This review by Rustgi elaborates on the known genetic syndromes that underlie familial pancreatic cancer. It aims to delineate the subtypes of syndromic hereditary pancreatic cancer in which germline genetic mutations have been identified and nonsyndromic familial pancreatic cancer in which genetic information is emerging.
Keywords: familial pancreatic cancer, hereditary pancreatitis, BRCA2, genetic testing
Abstract
Beset by poor prognosis, pancreatic ductal adenocarcinoma is classified as familial or sporadic. This review elaborates on the known genetic syndromes that underlie familial pancreatic cancer, where there are opportunities for genetic counseling and testing as well as clinical monitoring of at-risk patients. Such subsets of familial pancreatic cancer involve germline cationic trypsinogen or PRSS1 mutations (hereditary pancreatitis), BRCA2 mutations (usually in association with hereditary breast–ovarian cancer syndrome), CDKN2 mutations (familial atypical mole and multiple melanoma), or DNA repair gene mutations (e.g., ATM and PALB2, apart from those in BRCA2). However, the vast majority of familial pancreatic cancer cases have yet to have their genetic underpinnings elucidated, waiting in part for the results of deep sequencing efforts.
The most common type of pancreatic cancer is pancreatic ductal adenocarcinoma (abbreviated as PDA or PDAC), accounting for >90% of pancreatic cancers. Forty-four-thousand new PDAC cases occur in the United States every year. Nearly 40,000 deaths related to complications of PDAC occur annually in the United States. By 2020, PDAC will be the second leading cause of cancer-related mortality (Ma and Jemal 2013). Unfortunately, median survival is measured in months, and 5-yr survival is <5%–10%, attributable largely to presentation of disease at late stages. Recent genetic evidence suggests that preneoplastic cells may undergo epithelial–mesenchymal transition (EMT) and circulate in the blood to participate potentially in the formation of metastasis, which also may be a contributing factor to decreased survival (Rhim et al. 2012). The remaining subtypes include neuroendocrine tumors, which originate in islet cells, and mucinous cystadenocarcinomas arising from intraductal papillary mucinous neoplasms (IPMNs) and mucinous cystic neoplasms (MCNs). Rare pancreatic tumors include lymphomas, sarcomas, acinar cell carcinomas, adenosquamous carcinomas, colloid carcinomas, giant cell tumors, pancreatoblastomas, serous cystadenomas, signet ring cell carcinomas, solid and pseudopapillary tumors, and undifferentiated carcinomas.
As with some other cancers, PDAC may be classified further into either sporadic or hereditary. Sporadic PDAC reflects a combination of somatic genomic, genetic, and epigenetic alterations with a complex interplay with environmental factors (cigarette smoking, alcohol, and/or obesity). It is estimated that 5%–10% of PDAC has a hereditary basis with ∼80% penetrance (Permuth-Wey and Egan 2009). Some of these inherited conditions are well characterized for their incriminating germline gene mutations and recognizable clinical or phenotypic features, while others await identification of germline gene mutations through genomic or exomic sequencing efforts, building on other past approaches such as copy number variations (CNVs).
There is no uniform definition of nonsyndromic familial pancreatic cancer. It is likely that as more discoveries elucidate the genetics underlying familial pancreatic cancer, classification schemes will emerge, as has been evident for hereditary breast–ovarian cancer syndrome and Lynch syndrome, an inherited colorectal cancer predisposition syndrome. At the same time, clinical registries have led to “working” definitions. One such illustration is that familial pancreatic cancer is defined as a kindred with a pair of first-degree relatives with pancreatic cancer (Klein et al. 2004); in this context, the risk of developing pancreatic cancer in an at-risk family member is sixfold. This rises significantly more so if there are three or more first-degree relatives with pancreatic cancer (Hruban et al. 2010; Klein 2013). Other operational definitions take into account the age of onset of pancreatic cancer in a family with pancreatic cancer cases. Whereas risk assessment scores are used widely for hereditary breast cancer (National Comprehensive Cancer Network guidelines version 4.2013) and are emerging in Lynch syndrome (referred to as PREMM1,2,6) (Kastrinos et al. 2011), this approach (referred to as PancPRO) is in its nascency for potential widespread use (Wang et al. 2007).
The following sections delineate the subtypes of syndromic hereditary pancreatic cancer in which germline genetic mutations have been identified and nonsyndromic familial pancreatic cancer in which genetic information is emerging. Whenever possible, links to the pathogenesis of sporadic PDAC are emphasized.
Hereditary pancreatitis
The most appreciated and recognized risk factor for sporadic PDAC is chronic pancreatitis, which is triggered by acinar cell damage followed by autodigestion, a proinflammatory milieu, and fibrosis (Lowenfels et al. 1993; Yadav and Lowenfels 2013). Chronic pancreatitis results in manifestations of exocrine insufficiency and endocrine insufficiency, the latter typically following the former. An uncommon form of chronic pancreatitis is hereditary pancreatitis, which is an autosomal dominant inherited disorder and highly penetrant (>80%) and typically manifests prior to the age of 30 (Whitcomb 2013), although this may be dependent on ancestral origin in the family. Clinical suspicion is triggered by a family history of two or more first-degree relatives (or three or more second-degree relatives) with recurrent acute pancreatitis or chronic pancreatitis spanning two or more generations.
