Neural circuit assembly involves an initial overproduction of neurons and synaptic connections, followed by their selective elimination. Tasdemir-Yilmaz and Freeman show that Drosophila larval astrocytes are the primary phagocytic cell type in the pupal CNS neuropil. MB γ neuron axons are engulfed by astrocytes using the Draper and Crk/Mbc/dCed-12 signaling pathways, while Crk/Mbc/dCed-12, but not Draper, mediates the elimination of vCrz+ neurites. Eliminating Draper signaling delays early vCrz+ neurite degeneration, suggesting that glial cells promote neurite destruction through engulfment signaling.
Keywords: apoptosis, astrocyte, Drosophila, phagocytosis, pruning, synapse elimination
Abstract
Precise neural circuit assembly is achieved by initial overproduction of neurons and synapses, followed by refinement through elimination of exuberant neurons and synapses. Glial cells are the primary cells responsible for clearing neuronal debris, but the cellular and molecular basis of glial pruning is poorly defined. Here we show that Drosophila larval astrocytes transform into phagocytes through activation of a cell-autonomous, steroid-dependent program at the initiation of metamorphosis and are the primary phagocytic cell type in the pupal neuropil. We examined the developmental elimination of two neuron populations—mushroom body (MB) γ neurons and vCrz+ neurons (expressing Corazonin [Crz] neuropeptide in the ventral nerve cord [VNC])—where only neurites are pruned or entire cells are eliminated, respectively. We found that MB γ axons are engulfed by astrocytes using the Draper and Crk/Mbc/dCed-12 signaling pathways in a partially redundant manner. In contrast, while elimination of vCrz+ cell bodies requires Draper, elimination of vCrz+ neurites is mediated by Crk/Mbc/dCed-12 but not Draper. Intriguingly, we also found that elimination of Draper delayed vCrz+ neurite degeneration, suggesting that glia promote neurite destruction through engulfment signaling. This study identifies a novel role for astrocytes in the clearance of synaptic and neuronal debris and for Crk/Mbc/dCed-12 as a new glial pathway mediating pruning and reveals, unexpectedly, that the engulfment signaling pathways engaged by glia depend on whether neuronal debris was generated through cell death or local pruning.
In complex metazoans, neurons, neurites, and synapses are produced in excess early in development, and this is followed by a later phase of selective elimination of subsets of neurons or parts of neurons (termed pruning). The overproduction of neurons or the formation of excessive connections can be remarkable: Approximately 80% of neurons in the mesencephalic nucleus of the trigeminal nerve of chick embryos undergo cell death (Cowan et al. 1984; von Bartheld and Bothwell 1993), and in the mammalian retinogeniculate system, thalamic neurons prune ∼10 synaptic inputs for every successful input retained (Chen and Regehr 2000). Such early exuberant brain construction is thought to impart increased developmental plasticity through which trophic and activity-dependent mechanisms can be used to fine-tune neuronal numbers and connectivity for optimum circuit performance.
Neural circuit refinement can proceed by the wholesale elimination of a neuron and its projections or the selective destruction of specific axons, dendrites, or synapses. For instance, ∼50% of chick ciliary ganglion cells undergo cell death to remove entire cells after they have received afferent synaptic input and elaborated axonal projections (Landmesser and Pilar 1972, 1974). Similarly, ∼40% of mouse GABAergic inhibitory cortical neurons undergo cell death after they have migrated from their place of birth, acquired morphological features of cortical inhibitory neurons, and formed synaptic contacts with surrounding cells (Southwell et al. 2012). In contrast, other mammalian neurons selectively prune only specific parts of cells. For example motor neurons eliminate individual branches when neuromuscular junctions (NMJs) resolve states of polyinnervation to monoinnervation (Walsh and Lichtman 2003; Bishop et al. 2004), and branch-specific elimination occurs during eye-specific segregation of retinal ganglion cell inputs into the dorsal lateral geniculate nucleus (Chen and Regehr 2000; Hooks and Chen 2006).
Neural circuit refinement leads to the production of large amounts of neuronal debris in the form of neuronal cell corpses, fragmented axons and dendrites, and pruned synapses requiring disposal. Impressively, essentially all developmentally produced neuronal debris is rapidly eliminated from the nervous system through the activity of phagocytic glia. In the mammalian CNS, microglia are primarily responsible for clearing neural debris (Cuadros and Navascues 1998; Marin-Teva et al. 1999; Parnaik et al. 2000), and in the PNS, Schwann cells appear to play a key role in the elimination of exuberant motor neuron inputs (Bishop et al. 2004). The timely clearance of CNS debris is essential to avoid tissue inflammation and autoimmune responses (Savill et al. 2002). In addition, the nervous system appears especially sensitive to the presence of neural debris during synapse formation. For instance, suppressing the engulfing activity of glia and/or muscle at the Drosophila NMJ leads to accumulation of neural debris at the NMJ and in turn inhibits synaptic growth (Fuentes-Medel et al. 2009).
Molecular insights into the removal of cell corpses came first from work in Caenorhabditis elegans where a number of cell death abnormal (ced) mutants were found to delay or block the engulfment of dead cells (for review, see Reddien and Horvitz 2004). The CED-2/CED-5/CED-12 complex, which acts as a guanine nucleotide exchange factor (GEF) that activates Rac1 (Brugnera et al. 2002), was found to modulate cytoskeletal events during cell corpse internalization (Zhou et al. 2001a). A second pathway composed of CED-1 and CED-6 was also found to act genetically in parallel to the CED-2/CED-5/CED-12 pathway; loss of either pathway lead to a partial suppression of cell corpse engulfment, while simultaneous inactivation of both pathways suppressed clearance much more strongly (Ellis et al. 1991; Gumienny et al. 2001; Zhou et al. 2001b; Kinchen et al. 2005). CED-1 is a transmembrane receptor that acts in the engulfing cell and is thought to recognize cell corpses (Zhou et al. 2001b) and degrade them (Yu et al. 2008) through the PTB domain-containing protein CED-6 (Liu and Hengartner 1998). Similar roles for Drosophila and vertebrate orthologs of these genes have revealed conserved functions for these pathways in the elimination of cell corpses (Smits et al. 1999; Manaka et al. 2004; Etchegaray et al. 2012; Van Goethem et al. 2012; van Ham et al. 2012), including by glia in the nervous system (Freeman et al. 2003; Wu et al. 2009).
Draper, the Drosophila homolog of CED-1, is also required in glia for the removal of pruned mushroom body (MB) γ-axonal debris during metamorphosis (Awasaki et al. 2006), sensory neuron dendrites (Williams et al. 2006), presynaptic debris shed during larval NMJ growth (Fuentes-Medel et al. 2009), and axons that have undergone injury-induced degeneration in the adult (MacDonald et al. 2006). Likewise, we recently discovered a role for Crk/Mbc/dCed-12 (Drosophila CED-2/CED-5/CED-12) in the clearance of axonal debris after axotomy (Ziegenfuss et al. 2012). However, precisely which subtypes of Drosophila glia are responsible for neuronal pruning in the CNS remains unclear. Mammalian microglia have been ascribed a role in synaptic pruning (Stevens et al. 2007; Paolicelli et al. 2011; Schafer et al. 2012), but additional subtypes of glia may also sculpt neuronal circuits, and molecular pathways mediating glial pruning of neurons remain poorly defined. In this study, we describe a key role for astrocytes in the clearance of neural debris during neural circuit reorganization. We identify the Crk/Mbc/dCed-12 complex as a new signaling pathway required for glial pruning of neurites. Surprisingly, we found that the precise molecular pathways engaged by glia to internalize neurite debris are linked with the type of intrinsic molecular programs used to drive neurite degeneration.
Results
Astrocytes acquire phagocytic properties during neural circuit remodeling
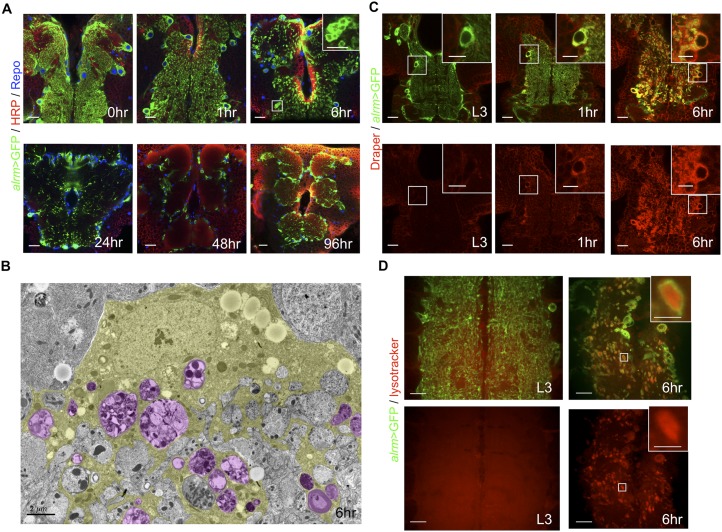
We wished to define the cellular and functional changes that occur in larval astrocytes during the larval-to-adult transition. We therefore examined the morphology of astrocytes in the Drosophila brain and ventral nerve cord (VNC) during larval and pupal stages using the alrm-Gal4 driver to express membrane-tethered GFP (alrm>GFP) (Doherty et al. 2009) specifically in astrocytes. At the third instar larval (L3) and white puparium stages (i.e., 0 h after puparium formation [APF]), astrocyte cell bodies resided at the edge of the neuropil (Fig. 1A; Supplemental Fig. 1A,B). Astrocyte processes extended exclusively into the neuropil and exhibited a tufted morphology, with very fine processes infiltrating the entire neuropil and associating closely with synapses (Figs. 1A, 3A [below]; Supplemental Fig. 1A,B; T Stork and M Freeman, unpubl.). Unexpectedly, astrocyte processes underwent a dramatic transformation at puparium formation such that at 1 h APF, fine processes became slightly enlarged and took on a vacuolated appearance (Fig. 1A). Vacuolization of astrocyte membranes became more extreme by 6 h APF; later, astrocyte processes became progressively sparser in the CNS; and by 48 h APF, astrocyte membranes were absent from the neuropil (Fig. 1A). Throughout metamorphosis, astrocyte cell bodies remained at the periphery of the neuropil, and their membranes appeared to reinfiltrate the neuropil (by 96 h APF) prior to eclosion as adult animals (Fig. 1A).
