The kinetics with which promoter-proximal paused Pol II undergoes premature termination versus productive elongation is key to understanding mechanisms of metazoan transcription regulation. To assess the fate of paused Pol II, Lis and colleagues measured the stability of paused Pol II at the Drosophila Hsp70 locus through a combination of optical studies and biochemical analysis. Heat shock drastically increases elongating Pol II without decreasing termination, indicating that regulation acts at the step of paused Pol II entry to productive elongation.
Keywords: RNA polymerase II, promoter-proximal pausing, termination, escape to productive elongation, photoactivation, nascent RNA
Abstract
The kinetics with which promoter-proximal paused RNA polymerase II (Pol II) undergoes premature termination versus productive elongation is central to understanding underlying mechanisms of metazoan transcription regulation. To assess the fate of Pol II quantitatively, we tracked photoactivatable GFP-tagged Pol II at uninduced Hsp70 on polytene chromosomes and showed that Pol II is stably paused with a half-life of 5 min. Biochemical analysis of short nascent RNA from Hsp70 reveals that this half-life is determined by two comparable rates of productive elongation and premature termination of paused Pol II. Importantly, heat shock dramatically increases elongating Pol II without decreasing termination, indicating that regulation acts at the step of paused Pol II entry to productive elongation.
Many metazoan genes have a high occupancy of transcriptionally engaged RNA polymerase II (Pol II) paused near their promoters (Bentley and Groudine 1986; Rougvie and Lis 1988; Muse et al. 2007; Zeitlinger et al. 2007; Core et al. 2008). Increasingly, studies indicate that the transition of this promoter-proximal Pol II into productive elongation is one of the major regulatory checkpoints of gene expression. Previous studies hypothesized that the accumulated Pol II at the promoter is either stably paused or iteratively terminating prematurely during early elongation (Bentley and Groudine 1986; Rougvie and Lis 1988). Interestingly, recent reports indicate that both pausing (DSIF and NELF) (Wu et al. 2003; Rahl et al. 2010) and termination (Dcp1a, Xrn2, and TTF2) (Brannan et al. 2012) factors are enriched at metazoan promoters, and their depletions can alter the distribution of Pol II. Therefore, the relative contribution of pausing or premature termination to promoter Pol II accumulation and its regulation is a central question that has yet to be quantitatively addressed in vivo.
The Drosophila Hsp70 heat-shock gene possesses a promoter-proximal Pol II that has been extensively characterized (Fuda et al. 2009). Under basal conditions, Pol II on Hsp70 pauses at sites 20–40 base pairs (bp) downstream from the transcription start site (Rasmussen and Lis 1993; Kwak et al. 2013), producing an accumulation of Pol II at the 5′ end of the gene and a basal distribution of Pol II along the gene body (Lis 1998). Upon heat-shock induction, the escape of paused Pol II to productive elongation as well as the initiation rate increase up to 100-fold, leading to the massive production of Hsp70 mRNA.
High-resolution live-cell imaging of the polytene chromosomes in Drosophila salivary glands provides one method to analyze the kinetics of Pol II induction and elongation during the heat-shock activation of Hsp70 (Darzacq et al. 2009). The heat-shock-activated endogenous Hsp70 loci (87A/C) can be easily located in living polytene nuclei because they produce a distinct doublet of intense GFP-Pol II (or RFP)-containing puffs after the activation. Fluorescence recovery after photobleaching (FRAP) can then be used to measure dynamics and elongation rates (Yao et al. 2007). However, the dynamics of Pol II under uninduced conditions has been difficult to assay due to the challenges of locating the endogenous Hsp70 loci without heat-shock activation.
Notably, many features of the paused Pol II at the uninduced Hsp70 are similar to the large number (∼70%) of active Drosophila genes containing paused Pol II (Core et al. 2012). In addition, many of the proteins identified to be involved in Hsp70 gene regulation have corresponding activities at other genes in various organisms (Fuda et al. 2009). These findings indicate that the mechanisms governing Hsp70 gene regulation are general. Therefore, in order to gain insights into the kinetic status of the promoter-proximal paused Pol II (paused Pol II that is stable vs. prematurely terminating), we measured the stability of paused Pol II at Drosophila Hsp70 through a combination of complementing optical and biochemical strategies that resolves previous challenges. We found that promoter-paused Pol II is relatively stable, and its entry to productive elongation, not its termination, is regulated by heat-shock activation.