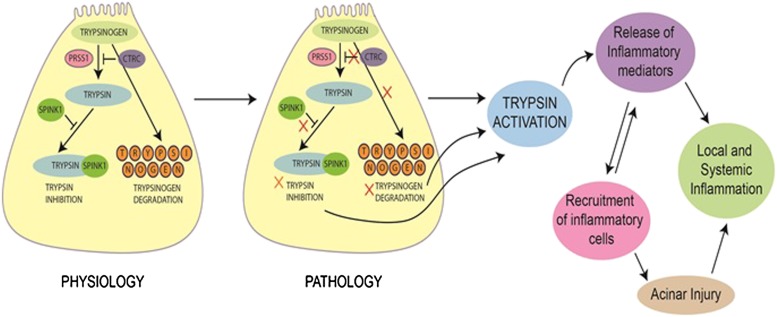
Germline mutations in PRSS1 (protease, Ser1 [chromosome 7q35]), which encodes the cationic trypsinogen protein, are associated with hereditary pancreatitis (Whitcomb et al. 1996). Trypsinogen is abundant in acinar cells. It hydrolyzes dietary proteins at lysine and arginine residues, thereby activating proenzymes. Premature activation of trypsinogen results in acute pancreatitis (Fig. 1). The original description of a germline mutation in PRSS1 was an arginine-to-histidine substitution at amino acid 117, referred to as R117H. Arg117 is sensitive to trypsin, and cleavage at this site allows for inactivation of trypsin. However, this mutation eliminates the trypsin hydrolysis site. Other mutations have also been described in cationic trypsinogen, some of which are listed here: N29I, A16V, D22G, K23R, A121T, and R122C. R116C is a rare mutation that may result in misfolding of the protein and present an alternative cause of activation (Kereszturi et al. 2009). Patients with hereditary pancreatitis have recurrent acute pancreatitis, which can often evolve into chronic pancreatitis. Their lifetime risk for PDAC is 35-fold (or more) by ages 70–75. Patients with hereditary pancreatitis may benefit from a low-fat (low-triglyceride) diet and abstinence from cigarette smoking and alcohol as a means to ameliorate progression to chronic pancreatitis.
Figure 1.
Trypsinogen–trypsin cascade. Under normal physiological conditions in the pancreatic acinar cell, trypsinogen activates trypsin, a serine protease, resulting in hydrolysis of proteins. This is held in balance by trypsinogen degradation and inhibition of trypsin. However, trypsinogen mutations can result in constitutive trypsin activation, establishing a cascade of events involving acinar cell injury, recurrent acute pancreatitis, and, as observed in hereditary pancreatitis, a progression to chronic pancreatitis and an increased risk for PDAC. This is augmented by mutations in CTRC in both hereditary pancreatitis and idiopathic chronic pancreatitis. Thus, SPINK1 mutations may be important in a modifier capacity in idiopathic chronic pancreatitis.
Modeling of hereditary pancreatitis has revealed that pancreata from Elastase-R122H (mPRSS1) transgenic mice display early-onset acinar cell injury and inflammatory cell infiltration (Archer et al. 2006). With age, the transgenic mice develop pancreatic fibrosis and display acinar cell dedifferentiation and an augmented response to cerulein-induced pancreatitis. Finally, activation of c-jun N-terminal kinase and extracellular signal-regulated kinase was observed, which may be linked to activation of inflammation in these mice.
Chymotrypsin C (CTRC) gene (chromosome 1p36.21) mutations can be found in chronic pancreatitis (Rosendahl et al. 2008). CTRC mutations appear to boost the effects of the mutant forms of cationic trypsinogen (Fig. 1; Szabo and Sahin-Toth 2012). Two alterations in the CTRC gene, R254W and K247_R254del, were significantly overrepresented in a study of cohorts of patients with idiopathic chronic pancreatitis and hereditary pancreatitis (Rosendahl et al. 2008). Functional analysis of the CTRC variants showed impaired activity and/or reduced secretion, which resulted in “removal” of the normal negative degradative influence of CTRC on trypsin levels.
The aforementioned mutations in hereditary pancreatitis, especially in PRSS1, need to be distinguished from other mutations found in sporadic idiopathic chronic pancreatitis. The serine protease inhibitors of the Kazal type (SPINK) are a family of genes in which SPINK1 mutations (e.g., N34S) are found to be associated in some patients who have idiopathic chronic pancreatitis (Fig. 1). Similarly, heterozygous mutations in the cystic fibrosis transmembrane receptor (CFTR) gene may be found in a subset of these patients (Cohn et al. 1998, 2000; Sharer et al. 1998). CFTR is an ion channel involved in the transport of chloride and thiocyanate. While CFTR gene mutations are traditionally associated with cystic fibrosis, they may also contribute to chronic pancreatitis. CFTR mutations lead to impaired chloride secretion and/or loss of bicarbonate secretion across ductal epithelial cells, which causes the accumulation of protein-rich materials, damage to acini, and subsequent fibrosis (Ooi et al. 2011).