Figure 1.
Astrocytes transform into phagocytes at the initiation of neural circuit remodeling. (A) Astrocytes in controls were labeled with GFP (alrm-Gal4, UAS-mCD8∷GFP; green) in A, C, and D, the neuropil was labeled with HRP-Cy3 (red), and glial nuclei were labeled with anti-Repo (blue). Time points are as indicated (APF). Thoracic and brain regions are shown. (Inset) High-magnification view of the boxed region. Single z-sections are shown for A, C, and D. Bars: 20 μm; insets, 10 μm. (B) TEM image from a cross-section of the abdominal VNC of a control animal. An astrocyte (outlined in yellow) at the edge of the neuropil, exhibiting cytoplasmic vacuoles (magenta). Genotype used was as follows: alrm-Gal4, UAS-mCD8∷GFP/+. Bar, 2 μm. (C) Draper was labeled in red. Time points are as indicated (APF). (Insets) High-magnification view of the boxed regions. Genotype used was as follows: alrm-Gal4, UAS-mCD8∷GFP. Bars: 20 μm; insets, 10 μm. (D) LysoTracker was labeled in red. Note that all LysoTracker+ puncta in the neuropil were surrounded by GFP+ membranes of astrocytes. (Inset) High-magnification view of vacuole filled with LysoTracker. Genotype used was as follows: alrm-Gal4, UAS-mCD8∷GFP. Bars: 20 μm; inset, 5 μm.
Figure 3.
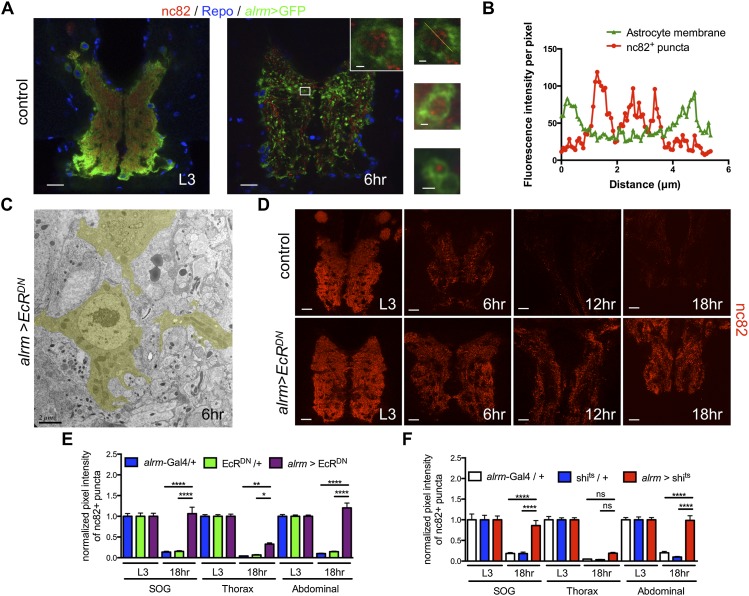
Astrocytes engulf and clear synaptic material from the neuropil. (A) In A and D, confocal images are single z-confocal slices. Astrocytes were labeled with GFP (alrm-Gal4, UAS-mCD8∷GFP/+; green), glial nuclei were labeled with anti-Repo (blue), and, in A and D, active zones were labeled with nc82 (antibody for Brp; red). Time points are as follows: L3 (third instar larva) and APF (hours APF). (Top inset) High-magnification view of the boxed region. (Middle and bottom insets) Other examples of vacuoles. Bars: 20 μm; insets, 1 μm. (B) The graph shows fluorescence intensity per pixel in each channel for the inset in A of the astrocyte membrane and nc82+ puncta along a line drawn through a vacuole. (C) TEM image from a cross-section of the abdominal VNC of an alrm>EcRDN animal. An astrocyte (outlined in yellow) at the edge of the neuropil. Time point is as indicated (APF). Genotype used was as follows: alrm>EcRDN (alrm-Gal4,UAS-mCD8∷GFP/UAS-EcRDN). Bars, 2 μm. (D) Time points are as indicated (APF). Genotypes used were as follows: control (alrm-Gal4/+) and alrm>EcRDN (alrm-Gal4/UAS-EcRDN). Bars, 20 μm. (E,F) Quantification of pixel intensity of nc82+ puncta at L3 and 18 h APF in SOG, thorax, and abdominal regions of the VNC. (E) Genotypes used were as follows: EcRDN/+ (UAS-EcRDN/+) and alrm>EcRDN (alrm-Gal4/UAS-EcRDN). Error bars represent ±SEM. N-values are as follows: alrm-Gal4/+, N ≥ 20; UAS-EcRDN/+, N ≥ 16; and alrm-Gal4/UAS-EcRDN, N ≥ 20 (N denotes number of measurements). (*) P < 0.05; (**) P < 0.01; (****) P < 0.0001. (F) Genotypes used were as follows: shits/+ (UAS-shits/+) and alrm>shits (alrm-Gal4/UAS-shits). Flies were raised at 18°C and then shifted to the restrictive temperature (30°C) at 0 h APF for 18 h. Error bars represent ±SEM; N ≥ 12; (****) P < 0.0001.
We performed transmission electron microscopy (TEM) to define the cellular basis of this astrocyte transformation in greater detail. We compared the morphology of astrocytes in the VNC of control animals at L3 (Supplemental Fig. 2) and 6 h APF (Fig. 1B). We defined astrocytes according to the following criteria: (1) the position of their cell bodies adjacent to the neuropil, (2) their electron-dense cytoplasm compared with surrounding cells, and (3) the fibrous morphology infiltrating to the neuropil. Consistent with our light microscopic observations, we found that L3 astrocyte membranes were fibrous and extended profusely into the synaptic neuropil (Supplemental Fig. 2). By 6 h APF, however, astrocyte morphology had changed dramatically: Astrocyte membranes, when compared with those of L3 animals, were less fibrous, and their cytoplasm was filled with many large vacuoles that appeared to be filled with cellular debris, suggesting that astrocytes were engulfing neuronal material.
We examined expression of Draper, a marker for engulfing cells in Drosophila (MacDonald et al. 2006; Ziegenfuss et al. 2008, 2012; Doherty et al. 2009), using Draper antibody and found that astrocytes at L3 expressed low to undetectable levels of Draper. However, at 1 h APF, Draper became detectable in astrocytes, and by 6 h APF, astrocytic Draper levels were significantly elevated (Fig. 1C) and specific to astrocytes in the neuropil (Supplemental Figs. 3, 4). dCed-6, which is required for Draper signaling, was expressed in astrocytes at L3 and 6 h APF, although in contrast to Draper, total dCed-6 levels did not change noticeably between L3 and 6 h APF (Supplemental Fig. 5). Finally, lysosomal activity (detected by LysoTracker) was not obvious in the neuropil at L3; however, robust lysosomal activity was observed in the neuropil by 6 h APF, and all LysoTracker+ staining was found within astrocytic vacuoles (Fig. 1D). Together, these data argue that astrocytes take on a phagocytic phenotype at pupariation and suggest that astrocytes may be the primary phagocytic cell type in the pupal neuropil.
Ecdysone receptor (EcR) cell-autonomously regulates transformation of astrocytes into phagocytes
The steroid hormone 20-hydroxyecdysone (ecdysone) is a major regulator of neuronal remodeling and cell death during metamorphosis (Robinow et al. 1993; Truman et al. 1994; Jiang et al. 1997; White et al. 1997; Schubiger et al. 1998). Given that astrocytes transform into phagocyte-like cells at this time of intense steroid-mediated developmental signaling, we wished to determine whether ecdysone signaling modulated astrocyte phagocytic phenotypes.
