.05). In addition, we found no significant difference in genotype frequencies between HCC and controls. Conclusions: Our data suggest that SNP rs1990172 and SNP rs975263 in the MACC1 gene may be potential genetic markers for HCC recurrence in LT patients.
Keywords: metastasis-associated in colon cancer-1, epithelial-to-mesenchymal transition
Introduction
Primary hepatocellular carcinoma (HCC), which mainly constitutes 70% to 85% of liver cancer 1, is the six most common malignancy and the third most common cause of cancer related death worldwide 1,2. However, most of the burdens are in developing countries, even half of these cases and deaths are estimated to occur in China 1. HCC is one of the most devastating neoplasms worldwide with very poor prognosis. The recurrence rate after resection is approximately 50% at 2 years and 75% at 5 years 3. Liver transplantation (LT) is a potential curative therapy for HCC patients 4. However, recurrence is the main obstacle for the long term survival of LT patients 5. For better clinical decision-making regarding therapy and better distribution of precious liver grafts, the stratification of outcome in HCC patients is mainly according to the clinicopathological characteristics, such as tumor size, tumor number, histopathologic grading, vascular invasion and preoperative serum alpha-fetoprotein (AFP)6. However, specific biomarkers for predicting recurrence and long term survival rates are still lacking.
Metastasis-associated in colon cancer-1 (MACC1) gene is located on chromosome 7 at position 7p21.1. The newly identified gene was initially discovered by genome-wide expression analysis in primary and metastatic colon cancer 7. In this study, Stein et al showed that MACC1 expression in tumor specimens was an independent prognostic indicator of metastasis formation and metastasis-free survival 7. Further study indicated that MACC1 was a key regulator of HGF-MET signaling pathway, whose function has been identified in tumor oncogenesis, invasion and metastasis 8,9. Thus, MACC1 promoted proliferation, invasion and HGF-induced scattering of colon cancer cells in cell culture and tumor growth and metastasis 7. Subsequently, MACC1 has also been identified in gastric cancer (GC) and contributed to a poor prognosis of GC by promoting tumor cell proliferation and invasion as well as the epithelial-to-mesenchymal transition (EMT) 10. Moreover, the overexpression of MACC1 was also related to poor disease-free survival (DFS) in lung adenocarcinoma 11. In pancreatic cancer, the overexpression of serum MACC1 may be a diagnostic marker and associate with tumor metastasis 12. High expression of MACC1 was related to reduced relapse-free survival in rectal cancer 13. In ovarian carcinoma, MACC1 also associated with metastasis of tumor cell and can be a target for antitumor therapy 14. Furthermore, MACC1 was more frequently expressed in vascular invasive HCC 15 and associated with overall survival (OS) and DFS 16. These findings suggested that MACC1 may play the prognostic role for a variety of malignancies.
In previous studies, genetic variation was significantly correlated with higher expression of MACC1, which could contribute to colorectal cancer (CRC) progression through mechanisms other than or additional to Met transcriptional upregulation 17. Moreover, the relationship between the single nucleotide polymorphisms (SNP) of MACC1 and OS or metastasis-free survival (MFS) of CRC patients has been detected 18,19. However, the effect of MACC1 SNPs on the clinical outcome of HCC has not been investigated yet.
In this study, we investigated whether tagSNPs of MACC1 were correlated with clinicopathological characteristics, recurrence and overall survival of HCC in patients treated with LT. Further, the relationship between the genotype of MACC1 and the risk to development of HCC was also examined.
Materials and methods
Patients and samples
The present study enrolled 187 HCC patients who underwent LT in our center from 2003 to 2012. The inclusion criteria were as follows: (a) HCC was diagnosed either before or following transplantation (as an incidental finding). The diagnoses were confirmed by two experienced pathologists; (b) all patients included in this study were Han Chinese; (c) clinicopathological variables, such as portal vein tumor thrombi (PVTT), preoperative α-fetoprotein (AFP) concentrations, age, tumor size, and gender were obtained before surgery and during follow-up; (d) absence of de novo HCC nodules in the transplanted liver. This study protocol was approved by the Ethical Review Committee of the First Affiliated Hospital, School of Medicine, Zhejiang University, and informed consent was obtained according to the Declaration of Helsinki. Among the 187 HCC patients, 183 cases had tumorous tissue sample, 117 cases had adjacent normal tissue sample. There were 113 matched-pairs among the 187 HCC patients. All the specimens from 187 patients were stored at -80°C before extracting genomic DNA. The study was conducted in blinded fashion so that all outcomes were unknown to investigators performing genotype.