Results and Discussion
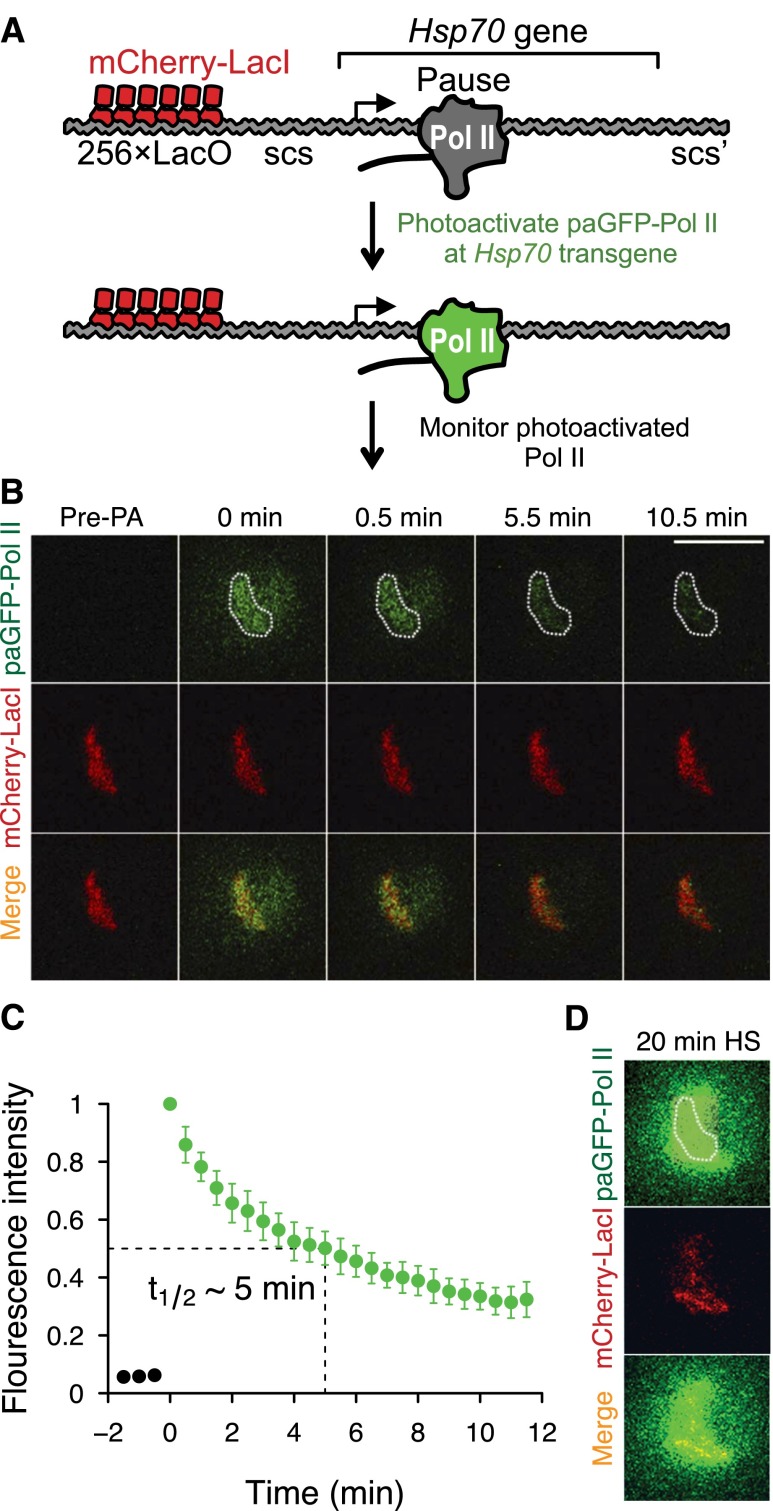
To examine the kinetic fate of paused Pol II, we used an optical approach to measure the stability of Pol II fused to the photoactivatable GFP (paGFP) (Patterson and Lippincott-Schwartz 2002) at the Hsp70 locus on Drosophila salivary gland polytene chromosomes (Fig. 1A). These interphase-like giant chromosomes have been used as a platform for high-resolution imaging of the dynamics of Pol II and other transcription factors by optical pulse-chase experiments at targeted genomic loci (Lis 2007). However, under uninduced basal conditions, identifying the endogenous Hsp70 gene loci is technically challenging, since the paused Pol II signal from Hsp70 is too weak to allow it to be easily distinguished from other Pol II signals (Supplemental Fig. 1a).
Figure 1.