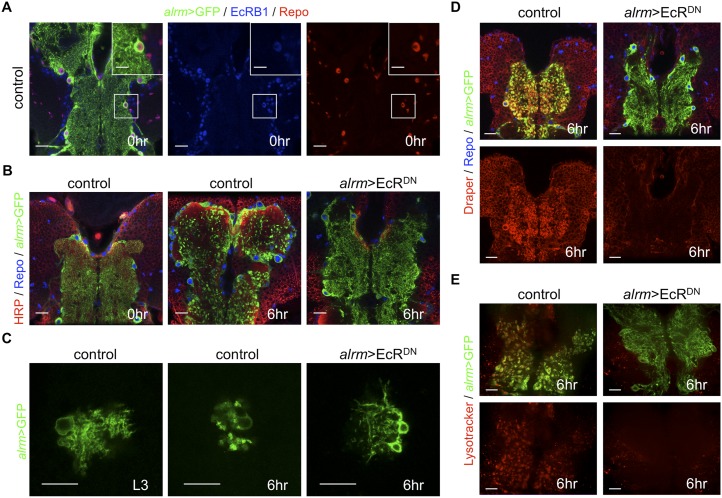
Ecdysone binds to a heterodimer receptor composed of the EcR and ultraspiracles (Usp) (Koelle et al. 1991; Yao et al. 1993). There are multiple isoforms of EcR, termed A, B1, and B2 (Talbot et al. 1993). EcR-B1 is expressed at high levels in neurons as they initiate pruning of larval projections (Truman et al. 1994) and is required for pruning (Schubiger et al. 1998, 2003; Lee et al. 2000; Kuo et al. 2005), while EcR-A appears to be up-regulated as neurons initiate outgrowth of adult-specific branches (Truman et al. 1994). Using antibodies specific for EcR-B1 and EcR-A, we found that astrocytes express EcR-B1 at 0 h APF (Fig. 2A) but not EcR-A (Supplemental Fig. 6).
Figure 2.
EcR signaling cell-autonomously regulates transformation of astrocytes into phagocytes. (A) Astrocytes in controls were labeled with GFP (alrm-Gal4, UAS-mCD8∷GFP/+; green) in A–E, and EcR-B1 (blue) and glial nuclei were labeled with anti-Repo (red). (Insets) High-magnification view of an astrocyte cell body. (A–E) All confocal images are single z-confocal slices. Bars: 20 μm; inset, 10 μm. (B) The neuropil was labeled with HRP-Cy3 (red) and, in B and D, glial nuclei were labeled with anti-Repo (blue). Time points are as indicated (APF). Note the lack of transformation of astrocytes at 6 h APF. (B,D,E) Genotypes used were as follows: control (alrm-Gal4,UAS-mCD8∷GFP/+) and alrm>EcRDN (alrm-Gal4,UAS-mCD8∷GFP/UAS-EcRDN). Bars, 20 μm. (C) Astrocyte MARCM clones at L3 and 6 h in controls (alrm-Gal4,UAS-mCD8∷GFP, repoflp6-2;FRT2A/FRT2A,tub-Gal80) or astrocytes expressing EcRDN (alrm-Gal4,UAS-mCD8∷GFP, repoflp6-2/UAS-EcRDN;FRT2A/FRT2A,tub-Gal80) at 6 h APF. Bars, 20 μm. (D) Draper was labeled in red. Time points are as indicated (APF). Note the strong knockdown of draper in astrocytes in alrm>EcRDN animals compared with controls at 6 h APF. Bars, 20 μm. (E) LysoTracker was labeled in red. Time points are as indicated (APF). LysoTracker+ puncta in astrocytes were blocked at 6 h APF in alrm>EcRDN animals. Bars, 20 μm.
We next assayed for EcR function in astrocytes by driving expression of two different dominant-negative versions of EcR: UAS-EcRDN F645A and UAS-EcRDN W650A (Cherbas et al. 2003). In animals expressing EcRDN constructs in astrocytes (alrm>EcRDN), we found that astrocyte morphology appeared normal at L3 (Supplemental Fig. 5), suggesting that EcR signaling is not critical before L3 for normal astrocyte development. However, astrocytic EcRDN expression completely blocked their morphological transformation into the highly vacuolated morphology apparent in controls at 6 h APF (Fig. 2B). Suppression of astrocyte morphological transformation was even accomplished when EcRDN was driven in one to two astrocytes rather than the entire population (Fig. 2C). Consistent with full suppression of the transformation of astrocytes into phagocytes, EcRDN expression in astrocytes also blocked the Draper up-regulation that normally occurs from 0 to 6 h APF (Fig. 2D) as well as induction of phagolysosomal activity (Fig. 2E).
Since Draper is a potent regulator of phagocytic status of glial cells in Drosophila, we next overexpressed Draper in astrocytes in a EcRDN background to determine whether Draper activation was sufficient to make astrocytes phagocytic. Despite high-level expression of Draper in EcRDN animals, astrocytes failed to transform into phagocytes at puparium formation (Supplemental Fig. 7), indicating that additional factors downstream from EcR are necessary for activation of the phagocyte program in astrocytes. We attempted to determine the developmental consequences of complete blockade of astrocyte phagocytic function throughout metamorphosis; however, we found that the effects of EcRDN were only transient: Despite continued expression of EcRDN in astrocytes, Draper activation was obvious by 48 h APF (Supplemental Fig. 8). Not all molecular components of the engulfment machinery are activated in astrocytes at pupariation. For instance, astrocytic dCed-6 levels at L3 are similar to those at 6 h APF, and we found no obvious change in dCed-6 in astrocytes expressing EcRDN (Supplemental Fig. 5). These results demonstrate that loss of EcR signaling is sufficient to cell-autonomously suppress the transformation of astrocytes into phagocytes at pupariation.
Astrocytes engulf synaptic material from the neuropil
Neuronal remodeling during metamorphosis results in the loss of nearly all synapses in the neuropil by 48 h APF, and adult-specific synapses are subsequently generated (A Muthukumar and M Freeman, unpubl.). We used antibodies to the presynaptic active zone marker Bruchpilot (nc82) to mark synapses and examine their fate during the first 48 h of metamorphosis. At L3, nc82 labeled synapses throughout the neuropil; by 6 h APF, we found that many nc82+ puncta were located within the astrocytic vacuoles (Fig. 3A,B; Supplemental Fig. 9A).
We next sought to determine whether astrocyte transformation into phagocytes or other events downstream from EcR in astrocytes was required for the elimination of CNS synapses. Based on nc82+ staining, it appeared that most synapses were eliminated by 6 h APF and were nearly gone by 12–18 h APF (Fig. 3D). Impressively, blockade of EcR signaling in astrocytes suppressed the clearance of nc82+ synapses from multiple brain regions at 18 h APF (Fig. 3E). Blockade of astrocytic endocytic function during early pupal stages by expression of Shibirets (Shits) resulted in a similar suppression of synaptic clearance from the neuropil (Fig. 3F). Finally, when we examined the ultrastructure of astrocytes at 6 h APF, we found that the large vacuoles filled with cellular debris normally present in control astrocytes (Fig. 1B) were absent from astrocytes expressing EcRDN (Fig. 3C). These data argue that some portion of internalized debris in astrocytes at 6 h APF is synaptic material.
Draper and Crk/Mbc/dCed-12 act in a partially redundant fashion to promote astrocyte transformation into phagocytes and synaptic clearance
The Draper signaling pathway and the Crk/Mbc/dCed-12 complex are essential for glial clearance of axonal debris after axotomy in Drosophila (MacDonald et al. 2006; Ziegenfuss et al. 2012). Loss of either pathway leads to a near-complete blockade of glial engulfment of degenerating olfactory receptor neurons in the adult (Ziegenfuss et al. 2012). Draper also has been shown previously to play a role in neuronal pruning in Drosophila (Awasaki et al. 2006), but in which subtype of glia Draper functions to engulf axonal debris remains unknown, and roles for Crk/Mbc/dCed-12 in synaptic, axonal, or dendritic pruning remain unexplored.
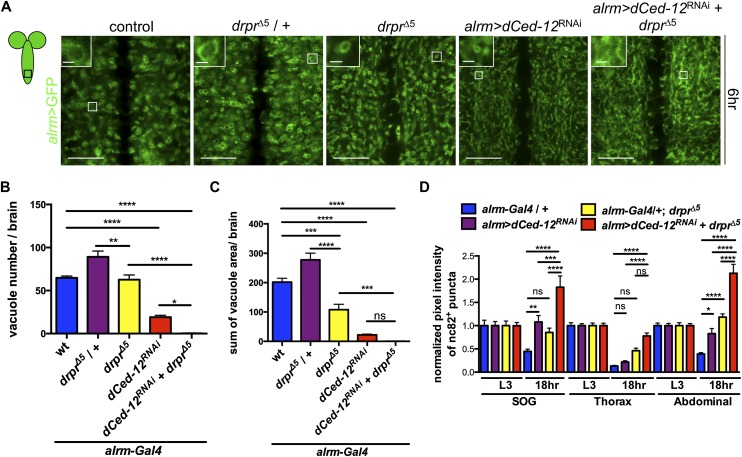
We assayed for requirements for Draper in the transformation of astrocytes into phagocytes. In the posterior abdominal region of the VNC at 6 h APF, drprΔ5-null mutants exhibited similar numbers of astrocytic vacuoles compared with controls, although vacuole size was significantly reduced (by ∼50% in drprΔ5-null mutants compared with controls) (Fig. 4A–C). In contrast, knockdown of dCed-12 in astrocytes (alrm>dCed-12RNAi) led to a very strong decrease in astrocytic vacuole number and size: Vacuole areas were reduced by 90%, and vacuole numbers were reduced by 70% compared with controls (Fig. 4A–C). Knockdown of Crk in astrocytes led to vacuole areas reduced by ∼20% compared with controls (Supplemental Fig. 10A–C), further supporting a requirement for the Crk/Mbc/dCed-12 complex in the induction of the astrocytic phagocyte program. To determine whether loss of both signaling pathways enhanced the observed changes, we knocked down dCed-12 in a drprΔ5-null background and found a near-complete loss of astrocytic vacuole formation (Fig. 4A–C). A similar phenotype was seen when we knocked down Crk in astrocytes in a drprΔ5-null background but to a lesser extent (Supplemental Fig. 10A–C).