Follow-up
Follow-up data were obtained after LT for 156 patients. The follow-up period was defined from the date of operation to the death or last follow up of the HCC patients. Tumor recurrence or metastasis was monitored by AFP, ultrasonography, chest X-ray, and emission computed tomography every 3 months for the first 2 years and semiannually thereafter. Recurrence was diagnosed by imaging techniques, either intrahepatically or extrahepatically. An increase of AFP without radiologic evidence of a new tumor was not diagnosed as recurrence till this became recognizable on imaging.
Genotyping
Genomic DNA was isolated from frozen tumor tissue and adjacent normal tissue using DNeasy Tissue Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. In this study, five tagSNPs were carefully selected in two ways: 1) by literature retrieval. 2) by analysis of HapMap genotyping data in Chinese (http://www.hapmap.org), with high frequencies[minor allelic frequency (MAF) >0.1] according to the method used by Carlson et al20. In this study, SNP of rs1990172 was successfully genotyped in 178 tumorous tissue samples and 114 adjacent normal tissue samples. SNP of rs975263 was successfully genotyped in 177 tumorous tissue samples and 117 adjacent normal tissue samples. SNP of rs3735615, rs4721888, rs2241056 were successfully genotyped in 183 tumorous tissue samples and 117 adjacent normal tissue samples, respectively. Genotyping of all selected SNPs were conducted using Applied Biosystems SNaP-Shot and TaqMan technology. To assess the reliability of genotyping, a random selection of >10% of samples were regenotyped with 100% concordance. PCR primers were listed in Supplementary Material: Table S1.
Statistical analysis
The allele and genotype frequencies between tumorous tissue groups and adjacent normal tissue groups as well as clinicopathological characteristics were tested using the Pearson χ2-test or Fisher exact test. RFS was defined from the date of transplantation to the time of first recurrence or the last follow-up assessment. OS was calculated from the date of transplantation to the death of patients or the last observation. Patients without recurrence at the time of last follow-up were treated as censored events. The relationship of MACC1 SNPs to RFS or OS was identified using Kaplan-Meier method and analyzed using log-rank test. Then, univariate Cox regression analysis was used in analyzing the association between the positive SNPs or clinicopathological characteristics and RFS or OS of HCC patients. Further, Cox's proportional hazard model were used for the multivariate analysis only when the variables were significant in the univariate analysis. Hazard ratios (HR) and 95% confidence intervals (95%CI) of the hazard ratios were derived from univariate and multivariate Cox proportional hazards models. All tests were two-tailed and a p<0.05 was considered statistically significant. Data analyses were performed using SPSS software (Version 20.0, SPSS Inc. Chicago, IL).
Results
Patient characteristics
In present study, we have obtained 156 HCC patients clinicopathological and survival data among all the 187 included HCC patients. Among these 156 HCC patients, there were 143 male (91.7%) and 13 female (8.3%), respectively. There were 81 patients (51.9%) with age <50 years old and 75 patients (48.1%) with age >50 years old. The follow-up time ranged from 1.8 to 116.7 months. Over a mean follow up period of 37.0 months, 86 deaths (55.1%) were recorded. The patient clinicopathological characteristics were presented in Table 1.
Table 1.
Patient characteristics with respect to recurrence and overall survival
| Variables | Recurrence-free n |
Recurrence n |
HR(95% CI) | P value | Survival n | Death n |
HR(95% CI) | P value |
|---|---|---|---|---|---|---|---|---|
| Age, years | ||||||||
| ≤50 | 29 | 52 | 0.67(0.44-1.01) | 0.053 | 34 | 47 | 0.82(0.54-1.26) | 0.372 |
| >50 | 35 | 40 | 36 | 39 | ||||
| Gender | ||||||||
| Female | 7 | 6 | 1.68(0.73-3.84) | 0.222 | 8 | 5 | 2.02(0.82-4.97) | 0.129 |
| Male | 57 | 86 | 62 | 81 | ||||
| Preoperative AFP, ng/mL | ||||||||
| ≤400 | 40 | 30 | 2.33(1.51-3.61) | <0.001 | 40 | 30 | 1.90(1.22-2.96) | 0.005 |
| >400 | 24 | 62 | 30 | 56 | ||||
| PVTT | ||||||||
| Negative | 53 | 47 | 2.85(1.88-4.31) | <0.001 | 55 | 45 | 2.31(1.51-3.53) | <0.001 |
| Positive | 11 | 45 | 15 | 41 | ||||
| Tumor size, cm | ||||||||
| ≤5 | 52 | 36 | 3.52(2.30-5.39) | <0.001 | 55 | 33 | 3.41(2.20-5.30) | <0.001 |
| >5 | 12 | 56 | 15 | 53 |
Hazard ratios (HR), 95% confidence intervals (95% CI) and P value were obtained from univariate Cox regression analysis.