Imaging the stability of paused Pol II at the uninduced Hsp70 transgene in living cells. (A) Schematic of the live-cell imaging experiment showing mCherry-LacI bound to the operator sites (red) and photoactivated Pol II (green). Scs and scs′ are the insulator elements flanking the Hsp70 gene. (B) Uninduced mCherry-marked Hsp70 transgene before and following the time course after paGFP-Pol II photoactivation. The region of interest is outlined by white dots. (C) Normalized fluorescence intensities of paGFP-Pol II fluorescence decay after photoactivation (FDAP) under the uninduced condition (n = 9). (D) After heat shock, images of mCherry-LacI, paGFP-Pol II (rephotoactivated after heat shock), and merge at the Hsp70 transgene. Error bars indicate SD. Bar, 10 μm.
To circumvent this problem, we generated a transgenic Hsp70 gene that can be easily identified on the polytene chromosomes for targeted analysis. The full-length Hsp70 gene is marked with 256 repeats of Escherichia coli Lac operator sites (LacO) and can be rapidly identified by coexpressing a fluorescently tagged Lac repressor (mCherry-LacI) in salivary gland nuclei (Fig. 1A). We tested that this LacO-tagged transgenic Hsp70 is functionally equivalent to the endogenous Hsp70 gene (Zobeck et al. 2010) by examining the recruitment and levels of Pol II intensity at puffs in response to heat shock (Supplemental Fig. 1b,c). We also confirmed that the paGFP-labeled Pol II subunit (Rpb9) reliably tracks Pol II at both the endogenous Hsp70 loci and the LacO-tagged Hsp70 transgene (Supplemental Figs. 2, 3).
The paGFP-Pol II is fluorescently inert at the Hsp70 transgene before photoactivation (Fig. 1B). To examine the dynamics of paused Pol II at the Hsp70 loci, we used laser-scanning confocal microscopy to specifically photoactivate paGFP-Pol II at the LacO-marked Hsp70 transgene under uninduced conditions (Fig. 1A). Figure 1, B and C, shows the time series of the fluorescence decay of paGFP-Pol II at the Hsp70 transgene. Importantly, the fact that the paGFP-Pol II signal is heat-shock-inducible validates that the locus examined at the single mCherry-LacI band was the Hsp70 transgene (Fig. 1D). Photobleaching is minimal during imaging (Supplemental Fig. 4), indicating that the decay of signal is due to the release of paGFP-Pol II from the transgene. The resulting decay approximates first-order kinetics with the half-life of ∼5 min (Fig. 1C). Because the main form of Pol II at the uninduced Hsp70 locus is paused Pol II (Core et al. 2012), this clearance half-life of 5 min indicates that paused Pol II is relatively stable but has a finite lifetime (Table 1).
Table 1.
Rate constants for the kinetics of promoter-proximal Pol II at uninduced Hsp70
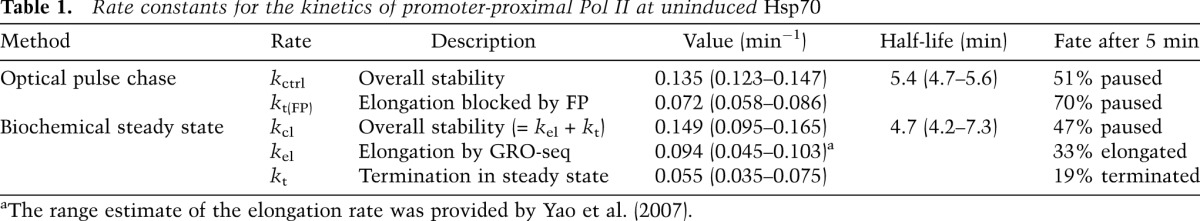
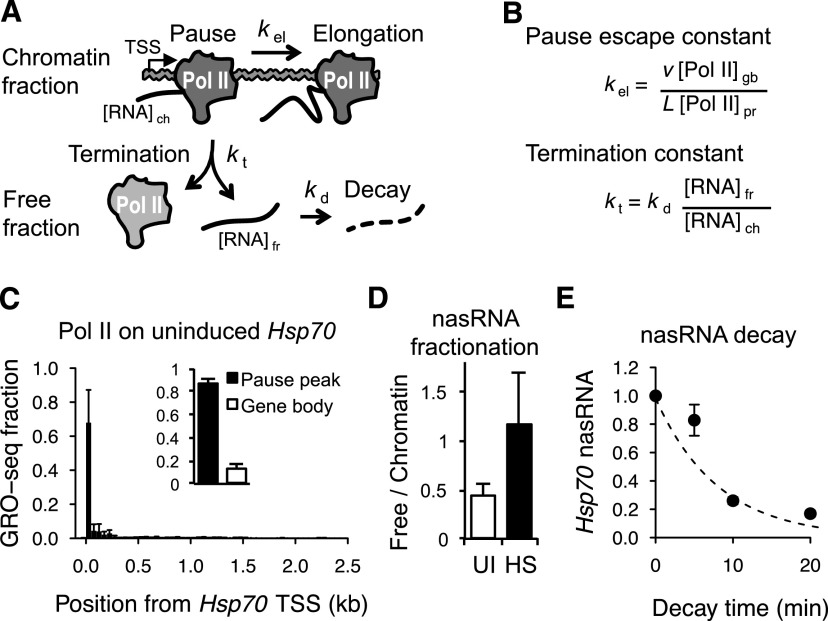