Figure 4.
Draper and Crk/Mbc/dCed-12 pathways are required for the formation of astrocytic vacuoles. (A) Astrocytes were labeled with GFP (alrm-Gal4, UAS-mCD8∷GFP/+; green). Images zoomed at the abdominal tip of the VNC at 6 h APF. (Insets) One representative vacuole. Images are single z-confocal slices. Bars: 20 μm; insets, 2 μm. Genotypes used were as follows: control (alrm-Gal4,UAS-mCD8∷GFP/+), drprΔ5/+ (alrm-Gal4,UAS-mCD8∷GFP/+; drprΔ5/+), drprΔ5 (alrm-Gal4,UAS-mCD8∷GFP/+; drprΔ5), alrm>dCed-12RNAi (alrm-Gal4, UAS-mCD8∷GFP/UAS-dCed-12RNAi), and alrm>dCed-12RNAi+drprΔ5 (alrm-Gal4, UAS-mCD8∷GFP/UAS-dCed-12RNAi/+; drprΔ5). (B) Quantification of number of vacuoles of figures in A. Error bars represent ±SEM; number of brains used was five or more; two hemisegments were measured in each brain. (***) P < 0.001; (****) P < 0.0001. (C) Quantification of a cross-section area of vacuoles of the figures in A. Error bars represent ±SEM; number of brains used was five or more; two hemisegments were measured in each brain. (*) P < 0.05; (**) P < 0.01; (****) P < 0.0001. (D) Quantification of pixel intensity of nc82+ puncta in SOG, thorax, and abdominal regions of the VNC at L3 and 18 h APF from Supplemental Figure 11. Genotypes used were as follows: alrm>dCed-12RNAi (alrm-Gal4/UAS-dCed-12RNAi) and alrm>dCed-12RNAi+drprΔ5 (alrm-Gal4/UAS-dCed-12RNAi; drprΔ5). Error bars represent ±SEM; N-values are as follows: alrm-Gal4/+, N ≥ 18; alrm>dCed-12RNAi, N ≥ 20; alrm-Gal4/+; drprΔ5, N ≥ 16; and alrm>dCed-12RNAi+drprΔ5, N ≥ 18. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001; (****) P < 0.0001.
We next assayed for clearance of synaptic structures at 18 h APF in drprΔ5 mutants and astrocytic dCed-12RNAi animals. We found that drprΔ5-null animals had a significant amount of nc82+ debris in the abdominal region of VNC (Fig. 4D; Supplemental Fig. 11). Astrocytic knockdown of dCed-12 resulted in a significant accumulation of nc82+ debris in both the SOG and abdominal region of the VNC. Astrocytic knockdown of Crk also gave a phenotype similar to dCed-12RNAi but to a lesser effect (Supplemental Fig. 12A,B). When we depleted both Draper and Crk/Mbc/dCed-12 signaling by astrocytic knockdown of dCed-12 in a drprΔ5-null background, we observed significantly elevated levels of nc82+ debris compared with knockdown of either pathway alone or controls (Fig. 4D; Supplemental Fig. 11). These data indicate that Draper and Crk/Mbc/dCed-12 act in a partially redundant fashion to transform astrocytes into phagocytes and promote engulfment of synaptic material during metamorphosis.
EcR, Draper, and Crk/Mbc/dCed-12 function in astrocytes to promote MB γ neuron clearance
In draper-null mutants, pruned MB γ neuron axon clearance is delayed by ∼2 d, but by adult stages, axonal material is ultimately pruned (Awasaki et al. 2006). Based on our analysis above, astrocytes are ideal candidates for clearing pruned axons in addition to synaptic material, since the timing of astrocyte transformation is coincident with MB γ neuron pruning, and their processes are close to the MB before clearance (Supplemental Fig. 13). In addition, Crk/Mbc/dCed-12 is a potential new signaling pathway functioning with Draper to clear pruned MB γ neuron axons and could account for the complete (although delayed) clearance in the absence of Draper.
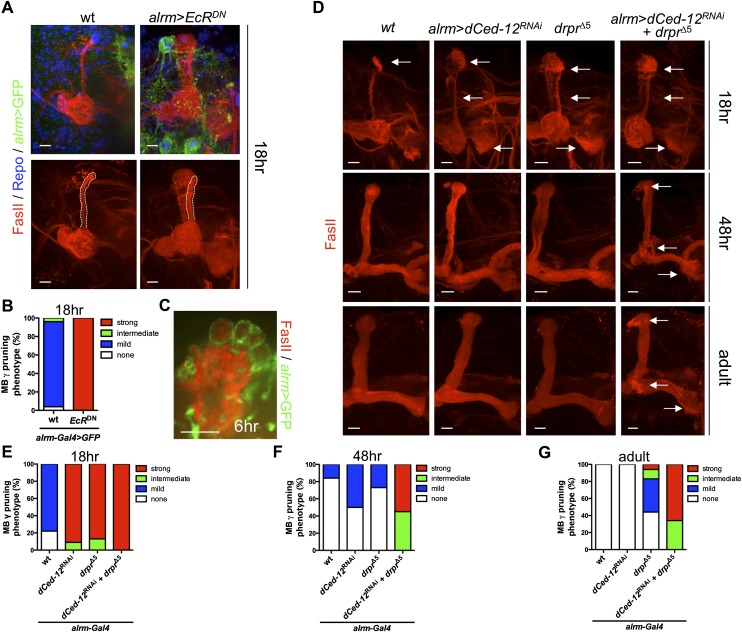
We first assayed MB γ neuron pruning in animals in which transformation into phagocytes was blocked by astrocytic EcRDN expression (Fig. 5A). FasII antibody labels MB α/β and γ neurons, which can be easily discriminated by position, morphology, and pruning: At L3, FasII labels the entire MB, but by 18 h APF, MB γ neurons are pruned and FasII staining is seen only in thin medial and dorsal processes, which are the developing adult-specific α/β lobes (Fig. 5A; Supplemental Fig. 14A–C; Awasaki and Ito 2004; Awasaki et al. 2006). We found that while the dorsal and medial lobes of MB γ neurons were largely pruned by 18 h APF in controls, pruning of these axonal branches was strongly inhibited by astrocytic EcRDN expression (Fig. 5A). Blockade of astrocytic endocytic function using Shits also resulted in a suppression of MB γ axon clearance but to a lesser extent than EcRDN (Supplemental Fig. 15A,B). To determine whether astrocytes phagocytose pruning MB γ neurons, we examined astrocytic phagolysosomes at an early puparium stage. At 6 h APF, we found FasII+ MB axonal material located within the astrocytic phagolysosomes and increased infiltration of the MB lobes by astrocyte processes (Fig. 5C; Supplemental Fig. 9B). These data indicate that transformation of astrocytes into phagocytes through EcR signaling is required for proper MB γ neuron pruning.
Figure 5.
EcR, Draper, and Crk/Mbc/dCed-12 function in astrocytes to promote MB γ neuron clearance. (A) Astrocytes were labeled with GFP (alrm-Gal4, UAS-mCD8∷GFP/+; green) in A and C, glial nuclei were labeled with anti-Repo (blue), and MB lobes were labeled with anti-FasII (red) in A, C, and D. The adult-specific dorsal lobe is outlined (white line). Time point is as indicated (APF). Genotypes used were as follows: wild type (wt; alrm-Gal4,UAS-mCD8∷GFP/+) and alrm>EcRDN (alrm-Gal4,UAS-mCD8∷GFP/UAS-EcRDN). (A–D) All confocal images are z-projections. Bars, 10 μm. (B) Quantification of MB γ neuron pruning phenotype from A using the categories shown in Supplemental Figure 14 (see the Materials and Methods). Wild type, N = 24; alrm>EcRDN, N = 20 hemisegments quantified. (C) Image of magnified dorsal MB lobe. Note the FasII+ debris inside astrocytic vacuoles. Time point is as indicated (APF). Genotype used was as follows: alrm-Gal4, UAS-mCD8∷GFP/+. Bars, 10 μm. (D) MB debris was scored at the time points indicated (APF). Arrows indicate extra MB γ debris. Bars, 10 μm. Genotypes used were as follows: Wild type (wt; alrm-Gal4/+), alrm>dCed-12RNAi (alrm-Gal4/UAS-dCed-12RNAi), drprΔ5 (alrm-Gal4/+; drprΔ5), and alrm>dCed-12RNAi+drprΔ5 (alrm-Gal4/UAS-dCed-12RNAi; drprΔ5). (E–G) Quantification of the MB γ neuron pruning phenotype from D. N-values are as follows: wild type, N = 36; alrm>dCed-12RNAi, N = 22; drprΔ5, N = 32; and alrm>dCed-12RNAi+drprΔ5, N = 23 hemisegments quantified at 18 h APF. N-values are as follows: wild type, N = 58; alrm>dCed-12RNAi, N = 30; drprΔ5, N = 26; and alrm>dCed-12RNAi + drprΔ5, N = 22 hemisegments quantified at 48 h APF. N-values are as follows: wild type, N = 38; alrm>dCed-12RNAi, N = 20; drprΔ5, N = 18; and alrm>dCed-12RNAi+drprΔ5, N = 32 hemisegments quantified at newly eclosed adults.