Analysis of SNP frequencies and HCC development as well as tumor recurrence and overall survival
Five SNPs in the MACC1 gene were genotyped in tumorous tissue samples and adjacent normal tissue samples of HCC patients with LT. The allele frequency and genotype frequency of each SNP was shown in Table 2. MAFs were similar to those given in HapMap SNP database for a CHB analysis panel and all five MACC1 SNP genotype frequencies were in Hardy-Weinberg equilibrium( P>0.05) (data not shown). The number of samples detected for each polymorphism was not exactly equal due to unsuccessful amplification of some samples. The difference of genotype or allele frequencies between tumorous tissue samples and adjacent normal tissue samples of HCC patients with LT was not statistically significant, indicating that these five SNPs were not involved in the susceptibility of HCC (Supplementary Material: Table S2).
Table 2.
Genotype and allele distributions in tumorous tissue and adjacent normal tissue of HCC patients treated by LT
| SNP | Adjacent normal tissues n (%) | tumorous tissues n (%) | P value | Recurrence-free n (%) | Recurrence n (%) | P value | Survival n (%) | Death n (%) | P value |
|---|---|---|---|---|---|---|---|---|---|
| rs1990172 | |||||||||
| GG | 83(72.2) | 135(75.8) | 0.310a | 52(85.3) | 61(68.5) | 0.001c | 54(80.6) | 59(71.1) | 0.072c |
| GT | 31(27.0) | 38(21.4) | 6(9.8) | 26(29.2) | 10(14.9) | 22(26.5) | |||
| TT | 1(0.8) | 5(2.8) | 3(4.9) | 2(2.3) | 3(4.5) | 2(2.4) | |||
| G allele | 197(85.7) | 308(86.5) | 0.807b | ||||||
| T allele | 33(14.3) | 48(13.5) | |||||||
| rs3735615 | |||||||||
| GG | 86(73.5) | 140(76.5) | 0.394a | 50(80.7) | 63(69.2) | 0.179c | 54(79.4) | 59(69.4) | 0.255c |
| CG | 31(26.5) | 41(22.4) | 11(17.7) | 27(29.7) | 13(19.1) | 25(29.4) | |||
| CC | 0(0.0) | 2(1.1) | 1(1.6) | 1(1.1) | 1(1.5) | 1(1.2) | |||
| G allele | 203(86.8) | 321(87.7) | 0.801b | ||||||
| C allele | 31(13.2) | 45(12.3) | |||||||
| rs4721888 | |||||||||
| GG | 75(64.1) | 119(65.0) | 0.932a | 38(61.3) | 61(67.0) | 0.741c | 44(64.7) | 55(64.7) | 0.610c |
| CG | 33(28.2) | 52(28.4) | 19(30.6) | 24(26.4) | 20(29.4) | 23(27.1) | |||
| CC | 9(7.7) | 12(6.6) | 5(8.1) | 6(6.6) | 4(5.9) | 7(8.2) | |||
| G allele | 183(78.2) | 290(79.2) | 0.760b | ||||||
| C allele | 51(21.8) | 76(20.8) | |||||||
| rs2241056 | |||||||||
| GG | 60(51.3) | 95(51.9) | 0.567a | 32(51.6) | 50(54.9) | 0.543c | 36(52.9) | 46(54.1) | 0.921c |
| AG | 53(45.3) | 77(42.1) | 25(40.3) | 38(41.8) | 29(42.7) | 34(40.0) | |||
| AA | 4(3.4) | 11(6.0) | 5(8.1) | 3(3.3) | 3(4.4) | 5(5.9) | |||
| G allele | 173(73.9) | 267(73.0) | 0.850b | ||||||
| A allele | 61(26.1) | 99(27.0) | |||||||
| rs975263 | |||||||||
| TT | 82(70.1) | 129(72.9) | 0.363a | 49(80.3) | 58(65.2) | 0.004c | 51(76.1) | 56(67.5) | 0.087c |
| CT | 34(29.1) | 43(24.3) | 8(13.1) | 30(33.7) | 12(17.9) | 26(31.3) | |||
| CC | 1(0.8) | 5(2.8) | 4(6.6) | 1(1.1) | 4(6.0) | 1(1.2) | |||
| T allele | 198(84.6) | 301(85.0) | 0.907b | ||||||
| C allele | 36(15.4) | 53(15.0) |
aχ2 test. (Pearson χ2 test). bχ2 test. (Fisher's exact test). cLog-rank test.
Effect of investigated MACC1 SNPs on RFS and OS in HCC patients treated with LT was evaluated by univariate Cox regression analysis using an additive model of inheritance in accordance with the distributions of the five SNPs (Table 2). Among selected SNPs, rs1990172 and rs975263 were significantly associated with an increased risk for recurrence (P=0.001 & P=0.004, respectively). Meanwhile, the other three SNPs didn't show an obvious impact on tumor recurrence. However, the association between the SNPs of rs1990172 or rs975263 and overall survival in HCC patients treated with LT did not reach the significance (P=0.072 & P=0.087, respectively), which might be a result of the limited sample size. Remaining three SNPs showed no significant impact on overall survival, either (Table 2).
