Left/right asymmetric features of the body are either randomly distributed on either the left or right side within a population (antisymmetries) or found on one particular side (directional asymmetries). Nervous systems display both types of asymmetries, but it is unknown whether shared regulators establish them. Here, Cochella et al. show that the transcription factor die-1 controls both antisymmetry and directional asymmetry in distinct sensory systems in C. elegans. This study uncovers the first molecular link between two different kinds of body plan asymmetries.
Keywords: asymmetry, transcriptional control, C. elegans
Abstract
Left/right asymmetric features of animals are either randomly distributed on either the left or right side within a population (“antisymmetries”) or found stereotypically on one particular side of an animal (“directional asymmetries”). Both types of asymmetries can be found in nervous systems, but whether the regulatory programs that establish these asymmetries share any mechanistic features is not known. We describe here an unprecedented molecular link between these two types of asymmetries in Caenorhabditis elegans. The zinc finger transcription factor die-1 is expressed in a directionally asymmetric manner in the gustatory neuron pair ASE left (ASEL) and ASE right (ASER), while it is expressed in an antisymmetric manner in the olfactory neuron pair AWC left (AWCL) and AWC right (AWCR). Asymmetric die-1 expression is controlled in a fundamentally distinct manner in these two neuron pairs. Importantly, asymmetric die-1 expression controls the directionally asymmetric expression of gustatory receptor proteins in the ASE neurons and the antisymmetric expression of olfactory receptor proteins in the AWC neurons. These asymmetries serve to increase the ability of the animal to discriminate distinct chemosensory inputs.
The body plan of most higher animals is largely bilaterally symmetric, yet there are deviations from bilaterality in visceral organ placement (Ludwig 1932) and nervous systems (Hobert et al. 2002; Rogers et al. 2013). Asymmetries come in two fundamentally distinct types. “Antisymmetries” refer to stochastic left/right asymmetries in which a specific feature is present on either the left or right side of an animal (Palmer 2005). For example, claw size in crustaceans and paw preference in many animals are antisymmetric. In contrast to antisymmetries, “directional asymmetries” refer to asymmetries that are stereotypically found on only one side of the animal (within at least 95% of animals in a population) (Palmer 2005). For example, language production in the left hemisphere of the human brain is directional.
Compared with developmental decisions along the anterior/posterior or dorsal/ventral axes, there are still fundamental gaps in our understanding of patterning along the left/right axis, particularly in the nervous system. Specifically, since the molecular and mechanistic basis of the establishment of asymmetries is generally poorly understood, it has not been possible to compare and perhaps find commonalities in the control of antisymmetries and directional asymmetries. Intriguingly, evolutionary tracing of left/right asymmetric features suggests that antisymmetries may be representative of an ancestral state that can become fixed to result in a directional asymmetry (Palmer 2004). However, the molecular basis for such an evolutionary progression is unclear.
The nervous system of the nematode Caenorhabditis elegans displays two striking examples of antisymmetry and directional asymmetry in distinct sensory systems. The AWC olfactory neuron pair expresses several G-protein-coupled receptor (GPCR)-type olfactory receptors in a stochastic, anti-correlated, left/right asymmetric manner (Fig. 1; Troemel et al. 1999; Bauer Huang et al. 2007). In contrast, the ASE gustatory neuron pair expresses putative chemoreceptors of the receptor-type guanylyl cyclase family (gcy genes) in a directionally asymmetric manner (Fig. 1; Yu et al. 1997; Ortiz et al. 2006). In both cases, asymmetric expression of chemoreceptors ensures that the left and right neurons are able to discriminate between distinct sensory cues (Pierce-Shimomura et al. 2001; Wes and Bargmann 2001; Ortiz et al. 2009). Such discrimination would not be possible if receptors were coexpressed in both left and right neurons.
Figure 1.
AWC and ASE asymmetry. Previously known components of genetic pathways that control AWC and ASE asymmetries. Not all genes known to be involved are shown. Black and gray gene names indicate whether a gene is more active or more expressed (black) in one neuron compared with the other neuron.
Genetic screens for mutants that affect left/right asymmetric expression of the putative olfactory receptor str-2 in AWC left/right (AWCL/R) have revealed a transient neural network formed via NSY-5 gap junctions between distinct sensory neurons, including AWC (Chuang et al. 2007). A calcium-triggered signaling pathway operating downstream from this gap junction network establishes AWC asymmetry (Fig. 1; Sagasti et al. 2001; Chuang and Bargmann 2005). However, none of these mechanisms operate in the establishment of ASE asymmetry (Chang et al. 2003). Left/right asymmetry of the two ASE neurons is rather conferred by a Notch-dependent signaling event in the early embryo that establishes a specific chromatin state at a microRNA (miRNA) locus, lsy-6, in the precursors of the left ASE neuron (Poole and Hobert 2006; Cochella and Hobert 2012). After birth of the ASE left (ASEL) neuron, this miRNA then operates through a complex gene regulatory network to determine left/right asymmetric gcy gene expression (Fig. 1; Hobert 2006). At the core of the gene regulatory network is the ASEL-restricted C2H2 zinc finger transcription factor DIE-1, whose loss results in a conversion of ASEL identity to ASE right (ASER) identity (“two-ASER phenotype”) (Fig. 1; Chang et al. 2004).
While investigating the directionally asymmetric regulation of die-1 expression in ASEL in more detail, we found that die-1 is also expressed in an antisymmetric manner in the AWC neuron pair. We show that this asymmetry is necessary and sufficient for antisymmetric expression of the GPCR-type chemoreceptors of the AWC neurons. Our studies provide the first molecular link between two completely different kinds of asymmetries in an animal's body plan.