The lifetime of the paused Pol II can be a consequence of the escape into productive elongation, premature termination, or both. To measure the contribution of each to the stability of paused Pol II, we developed an independent biochemical kinetic strategy in Drosophila S2 cells (Fig. 2A). First, we evaluated the rate of escape into productive elongation. If a certain fraction of paused Pol II escape into productive elongation every minute (kel), these escaped Pol II will be distributed in the gene body region defined by the speed of elongation. Therefore, the rate of escaping Pol II can then be derived from the speed of Pol II elongation and the relative ratio between gene body and paused Pol II density (Fig. 2B,; Supplemental Material). For this measurement, we used pre-existing nuclear run-on sequencing (GRO-seq or PRO-seq) data sets in S2 cells (Fig. 2C; Core et al. 2012; Kwak et al. 2013), and the elongation speed of 1.5 kb/min for Hsp70 from previous studies (Yao et al. 2007; Ardehali and Lis 2009). The run-on sequencing results show that at Hsp70, an average of 86% of engaged Pol IIs are restricted to the promoter-proximal region, and 13% are distributed in the 2.4-kb gene body region (Fig. 2C). From these estimates, we calculated that kel = 0.094 min−1; that is, 33% of paused Pol II escape into elongation every 5 min (Table 1).
Figure 2.
Biochemical analysis of steady-state paused Pol II kinetics. (A) Schematic showing the kinetic fates of paused Pol II and short nascent RNA (nasRNA). (kel) Kinetic constant of paused Pol II elongation; (kt) kinetic constant of paused Pol II termination; (kd) short nascent RNA decay constant; ([RNA]fr,) free Hsp70 short nascent RNA; ([RNA]ch) chromatin-bound Hsp70 short nascent RNA. (B) The equations used to calculate the pause Pol II elongation and termination rate constants (see the Supplemental Material). (v) Elongation rate; (L) gene length; ([Pol II]pr) Pol II fraction in pause region; ([Pol II]gb) Pol II fraction in gene body region. (C) Estimation of kel from GRO-seq (and PRO-seq) read fractions in Hsp70 (50-bp bins; n = 14 independent data sets). The inset shows read fractions in pause region (−50 to +250 from the transcription start site [TSS]) and gene body (+300 to 2.4 kb). (D) Measuring the ratio between free and chromatin-bound short nascent RNA by qRT–PCR in uninduced (UI; n = 9) and 15-min heat-shock (HS; n = 8) conditions. (E) Estimation of kd from free Hsp70 short nascent RNA decay after Triptolide (10 μM) addition (qRT–PCR). Each time point is normalized to the pretreatment level. (C–E) Error bars indicate the SEM.
Having measured the amount of paused Pol II that escapes into productive elongation, we then devised a biochemical measurement of the Pol II that terminates prematurely. Paused Pol II that terminates will dissociate from chromatin, and the engaged short nascent RNA will be released (Fig. 2A). Therefore, the amount of free short nascent RNA relative to the chromatin-associated (paused) short nascent RNA will reflect the rate of premature termination (Fig. 2B). We measured the amount of short nascent Hsp70 RNA from chromatin-associated (paused) and free (terminated) Pol II using biochemical fractionation (Wuarin and Schibler 1994). Short nascent RNA was quantitatively measured by ligation-mediated quantitative RT–PCR (qRT–PCR) targeting most of the Hsp70 pausing region within 25–40 bp downstream from the transcription start site (Supplemental Fig. 5). The nascent RNA of the free fraction was, on average, 45% of the chromatin-bound fraction level in uninduced Hsp70 (Fig. 2D, UI), suggesting that a significant amount of Pol II may be terminating from the uninduced Hsp70 pause sites.