Then, further assessment was performed by univariate and multivariate Cox regression analysis for the impact of SNP rs1990172 or SNP rs975263 on recurrence and overall survival in HCC patients treated with LT on the basis of an additive, dominant, recessive and overdominant model of inheritance, respectively. Results were shown in table 3. In the dominant model of inheritance, patients who have the genotype of GT or TT for SNP rs1990172 or patients who have the genotype of CT or CC for SNP rs975263 may confront a high risk of tumor recurrence. In the overdominant model, genotype of GT for SNP rs1990172 or genotype of CT for SNP rs975263 indicated a shorter recurrence-free survival time. Though, in the overdominant of inheritance, genotype of GT for SNP rs1990172 showed a significantly increased risk for reduced overall survival in univariate Cox regression analysis (P=0.028), but it did not reach a significance in multivariate Cox regression analysis (P=0.198). The results showed that in additive and recessive model of inheritance, there was no significance for the impact of investigated SNPs on tumor recurrence and overall survival of HCC patients treated with LT (Table 3). Further, in the multivariate Cox regression analysis, tumor size, preoperative AFP level, PVTT and SNP rs1990172 or rs975263 were four independent prognostic factor for recurrence in both dominant and overdominant model of inheritance (Table 4).
Table 3.
Impact of MACC1 SNP rs1990172 and rs975263 on recurrence and overall survival, shown for additive, dominant, recessive, and overdominant genetic models of inheritance
| Model | Genotype | Recurrence-free n (%) | Recurrence n (%) | HR(95% CI) | P value | Survival n (%) | Death n (%) |
HR(95% CI) | P value |
|---|---|---|---|---|---|---|---|---|---|
| rs1990172 | |||||||||
| Additive | GG | 52(85.3) | 61(68.5) | 1.36(0.96-1.91)a | 0.082a | 54(80.6) | 59(71.1) | 1.23(0.84-1.79)a | 0.284a |
| GT | 6(9.8) | 26(29.2) | 1.32(0.93-1.86)b | 0.123b | 10(14.9) | 22(26.5) | 1.11(0.75-1.64)b | 0.595b | |
| TT | 3(4.9) | 2(2.2) | 3(4.5) | 2(2.4) | |||||
| Dominant | GG | 52(85.3) | 61(68.5) | 1.87(1.20-2.94)a | 0.006a | 54(80.6) | 59(71.1) | 1.51(0.94-2.43)a | 0.090a |
| GT&TT | 9(14.8) | 28(31.5) | 1.80(1.14-2.85)b | 0.011b | 13(19.4) | 24(28.9) | 1.26(0.78-2.03)b | 0.353b | |
| Recessive | GG> | 58(95.1) | 87(97.8) | 0.51(0.12-2.06)a | 0.341a | 64(95.5) | 81(97.6) | 0.60(0.15-2.43)a | 0.471a |
| TT | 3(4.9) | 2(2.2) | 0.48(0.12-1.97)b | 0.308b | 3(4.5) | 2(2.4) | 0.64(0.16-2.62)b | 0.532b | |
| Overdominant | GG&TT | 55(90.2) | 63(70.8) | 2.30(1.45-3.64)a | 0.0004a | 57(85.1) | 61(73.5) | 1.73(1.06-2.830)a | 0.028a |
| GT | 6(9.8) | 26(29.2) | 2.27(1.41-3.66)b | 0.001b | 10(14.9) | 22(26.5) | 1.39(0.84-2.29)b | 0.198b | |
| rs975263 | |||||||||
| Additive | TT | 49(80.3) | 58(65.2) | 1.23(0.87-1.74)a | 0.248a | 51(76.1) | 56(67.5) | 1.09(0.75-1.60)a | 0.642a |
| CT | 8(13.1) | 30(33.7) | 1.25(0.88-1.77)b | 0.215b | 12(17.9) | 26(31.3) | 1.05(0.71-1.54)b | 0.825b | |
| CC | 4(6.6) | 1(1.1) | 4(6.0) | 1(1.2) | |||||
| Dominant | TT | 49(80.3) | 58(65.2) | 1.62(1.05-2.51)a | 0.031a | 51(76.1) | 56(67.5) | 1.32(0.83-2.09)a | 0.241a |
| CT&CC | 12(19.7) | 31(34.8) | 1.69(1.09-2.63)b | 0.020b | 16(23.9) | 27(32.5) | 1.21(0.76-1.93)b | 0.413b | |
| Recessive | TT&CT | 57(93.4) | 88(98.9) | 0.25(0.03-1.76)a | 0.162a | 63(94.0) | 82(98.8) | 0.28(0.04-2.03)a | 0.209a |
| CC | 4(6.6) | 1(1.1) | 0.23(0.03-1.66)b | 0.145b | 4(6.0) | 1(1.2) | 0.29(0.04-2.12)b | 0.224b | |
| Overdominant | TT&CC | 53(86.9) | 59(66.3) | 2.00(1.28-3.11)a | 0.002a | 55(82.1) | 57(68.7) | 1.56(0.98-2.49)a | 0.061a |
| CT | 8(13.1) | 30(33.7) | 2.16(1.37-3.39)b | 0.001b | 12(17.9) | 26(31.3) | 1.42(0.88-2.27)b | 0.148b |
a Hazard ratios (HR) and 95% confidence intervals (95% CI) were obtained from univariate Cox regression analysis.