Results
Regulation of directionally asymmetric die-1 expression in the ASE neurons
Our initial goal was to examine the regulation of asymmetric expression of the die-1 locus in the ASE neurons in more detail on the level of both cis-regulatory elements and trans-acting factors. Our starting point for this analysis was the previously reported finding that the 3′ untranslated region (UTR) of the die-1 locus contains cis-regulatory elements that are sufficient to repress gene expression in ASER, thereby imposing left/right asymmetric gene expression onto the two ASE neurons (Chang et al. 2004; Didiano et al. 2010). However, even though these likely post-transcriptional cis-regulatory elements are sufficient to direct asymmetric gene expression, they are not required. If the 3′ UTR of the die-1 gene is deleted in the context of a 40-kb fosmid-based reporter construct containing the full die-1 locus, die-1 expression remains left/right asymmetric (Didiano et al. 2010). This 3′ UTR-independent asymmetry could be ascribed to either differential post-translational control of DIE-1 protein levels in ASEL versus ASER or differential transcription of the die-1 gene in ASEL versus ASER. If the latter case were true, one would expect to identify cis-regulatory elements outside the coding sequences and UTRs that direct asymmetric gene expression.
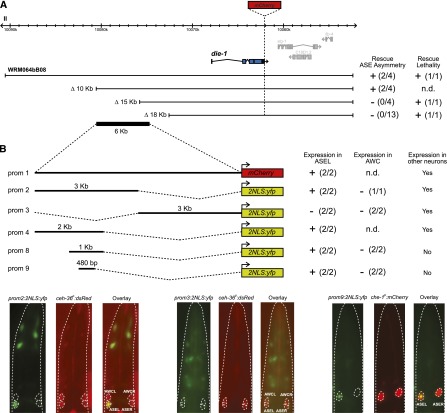
The unusually large size of the die-1 locus (∼27-kb 5′ intergenic region, first introns ∼3.3 kb) (Fig. 2A) hampered initial attempts to identify such elements in proximity to the die-1 gene. We set out to assay for the existence of transcriptional regulatory elements using a strategy in which we made use of the ability of a mCherry-tagged die-1 fosmid reporter to rescue the ASE asymmetry defect of die-1 mutant animals. We generated deletion derivatives of this fosmid reporter and asked how much of the genomic sequence upstream of the protein-coding part of the die-1 locus is required for rescue of the ASE asymmetry defect. This mapping led to the identification of a 5-kb region located ∼8 kb upstream of the first exon of die-1 that is required for rescue of ASE asymmetry defects (Fig. 2A; Supplemental Table S1). Constructs that do not rescue the ASE asymmetry defects still rescued the lethality of die-1 caused by hypodermal patterning defects, demonstrating that hypodermal and neuronal regulatory elements can be separated (Fig. 2A; Supplemental Table S1). Further analysis showed that the distal upstream region contains a 480-base-pair (bp) regulatory element that drives expression exclusively in ASEL and no other neurons (Fig. 2B). We conclude that the die-1 locus contains two distinct types of cis-regulatory elements that have the ability to direct left/right asymmetric gene expression: one located in the 3′ UTR and likely conferring post-transcriptional repression in ASER and one upstream of the locus and likely conferring transcriptional restriction to ASEL.
Figure 2.
Cis-regulatory analysis of asymmetric die-1 expression in the ASE neurons. (A) Rescue of die-1 fosmid derivatives (representative lines otEx4144, otEx4300, otEx4299, and otEx4342). Each construct was analyzed for its ability to rescue the loss of ASE neuron asymmetry observed in die-1(ot26) as well as the lethality observed in die-1(w34) due to the epidermal enclosure failure. The number of independent lines that showed rescue/number of lines analyzed are shown in parentheses. For the ASE asymmetry scoring, lines considered as providing rescue did so in >80% of the observed animals; lines considered as nonrescuing had 0% of observed animals with the wild-type phenotype. L4 and young adult animals were scored. (B) Defining an ASEL-specific cis-regulatory element in the die-1 locus. The number of independent lines that showed expression/the number of lines analyzed are shown in parentheses. Representative images of larvae carrying three different promoter fusions are shown. die-1prom2 (otEx4349) drives expression in ASEL but not in ASER or the AWC neurons. die-1prom3 (otEx4352) drives expression in a few neurons as well as in some hypodermal cells but not in the ASE or AWC neurons. die-1prom9 drives expression solely in ASEL.