For a quantitative estimation of the paused Pol II termination rate, we analyzed the steady-state kinetics of short Hsp70 nascent RNA termination and decay (Fig. 2A; Supplemental Material). At steady state, the decay rate of the free nascent RNA should be equal to its production rate from the terminating Pol II. Therefore, the termination rate can be derived from the free RNA decay rate and the ratio between free and chromatin-bound short nascent RNA (Fig. 2B). To estimate the decay rate, we blocked the RNA production at the initiation step by using Triptolide, a potent chemical inhibitor of the TFIIH helicase XPB (Titov et al. 2011), and measured the time course of free nascent RNA decay. The time course showed a decay half-life of ∼6 min and the decay constant kd = 0.123 min−1 (Fig. 2E). This decay constant, when combined with the ratio between free and chromatin-bound short nascent RNA (described above), allows the calculation of the termination constant kt = 0.055 min−1, meaning that 19% of paused Pol II terminate prematurely every 5 min (Table 1). The rate of clearing of paused Pol II is the sum of termination and escape to elongation; adding these rates gives a Pol II that has a clearance kinetic constant (kcl) of 0.149 min−1, which corresponds to the half-life of 4.7 min (Table 1). Therefore, the two independent methods—biochemical steady-state kinetics and optical pulse-chase measurements—are in close agreement with each other.
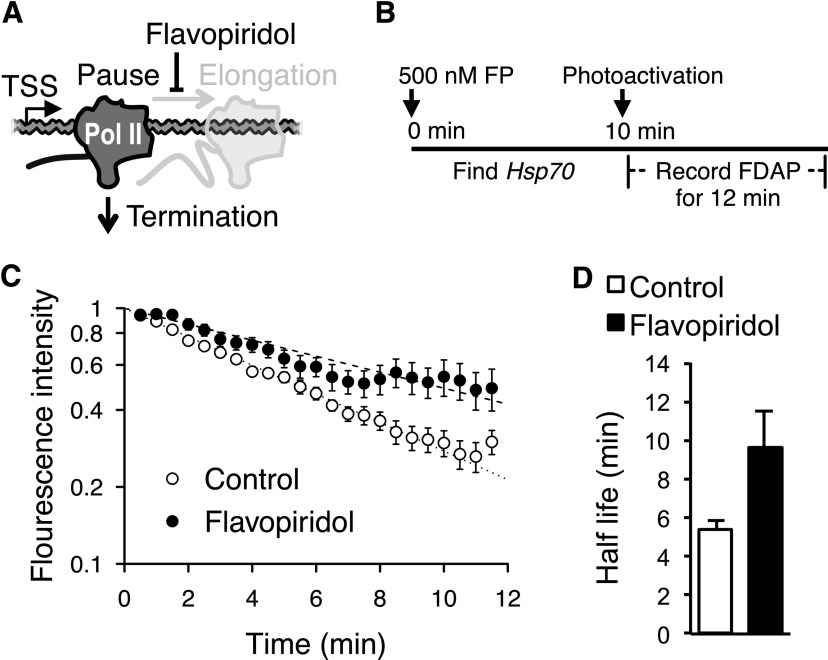
P-TEFb (positive transcription elongation factor b) kinase activity is required for the escape of paused Pol II into productive elongation at most genes, including Drosophila Hsp70 (Lis et al. 2000; Chao and Price 2001; Ni et al. 2008; Rahl et al. 2010). To obtain an independent estimate of the early termination rate of paused Pol II in vivo, salivary glands were treated with the P-TEFb kinase inhibitor Flavopiridol (Chao and Price 2001; Ni et al. 2008) to block transcription elongation (Fig. 3A). Assaying the fluorescence decay after photoactivation (FDAP) of paGFP-Pol II at the uninduced Hsp70 transgene then reveals the loss of Pol II by mechanisms that are independent of P-TEFb activity, and we assume this is mainly by termination (Fig. 3B). We further analyzed the fluorescence using a quantitative decay model in both control and Flavopiridol treatment (Supplemental Fig. 6). The decay curves show the stabilization of Pol II (Fig. 3C; Supplemental Fig. 7) and near doubling of the half-life with Flavopiridol treatment (Fig. 3D; Table 1), indicating that the elongation and termination can also be distinctly measured using optical methods in vivo. Together, the optical and biochemical analyses provide a strong indication that paused Pol II is relatively stable in uninduced cells but does undergo slow transitions to both productive elongation or premature termination in living cells. This is additionally supported by a very recent independent and different approach showing that many other active Drosophila genes also have stably paused Pol II (Henriques et al. 2013).