b Hazard ratios (HR) and 95% confidence intervals (95% CI) were obtained from multivariate Cox regression analysis.
Table 4.
Multivariate Cox regression analysis of variables related to tumor recurrence at univariate analysis
| Model | variable | HR(95% CI) | P value |
|---|---|---|---|
| rs1990172 | |||
| Dominant | GT&TT genotype | 1.80(1.14-2.85) | 0.011 |
| Tumor size>5cm | 2.46(1.58-3.83) | <0.001 | |
| AFP >400 ng/mL | 1.83(1.17-2.88) | 0.009 | |
| PVTT | 2.44(1.59-3.75) | <0.001 | |
| Overdominant | GT genotype | 2.27(1.41-3.66) | 0.001 |
| Tumor size>5cm | 2.30(1.47-3.61) | <0.001 | |
| AFP >400 ng/mL | 1.95(1.24-3.07) | 0.004 | |
| PVTT | 2.50(1.62-3.85) | <0.001 | |
| rs975263 | |||
| Dominant | CT&CC genotype | 1.69(1.09-2.63) | 0.02 |
| Tumor size>5cm | 2.58(1.65-4.03) | <0.001 | |
| AFP >400 ng/mL | 1.83(1.16-2.88) | 0.01 | |
| PVTT | 2.36(1.54-3.63) | <0.001 | |
| Overdominant | CT genotype | 2.16(1.37-3.39) | 0.001 |
| Tumor size>5cm | 2.49(1.59-3.90) | <0.001 | |
| AFP >400 ng/mL | 1.94(1.23-3.07) | 0.004 | |
| PVTT | 2.41(1.56-3.72) | <0.001 |
AFP, α-fetoprotein; PVTT, portal vein tumor thrombi; HR, hazard ratio; CI, confidence interval.
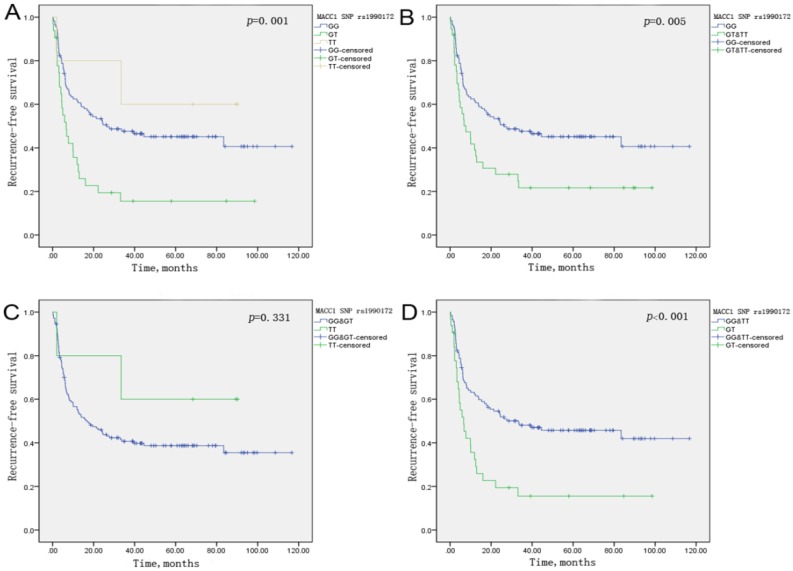
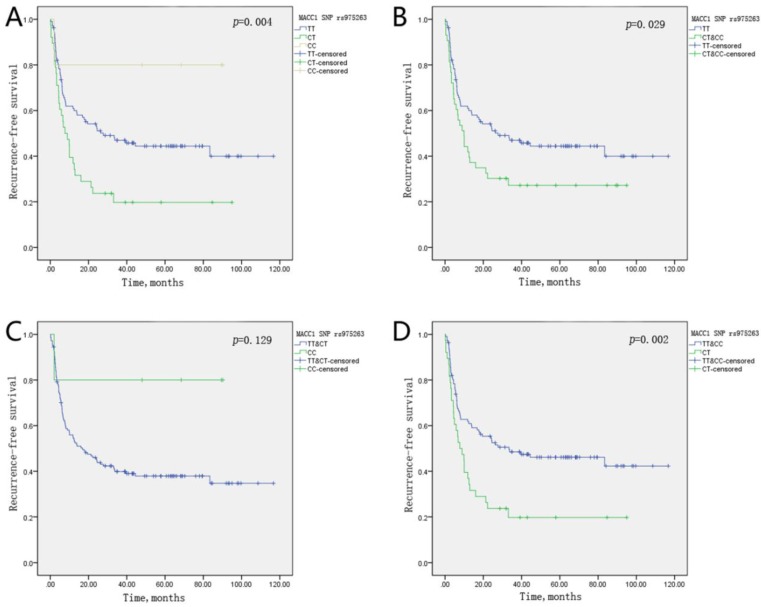
Moreover, Kaplan-Meier survival curves were presented in accordance with an additive, dominant, recessive and overdominant model of inheritance for SNP rs1990172 (Figure 1) and SNP rs975263 (Figure 2) on tumor recurrence of HCC patients treated with LT. From Figure 1, we can see that SNP rs1990172 obviously correlated with recurrence-free survival in a dominant and overdominant model of inheritance. The same conclusion can be drawn for SNP rs975263 from Figure 2.