Different phases of asymmetric die-1 expression in ASEL are controlled by distinct trans-acting factors
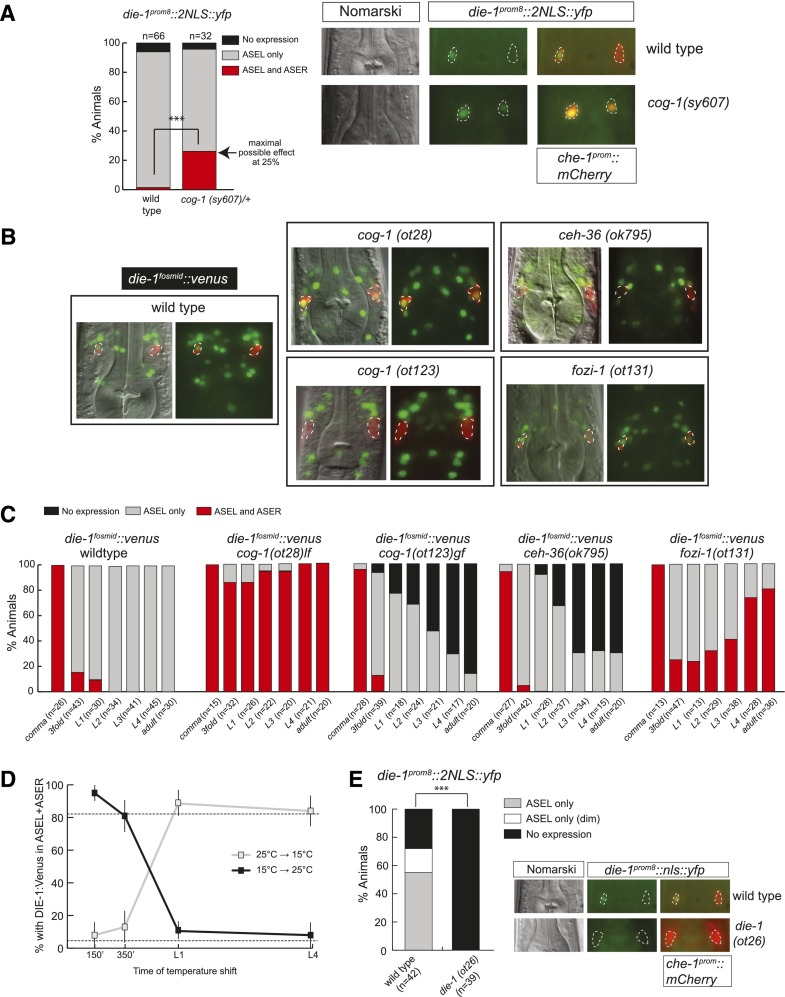
Our previous genetic analysis of die-1 regulation has identified the homeobox gene cog-1 as a regulator—most likely indirect—of asymmetric expression of the die-1 3′ UTR (Johnston et al. 2005; Didiano et al. 2010). We found that expression of the die-1 transcriptional reporter that we described above also depends on cog-1; it is derepressed in ASER in cog-1 mutant animals (Fig. 3A). We found that cog-1 also affects the expression of the DIE-1 protein expressed from the entire die-1 locus; the reporter is also derepressed in the ASER neurons of cog-1(−) mutant animals (ot28) (Fig. 3B,C). Conversely, a gain-of-function allele of cog-1, ot123, which is ectopically expressed in the ASEL neuron (due to a 3′ UTR deletion that renders it refractory to lsy-6-mediated repression in ASEL) (Sarin et al. 2007), results in repression of die-1 expression in ASEL (Fig. 3B,C).
Figure 3.
Different phases of asymmetric die-1 expression in ASEL are controlled by distinct trans-acting factors. (A) cog-1 restricts die-1 expression at the transcriptional level. The die-1 prom8∷2nls∷yfp transcriptional reporter (otEx4359) (Fig. 1C) shows derepression in ASER in the background of a strong loss-of-function allele of cog-1(sy607). This allele is maintained as a heterozygote, as it has very low viability in homozygosity. Thus, the progeny of heterozygous mothers were analyzed. These should produce one-fourth of homozygous cog-1(sy607) progeny, and thus 25% of animals showing expression in ASEL+ASER suggest a fully penetrant derepression of die-1 in ASER in cog-1(sy607). Representative images are shown in which the ASE neurons are marked by che-1prom∷mCherry. (B) Representative images of L4/young adult animals with die-1∷2xFlag∷Venus (otIs274) fosmid expression in different ASE asymmetry mutant backgrounds. Animals contain che-1prom∷mCherry in the background to facilitate identification of the ASE neurons. Images were derived by Z-projections of stacks with focal planes showing die-1fosmid∷ 2xFlag∷Venus and che-1prom∷mCherry signals. The identity of the ASE neurons was confirmed by combining Nomarski and ASE-specific che-1prom∷mCherry signals. The die-1∷2xFlag∷Venus was carefully assessed to be in the corresponding ASE neuron nucleus. Due to the aggregation of the mCherry protein, some neurons have low or little mCherry in the nucleus. The 2xFlag∷Venus-labeled functional DIE-1 transcription factor (derived from the fosmid-based die-1∷2xFlag∷Venus reporter) is located exclusively in the nucleus. Some neurons with little or no mCherry (derived from the transcriptional che-1prom∷mCherry reporter) in the nucleus show no or little “yellow” staining in the merged fluorescence channels. See C for quantification. (C) Time course of die-1∷2xFlag∷Venus (otIs274) expression in different ASE asymmetry mutant backgrounds during early development until adulthood. Due to the difficulty of handling sterile cog-1(sy607)-null mutants, we scored the cog-1(ot28) hypomorphic allele, which also shows fully penetrant defects. (D) cog-1 is only required shortly after the birth of the ASE neurons to restrict die-1 expression to ASEL. The die-1∷2xFlag∷Venus (otIs274) reporter was analyzed in the background of a temperature-sensitive allele of cog-1(ot221). All animals were scored as L4 or young adults. Animals were grown at either the permissive temperature (15°C) or the restrictive temperature (25°C) and shifted to the opposite temperature at the indicated times. For synchronization, embryos were extracted with bleach, and two-cell embryos were isolated and allowed to develop for 150 or 350 min. The remaining animals were arrested as L1 by hatching in the absence of food and then allowed to continue development until L4 in the presence of food. (E) die-1 is required for its own expression. The die-1 prom8∷nls∷yfp transcriptional reporter (otEx4359) (Fig. 1C) loses expression in ASEL in the background of a loss-of-function allele of die-1(ot26). Representative images are shown in which the ASE neurons are marked by che-1prom∷mCherry. Animals were scored as L4 or young adults.
We next examined temporal aspects of die-1 regulation by cog-1. Confirming previous antibody staining results (Heid et al. 2001), we found that the rescuing die-1 fosmid reporter is initially expressed very broadly throughout the developing embryo, including the hypodermis, where die-1 acts to ensure ventral enclosure during gastrulation (Supplemental Fig. S1). Again mirroring antibody staining, reporter expression of the fosmid reporter vanishes at ∼460 min of embryonic development (Supplemental Fig. S1). die-1 expression then reinitiates in many neurons, including both ASEL and ASER; shortly thereafter, expression in ASER becomes undetectable (Fig. 3B,C, Supplemental Fig. S1). ASEL-restricted expression is maintained throughout the embryonic, larval, and adult stages (Figure 3B,C, Supplemental Fig. S1). ASEL restriction is paralleled by the onset of cog-1 expression exclusively in ASER, as assessed with a cog-1 fosmid reporter (Supplemental Fig. S1).