Figure 3.
Optical measurement of the termination rate of paused Pol II at the Hsp70 transgene in living cells. (A) Schematic diagram outlining overall logic of using Flavopiridol to measure paused Pol II termination kinetics. (B) Illustration of the experimental scheme. (C) Semi-log plot of normalized fluorescence intensities of paGFP-Pol II under uninduced condition in control (n = 9) and with Flavopiridol treatment (n = 7). Data are corrected for background Pol II signal (see the Materials and Methods). Error bars indicate SD. (D) Half-lives of paused Pol II decay with Flavopiridol treatment and control. (C,D) Error bars indicate SEM.
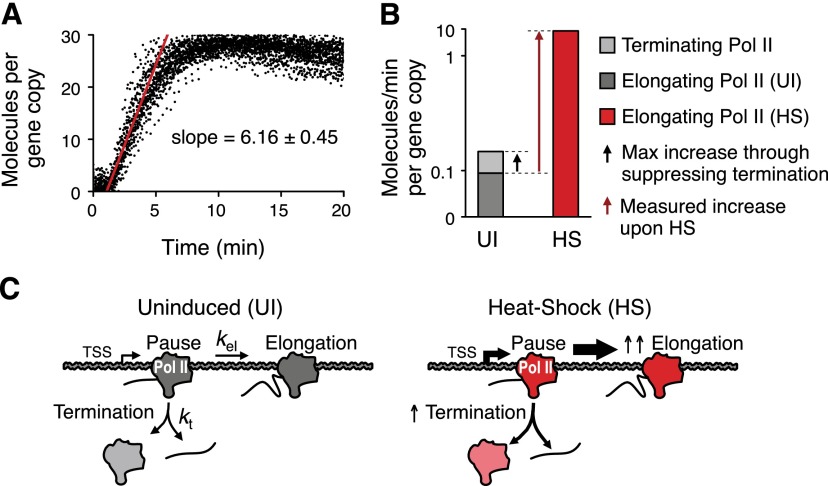
The fact that paused Pol II can terminate raises the intriguing possibility that premature termination may play a role in Hsp70 gene regulation, as demonstrated in the examples of prokaryotic or viral promoters (Kao et al. 1987; Gollnick and Babitzke 2002). We assessed the contribution of premature termination to the regulation of Hsp70 expression level upon heat-shock induction based on our kinetic findings. The rate of Pol II escape into elongation rapidly increases after the heat-shock induction. This increase is equal to the initial rate of Pol II recruitment to the activated Hsp70, which was measured previously using a high-temporal-resolution recruitment study of Pol II in living cells (Zobeck et al. 2010), and here we estimate this rate to be about six molecules per minute per promoter (Fig. 4A). If all prematurely terminating Pol II are converted to productively elongating Pol II, the expected increase would be 0.055 molecules per minute per promoter (Table 1), which explains only a small fraction of the elongating Pol II (Fig. 4B). In addition, we observed an increase rather than the decrease of terminated nascent RNA fractions from paused Pol II upon heat shock (Fig. 2D, HS), opposite to the expectation if down-regulation of premature termination contributes to the heat-shock induction. Collectively, these findings provide a comprehensive assessment of paused Pol II kinetics of elongation and termination on Hsp70 (Fig. 4C). Although cycles of initiation and premature termination occur at the promoter-proximal Pol II on Hsp70, this process is slow, and Pol II at the pause is relatively stable; the changes in premature termination do not appear to contribute to Hsp70 activation. Rather, the regulation of the Hsp70 gene upon heat shock occurs mainly through stimulation of escape from the stable pause.
Figure 4.
Kinetics of early elongating Pol II at Hsp70 during heat-shock induction. (A) Estimation of initial Pol II escape rate using live-cell imaging at the endogenous Hsp70 upon heat shock. The imaging data from a previous study (Zobeck et al. 2010) were analyzed (n = 33). (B) Contribution of premature termination to Pol II escape rate upon heat-shock induction. (UI) Uninduced condition; (HS) heat-shock induction. The Y-axis is in a nonlinear scale. (C) Model of early elongating Pol II kinetics under uninduced and heat-shock-activated conditions. The width of an arrow reflects the magnitude of the rate.