Figure 1.
Kaplan-Meier curve of MACC1 SNP rs1990172 on recurrence-free survival for HCC patients undergoing LT in an additive(A), dominant(B), recessive(C), and overdominant model(D) of inheritance, respectively. P-value was calculated by Log-rank test.
Figure 2.
Kaplan-Meier curve of MACC1 SNP rs975263 on recurrence-free survival for HCC patients undergoing LT in an additive(A), dominant(B), recessive(C), and overdominant model(D) of inheritance, respectively. P-value was calculated by Log-rank test.
Association between genetic polymorphisms and clinicopathological variables
To avoid a false association and demonstrate the relevant relationship of the recurrence and overall survival with polymorphisms, the associations of SNP rs1990172 and SNP rs975263 with clinicopathological characteristics were analyzed. There was no statistical significance between these two investigated SNPs and HCC patient age, gender, tumor size, preoperative AFP level, PVTT (Table 5).
Table 5.
Association of rs1990172 and rs975263 genotypes with clinicopathological variables.
| Variables | rs1990172 | P value | rs975263 | P value | ||||
|---|---|---|---|---|---|---|---|---|
| GG n (%) | GT n (%) | TT n (%) | TT n (%) | CT n (%) | CC n (%) | |||
| Age, years | ||||||||
| ≤50 | 59(75.6) | 17(21.8) | 2(2.6) | 0.858a | 57(73.1) | 19(24.4) | 2(2.5) | 0.811a |
| >50 | 54(75.0) | 15(20.8) | 3(4.2) | 50(69.4) | 19(26.4) | 3(4.2) | ||
| Gender | ||||||||
| Female | 11(84.6) | 2(15.4) | 0(0) | 0.646a | 9(69.2) | 3(23.1) | 1(7.7) | 0.654a |
| Male | 102(74.5) | 30(21.9) | 5(3.6) | 98(71.5) | 35(25.5) | 4(3.0) | ||
| Preoperative AFP, ng/mL | ||||||||
| ≤400 | 51(76.1) | 15(22.4) | 1(1.5) | 0.521a | 50(75.8) | 15(22.7) | 1(1.5) | 0.405a |
| >400 | 62(74.7) | 17(20.5) | 4(4.8) | 57(67.9) | 23(27.4) | 4(4.8) | ||
| PVTT | ||||||||
| Negative | 73(76.1) | 20(20.8) | 3(3.1) | 0.959a | 68(70.8) | 25(26.1) | 3(3.1) | 0.953a |
| Positive | 40(74.1) | 12(22.2) | 2(3.7) | 39(72.2) | 13(24.1) | 2(3.7) | ||
| Tumor size, cm | ||||||||
| ≤5 | 67(79.8) | 13(15.5) | 4(4.8) | 0.094a | 63(75.0) | 17(20.2) | 4(4.8) | 0.175a |
| >5 | 46(69.7) | 19(28.8) | 1(1.5) | 44(66.7) | 21(31.8) | 1(1.5) | ||
aχ2 test. (Pearson χ2 test)
Discussion
Metastasis-associated in colon cancer-1(MACC1) gene is a novel oncogene located on chromosome 7p21.1. In this study, we examined whether genetic variations of MACC1 gene had predictive value for recurrence and overall survival of HCC patients treated with LT. Notably, our results showed that polymorphisms of rs1990172 and rs975263 were significantly associated with tumor recurrence of HCC patients undergoing LT. As far as we know, this is the first investigation of MACC1 gene polymorphisms on the prognosis of HCC.