Using a temperature-sensitive cog-1 allele, ot221 (Sarin et al. 2007), we asked when cog-1 acts to control die-1 expression. Through temperature shifts at different embryonic and post-embryonic stages, we found that cog-1 is only transiently required in the embryo to repress die-1 in ASER, even though cog-1 is continuously expressed in ASER (Fig.3D). If cog-1 gene activity is removed post-embryonically, die-1 expression remains asymmetric (Fig. 3D). These data demonstrate that cog-1 is required to establish asymmetric expression of die-1 but that this laterality is maintained independently of cog-1 later on.
The sustained post-embryonic expression of die-1 in ASEL is required to maintain the expression of terminal markers of ASE laterality (O'Meara et al. 2010). Since the transient need for cog-1 to ensure die-1 repression in ASER suggests that other factors are required to maintain die-1 expression and hence ASE asymmetry, we examined how die-1 expression is maintained in ASEL. We found that ASEL-specific expression of the die-1 transcriptional reporter is abolished in die-1 mutants (Fig. 3E), suggesting that die-1 autoregulates its expression. In addition to autoregulation, we identified two more genes that are required to maintain die-1 expression in ASEL. Animals that lack the ceh-36 homeobox gene, a previously identified regulator of ASEL identity (Chang et al. 2003) that is expressed in both ASEL and ASER (Supplemental Fig. S1), show normal ASEL-restricted expression of die-1 in early developmental stages but lose die-1 expression in later stages (Fig. 3B,C). We corroborated the maintenance role of ceh-36 by finding that the conversion of terminal ASEL identity to ASER identity in ceh-36 mutants is indeed much more pronounced in older animals than in younger animals (Supplemental Table S2).
We found that fozi-1 is another factor required for maintenance of asymmetric die-1 expression in ASEL. fozi-1 is a zinc finger transcription factor expressed in the ASER neurons from threefold embryos throughout larval and adult stages (Supplemental Fig. S1; Johnston et al. 2006). We found that in fozi-1 mutants, die-1 fosmid reporter expression becomes derepressed in the ASER neuron but only in later larval and adult stages (Fig. 3B,C). This is a double-negative feedback control mechanism, as die-1 also represses fozi-1 in ASEL (Johnston et al. 2006).
Taken together, die-1 expression passes through several distinct phases: an early bilateral phase, likely controlled by bilateral ASE fate determinants such as che-1 and nhr-67 (Etchberger et al. 2007; Sarin et al. 2009); a restriction to ASEL that requires transient activity of the cog-1 homeobox gene; and a maintenance phase that requires ceh-36 and die-1 itself to promote expression in ASEL and fozi-1 to repress expression in ASER.
Antisymmetric expression of die-1 in the AWC neurons
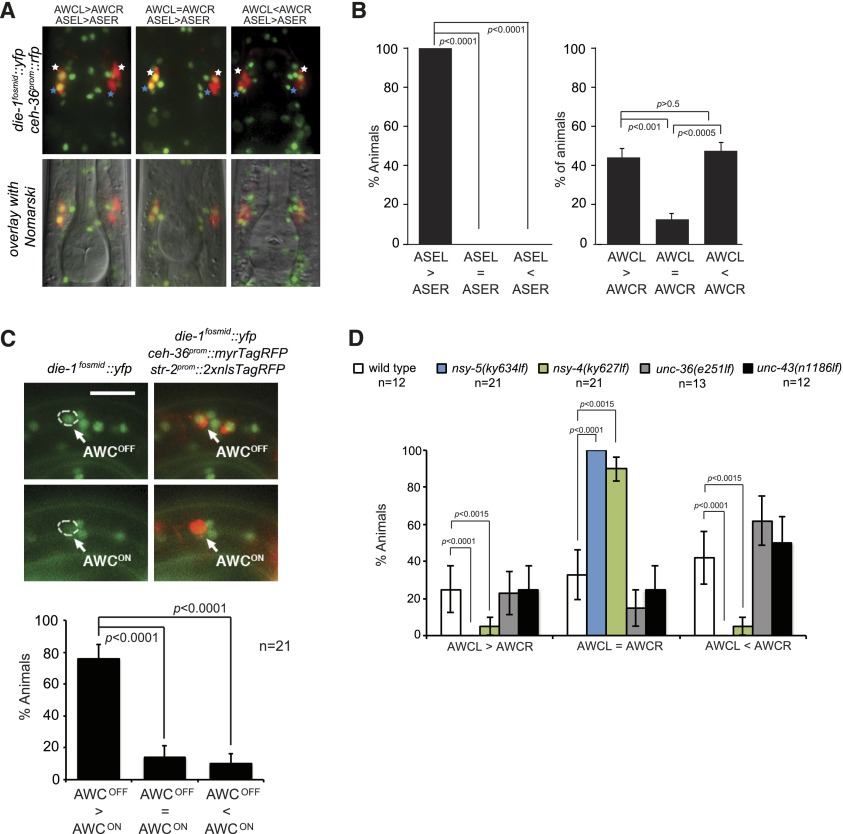
In the course of examining expression of the die-1 fosmid-based reporter in the ASE neurons, we noted that die-1 is also expressed in an asymmetric manner in the AWC olfactory neuron pair (Fig. 4A) but in a manner fundamentally distinct from the directionally asymmetric expression in the ASE neuron pair (Fig. 4A,B). Most adult animals show a strong bias of expression of die-1 to either the left or right AWC neuron (Fig. 4B). This antisymmetric die-1 expression is already evident in late embryos when the initial decision of AWC asymmetry control is made (Supplemental Fig. S2). ASEL-specific expression of die-1 can be observed before any AWC expression of die-1 is observed (data not shown).
Figure 4.