Familial atypical mole and multiple melanoma (FAMMM) syndrome
An autosomal dominant disorder, FAMMM is characterized by the following in a family: melanomas in more than one first- or second-degree relative, high total body nevi count (often >50), and nevi with certain histopathological features. The melanomas can arise from the atypical moles or de novo (superficially spreading melanoma and/or nodular melanoma). Three original descriptions in different kindreds implicated germline mutations or microdeletions in CDNK2 (chromosome 9p21.3), in particular restricted to the p16INK4a isoform (chromosome 9p21.3), as causative for FAMMM (Gruis et al. 1995; Ranade et al. 1995). These mutant p16 proteins cannot inhibit the cyclin D1/CDK4 or cyclin D1/CDK6 complex. CDNK2 mutations are associated with 60%–90% melanoma risk by age 80 as well as an increased pancreatic cancer risk of up to 20% by age 75. Other studies have led to equally worrisome correlations. In one study, there was a 25% increased risk of pancreatic cancer in FAMMM kindreds, and in another study, a cohort showed a 13-fold to 22-fold increase in pancreatic cancer (Lynch et al. 2008). The age of onset of pancreatic cancer in FAMMM is quite variable, perhaps reflecting incomplete penetrance. Importantly, nearly 90% of sporadic PDACs harbor alterations (promoter hypermethylation, mutations, and microdeletions) in CDKN2, providing insights between the pathogenesis of FAMMM and the pathogenesis of sporadic PDAC. It should be noted that while melanoma and pancreatic cancer constitute the two most frequent cancers, respectively, in FAMMM, anecdotal reports note a variety of other cancers in association with FAMMM, but a clear pattern is lacking. Interestingly, CDK4 mutations that occur in the CDKN2-binding domain have been reported in FAMMM kindreds (Zuo et al. 1996). Abnormal cell cycle progression and proliferation in FAMMM is evident also in sporadic PDAC. The reasons underlying the tissue-specific functional consequences of mutant p16 protein in FAMMM are not clear. It is likely that mutant p16 protein promotes genomic instability in sporadic PDAC by virtue of its contribution to uncontrolled cell division (Campbell et al. 2010).
Upon determination that FAMMM exists in a kindred, genetic testing and counseling should be explored, complemented by careful dermatological examinations and consideration of noninvasive or invasive screening of the pancreas (see below).
Peutz-Jeghers (PJ) syndrome and other colon cancer syndromes: associations with pancreatic cancer
As an autosomal dominant disorder characterized by germline STK11 (LKB1) gene mutations, patients have early onset of large, multiple small intestinal and colonic polyps with increased risk of cancers at these sites (Rustgi 2007). As patients age, they have an increased risk of breast, lung, pancreatic, cervical, and other cancers. The types of pancreatic cancers include PDAC, IPMN, and serous cystadenocarcinomas (Sato et al. 2001). One comprehensive Dutch study of PJ syndrome patients revealed a 26% increased risk of pancreatic cancer by age 70 yr and relative risk of 76 (Korsse et al. 2013). Patients may also present with ampullary cancer or distal bile duct cancer. Conditional deletion of Lkb1 in the mouse pancreas results in postnatal acinar cell degeneration, acinar-to-ductal metaplasia, and serous cystadenomas (Hezel et al. 2008), suggesting some discordance between the pancreatic phenotype observed in PJ syndrome patients and this particular mouse model. In that vein, patients with familial adenomatous polyposis (FAP), characterized by germline APC gene mutations resulting in derangement of Wnt signaling, do not typically have pancreatic cancer but may have ampullary cancer or duodenal cancer. This suggests that APC mutations are not critical in sporadic PDAC. Lynch syndrome is an inherited form of colon cancer that is associated with germline mismatch repair (MMR) gene mutations (e.g., hMLH1, hMSH2, hMSH6, and hPMS2), resulting in microsatellite instability (MSI) in the colon cancers (and extracolonic cancers that are associated with Lynch syndrome). In a study of 147 families with germline MMR gene mutations, the cumulative risk of pancreatic cancer was 1.31% up to age 50 yr and 3.68% up to age 70 yr, an 8.6-fold increase compared with the general population (Kastrinos et al. 2009). However, MSI is not common in sporadic PDAC.