In previous studies, the prognostic role of MACC1 gene for tumor recurrence and overall survival in HCC patients has already been identified 16,21,22. The predictive value of MACC1 overexpression has also been noticed in CRC 7,23, GC 10, lung carcinoma 11, pancreatic cancer 12. Moreover, Galimi F et al 17 has demonstrated that Chromosome 7 polysomy and gain of the p-arm were significantly correlated with MACC1 overexpression leading to a poor prognosis of patients, which meant that genetic variation might affect the prognosis of patients through regulating the expression of MACC1 gene. As known to all that SNP is an important form of genetic variation, so whether SNP of MACC1 gene has an impact on the prognosis of malignancy needed to be further confirmed. Thus, Lang AH et al 18 has demonstrated that SNP rs1990172 can be a predictor for reduced overall survival in colorectal cancer patients. Moreover, Schmid F et al declared that CT genotype of SNP rs975263 might be associated with a reduced survival for younger colon cancer patients in early stages 19.
Actually, these two SNPs mentioned above were just the very significant SNPs covered in our analysis. SNP rs1990172 is situated in an intronic region of the MACC1 gene, which has no impact on coding exon of MACC1 gene. While SNP rs975263 is a nonsynonymous one with an exchange of leucine to serine at codon 515, fifth exon. In fact, these two SNPs are in strong linkage disequilibrium with each other (r2=0.858; http://www.hapmap.org). However, the substitution of leucine to serine at codon 515, fifth exon demonstrated no adverse effect on the structure or function of the MACC1 protein in silico analysis using software tools PolyPhen-2 and SIFT, respectively 18. Moreover, the exchange of leucine to serine in the MACC1 gene was also demonstrated no impact on expression level of MACC1 mRNA in the CRC in vitro experiments 19. Thus, further investigations need to be conducted to demonstrate the relationship between SNPs and poor prognosis of HCC patients.
In addition to evaluating the impact of SNPs within MACC1 on tumor recurrence and overall survival in HCC patients, we simultaneously evaluated potential correlation between MACC1 polymorphisms and HCC susceptibility. The results showed that there was no significant difference in the allele or genotype distribution of the polymorphisms in MACC1 gene between tumor tissue and adjacent normal tissue, which indicated that these five SNPs might not influence the risk of development of HCC. To our knowledge, no other studies on the impact of MACC1 polymorphisms on HCC development have been published.
It's worth mentioning that there were some inevitable limitations in present study. First, metastasis associated in colon cancer 1 (MACC1) gene, just as its name implies, is mainly correlated with the process of tumor metastasis. Thus, metastasis-free survival would be preferred as a more accurate endpoint. However, tumor recurrence and poor overall survival come hand in hand with tumor metastasis 24. Thus, RFS and OS may also act as reasonable observation endpoints. Second, most of the included patients are unavoidably infected with hepatitis-B virus. Therefore, further studies are necessary to demonstrate the association between the polymorphisms and the prognosis of HCC patients caused by other diseases. Further, the sample size in our study is relative small. Further investigations are warranted to validate the prognostic role in large retrospective and prospective cohorts. Moreover, genomic DNA used in this study was extracted from liver tissue after LT. However, it is preferred to isolate genomic DNA from peripheral blood in our further studies, which is convenient and acceptable for the patients before operation.
In conclusion, this is the first study to demonstrate the correlation between polymorphisms of MACC1 gene and the prognosis of HCC patients undergoing LT. Our results suggest the potential role of MACC1 polymorphisms as a genetic marker for tumor recurrence in HCC patients treated with LT, which may help to stratify high risk HCC patients before LT. However, these findings need to be further confirmed through more large retrospective and prospective studies.
Supplementary Material
Table S1 - Table S2.
Acknowledgments
The authors gratefully acknowledge the study participants, the many surgeons and nurses who kindly facilitate the recruitment and collection of patient information. We also thank for the support of National Natural Science Foundation of China (No:81101840 & 81302074), National S&T Major Project (No.2012ZX10002-017), National Basic Research Program of China (973 Program) (No.2009CB522407) and Education Department of Zhejiang Province Research Project (No.Y201223762).