Antisymmetric expression of die-1 in the AWC neurons. (A) Representative images of die-1fosmid∷ 2xFlag∷Venus for animals showing asymmetric expression in AWC and ASE neurons labeled by expression of ceh-36 promTagRFP. Images were derived by Z-projections of stacks with focal planes showing die-1fosmid∷ 2xFlag∷Venus signals. White stars indicate AWCs, and blue stars indicate ASE neurons. (B) Quantification of die-1 asymmetry in AWC and ASE neurons. Acquired image stacks of die-1fosmid∷2xFlag∷Venus, ceh-36prom∷TagRFP (otIs264)-expressing animals were analyzed for expression in left or right AWC and ASE neurons. P-values were calculated using a Z-test. Error bars represent standard error of proportion. n = 45 for both quantifications. (C) die-1 is expressed in AWCOFF. Representative images of die-1fosmid∷yfp (vyEx1506), ceh-36prom∷myrTagRFP(vyEx792), and str-2prom∷2xnlsTagRFP(vyIs51) in a first stage larva. (Top panels) The AWCOFF cell was defined as str-2prom∷2xnlsTagRFP-negative and ceh-36prom∷myrTagRFP-positive. (Bottom panels) The AWCON cell was identified as str-2prom∷2xnlsTagRFP-positive nuclei and ceh-36prom∷myrTagRFP-positive. Bar, 10 μm. (Bottom graph) To quantify die-1fosmid∷yfp expression in AWCON and AWCOFF cells, the single focal plane with the brightest fluorescence in each AWC was selected from the acquired image stack and measured for fluorescence intensity. Each animal was categorized into one of three categories (AWCOFF > AWCON, AWCON = AWCOFF, and AWCOFF < AWCON) based on the comparison of YFP intensities between AWCON and AWCOFF cells of the same animal. P-values were calculated using a Z-test. Error bars represent standard error of proportion. (D) Antisymmetric die-1 expression, assayed with die-1fosmid∷yfp (otIs274), in AWCL/R is regulated by a nsy-5-dependent gap junction neural network but is independent of the calcium-triggered MAPK pathway. First larval stage animals were scored. P-values were calculated using Fisher's exact test. Error bars represent standard error of proportion.
Previous work had shown anti-correlated expression of the GPCRs str-2 and srsx-3; the str-2-expressing cell has been termed “AWCON,” while the srsx-3-expressing cell has been termed “AWCOFF” (Bauer Huang et al. 2007). Using a transgenic reporter strain in which AWCON is labeled with str-2prom∷2xnlsTagRFP, we found that expression of the die-1 fosmid reporter correlates with the AWCOFF state (Fig. 4C).
die-1 affects antisymmetric chemoreceptor expression in the AWC neuron pair
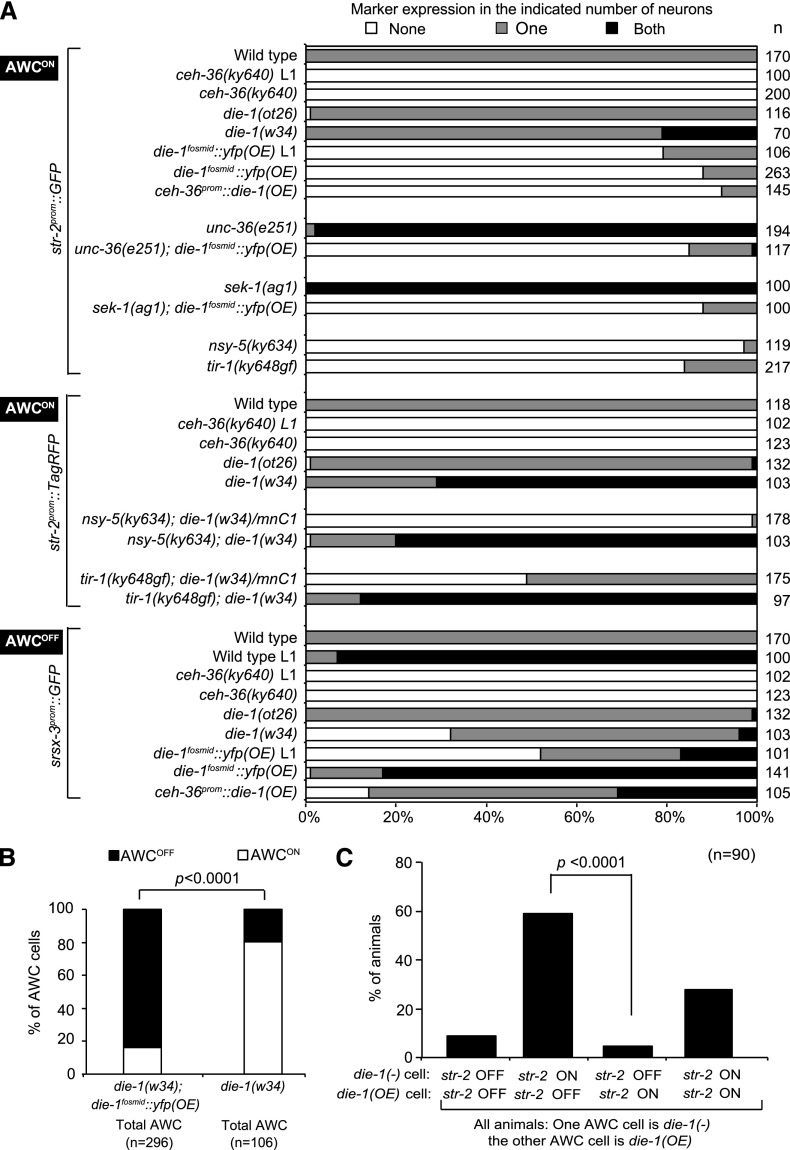
We next asked whether asymmetric die-1 expression in the AWC neurons is required for antisymmetric expression of the GPCRs in the AWC neuron pair. We found that in die-1(w34)-null mutants, str-2prom∷GFP (AWCON marker) is more frequently expressed in both AWC neurons (Fig. 5A). The expected opposite effect is observed on the antisymmetric expression of srsx-3 (AWCOFF marker), which fails to be expressed in a substantial number of die-1(−) animals (Fig. 5A). In animals that carry a die-1 missense mutation allele, ot26, AWC asymmetry is not affected at all (Fig. 5A). This is notable because in die-1(ot26) animals, ASE asymmetry is as severely affected as in die-1(w34)-null mutants (Supplemental Table S1). The single amino acid change in the C-terminal region of the DIE-1 protein in die-1(ot26) animals may reveal a distinct requirement for a specific part of the protein in controlling ASE versus AWC asymmetry. Alternatively, if the missense mutation merely reduces overall gene activity, these data may indicate that AWC and ASE require distinct levels of die-1 activity to control their respective asymmetries.