DNA repair defects: a potential unifying theme in a subset of familial pancreatic cancer
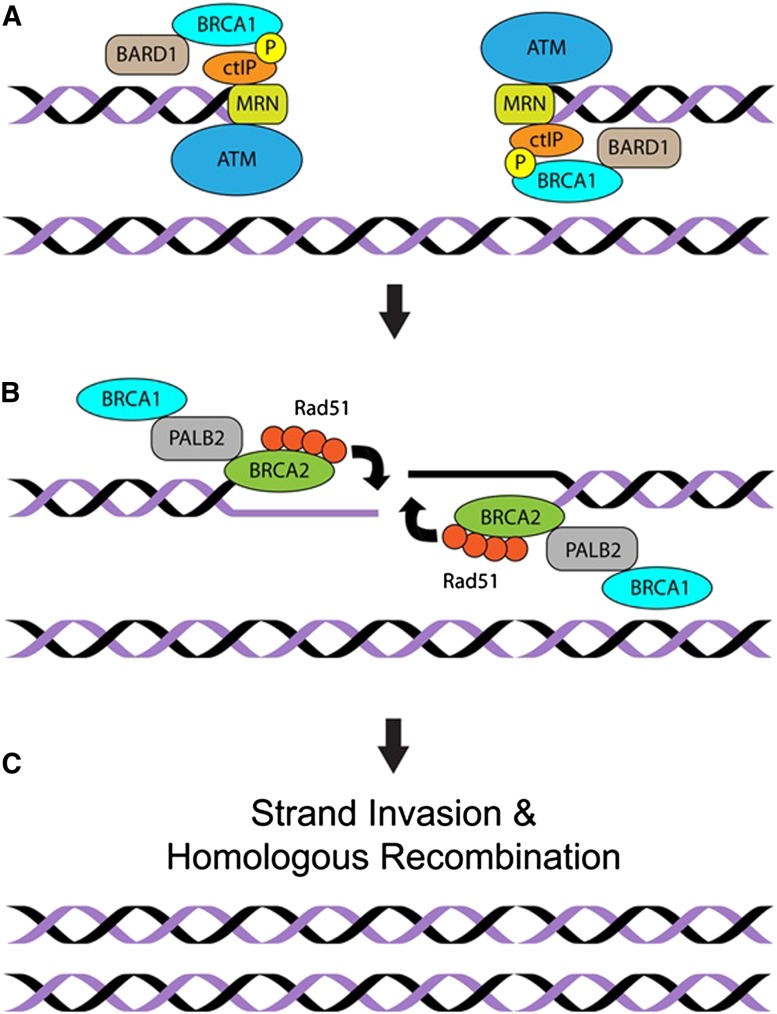
DNA repair processes respond to DNA damage, which can occur through abnormal endogenous cellular processes (e.g., mismatch of nucleotide bases, depurination, β elimination, and hydroxyl radical damage) or exposure to exogenous agents (ultraviolet light, ionizing radiation, and chemical carcinogens). One particularly cancer-causing type of DNA damage, based on the ability to promote loss of heterozygosity and translocations, is double-stranded breaks (DSBs). Repair of DSBs can involve nonhomologous end-joining (NHEJ), microhomology-mediated end-joining (MMEJ), and homologous recombination (HR). FANCC and FANCJ, as part of the Fanconi anemia complement of repair proteins, help preserve the integrity of replication forks during S phase. When replication forks collapse into DSBs, repair is then completed with assistance from ATM, BRCA2, and PALB2 (Fig. 2).
Figure 2.
DNA repair and HR. Mutations in the DNA repair cascades provide clues in the pathogenesis of familial pancreatic cancer. dsDNA breaks are recognized first by several protein complexes: BRCA1–BARD1; CTIP (C-terminal-binding protein, interacting protein), which associates with MRN (consisting of MRE11, RAD50, and NBS1, also known as Nibrin); and the ATM protein kinase. (A) These enzyme complexes are needed to perform the exonucleolytic resection of one of strand of DNA at the DSB, generating a 3′ overhang. BRCA1 fosters the accumulation of PALB2, a functional partner of BRCA2. (B) BRCA2 serves as a reservoir of RAD51, which is loaded onto the newly created ssDNA. RAD51 promotes invasion of this ssDNA into the intact sister chromatid to complete the repair of the DSB though replication of the sister (not shown). (C) The copied DNA is then resolved into two intact sister chromatids, completing the repair process.
The ATM kinase is activated by DSBs to delay cell cycle progression and promote DSB resection. This resection of one strand of DNA creates a single-stranded element onto which RAD51 is loaded by BRCA2 and PALB2 (Zhang et al. 2009). RAD51 then helps pair this strand with the template on the double-strand sister chromatid to replace damaged segments through replication and recombination (Fig. 2). Germline mutations in some of the DNA repair genes have been discovered in familial pancreatic cancer. The relevance of this is that accumulation of damaged DNA can trigger different states—senescence, apoptosis, or unregulated cell division—that may lead to formation of cancer.