We next examined MB γ neuron pruning in controls, draper mutants, dCed-12RNAi animals, and animals in which both Draper and dCed-12 signaling had been depleted. We scored pruning phenotypes at multiple time points, including 18 and 48 h APF and in newly eclosed adults (Fig. 5D–G). Consistent with previous observations, in drprΔ5-null mutants, we found a strong suppression of MB γ neuron pruning at 18 h APF and a mild phenotype in adults (Fig. 5D–G). This defect in clearance represented astrocytic Draper function, as expression of draperRNAi in astrocytes resulted in a similar phenotype (Supplemental Fig. 16A,B). Astrocytic knockdown of dCed-12 also resulted in a strong suppression of MB γ neuron pruning at 18 h APF and a mild phenotype at 48 h APF, although pruning in adults was similar to that in controls (Fig. 5D–G). Similar results were found with astrocytic knockdown of Crk or mbc, although the observed phenotypes were slightly weaker than those observed with dCed-12RNAi (Supplemental Fig. 17A,B). These observations identify the Crk/Mbc/dCed-12 complex as a new glial signaling pathway required for MB axonal pruning.
Interestingly, astrocytic knockdown of dCed-12 in a drprΔ5-null mutant background resulted in an additive phenotype, with dCed-12RNAi, drprΔ5 animals exhibiting a stronger suppression of MB γ axon pruning at 48 h APF and into adult stages (Fig. 5D–G). Similar results were found with astrocytic knockdown of Crk in a drprΔ5-null mutant background (Supplemental Fig. 17C,D). We conclude that Draper and Crk/Mbc/dCed-12 act in a partially redundant fashion during neuronal pruning events.
Loss of Draper and Crk/Mbc/dCed-12 delay clearance of vCrz+ neurons (expressing Corazonin [Crz] neuropeptide in the VNC)
Astrocyte membranes are present throughout the larval neuropil (except MB lobes, as stated previously) at late larval stages, and all appear to transform into phagocytes. This observation suggests that astrocytes have widespread roles in neuronal pruning. To explore this possibility, we examined the peptidergic vCrz neurons that undergo apoptosis during early metamorphosis between 0 and 6 h APF and whose neurites are eliminated (Choi et al. 2006; Lee et al. 2011).
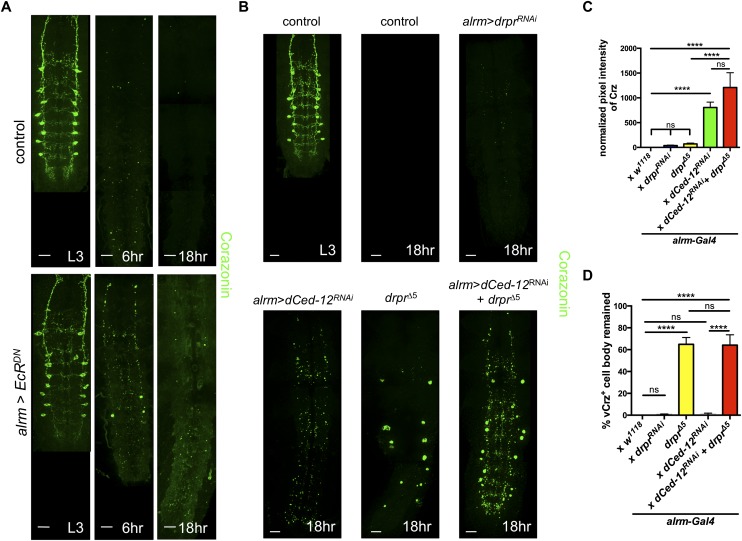
In controls at L3, the α-Crz antibody labels eight pairs of vCrz neurons and a few neurons in the brain, and by 6 h APF, almost all of the vCrz+ neuronal debris was cleared from the neuropil (Choi et al. 2006). We blocked astrocyte phagocytic function by expressing EcRDN in astrocytes and found that many vCrz+ cell bodies and neurite debris remained at 6 h APF and that neurite debris lingered until 18 h APF (Fig. 6A). We next examined animals depleted for Draper, dCed-12, or both and quantified the clearance of vCrz+ cell bodies and neurites. We found a striking difference in the requirements for Draper and dCed-12 in the clearance of vCrz+ cell bodies versus neurites. In drprΔ5-null mutants at 18 h APF, nearly all vCrz+ neurites were eliminated from the neuropil, while the majority of vCrz+ cell bodies remained (Fig. 6B–D). In contrast, astrocytic knockdown of dCed-12 resulted in a lack of vCrz+ cell bodies, but neurite debris was significantly retained (Fig. 6B–D). Pan-glial knockdown of dCed-12 also resulted in a lack of vCrz+ cell bodies and a partial suppression of clearance of neurites (Supplemental Fig. 18A–C). Preservation of vCrz+ neurite debris at 18 h APF (rather than complete loss of Crz+ signal) indicates that the Crz peptide is stable for at least 18 h APF and is a useful marker for unengulfed neurite debris.
Figure 6.
EcR, Draper, and Crk/Mbc/dCed-12 function in astrocytes to promote clearance of vCrz+ neuronal debris. (A) vCrz+ neurons were labeled with anti-Crz (green) in A and B. Time points used were as follows: L3 (third instar larva). Genotypes used were as follows: control (alrm-Gal4, UAS-mCD8∷GFP/+) and alrm>EcRDN (alrm-Gal4, UAS-mCD8∷GFP/UAS-EcRDN). Confocal images in A and B are z-projection images. Bars, 20 μm. (B) Time points are as indicated in A. Bars, 20 μm. Genotypes used were as follows: control (alrm-Gal4/+), alrm>drprRNAi (alrm-Gal4/drprRNAi), drprΔ5 (alrm-Gal4/+; drprΔ5), alrm>dCed-12RNAi (alrm-Gal4/dCed-12RNAi), and alrm>dCed-12RNAi+drprΔ5 (alrm-Gal4/dCed-12RNAi; drprΔ5). (C) Quantification of pixel intensity of vCrz+ debris from B. (C,D) N-values are as follows: alrm-Gal4/+, N = 32; alrm-Gal4/drprRNAi, N = 11; alrm-Gal4/+; drprΔ5, N = 11; alrm-Gal4/dCed-12RNAi, N = 14; and alrm-Gal4/dCed-12RNAi; drprΔ5, N = 12 brains quantified. Error bars represent ±SEM. (****) P < 0.0001. (D) Quantification of the percentage of remaining vCrz+ cell bodies from B. Error bars represent ±SEM. (****) P < 0.0001.
Glial clearance of cell bodies and neurites appears to be a separable and additive function for Draper and dCed-12. Elimination of both pathways did not increase the number of cell bodies or amount of neurite debris when compared with elimination of each pathway alone (Fig. 6B–D). In addition, these pathways appear to be acting in different subsets of glial cells. Draper appears to function in nonastrocyte glia (e.g., ensheathing or cortex glia) to clear vCrz+ cell bodies, since astrocytic knockdown of draper had no effect on cell body clearance (Fig. 6B–D), and nonastrocyte glial depletion of draper led to similar levels of remaining vCrz+ cell bodies compared with depletion from all glia (Supplemental Fig. 19A,B). Clearance is likely not mediated by neurons themselves, since vCrz+ neurons did not express Draper at 6 h APF (Supplemental Fig. 20), and vCrz+ neuronal depletion of draper had no effect on neurite or cell body clearance (Supplemental Fig. 21A–C).
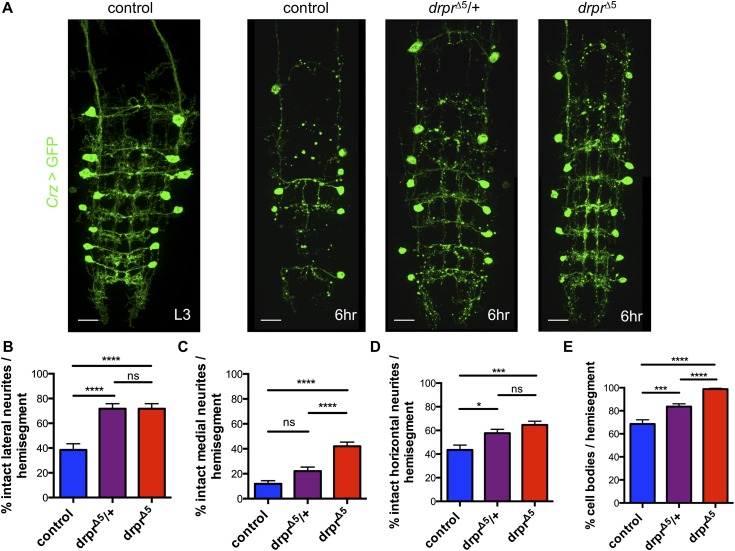
Engulfing cells such as microglia can promote apoptotic death of target cells, but this has generally been examined before neurite elaboration has occurred (Marin-Teva et al. 2004; Cunningham et al. 2013). Can engulfing activity of glia promote neurite destruction? To answer this question, we labeled vCrz+ neuronal membranes with membrane-tethered GFP (Crz-Gal4; UAS-mCD8∷GFP) and assayed clearance in controls and draper mutants. By 6 h APF, the majority of cell bodies and neurites were cleared in controls (Fig. 7A–E). However, removing only a single copy of draper (drprΔ5/+) resulted in dominant suppression of clearance of both vCrz+ cell bodies and neurites at 6 h APF (Fig. 7A–E), which was enhanced in drprΔ5 homozygous mutants (Fig. 7A–E). This observation argues that the fragmentation of vCrz+ neurons is highly sensitive to draper gene dosage and suggests that engulfing glia can actively promote the destruction of targets during clearance.