Figure 5.
die-1 affects antisymmetry of AWC neurons. (A) Expression phenotypes of integrated transgenes of str-2prom∷GFP(kyIs140), str-2prom∷TagRFP (vyIs68), and srsx-3prom∷GFP (vyIs68). (OE) Overexpression. die-1fosmid∷yfp(OE) data in the sixth and seventh rows are combined from both vyEx1462 and vyEx1476, showing qualitatively similar results. (B,C) Only vyEx1462 was used in double-mutant analysis with unc-36(e251) and sek-1(ag1) and in mosaic analysis. Since the ceh-36 promoter is active in not only AWCL/R but also ASEL/R, we also examined and observed the expected ASER-to-ASEL transformation in ceh-36prom∷die-1 animals (vyEx1510, 1511) (Supplemental Fig. S3). (B) Expression of an integrated transgene of str-2prom∷GFP (kyIs140) in die-1(w34) animals with or without the extrachromosomal array die-1fosmid∷yfp; odr-1prom∷DsRed (vyEx1462) in total AWC cells. A Z-test was used to calculate P-values. (C) A subset of the data from B is presented here. Shown are only those animals in which die-1 activity is present in one, but not the other, AWC neuron (as assessed with the odr-1prom∷DsRed array marker), revealing again that die-1 activity correlates with the absence of str-2 expression. A Z-test was used to calculate P-values.
We examined whether expression of die-1 is not only required for the establishment of the AWCOFF state but also sufficient to convert the AWCON cell to an AWCOFF cell. To this end, we used two different approaches. First, we generated animals that express multiple copies of the die-1 fosmid reporter. Consistent with the loss-of-function phenotype (two AWCON neurons) and the expression of die-1 in AWCOFF, we found that in these transgenic animals [die-1fosmid(OE)], both AWC neurons adopt the AWCOFF identity (Fig. 5A). To confirm that this effect is indeed due to ectopic expression of die-1 in both AWC neurons, we also engineered transgenic animals that express die-1 under the control of the ceh-36 promoter, which is bilaterally expressed in the AWC neurons. These animals also display a two-AWCOFF phenotype (Fig. 5A).
The die-1 loss-of-function and gain-of-function phenotypes and the AWCOFF-biased die-1 expression suggested that die-1 operates in a cell-autonomous manner. We corroborated this notion using genetic mosaic analysis. To this end, we generated transgenic die-1(w34)-null mutants that carry an extrachromosomal array containing the rescuing die-1 fosmid reporter and the AWCL+AWCR-expressed odr-1prom∷DsRed marker. We scored expression of the AWCON marker str-2prom∷GFP, contained on a separate, chromosomally integrated array. We found that the presence of the die-1 array in an AWC neuron results in an increased probability of the AWCOFF state compared with AWC neurons that have lost the array (Fig. 5B). Furthermore, we specifically examined animals that contain die-1(+) gene activity on a rescuing array (as assessed with the expression of the odr-1 marker) in one AWC neuron but not the contralateral homolog. We found that in these animals, the presence of the die-1 array tracks with the adoption of the AWCOFF phenotype (Fig. 5C). Taken together, these data suggest that die-1 acts cell-autonomously in AWCOFF.
Regulation of antisymmetric die-1 expression in AWCL/R
AWC antisymmetry has been shown to be controlled by a calcium-triggered MAPK signaling cascade, which is active in the AWCOFF neuron, resulting in asymmetric expression of the GPCRs str-2 and srsx-3 (Fig. 1; Sagasti et al. 2001; Chuang and Bargmann 2005). Surprisingly, we found that loss-of-function mutations in this pathway do not have a significant impact on the antisymmetric expression of die-1 (Fig. 4D). However, through genetic epistasis analysis, we found that this calcium-triggered MAPK pathway requires die-1. Specifically, we found that the “two-AWCON” phenotype of unc-36 calcium channel mutants or sek-1 MAPKK mutants is suppressed by the die-1 overexpression transgene, which induces a “two-AWCOFF” phenotype (Fig. 5A). Similarly, we found that the “two-AWCOFF” phenotype of animals that either lack the nsy-5 gene (which encodes a gap junction protein) or carry a gain-of-function mutation in the tir-1 gene (which encodes an adaptor protein in the MAPK cascade) is suppressed in the context of a die-1-null mutant background (Fig. 5A). In light of the independence of asymmetric die-1 expression on the calcium/MAPK pathway (Fig. 4D), these data suggest that an upstream event triggers two downstream events in parallel: asymmetric activation of the calcium/MAPK pathway and asymmetric expression of die-1. Both events are required to subsequently determine asymmetric GPCR expression.
We tested whether the transient neural network formed via the NSY-5 innexin gap junction protein and controlled by the NSY-4 claudin-like protein may provide the upstream asymmetry trigger for antisymmetric die-1 expression. We found that in nsy-4 and nsy-5 mutants, die-1 expression in AWC becomes completely symmetrized (Fig. 4D). We conclude that two separate pathways, both triggered by nsy-4 and nsy-5, converge to control AWC antisymmetry.