Approximately 5%–10% of breast cancers are believed to be hereditary in nature, many of which are associated with germline BRCA1 or BRCA2 mutations. Of note, up to 10% of sporadic PDACs are believed to harbor BRCA2 mutations. Compared with BRCA1 mutation carriers, BRCA2 mutation carriers are at increased risk for pancreatic cancer. In a study of 173 families with germline BRCA2 mutations and breast/ovarian cancers, representing a cohort of 3728 individuals, the relative risk of pancreatic cancer was 3.51 (Goldgar 1995). This risk is particularly evident in families of Ashkenazi Jewish descent, where a germline 6174delT BRCA2 mutation has been reported (Ozçelik et al. 1997). Some estimates indicate that up to 10% of familial pancreatic cancer may be attributable to germline BRCA2 mutations, although the age of onset may not be particularly early in life. In a separate study, the relative risk of pancreatic cancer was nearly 2.3 in BRCA1 mutation carriers (Thompson et al. 2002). Analysis of conditional knockouts of Brca2 in the mouse pancreas has produced conflicting results on KRASG12D-driven pancreatic carcinogenesis. In one setting, Brca2 accelerates lesions in KRASG12D-expressing mice irrespective of Trp53 status, with evidence of only loss of one allele of Brca2 being necessary (Skoulidis et al. 2010). In another mouse model, Brca2 loss decelerates KRASG12D-driven pancreatic carcinogenesis but accelerates it when combined with Trp53 mutation (Rowley et al. 2011), again underscoring the careful interpretation that is necessary of some mouse models to correlate with human pancreatic cancer.
In a genome-wide sequencing analysis of nearly 170 families with familial pancreatic cancer, two families were identified as having germline heterozygous ATM mutations, and four families had deleterious homozygous ATM mutations (Roberts et al. 2012). PALB2 performs its tumor suppressor role at least in part by supporting HR-type DSB repair (HR-DSBR) through physical interactions with BRCA1, BRCA2, and RAD51 (Zhang et al. 2009). In an analysis of nearly 100 families with familial pancreatic cancer, four families had evidence of protein-truncating mutations in PALB2 (Jones et al. 2009), confirmed in another study (Tischkowitz et al. 2009). The role of the Palb2 mutation in the initiation of pancreatic cancer via loss of its chromosome integrity maintenance role has not been investigated to date in mouse pancreata. However, Palb2 is known to play an important cancer-suppressive role in another tissue (namely, the breast), as it synergizes with Trp53 to limit mammary tumors in mice (Bowman-Colin et al. 2013). It is likely that germline BRCA2, ATM, or PALB2 mutations in human familial pancreatic cancer contribute to defective DNA repair, accumulation of damaged DNA, genomic instability, and eventual cancer formation. Genomic instability is a hallmark feature of sporadic PDAC (Campbell et al. 2010), and understanding defective DNA repair mechanisms may shed light on the molecular pathogenesis of sporadic PDAC.
Approaches to discovery of new gene mutations
Although advances have been made in associating germline gene mutations with some forms of familial pancreatic cancer, our full realization of the degree to which pancreatic cancer is driven by underlying genetic predisposition requires further elucidation. Initial clues were provided by an analysis of CNVs. CNVs correspond to large regions of the genome that have been duplicated, deleted, or inverted or undergone translocation and can provide clues as to potential genes that contribute to pathogenesis of inherited and/or sporadic cancer. In an initial study of 57 familial pancreatic cancer kindreds, a total of 56 unique genomic regions with CNVs were identified that were not present in controls, including 31 amplifications and 25 deletions (Lucito et al. 2007). In a subsequent study of 120 familial pancreatic cancer cases (and 1194 controls), a total of 93 nonredundant familial pancreatic cancer-specific CNVs (53 losses and 40 gains) were identified in 50 cases, with each CNV present in a single individual (Al-Sukhni et al. 2012).
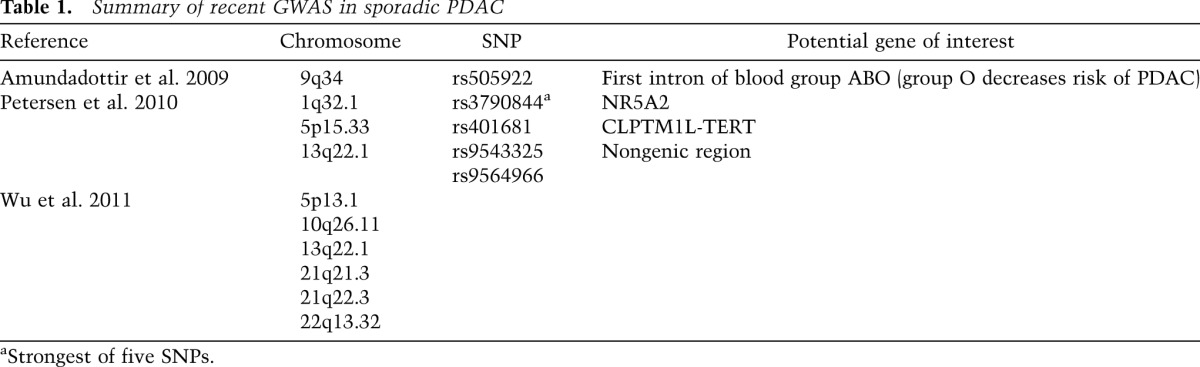
Genome-wide associations studies (GWASs) in patients with sporadic PDAC may highlight gene loci that could be tested for linkage in familial pancreatic cancer (Petersen et al. 2010; Wu et al. 2011). To that end, susceptibility loci have been identified, although differences in results may be attributable to the evaluation of different populations (Table 1). Deep DNA sequencing efforts, which are ongoing, in familial pancreatic cancer should shed new insights into new genes, genotypic–phenotypic correlations, pathways in familial and sporadic PDAC pathogenesis, and, hopefully, opportunities for diagnosis and therapy. An example of the latter is the potential application of poly-ADP-ribose polymerase (PARP) inhibitors or mitomycin C in BRCA2+ mutation carriers with pancreatic cancer.