Figure 7.
Loss of Draper delays neurite degeneration and clearance of vCrz+ neuronal cell bodies. (A) vCrz+ neurons were labeled with GFP (Crz-Gal4, UAS-mCD8∷GFP; green). Time points are as indicated (APF). Bars, 20 μm. Genotypes used were as follows: control (Crz-Gal4, UAS-mCD8∷GFP; +), drprΔ5/+ (Crz-Gal4, UAS-mCD8∷GFP; drprΔ5/+), and drprΔ5 (Crz-Gal4, UAS-mCD8∷GFP; drprΔ5). (B–D) Quantification of the percentage of intact lateral (B), medial (C), and horizontal (D) axons of vCrz+ neurons per hemisegment from A. (B–E) Control, N = 30; drprΔ5/+, N = 30; and drprΔ5, N = 32 hemisegments quantified. Error bars represent ±SEM. (*) P < 0.05; (***) P < 0.001; (****) P < 0.0001. (E) Quantification of the percentage of cell bodies of vCrz+ neurons per hemisegment from A.
Discussion
Widespread cell death and pruning occur in developing nervous systems of complex metazoans. Roles for astrocytes in the clearance of neuronal debris remain undefined. In this study, we showed that Drosophila larval astrocytes transform from supportive cells into phagocytes that engulf significant amounts of pruned synapses and neural debris. Astrocytic engulfment of pruned synapses and MB γ neural debris requires the Draper signaling pathway, which acts in a partially redundant fashion with the Crk/Mbc/dCed-12 complex, a pathway that we implicate for the first time in neural pruning events. Detailed genetic analysis of these pathways revealed unexpected, context-dependent usage of these signaling pathways to clear subcompartments of neurons during nervous system reorganization.
Drosophila larval astrocytes transform into phagocytes at the initiation of metamorphosis
Astrocytes serve as regulators of synapse formation and function and are generally supportive of neural circuits. Based on several lines of evidence, we showed that astrocytes transform morphologically and functionally into phagocytes at pupariation and engulf significant amounts of neural debris. Phagocytic astrocytes take on a highly vacuolated appearance, up-regulate the engulfment molecule Draper, contain cytoplasmic vacuoles filled with debris that stains for synaptic or axonal markers, and exhibit high levels of lysosomal activity. Activation of this phagocytic program depends on cell-autonomous signaling through the EcR, since blockade of EcR in even single astrocytes suppressed their transformation by all morphological and molecular criteria. Blockade of astrocyte phagocytic function by multiple methods (e.g., EcRDN or Shits) suppressed the clearance of synapses throughout the CNS, axons of MB γ neurons in the central brain, and the neurites of vCrz+ cells in the VNC. This latter observation defines vCrz+ neurons as a new system to explore astrocyte–neuron interactions during neuronal apoptosis and neurite or synapse elimination in the CNS.
While roles for Drosophila glia in the engulfment of pruned MB γ axons have been previously described (Awasaki and Ito 2004; Watts et al. 2004; Awasaki et al. 2006, 2011), it was unexpected that astrocytes would be the phagocytic cell type. In the adult Drosophila brain, a second type of neuropil glial cells, ensheathing glia, is responsible for engulfing injured axonal debris, while astrocytes fail to respond in a detectable way morphologically or molecularly (e.g., by Draper up-regulation) (Doherty et al. 2009). The stark difference in glial subtypes executing phagocytic function in the pupa versus the adult could result from differences in glial genetic programs during development versus in mature glia or indicate a key difference in the molecular nature of pruned neurites compared with those undergoing injury-induced axon degeneration.
The extent to which mammalian astrocytes are phagocytes in vivo has remained unclear until recently. In culture, astrocytes can be phagocytic (Roldan et al. 1997; Tansey and Cammer 1998) and engulf apoptotic cells (Chang et al. 2000) or amyloid-β (Wyss-Coray et al. 2003). In vivo, the post-laminar optic nerve head myelination transition zone (MTZ) astrocytes express Mac-2, a molecule implicated in phagocytic activity, and internalize axonal evulsions, and this event appears to be increased in frequency in glaucoma models (Nguyen et al. 2011). Most impressively, Chung et al. (2013) recently directly demonstrated that astrocytes engulf synaptic material in vivo and that this event is mediated by MEGF10 (mouse Draper) and the engulfment receptor MERTK. Thus, Draper/MEGF10-dependent engulfment of pruned synapses appears to be an ancient astrocytic mechanism; whether the same is true for the Crk/Mbc/dCed-12 pathway remains to be determined. Transcriptome data from purified mouse forebrain astrocytes support this notion, since molecular pathways for engulfment are highly enriched, including, in addition to MEGF10, Gulp1 (dCed-6), and in addition to Crk, Dock1 (Mbc) and Elmo (dCed-12) (Cahoy et al. 2008). It will be important to explore whether these additional pathways have similar roles in astrocytes for the pruning of mammalian neural circuits during development and whether astrocyte engulfment activity is modified in neurological diseases involving axonal, dendritic, or synaptic loss.
Activation of astrocyte phagocytic function requires Draper and the Crk/Mbc/dCed-12 signaling pathway
We examined two types of neurons that undergo different types of developmental reorganization. MB γ neurons prune their medial and dorsal axon branches and dendrites, their cell bodies remain viable, and, at midpupal stages, they re-extend medial axon branches to establish adult-specific connectivity (Lee et al. 2000; Watts et al. 2004). In contrast, vCrz+ neurons exhibit complete neurite degeneration, and cell bodies undergo apoptotic death and eventually are completely eliminated (Choi et al. 2006). We found that in addition to there being remarkable differences in the patterns of fragmentation exhibited by these subsets of neurons, there are also critical differences in the engulfment signaling pathways used to promote their initial destruction and clearance.
Previous work has revealed a role for Draper in MB γ neuron pruning, with draper-null mutants exhibiting a delay of ∼2 d in the clearance of pruned MB γ neuron axonal debris (Awasaki et al. 2006). We provide evidence supporting a key role for the Crk/Mbc/dCed-12 complex in the clearance of MB γ neuron axonal debris and demonstrate that Crk/Mbc/dCed-12 knockdown and draper mutants have additive phenotypes, indicating that these signaling pathways act in a partially redundant fashion in astrocytes to promote clearance of pruned MB γ neuron axons. While these phenotypes are additive, we note that draper mutants exhibit a stronger delay in MB γ neuron pruning, suggesting that Draper signaling plays a more prominent role in this brain region than signaling through the Crk/Mbc/dCed-12 complex. However, we cannot exclude the possibility that a Crk, mbc, or dCed-12 mutant might have a stronger phenotype than the RNAi lines used in this study.
vCrz+ neuronal clearance appears to involve both Draper signaling and the Crk/Mbc/dCed-12 complex, with the former primarily promoting clearance of vCrz+ cell bodies, and the latter driving clearance of degenerating vCrz+ neurites. In draper-null mutants at 18 h APF, the majority of neuronal cell bodies remained at the edge of the neuropil, while neurite debris was largely cleared. Reciprocally, astrocytic knockdown of dCed-12 suppressed neurite clearance, while vCrz+ cell bodies were promptly eliminated. The lack of additivity of the phenotypes for either neurite or cell body clearance in draper-null mutants with astrocytic dCed-12RNAi suggests that elimination of cell bodies is primarily driven by the Draper pathway, while neurite clearance is largely accomplished by signaling through the Crk/Mbc/dCed-12 complex.
Our study reveals that the genetic pathways engaged by glia to engulf pruned neuronal material versus apoptotic neurons are context-dependent and correlate with the type of destructive program. Interestingly, the molecular pathways that mediate axonal degeneration during axon pruning versus apoptosis are also distinct in mammalian cultured neurons. Local withdrawal of NGF in the axon activates Caspase 6-dependent axon degeneration, which is not sensitive to XIAP1 inhibition. In contrast, whole-cell NGF withdrawal leads to Caspase 6-independent apoptotic cell death and degeneration of axons, which is then sensitive to XIAP1 (Cusack et al. 2013). These observations argue that context matters when neurites and cell bodies are being destroyed. We speculate that neuron–glia signaling during engulfment events might also be compartmentalized, with neurites and cell bodies generating different types of “eat me” cues for clearance by glia.
Do engulfing glial cells actively promote the destruction of target neurons? We made the intriguing observation that loss of a single copy of draper is sufficient to dominantly suppress the elimination of vCrz+ cell bodies and neurites at 6 h APF. Moreover, loss of two copies results in the retention of nearly all vCrz+ cell bodies and significant parts of the vCrz+ scaffold, and many regions of the scaffold appeared morphologically intact, suggesting a delay of neurite fragmentation. Previous work has shown that expression of the anti-apoptotic molecule P35 is sufficient to suppress the pruning of vCrz+ neurites for at least 1 d (Choi et al. 2006); we extended this observation and found this to be true for at least 2 d (data not shown). Thus, blocking apoptosis in vCrz+ is sufficient to significantly delay neurite degeneration. This observation, coupled with the known role for CED-1 in actively promoting the death of engulfment targets (Hoeppner et al. 2001; Reddien et al. 2001), suggests that engulfing nonastrocyte glia may promote neuronal apoptosis through Draper signaling. This in turn would promote degeneration of the neurite scaffold after cell body death. If this underlies the delay that we observed in neurite degeneration in draper mutants, it would argue that glia actively sculpt neural networks by promoting the destruction of selected subsets of neurons. Subsequent work exploring the survival of cell bodies preserved in draper-null animals will be essential to explore this exciting possibility.