References
- 1.Jemal A, Bray F, Center MM. et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Shin HR, Bray F. et al. Estimates of worldwide burden of cancer in 2008:GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin. 2012;62:394–9. doi: 10.3322/caac.21161. [DOI] [PubMed] [Google Scholar]
- 4.Mazzaferro V, Regalia E, Doci R. et al. Liver transplantation for the treatment of small hepatocellular carcinomas in patients with cirrhosis. N Engl J Med. 1996;334:693–9. doi: 10.1056/NEJM199603143341104. [DOI] [PubMed] [Google Scholar]
- 5.Zimmerman MA, Ghobrial RM, Tong MJ. et al. Recurrence of hepatocellular carcinoma following liver transplantation: a review of preoperative and postoperative prognostic indicators. Arch Surg. 2008;143:182–8. doi: 10.1001/archsurg.2007.39. [DOI] [PubMed] [Google Scholar]
- 6.Zheng SS, Xu X, Wu J. et al. Liver transplantation for hepatocellular carcinoma: Hangzhou experiences. Transplantation. 2008;85:1726–32. doi: 10.1097/TP.0b013e31816b67e4. [DOI] [PubMed] [Google Scholar]
- 7.Stein U, Walther W, Arlt F. et al. MACC1, a newly identified key regulator of HGF-MET signaling, predicts colon cancer metastasis. Nat Med. 2009;15:59–67. doi: 10.1038/nm.1889. [DOI] [PubMed] [Google Scholar]
- 8.Trusolino L, Bertotti A, Comoglio PM. MET signalling: principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol. 2010;11:834–48. doi: 10.1038/nrm3012. [DOI] [PubMed] [Google Scholar]
- 9.Gherardi E, Birchmeier W, Birchmeier C. et al. Targeting MET in cancer: rationale and progress. Nat Rev Cancer. 2012;12:89–103. doi: 10.1038/nrc3205. [DOI] [PubMed] [Google Scholar]
- 10.Wang L, Wu Y, Lin L. et al. MACC1 upregulation predicts a poor prognosis of gastric cancer, and promotes tumor cell proliferation and invasion. Int J Cancer. 2013 doi: 10.1002/ijc.28140. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 11.Shimokawa H, Uramoto H, Onitsuka T. et al. Overexpression of MACC1 mRNA in lung adenocarcinoma is associated with postoperative recurrence. J Thorac Cardiovasc Surg. 2011;141:895–8. doi: 10.1016/j.jtcvs.2010.09.044. [DOI] [PubMed] [Google Scholar]
- 12.Wang G, Kang MX, Lu WJ. et al. MACC1: A potential molecule associated with pancreatic cancer metastasis and chemoresistance. Oncol Lett. 2012;4:783–791. doi: 10.3892/ol.2012.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawamura M, Saigusa S, Toiyama Y. et al. Correlation of MACC1 and MET expression in rectal cancer after neoadjuvant chemoradiotherapy. Anticancer Res. 2012;32:1527–31. [PubMed] [Google Scholar]
- 14.Zhang R, Shi H, Chen Z. et al. Effects of metastasis-associated in colon cancer 1 inhibition by small hairpin RNA on ovarian carcinoma OVCAR-3 cells. J Exp Clin Cancer Res. 2011;30:83. doi: 10.1186/1756-9966-30-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shirahata A, Fan W, Sakuraba K. et al. MACC 1 as a marker for vascular invasive hepatocellular carcinoma. Anticancer Res. 2011;31:777–80. [PubMed] [Google Scholar]
- 16.Qiu J, Huang P, Liu Q. et al. Identification of MACC1 as a novel prognostic marker in hepatocellular carcinoma. J Transl Med. 2011;9:166. doi: 10.1186/1479-5876-9-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galimi F, Torti D, Sassi F. et al. Genetic and expression analysis of MET, MACC1, and HGF in metastatic colorectal cancer: response to met inhibition in patient xenografts and pathologic correlations. Clin Cancer Res. 2011;17:3146–56. doi: 10.1158/1078-0432.CCR-10-3377. [DOI] [PubMed] [Google Scholar]
- 18.Lang AH, Geller-Rhomberg S, Winder T. et al. A common variant of the MACC1 gene is significantly associated with overall survival in colorectal cancer patients. BMC Cancer. 2012;12:20. doi: 10.1186/1471-2407-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmid F, Burock S, Klockmeier K. et al. SNPs in the coding region of the metastasis-inducing gene MACC1 and clinical outcome in colorectal cancer. Mol Cancer. 2012;11:49. doi: 10.1186/1476-4598-11-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carlson CS, Eberle MA, Rieder MJ. et al. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–20. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang YP, Qu JH, Chang XJ. et al. High intratumoral metastasis-associated in colon cancer-1 expression predicts poor outcomes of cryoablation therapy for advanced hepatocellular carcinoma. J Transl Med. 2013;11:41. doi: 10.1186/1479-5876-11-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qu JH, Chang XJ, Lu YY. et al. Overexpression of metastasis-associated in colon cancer 1 predicts a poor outcome of hepatitis B virus-related hepatocellular carcinoma. World J Gastroenterol. 2012;18:2995–3003. doi: 10.3748/wjg.v18.i23.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stein U, Burock S, Herrmann P. et al. Circulating MACC1 transcripts in colorectal cancer patient plasma predict metastasis and prognosis. PLoS One. 2012;7:e49249. doi: 10.1371/journal.pone.0049249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marx V. Tracking metastasis and tricking cancer. Nature. 2013;494:133–6. doi: 10.1038/494131a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 - Table S2.