Table 1.
Summary of recent GWAS in sporadic PDAC
Translational applications
Certainly, in familial pancreatic cancer where the gene mutation is known, genetic counseling and testing should be offered. This has important clinical implications in the affected individual as well as for at-risk individuals in the family. There is no consensus as to how to screen clinically individuals with known genetic mutations or those who have a strong family history absent a known genetic mutation for the possibility of incipient early pancreatic cancer. Clinical screening approaches segregate into noninvasive imaging of the pancreas (magnetic resonance imaging [MRI] or computerized tomography [CT]) or invasive methodologies, such as endoscopic ultrasound (Canto et al. 2012). One study found that in asymptomatic patients with familial pancreatic cancer, 7% had pancreatic cancer, and 16% had IPMN (Poley et al. 2009), suggesting that screening may be effective to detect preneoplastic or early stage neoplastic lesions. However, the age at which to start (and stop) screening, the subsequent frequency of screening, and how to manage ambiguous lesions are not clear (Canto et al. 2013).
New approaches under investigation (not yet established for implementation) would involve strategies for the detection of circulating pancreatic cells in the blood of at-risk familial pancreatic cancer patients, especially those with small lesions (e.g., IPMN). Furthermore, developments in imaging with molecular probes might improve on the threshold of detection provided by either MRI or CT. As more genomic and genetic information emerges to unravel the mechanistic underpinnings of familial pancreatic cancer, undoubtedly new translational applications will evolve.
Acknowledgements
I thank Dr. Eric Brown for critical discussions. The National Institutes of Health (NIH)/National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) R01 DK060694, NIH/NIDDK P30-DK050306 Center for Molecular Studies in Digestive and Liver Diseases, the American Association for Cancer Research-Research Acceleration Network (AACR-RAN), the University of Pennsylvania Abramson Cancer Center's Translational Center of Excellence in Pancreas Cancer, and the University of Pennsylvania Basser Research Center for BRCA supported this work.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.228452.113.
References
- Al-Sukhni W, Joe S, Lionel AC, Zwingerman N, Zogopoulos G, Marshall CR, Borgida A, Holter S, Gropper A, Moore S, et al. 2012. Identification of germline genomic copy number variation in familial pancreatic cancer. Hum Genet 131: 1481–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, Fuchs CS, Petersen GM, Arslan AA, Bueno-de-Mesquita HB, Gross M, Helzlsouer K, Jacobs EJ, et al. 2009. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet 41: 986–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer H, Jura N, Keller J, Jacobson M, Bar-Sagi D 2006. A mouse model of hereditary pancreatitis generated by transgenic expression of R122H trypsinogen. Gastroenterology 131: 1844–1855 [DOI] [PubMed] [Google Scholar]
- Bowman-Colin C, Xia B, Bunting S, Klijn C, Drost R, Bouwman P, Fineman L, Chen X, Culhane AC, Cai H, et al. 2013. Palb2 synergizes with Trp53 to suppress mammary tumor formation in a model of inherited breast cancer. Proc Natl Acad Sci 110: 8632–8637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, Morsberger LA, Latimer C, McLaren S, Lin ML, et al. 2010. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 467: 1109–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto MI, Hruban RH, Fishman EK, Kamel IR, Schulick R, Zhang Z, Topazian M, Takahashi N, Fletcher J, Petersen G, et al. 2012. Frequent detection of pancreatic lesions in asymptomatic high-risk individuals. Gastroenterology 142: 796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto MI, Harinck F, Hruban RH, Offerhaus GJ, Poley JW, Kamel I, Nio Y, Schulick RS, Bassi C, Kluijt I, et al. 2013. International Cancer of the Pancreas Screening (CAPS) Consortium summit on the management of patients with increased risk for familial pancreatic cancer. Gut 62: 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn JA, Friedman KJ, Noone PG, Knowles MR, Silverman LM, Jowell PS 1998. Relation between mutations of the cystic fibrosis gene and idiopathic pancreatitis. N Engl J Med 339: 653–658 [DOI] [PubMed] [Google Scholar]
- Cohn JA, Bornstein JD, Jowell PS 2000. Cystic fibrosis mutations and genetic predisposition to idiopathic chronic pancreatitis. Med Clin N Am 84: 621–631 [DOI] [PubMed] [Google Scholar]