Our optical approach addresses the long-standing question of paused Pol II stability. One possible interpretation of the optical analysis is that the photoactivated paused Pol II in uninduced cells may be frequently terminated and recycled at the same locus. However, the paused Pol II is known to be phosphorylated at Ser5, and it is the unphosphorylated form of Pol II that is used in the reinitiation step (Laybourn and Dahmus 1990). Additionally, we observed previously in FRAP assays that, during the early phases of induction of the Hsp70 gene, Pol II dissociates from the locus rather than recycling and only partially recycles after accumulating at the locus at high concentrations (Yao et al. 2007; Zobeck et al. 2010). In addition, we further provide orthogonal, biochemical evidence of the stability of paused Pol II using nascent RNA, which can be assessed independently from recycling.
The molecular mechanism of premature termination still remains to be investigated. 3′ RNA processing and termination factors may have a role in this termination by targeting the nascent RNA engaged with paused Pol II near the promoter (Brannan et al. 2012). However, nascent RNA extending outside of the Pol II complex may not have enough accessibility for RNA processing factors, since the positions of promoter-proximal pausing in Hsp70 are, like many other paused genes in Drosophila, relatively close to the transcription start site (Rasmussen and Lis 1993; Kwak et al. 2013). Nonetheless, this mechanism may still exist if additional premature termination takes place after Pol II escapes pausing in nearby downstream regions or for other Drosophila genes shown to have more distal pausing (Kwak et al. 2013).
The prolonged pausing itself may also lead to spontaneous Pol II termination, similar to what was observed in Pol II pausing at DNA damage sites (Somesh et al. 2005; Anindya et al. 2007). In addition, it is possible that a trailing or a newly initiating Pol II molecule may collide with the paused Pol II and result in the termination of the leading Pol II molecule, as seen in in vitro studies (Saeki and Svejstrup 2009). This may explain our observation of increased short terminated transcripts under heat-shock induction, where initiation is much more frequent. Thus, premature termination appears to be a consequence of the promoter dynamics rather than a control mechanism for Pol II productive elongation.
Materials and methods
FDAP of polytene nuclei
Intact Drosophila salivary glands were dissected from third instar larvae and transferred to Grace's medium. For drug experiments, glands were transferred to 500 nM Flavopiridol (Sigma-Aldrich) diluted in medium. Laser-scanning confocal microscopy of salivary glands was carried out on a Zeiss 710 microscope. The mCherry-LacI-tagged Hsp70 transgene was identified using a 561-nm laser. Samples were photoactivated using a circular region of interest limited to the dimensions of the mCherry-LacI spot using a 405-nm laser. The fluorescence of both the mCherry-LacI and Rpb9-paGFP was imaged using 561- and 488-nm lasers every 30 sec for 12 min. Photoactivation curves were normalized for the first image to equal 1. Half-lives were obtained by fitting the FDAP data to an exponential component using a grid-search regression in a mixed linear decay model. To confirm that the Hsp70 gene was targeted, an objective preheated to 37°C (Bioptechs) was used to heat-shock samples for 20 min, and the locus was rephotoactivated.
Biochemical steady-state kinetic analysis
The rate constant of elongating Pol II from pausing (kel) was derived from GRO-seq and PRO-seq data (Core et al. 2012; Kwak et al. 2013) in Drosophila S2 cells at the Hsp70 gene: kel = vλ/[Pol II]pr, where v is the Pol II elongation speed (in kilobases per minute), λ is the gene body Pol II density (in reads per kilobase), and [Pol II]pr is the level of promoter-proximal Pol II (in reads). The kinetic constant of Pol II termination (kt) was determined from nascent RNA fractionation in S2 cells: kt = kd ([RNA]fr/[RNA]ch), where [RNA]ch is the nascent RNA in chromatin fraction, [RNA]fr is the nascent RNA in free fraction, and kd is the free nascent RNA decay constant. Nascent RNA fractionations were carried out as described previously (Wuarin and Schibler 1994), and a short spike-in RNA sequence from an Arabidopsis gene (RCP1) was added for the normalization after ligation-mediated qRT–PCR analysis.
Acknowledgments
We thank Jie Yao (Yale University) for providing the range estimates of the elongation rate on Hsp70. This work was supported by a grant from the National Institute of Health (NIH; GM25232 to J.T.L.) and fellowships from NIH (GM087003 to M.S.B.) and the Howard Hughes Medical Institute (to H.K.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.231886.113.