Discussion
We showed here that the nematode-specific die-1 gene has a versatile role in controlling distinct neuronal asymmetries. die-1 is asymmetrically expressed in both the ASE and AWC neurons but in a strikingly distinct manner. In ASE, it is expressed in a directionally asymmetric manner, while in AWC, it is expressed in an antisymmetric manner. Moreover, we showed that die-1 is required in both the ASE and AWC neurons to control their respective directional and stochastic asymmetries in chemoreceptor gene expression. Asymmetric chemoreceptor expression is critical for asymmetric functions of both ASE and AWC neurons. Our data provide an unprecedented link between the control mechanisms of two types of asymmetries: antisymmetry and directional asymmetry. die-1 can be considered a “node” that integrates two completely separate types of asymmetric regulatory inputs and then triggers two distinct types of transcriptional outputs, resulting in asymmetric expression of two distinct types of chemosensory systems.
The distinct nature of regulatory inputs into the die-1 locus in ASE and AWC is striking. All of the genes that we described here as regulators of specific phases of die-1 expression in ASE have either no function at all in AWC or display a completely distinct function in AWC. cog-1, which represses die-1 in ASER, is not expressed in AWC (based on the cog-1fosmid reporter) (data not shown), and loss of cog-1 does not affect AWC asymmetry (Chang et al. 2003). ASER-expressed fozi-1, which maintains die-1 restriction to ASEL and controls ASE terminal asymmetry (gcy expression), is expressed in both AWC neurons (based on the fozi-1fosmid reporter) (data not shown) but has no impact on terminal AWC asymmetry (data not shown). The die-1 maintenance factor in ASEL, ceh-36, does function in AWC but, unlike its asymmetric function in ASEL, has no role in asymmetry control of AWC per se; it rather acts as a terminal selector to induce all known aspects of AWC differentiation (Kim et al. 2010). Further reflecting distinct regulatory inputs, a discrete cis-regulatory element in the die-1 locus drives ASEL expression but not AWC expression, and we observed that directionally asymmetric expression of die-1 in ASEL precedes antisymmetric expression in AWCL/R in the embryo.
Directional asymmetry is thought to evolve from an antisymmetric state that became subsequently fixed in one specific configuration (Palmer 2004). Perhaps ASE directional asymmetry evolved from an antisymmetric state in which die-1 was, like in AWCL/R, antisymmetrically expressed in ASEL/R. The antisymmetric control of die-1 expression in ASE may then have become fixed because the distinct lineage histories of the ASEL and ASER neurons afforded an opportunity to employ a lineage-based mechanism to lock die-1 expression into ASEL (Poole and Hobert 2006; Cochella and Hobert 2012). The symmetric lineage histories of AWCL and AWCR did not permit the superimposition of such a lineage-based mechanism, and therefore the antisymmetric nature of die-1 expression was retained in AWCL/R. It will be interesting to trace the type of terminal ASE asymmetry and the expression of die-1 in highly diverse nematode species.
Materials and methods
Strains and transgenes
A list of strains and transgenes can be found in the Supplemental Material.
Reporter constructs
Fosmid reporter constructs were generated by recombineering using a previously described recombineering protocol (Tursun et al. 2009), with 2xFlag∷Venus or mCherry at the C terminus of die-1 (fosmid ID, WRM064bB08) or yfp at the C terminus of cog-1 (WRM067cF11), ceh-36 (WRM065bE05), fozi-1 (WRM062aF03), and lim-6 (WRM065aD03).
The deletions in the die-1∷mCherry fosmid reporter were generated also by recombineering, replacing the region to be deleted with a kanamycin resistance cassette described in Tursun et al. (2009) (pBALU Ext). Smaller promoter constructs were generated by cloning the specified fragments of the die-1 locus into pPD95.67 (containing the classic Fire vector MCS followed by 2nls∷yfp∷unc-54 3′ UTR). All fragments were amplified, including 5′ SalI and 3′ KpnI linkers for subcloning. The sequences of the primers used are listed in the Supplemental Material.
Fosmid reporters were typically injected as complex arrays (except for the overexpression experiments in Fig. 4). Injection mixes contained 10–15 ng/μL linearized fosmid, 2–5 ng/μL linearized coinjection marker, and 100–120 ng/μL fragmented genomic DNA from Escherichia coli (OP50). The promoter fusion constructs were all injected as simple arrays containing 50 ng/μL plasmid construct, 6 ng/μL elt-2 prom∷DsRed plasmid, and 70 ng/μL Bluescript.
Acknowledgments
We thank Feifan Zhang for the initial observation of fozi-1 affecting die-1 expression, and Qi Chen for expert assistance in generating transgenic strains. L.C. was supported by a Helen Hay Whitney Foundation Post-doctoral Fellowship, B.T. was supported by a Francis Goelet Post-doctoral Fellowship, Y.W.H. was supported by a National Institutes of Health (NIH) Organogenesis Training Grant, C.F.C. was supported by the NIH (R01GM098026) and an Alfred Sloan Research Fellowship, and O.H. was supported by the NIH (R01NS039996-05) and the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.233643.113.