- Goldgar DE 1995. Analysis of familial breast cancer in genetic analysis workshop 9: Summary of findings. Genet Epidemiol 12: 833–836 [DOI] [PubMed] [Google Scholar]
- Gruis NA, van der Velden PA, Sandkuijl LA, Prins DE, Weaver-Feldhaus J, Kamb A, Bergman W, Frants R 1995. Homozygotes for CDKN2 (p16) germline mutation in Dutch familial melanoma kindreds. Nat Genet 10: 351–353 [DOI] [PubMed] [Google Scholar]
- Hezel AF, Gurumurthy S, Granot Z, Swisa A, Chu GC, Bailey G, Dor Y, Bardeesy N, Depinho RA 2008. Pancreatic LKB1 deletion leads to acinar polarity defects and cystic neoplasms. Mol Cell Biol 28: 2414–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruban RH, Canto M, Goggins M, Schulick R, Klein AP 2010. Update on familial pancreatic cancer. Adv Surg 44: 293–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Hruban RH, Kamiyama M, Borges M, Zhang X, Parsons DW, Lin JC, Palmisano E, Brune K, Jaffee EM, et al. 2009. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science 324: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastrinos F, Mukherjee B, Tayob N, Wang F, Sparr J, Raymond VM, Bandipalliam P, Stoffel EM, Gruber SB, Syngal S 2009. Risk of pancreatic cancer in families with Lynch syndrome. JAMA 302: 1790–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastrinos F, Steyerberg EW, Mercado R, Balmaña J, Holter S, Gallinger S, Siegmund KD, Church JM, Jenkins MA, Lindor NM, et al. 2011. The PREMM(1,2,6) model predicts risk of MLH1, MSH2, and MSH6 germline mutations based on cancer history. Gastroenterology 140: 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kereszturi E, Szmola R, Kukor Z, Simon P, Weiss FU, Lerch MM, Sahin-Tóth M 2009. Hereditary pancreatitis caused by mutation-induced misfolding of human cationic trypsinogen: A novel disease mechanism. Hum Mutat 30: 575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AP 2013. Identifying people at a high risk of developing pancreatic cancer. Nat Rev Cancer 13: 66–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AP, Brune KA, Petersen GM, Goggins M, Tersmette AC, Offerhaus GJ, Griffin C, Cameron JL, Yeo CJ, Kern S, et al. 2004. Prospective risk of pancreatic cancer in familial pancreatic cancer kindreds. Cancer Res 64: 2634–2638 [DOI] [PubMed] [Google Scholar]
- Korsse SE, Harinck F, van Lier MGF, Biermann K, Offerhaus GJA, Krak N, Looman CWN, van Veelen W, Kuipers EJ, Wagner A, et al. 2013. Pancreatic cancer risk in Peutz-Jeghers syndrome patients: A large cohort study and implications for surveillance. J Med Genet 50: 59–64 [DOI] [PubMed] [Google Scholar]
- Lowenfels AB, Maisonneuve P, Cavallini G, Ammann RW, Lankisch PG, Andersen JR, Dimagno EP, Andrén-Sandberg A, Domellöf L 1993. Pancreatitis and the risk of pancreatic cancer: International Pancreatitis Study Group. N Engl J Med 328: 1433–1437 [DOI] [PubMed] [Google Scholar]
- Lucito R, Suresh S, Walter K, Pandey A, Lakshmi B, Krasnitz A, Sebat J, Wigler M, Klein AP, Brune K, et al. 2007. Copy-number variants in patients with a strong family history of pancreatic cancer. Cancer Biol Ther 6: 1592–1599 [DOI] [PubMed] [Google Scholar]
- Lynch HT, Fusaro RM, Lynch JF, Brand R 2008. Pancreatic cancer and the FAMMM syndrome. Fam Cancer 7: 103–112 [DOI] [PubMed] [Google Scholar]
- Ma J, Jemal A 2013. The rise and fall of cancer mortality in the USA: Why does pancreatic cancer not follow the trend? Future Oncol 9: 917–919 [DOI] [PubMed] [Google Scholar]
- Ooi CY, Dorfman R, Cipolli M, Gonska T, Castellani C, Keenan K, Freedman SD, Zielenski J, Berthiaume Y, Corey J, et al. 2011. Type of CFTR mutation determines risk of pancreatitis in patients with cystic fibrosis. Gastroenterology 140: 153–161 [DOI] [PubMed] [Google Scholar]
- Ozçelik H, Schmocker B, Di Nicola N, Shi XH, Langer B, Moore M, Taylor BR, Narod SA, Darlington G, Andrulis IL, et al. 1997. Germline BRCA2 6174delT mutations in Ashkenazi Jewish pancreatic cancer patients. Nat Genet 16: 17–18 [DOI] [PubMed] [Google Scholar]
- Permuth-Wey J, Egan KM 2009. Family history is a significant risk factor for pancreatic cancer: Results from a systematic review and meta-analysis. Fam Cancer 8: 109–117 [DOI] [PubMed] [Google Scholar]
- Petersen GM, Amundadottir L, Fuchs CS, Kraft P, Stolzenberg-Solomon RZ, Jacobs KB, Arslan AA, Bueno-de-Mesquita HB, Gallinger S, Gross M 2010. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet 42: 224–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poley JW, Kluijt I, Gouma DJ, Harinck F, Wagner A, Aalfs C, van Eijck CH, Cats A, Kuipers EJ, Nio Y, et al. 2009. The yield of first-time endoscopic ultrasonography in screening individuals at a high risk of developing pancreatic cancer. Gastroenterology 104: 2175–2181 [DOI] [PubMed] [Google Scholar]