Materials and methods
Drosophila strains
The Drosophila strains used were as follows: alrm-Gal4 (Doherty et al. 2009), pUAST-mCD8∷GFP (Lee and Luo 2001), draperΔ5 (MacDonald et al. 2006), UAS-shibirets, w1118, UAS-EcR.B1-ΔC655.F645A and UAS-EcR.B1-ΔC655.W650A (Cherbas et al. 2003), UAS-CrkRNAi (Vienna Drosophila RNAi Center [VDRC] transformant 19061), UAS-dCed12RNAi (VDRC transformant 107590), UAS-mbcRNAi (VDRC transformant 10644), Crz-Gal4 (Choi et al. 2006), repo-flp6-2 (a gift from T. Stork), pWiz-drprRNAi7b (MacDonald et al. 2006), repo-Gal4 (Sepp et al. 2001), tub-Gal80, and UAS-draperI (Logan et al. 2012). The alrm-Gal80 construct was generated by amplifying a 5-kb region of the alrm promoter using the following primers: 5′-GATCAACCGCGGACTACGCACAGATGTGGTCATCTGAATAGGTTTC-3′ and 5′-AAATTTGTCTAGATAGTGGCGATCCTTTCGCTCGGGAGCC-3′. The resulting fragment was cloned into the Cas40-GAL80 vector, and transgenic flies were generated using standard methods by BestGene, Inc.
Acknowledgments
We thank all members of the Freeman laboratory for helpful advice and discussions, and Tobi Stork, Lukas Neukomm, Megan Corty, Tsai-Yi Lu, Jaeda Coutinho-Budd, and A. Nicole Fox for critical reading of the manuscript. We thank Amy Sheehan for production of the alrm-Gal80 construct, and Takeshi Awasaki, Tobi Stork, and Jae Park for generously providing reagents. nc82 (developed by E. Buchner), anti-Repo (8D12; developed by C. Goodman), and 1D4 (developed by C. Goodman) were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by Department of Biology at The University of Iowa (Iowa City, IA). The EM portion of the project was supported by award number S10RR027897 from the National Center For Research Resources. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center For Research Resources or the National Institutes of Health (NIH). This work was supported by NIH grant NS053538 (to M.R.F.), and M.R.F. is an Investigator of the Howard Hughes Medical Institute.
Note added in proof
While this manuscript was submitted, a new study was also being published at the same time elucidating astrocytes' phagocytic role during MB γ neuronal pruning (Hakim et al. 2014).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.229518.113.
References
- Awasaki T, Ito K 2004. Engulfing action of glial cells is required for programmed axon pruning during Drosophila metamorphosis. Curr Biol 14: 668–677 [DOI] [PubMed] [Google Scholar]
- Awasaki T, Tatsumi R, Takahashi K, Arai K, Nakanishi Y, Ueda R, Ito K 2006. Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron 50: 855–867 [DOI] [PubMed] [Google Scholar]
- Awasaki T, Huang Y, O'Connor MB, Lee T 2011. Glia instruct developmental neuronal remodeling through TGF-β signaling. Nat Neurosci 14: 821–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DL, Misgeld T, Walsh MK, Gan WB, Lichtman JW 2004. Axon branch removal at developing synapses by axosome shedding. Neuron 44: 651–661 [DOI] [PubMed] [Google Scholar]
- Brugnera E, Haney L, Grimsley C, Lu M, Walk SF, Tosello-Trampont AC, Macara IG, Madhani H, Fink GR, Ravichandran KS 2002. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat Cell Biol 4: 574–582 [DOI] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, Xing Y, Lubischer JL, Krieg PA, Krupenko SA et al. 2008. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J Neurosci 28: 264–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang GH, Barbaro NM, Pieper RO 2000. Phosphatidylserine-dependent phagocytosis of apoptotic glioma cells by normal human microglia, astrocytes, and glioma cells. Neuro Oncol 2: 174–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Regehr WG 2000. Developmental remodeling of the retinogeniculate synapse. Neuron 28: 955–966 [DOI] [PubMed] [Google Scholar]
- Cherbas L, Hu X, Zhimulev I, Belyaeva E, Cherbas P 2003. EcR isoforms in Drosophila: Testing tissue-specific requirements by targeted blockade and rescue. Development 130: 271–284 [DOI] [PubMed] [Google Scholar]
- Choi YJ, Lee G, Park JH 2006. Programmed cell death mechanisms of identifiable peptidergic neurons in Drosophila melanogaster. Development 133: 2223–2232 [DOI] [PubMed] [Google Scholar]
- Chung WS, Clarke LE, Wang GX, Stafford BK, Sher A, Chakraborty C, Joung J, Foo LC, Thompson A, Chen C, et al. 2013. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature doi: 10.1038/nature12776 [DOI] [PMC free article] [PubMed]
- Cowan WM, Fawcett JW, O'Leary DD, Stanfield BB 1984. Regressive events in neurogenesis. Science 225: 1258–1265 [DOI] [PubMed] [Google Scholar]
- Cuadros MA, Navascues J 1998. The origin and differentiation of microglial cells during development. Prog Neurobiol 56: 173–189 [DOI] [PubMed] [Google Scholar]
- Cunningham CL, Martinez-Cerdeno V, Noctor SC 2013. Microglia regulate the number of neural precursor cells in the developing cerebral cortex. J Neurosci 33: 4216–4233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack CL, Swahari V, Hampton Henley W, Michael Ramsey J, Deshmukh M 2013. Distinct pathways mediate axon degeneration during apoptosis and axon-specific pruning. Nat Commun 4: 1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty J, Logan MA, Tasdemir OE, Freeman MR 2009. Ensheathing glia function as phagocytes in the adult Drosophila brain. J Neurosci 29: 4768–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis RE, Jacobson DM, Horvitz HR 1991. Genes required for the engulfment of cell corpses during programmed cell death in Caenorhabditis elegans. Genetics 129: 79–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchegaray JI, Timmons AK, Klein AP, Pritchett TL, Welch E, Meehan TL, Li C, McCall K 2012. Draper acts through the JNK pathway to control synchronous engulfment of dying germline cells by follicular epithelial cells. Development 139: 4029–4039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman MR, Delrow J, Kim J, Johnson E, Doe CQ 2003. Unwrapping glial biology: Gcm target genes regulating glial development, diversification, and function. Neuron 38: 567–580 [DOI] [PubMed] [Google Scholar]
- Fuentes-Medel Y, Logan MA, Ashley J, Ataman B, Budnik V, Freeman MR 2009. Glia and muscle sculpt neuromuscular arbors by engulfing destabilized synaptic boutons and shed presynaptic debris. PLoS Biol 7: e1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumienny TL, Brugnera E, Tosello-Trampont AC, Kinchen JM, Haney LB, Nishiwaki K, Walk SF, Nemergut ME, Macara IG, Francis R, et al. 2001. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell 107: 27–41 [DOI] [PubMed] [Google Scholar]
- Hakim Y, Yaniv SP, Schuldiner O 2014. Astrocytes play a key role in Drosophila mushroom body axon pruning. PLoS One (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeppner DJ, Hengartner MO, Schnabel R 2001. Engulfment genes cooperate with ced-3 to promote cell death in Caenorhabditis elegans. Nature 412: 202–206 [DOI] [PubMed] [Google Scholar]
- Hooks BM, Chen C 2006. Distinct roles for spontaneous and visual activity in remodeling of the retinogeniculate synapse. Neuron 52: 281–291 [DOI] [PubMed] [Google Scholar]
- Jiang C, Baehrecke EH, Thummel CS 1997. Steroid regulated programmed cell death during Drosophila metamorphosis. Development 124: 4673–4683 [DOI] [PubMed] [Google Scholar]
- Kinchen JM, Cabello J, Klingele D, Wong K, Feichtinger R, Schnabel H, Schnabel R, Hengartner MO 2005. Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C. elegans. Nature 434: 93–99 [DOI] [PubMed] [Google Scholar]
- Koelle MR, Talbot WS, Segraves WA, Bender MT, Cherbas P, Hogness DS 1991. The Drosophila EcR gene encodes an ecdysone receptor, a new member of the steroid receptor superfamily. Cell 67: 59–77 [DOI] [PubMed] [Google Scholar]
- Kuo CT, Jan LY, Jan YN 2005. Dendrite-specific remodeling of Drosophila sensory neurons requires matrix metalloproteases, ubiquitin-proteasome, and ecdysone signaling. Proc Natl Acad Sci 102: 15230–15235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landmesser L, Pilar G 1972. The onset and development of transmission in the chick ciliary ganglion. J Physiol 222: 691–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landmesser L, Pilar G 1974. Synaptic transmission and cell death during normal ganglionic development. J Physiol 241: 737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L 2001. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci 24: 251–254 [DOI] [PubMed] [Google Scholar]