References
- Anindya R, Aygün O, Svejstrup JQ 2007. Damage-induced ubiquitylation of human RNA polymerase II by the ubiquitin ligase Nedd4, but not Cockayne syndrome proteins or BRCA1. Mol Cell 28: 386–397 [DOI] [PubMed] [Google Scholar]
- Ardehali MB, Lis JT 2009. Tracking rates of transcription and splicing in vivo. Nat Struct Mol Biol 16: 1123–1124 [DOI] [PubMed] [Google Scholar]
- Bentley DL, Groudine M 1986. A block to elongation is largely responsible for decreased transcription of c-myc in differentiated HL60 cells. Nature 321: 702–706 [DOI] [PubMed] [Google Scholar]
- Brannan K, Kim H, Erickson B, Glover-Cutter K, Kim S, Fong N, Kiemele L, Hansen K, Davis R, Lykke-Andersen J, et al. 2012. mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol Cell 46: 311–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao SH, Price DH 2001. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem 276: 31793–31799 [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Gilchrist DA, Fargo DC, Kwak H, Adelman K, Lis JT 2012. Defining the status of RNA polymerase at promoters. Cell Rep 2: 1025–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Yao J, Larson DR, Causse SZ, Bosanac L, de Turris V, Ruda VM, Lionnet T, Zenklusen D, Guglielmi B, et al. 2009. Imaging transcription in living cells. Annu Rev Biophys 38: 173–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuda NJ, Ardehali MB, Lis JT 2009. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 461: 186–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollnick P, Babitzke P 2002. Transcription attenuation. Biochim Biophys Acta 1577: 240–250 [DOI] [PubMed] [Google Scholar]
- Henriques T, Gilchrist DA, Nechaev S, Bern M, Muse GW, Burkholder A, Fargo DC, Adelman K 2013. Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol Cell 52: 517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao SY, Calman AF, Luciw PA, Peterlin BM 1987. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 330: 489–493 [DOI] [PubMed] [Google Scholar]
- Kwak H, Fuda NJ, Core LJ, Lis JT 2013. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339: 950–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laybourn PJ, Dahmus ME 1990. Phosphorylation of RNA polymerase IIA occurs subsequent to interaction with the promoter and before the initiation of transcription. J Biol Chem 265: 13165–13173 [PubMed] [Google Scholar]
- Lis JT 1998. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harb Symp Quant Biol 63: 347–356 [DOI] [PubMed] [Google Scholar]
- Lis JT 2007. Imaging Drosophila gene activation and polymerase pausing in vivo. Nature 450: 198–202 [DOI] [PubMed] [Google Scholar]
- Lis JT, Mason P, Peng J, Price DH, Werner J 2000. P-TEFb kinase recruitment and function at heat shock loci. Genes Dev 14: 792–803 [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K 2007. RNA polymerase is poised for activation across the genome. Nat Genet 39: 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Z, Saunders A, Fuda NJ, Yao J, Suarez J-R, Webb WW, Lis JT 2008. P-TEFb is critical for the maturation of RNA polymerase II into productive elongation in vivo. Mol Cell Biol 28: 1161–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson GH, Lippincott-Schwartz J 2002. A photoactivatable GFP for selective photolabeling of proteins and cells. Science 297: 1873–1877 [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA 2010. c-Myc regulates transcriptional pause release. Cell 141: 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen EB, Lis JT 1993. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Natl Acad Sci 90: 7923–7927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AEA, Lis JTJ 1988. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54: 795–804 [DOI] [PubMed] [Google Scholar]
- Saeki H, Svejstrup JQ 2009. Stability, flexibility, and dynamic interactions of colliding RNA polymerase II elongation complexes. Mol Cell 35: 191–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somesh BP, Reid J, Liu W-F, Søgaard TMM, Erdjument-Bromage H, Tempst P, Svejstrup JQ 2005. Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell 121: 913–923 [DOI] [PubMed] [Google Scholar]
- Titov DV, Gilman B, He Q-L, Bhat S, Low W-K, Dang Y, Smeaton M, Demain AL, Miller PS, Kugel JF, et al. 2011. XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nat Chem Biol 7: 182–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-H, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, Enerly E, Larsson J, Lambertsson A, Handa H, Gilmour D 2003. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev 17: 1402–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuarin J, Schibler U 1994. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: Evidence for cotranscriptional splicing. Mol Cell Biol 14: 7219–7225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Ardehali MB, Fecko CJ, Webb WW, Lis JT 2007. Intranuclear distribution and local dynamics of RNA polymerase II during transcription activation. Mol Cell 28: 978–990 [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong J-W, Nechaev S, Adelman K, Levine M, Young RA 2007. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet 39: 1512–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zobeck KL, Buckley MS, Zipfel WR, Lis JT 2010. Recruitment timing and dynamics of transcription factors at the Hsp70 loci in living cells. Mol Cell 40: 965–975 [DOI] [PMC free article] [PubMed] [Google Scholar]