- Ranade K, Hussussian CJ, Sikorski RS, Varmus HE, Goldstein AM, Tucker MA, Serrano M, Hannon GJ, Beach D, Dracopoli NC 1995. Mutations associated with familial melanoma impair p16INK4 function. Nat Genet 10: 114–116 [DOI] [PubMed] [Google Scholar]
- Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, Reichert M, Beatty GL, Rustgi AK, Vonderheide RH, et al. 2012. EMT and dissemination precede pancreatic tumor formation. Cell 148: 349–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts NJ, Jiao Y, Yu J, Kopelovich L, Petersen GM, Bondy ML, Gallinger S, Schwartz AG, Syngal S, Cote ML, et al. 2012. ATM mutations in patients with hereditary pancreatic cancer. Cancer Discov 2: 41–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosendahl J, Witt H, Szmola R, Bhatia E, Ozsvári B, Landt O, Schulz HU, Gress TM, Pfützer R, Löhr M, et al. 2008. Chymotrypsin C (CTRC) variants that diminish activity or secretion are associated with chronic pancreatitis. Nat Genet 40: 78–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley M, Ohashi A, Mondal G, Mills L, Yang L, Zhang L, Sundsbak R, Shapiro V, Muders MH, Smyrk T, et al. 2011. Inactivation of Brca2 promotes Trp53-associated but inhibits KrasG12D-dependent pancreatic cancer development in mice. Gastroenterology 140: 1303–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustgi AK 2007. The genetics of hereditary colon cancer. Genes Dev 21: 2525–2538 [DOI] [PubMed] [Google Scholar]
- Sato N, Rosty C, Jansen M, Fukushima N, Ueki T, Yeo CJ, Cameron JL, Iacobuzio-Donahue CA, Hruban RH, Goggins M 2001. STK11/LKB1 Peutz-Jeghers gene inactivation in intraductal papillary-mucinous neoplasms of the pancreas. Am J Pathol 159: 2017–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharer N, Schwarz M, Malone G, Howarth A, Painter J, Super M, Braganza J 1998. Mutations of the cystic fibrosis gene in patients with cystic fibrosis. N Engl J Med 339: 645–652 [DOI] [PubMed] [Google Scholar]
- Skoulidis F, Cassidy LD, Pisupati V, Jonasson JG, Bjarnason H, Eyfjord JE, Karreth FA, Lim M, Barber LM, Clatworthy SA, et al. 2010. Germline Brca2 heterozygosity promotes Kras(G12D)-driven carcinogenesis in a murine model of familial pancreatic cancer. Cancer Cell 18: 499–509 [DOI] [PubMed] [Google Scholar]
- Szabo A, Sahin-Toth M 2012. Increased activation of hereditary pancreatitis-associated human cationic trypsinogen mutants in the presence of chymotrypsin C. J Biol Chem 287: 20701–20710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson D, Easton DF, Breast Cancer Linkage Consortium 2002. Cancer incidence in BRCA1 mutation carriers. J Natl Cancer Inst 94: 1358–1365 [DOI] [PubMed] [Google Scholar]
- Tischkowitz MD, Sabbaghian N, Hamel N, Borgida A, Rosner C, Taherian N, Srivastava A, Holter S, Rothenmund H, Ghadirian P, et al. 2009. Analysis of the gene coding for the BRCA2-interacting protein PALB2 in familial and sporadic pancreatic cancer. Gastroenterology 137: 1183–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Chen S, Brune KA, Hruban RH, Parmigiani G, Klein AP 2007. PancPRO: Risk assessment for individuals with a family history of pancreatic cancer. J Clin Oncol 25: 1417–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitcomb DC 2013. Genetic risk factors for pancreatic disorders. Gastroenterology 144: 1292–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitcomb DC, Gorry MC, Preston RA, Furey W, Sossenheimer MJ, Ulrich CD, Martin SP, Gates LK Jr, Amann ST, Toskes PP, et al. 1996. Hereditary pancreatitis is caused by a mutation in the cationic trypsinogen gene. Nat Genet 14: 141–145 [DOI] [PubMed] [Google Scholar]
- Wu C, Miao X, Huang L, Che X, Jiang G, Yu D, Yang X, Cao G, Hu Z, Zhou Y, et al. 2011. Genome-wide association study identifies five loci associated with susceptibility to pancreatic cancer in Chinese populations. Nat Genet 44: 62–66 [DOI] [PubMed] [Google Scholar]
- Yadav D, Lowenfels AB 2013. The epidemiology of pancreatitis and pancreatic cancer. Gastroenterology 144: 1252–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Ma J, Wu J, Ye L, Cai H, Xia B, Xiaochun Yu X 2009. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol 19: 524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Weger J, Yang Q, Goldstein AM, Tucker MA, Walker GJ, Hayward N, Dracopoli NC 1996. Germline mutations in the p16INK4a binding domain of CDK4 in familial melanoma. Nat Genet 12: 97–99 [DOI] [PubMed] [Google Scholar]