References
- Bauer Huang SL, Saheki Y, VanHoven MK, Torayama I, Ishihara T, Katsura I, van der Linden A, Sengupta P, Bargmann CI 2007. Left-right olfactory asymmetry results from antagonistic functions of voltage-activated calcium channels and the Raw repeat protein OLRN-1 in C. elegans. Neural Dev 2: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Johnston RJ Jr, Hobert O 2003. A transcriptional regulatory cascade that controls left/right asymmetry in chemosensory neurons of C. elegans. Genes Dev 17: 2123–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Johnston RJ, Frokjaer-Jensen C, Lockery S, Hobert O 2004. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature 430: 785–789 [DOI] [PubMed] [Google Scholar]
- Chuang CF, Bargmann CI 2005. A Toll-interleukin 1 repeat protein at the synapse specifies asymmetric odorant receptor expression via ASK1 MAPKKK signaling. Genes Dev 19: 270–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang CF, Vanhoven MK, Fetter RD, Verselis VK, Bargmann CI 2007. An innexin-dependent cell network establishes left-right neuronal asymmetry in C. elegans. Cell 129: 787–799 [DOI] [PubMed] [Google Scholar]
- Cochella L, Hobert O 2012. Embryonic priming of a miRNA locus predetermines postmitotic neuronal left/right asymmetry in C. elegans. Cell 151: 1229–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiano D, Cochella L, Tursun B, Hobert O 2010. Neuron-type specific regulation of a 3′ UTR through redundant and combinatorially acting cis-regulatory elements. RNA 16: 349–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchberger JF, Lorch A, Sleumer MC, Zapf R, Jones SJ, Marra MA, Holt RA, Moerman DG, Hobert O 2007. The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes Dev 21: 1653–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid PJ, Raich WB, Smith R, Mohler WA, Simokat K, Gendreau SB, Rothman JH, Hardin J 2001. The zinc finger protein DIE-1 is required for late events during epithelial cell rearrangement in C. elegans. Dev Biol 236: 165–180 [DOI] [PubMed] [Google Scholar]
- Hobert O 2006. Architecture of a microRNA-controlled gene regulatory network that diversifies neuronal cell fates. Cold Spring Harb Symp Quant Biol 71: 181–188 [DOI] [PubMed] [Google Scholar]
- Hobert O, Johnston RJ Jr, Chang S 2002. Left-right asymmetry in the nervous system: The Caenorhabditis elegans model. Nat Rev Neurosci 3: 629–640 [DOI] [PubMed] [Google Scholar]
- Johnston RJ Jr, Chang S, Etchberger JF, Ortiz CO, Hobert O 2005. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc Natl Acad Sci 102: 12449–12454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ Jr, Copeland JW, Fasnacht M, Etchberger JF, Liu J, Honig B, Hobert O 2006. An unusual Zn-finger/FH2 domain protein controls a left/right asymmetric neuronal fate decision in C. elegans. Development 133: 3317–3328 [DOI] [PubMed] [Google Scholar]
- Kim K, Kim R, Sengupta P 2010. The HMX/NKX homeodomain protein MLS-2 specifies the identity of the AWC sensory neuron type via regulation of the ceh-36 Otx gene in C. elegans. Development 137: 963–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W. 1932. Das Rechts-Links Problem im Tierreich und beim Menschen. Springer, Berlin. [Google Scholar]
- O'Meara MM, Zhang F, Hobert O 2010. Maintenance of neuronal laterality in Caenorhabditis elegans through MYST histone acetyltransferase complex components LSY-12, LSY-13 and LIN-49. Genetics 186: 1497–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz CO, Etchberger JF, Posy SL, Frokjaer-Jensen C, Lockery S, Honig B, Hobert O 2006. Searching for neuronal left/right asymmetry: Genomewide analysis of nematode receptor-type guanylyl cyclases. Genetics 173: 131–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz CO, Faumont S, Takayama J, Ahmed HK, Goldsmith AD, Pocock R, McCormick KE, Kunimoto H, Iino Y, Lockery S, et al. 2009. Lateralized gustatory behavior of C. elegans is controlled by specific receptor-type guanylyl cyclases. Curr Biol 19: 996–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AR 2004. Symmetry breaking and the evolution of development. Science 306: 828–833 [DOI] [PubMed] [Google Scholar]
- Palmer AR. 2005. Antisymmetry. In Variation: A central concept in biology (ed. B Hallgrimsson B, Hall BK), pp. 359–398. Elsevier Academic Press Publications, San Diego. [Google Scholar]
- Pierce-Shimomura JT, Faumont S, Gaston MR, Pearson BJ, Lockery SR 2001. The homeobox gene lim-6 is required for distinct chemosensory representations in C. elegans. Nature 410: 694–698 [DOI] [PubMed] [Google Scholar]
- Poole RJ, Hobert O 2006. Early embryonic programming of neuronal left/right asymmetry in C. elegans. Curr Biol 16: 2279–2292 [DOI] [PubMed] [Google Scholar]
- Rogers LJ, Vallortigara G, Andrew RJ 2013. Divided brains: The biology and behavior of brain asymmetries. Cambridge University Press, New York [Google Scholar]
- Sagasti A, Hisamoto N, Hyodo J, Tanaka-Hino M, Matsumoto K, Bargmann CI 2001. The CaMKII UNC-43 activates the MAPKKK NSY-1 to execute a lateral signaling decision required for asymmetric olfactory neuron fates. Cell 105: 221–232 [DOI] [PubMed] [Google Scholar]
- Sarin S, O'Meara MM, Flowers EB, Antonio C, Poole RJ, Didiano D, Johnston RJ Jr, Chang S, Narula S, Hobert O 2007. Genetic screens for Caenorhabditis elegans mutants defective in left/right asymmetric neuronal fate specification. Genetics 176: 2109–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin S, Antonio C, Tursun B, Hobert O 2009. The C. elegans Tailless/TLX transcription factor nhr-67 controls neuronal identity and left/right asymmetric fate diversification. Development 136: 2933–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troemel ER, Sagasti A, Bargmann CI 1999. Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell 99: 387–398 [DOI] [PubMed] [Google Scholar]
- Tursun B, Cochella L, Carrera I, Hobert O 2009. A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS ONE 4: e4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wes PD, Bargmann CI 2001. C. elegans odour discrimination requires asymmetric diversity in olfactory neurons. Nature 410: 698–701 [DOI] [PubMed] [Google Scholar]
- Yu S, Avery L, Baude E, Garbers DL 1997. Guanylyl cyclase expression in specific sensory neurons: A new family of chemosensory receptors. Proc Natl Acad Sci 94: 3384–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]