- Lee T, Marticke S, Sung C, Robinow S, Luo L 2000. Cell-autonomous requirement of the USP/EcR-B ecdysone receptor for mushroom body neuronal remodeling in Drosophila. Neuron 28: 807–818 [DOI] [PubMed] [Google Scholar]
- Lee G, Wang Z, Sehgal R, Chen CH, Kikuno K, Hay B, Park JH 2011. Drosophila caspases involved in developmentally regulated programmed cell death of peptidergic neurons during early metamorphosis. J Comp Neurol 519: 34–48 [DOI] [PubMed] [Google Scholar]
- Liu QA, Hengartner MO 1998. Candidate adaptor protein CED-6 promotes the engulfment of apoptotic cells in C. elegans. Cell 93: 961–972 [DOI] [PubMed] [Google Scholar]
- Logan MA, Hackett R, Doherty J, Sheehan A, Speese SD, Freeman MR 2012. Negative regulation of glial engulfment activity by Draper terminates glial responses to axon injury. Nat Neurosci 15: 722–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald JM, Beach MG, Porpiglia E, Sheehan AE, Watts RJ, Freeman MR 2006. The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons. Neuron 50: 869–881 [DOI] [PubMed] [Google Scholar]
- Manaka J, Kuraishi T, Shiratsuchi A, Nakai Y, Higashida H, Henson P, Nakanishi Y 2004. Draper-mediated and phosphatidylserine-independent phagocytosis of apoptotic cells by Drosophila hemocytes/macrophages. J Biol Chem 279: 48466–48476 [DOI] [PubMed] [Google Scholar]
- Marin-Teva JL, Cuadros MA, Calvente R, Almendros A, Navascues J 1999. Naturally occurring cell death and migration of microglial precursors in the quail retina during normal development. J Comp Neurol 412: 255–275 [PubMed] [Google Scholar]
- Marin-Teva JL, Dusart I, Colin C, Gervais A, van Rooijen N, Mallat M 2004. Microglia promote the death of developing Purkinje cells. Neuron 41: 535–547 [DOI] [PubMed] [Google Scholar]
- Nguyen JV, Soto I, Kim KY, Bushong EA, Oglesby E, Valiente-Soriano FJ, Yang Z, Davis CH, Bedont JL, Son JL, et al. 2011. Myelination transition zone astrocytes are constitutively phagocytic and have synuclein dependent reactivity in glaucoma. Proc Natl Acad Sci 108: 1176–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, et al. 2011. Synaptic pruning by microglia is necessary for normal brain development. Science 333: 1456–1458 [DOI] [PubMed] [Google Scholar]
- Parnaik R, Raff MC, Scholes J 2000. Differences between the clearance of apoptotic cells by professional and non-professional phagocytes. Curr Biol 10: 857–860 [DOI] [PubMed] [Google Scholar]
- Reddien PW, Horvitz HR 2004. The engulfment process of programmed cell death in Caenorhabditis elegans. Annu Rev Cell Dev Biol 20: 193–221 [DOI] [PubMed] [Google Scholar]
- Reddien PW, Cameron S, Horvitz HR 2001. Phagocytosis promotes programmed cell death in C. elegans. Nature 412: 198–202 [DOI] [PubMed] [Google Scholar]
- Robinow S, Talbot WS, Hogness DS, Truman JW 1993. Programmed cell death in the Drosophila CNS is ecdysone-regulated and coupled with a specific ecdysone receptor isoform. Development 119: 1251–1259 [DOI] [PubMed] [Google Scholar]
- Roldan A, Gogg S, Ferrini M, Schillaci R, De Nicola AF 1997. Glucocorticoid regulation of in vitro astrocyte phagocytosis. Biocell 21: 83–89 [PubMed] [Google Scholar]
- Savill J, Dransfield I, Gregory C, Haslett C 2002. A blast from the past: Clearance of apoptotic cells regulates immune responses. Nat Rev Immunol 2: 965–975 [DOI] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, Stevens B 2012. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74: 691–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubiger M, Wade AA, Carney GE, Truman JW, Bender M 1998. Drosophila EcR-B ecdysone receptor isoforms are required for larval molting and for neuron remodeling during metamorphosis. Development 125: 2053–2062 [DOI] [PubMed] [Google Scholar]
- Schubiger M, Tomita S, Sung C, Robinow S, Truman JW 2003. Isoform specific control of gene activity in vivo by the Drosophila ecdysone receptor. Mech Dev 120: 909–918 [DOI] [PubMed] [Google Scholar]
- Sepp KJ, Schulte J, Auld VJ 2001. Peripheral glia direct axon guidance across the CNS/PNS transition zone. Dev Biol 238: 47–63 [DOI] [PubMed] [Google Scholar]
- Smits E, Van Criekinge W, Plaetinck G, Bogaert T 1999. The human homologue of Caenorhabditis elegans CED-6 specifically promotes phagocytosis of apoptotic cells. Curr Biol 9: 1351–1354 [DOI] [PubMed] [Google Scholar]
- Southwell DG, Paredes MF, Galvao RP, Jones DL, Froemke RC, Sebe JY, Alfaro-Cervello C, Tang Y, Garcia-Verdugo JM, Rubenstein JL, et al. 2012. Intrinsically determined cell death of developing cortical interneurons. Nature 491: 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B, et al. 2007. The classical complement cascade mediates CNS synapse elimination. Cell 131: 1164–1178 [DOI] [PubMed] [Google Scholar]
- Talbot WS, Swyryd EA, Hogness DS 1993. Drosophila tissues with different metamorphic responses to ecdysone express different ecdysone receptor isoforms. Cell 73: 1323–1337 [DOI] [PubMed] [Google Scholar]
- Tansey FA, Cammer W 1998. Differential uptake of dextran beads by astrocytes, macrophages and oligodendrocytes in mixed glial-cell cultures from brains of neonatal rats. Neurosci Lett 248: 159–162 [DOI] [PubMed] [Google Scholar]
- Truman JW, Talbot WS, Fahrbach SE, Hogness DS 1994. Ecdysone receptor expression in the CNS correlates with stage-specific responses to ecdysteroids during Drosophila and Manduca development. Development 120: 219–234 [DOI] [PubMed] [Google Scholar]
- Van Goethem E, Silva EA, Xiao H, Franc NC 2012. The Drosophila TRPP cation channel, PKD2 and Dmel/Ced-12 act in genetically distinct pathways during apoptotic cell clearance. PLoS ONE 7: e31488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ham TJ, Kokel D, Peterson RT 2012. Apoptotic cells are cleared by directional migration and elmo1- dependent macrophage engulfment. Curr Biol 22: 830–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bartheld CS, Bothwell M 1993. Development of the mesencephalic nucleus of the trigeminal nerve in chick embryos: Target innervation, neurotrophin receptors, and cell death. J Comp Neurol 328: 185–202 [DOI] [PubMed] [Google Scholar]
- Walsh MK, Lichtman JW 2003. In vivo time-lapse imaging of synaptic takeover associated with naturally occurring synapse elimination. Neuron 37: 67–73 [DOI] [PubMed] [Google Scholar]
- Watts RJ, Schuldiner O, Perrino J, Larsen C, Luo L 2004. Glia engulf degenerating axons during developmental axon pruning. Curr Biology 14: 678–684 [DOI] [PubMed] [Google Scholar]
- White KP, Hurban P, Watanabe T, Hogness DS 1997. Coordination of Drosophila metamorphosis by two ecdysone-induced nuclear receptors. Science 276: 114–117 [DOI] [PubMed] [Google Scholar]
- Williams DW, Kondo S, Krzyzanowska A, Hiromi Y, Truman JW 2006. Local caspase activity directs engulfment of dendrites during pruning. Nat Neurosci 9: 1234–1236 [DOI] [PubMed] [Google Scholar]
- Wu HH, Bellmunt E, Scheib JL, Venegas V, Burkert C, Reichardt LF, Zhou Z, Farinas I, Carter BD 2009. Glial precursors clear sensory neuron corpses during development via Jedi-1, an engulfment receptor. Nat Neurosci 12: 1534–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss-Coray T, Loike JD, Brionne TC, Lu E, Anankov R, Yan F, Silverstein SC, Husemann J 2003. Adult mouse astrocytes degrade amyloid-β in vitro and in situ. Nat Med 9: 453–457 [DOI] [PubMed] [Google Scholar]
- Yao TP, Forman BM, Jiang Z, Cherbas L, Chen JD, McKeown M, Cherbas P, Evans RM 1993. Functional ecdysone receptor is the product of EcR and ultraspiracle genes. Nature 366: 476–479 [DOI] [PubMed] [Google Scholar]
- Yu X, Lu N, Zhou Z 2008. Phagocytic receptor CED-1 initiates a signaling pathway for degrading engulfed apoptotic cells. PLoS Biol 6: e61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Caron E, Hartwieg E, Hall A, Horvitz HR 2001a. The C. elegans PH domain protein CED-12 regulates cytoskeletal reorganization via a Rho/Rac GTPase signaling pathway. Dev Cell 1: 477–489 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Hartwieg E, Horvitz HR 2001b. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell 104: 43–56 [DOI] [PubMed] [Google Scholar]
- Ziegenfuss JS, Biswas R, Avery MA, Hong K, Sheehan AE, Yeung YG, Stanley ER, Freeman MR 2008. Draper-dependent glial phagocytic activity is mediated by Src and Syk family kinase signalling. Nature 453: 935–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegenfuss JS, Doherty J, Freeman MR 2012. Distinct molecular pathways mediate glial activation and engulfment of axonal debris after axotomy. Nat Neurosci 15: 979–987 [DOI] [PMC free article] [PubMed] [Google Scholar]