c-MYC is abnormally overexpressed in many human cancers. Kelly et al. discover that MCL-1 is essential to sustain c-MYC-driven mouse lymphomas. MCL-1 blockade killed c-MYC-driven human Burkitt lymphoma cells, even those bearing mutations in p53, but was well tolerated in healthy tissues. These findings identify MCL-1 as an attractive therapeutic target for MYC-driven cancers.
Keywords: cancer, apoptosis, MCL-1, p53, BCL-2, MYC
Abstract
The transcriptional regulator c-MYC is abnormally overexpressed in many human cancers. Evasion from apoptosis is critical for cancer development, particularly c-MYC-driven cancers. We explored which anti-apoptotic BCL-2 family member (expressed under endogenous regulation) is essential to sustain c-MYC-driven lymphoma growth to reveal which should be targeted for cancer therapy. Remarkably, inducible Cre-mediated deletion of even a single Mcl-1 allele substantially impaired the growth of c-MYC-driven mouse lymphomas. Mutations in p53 could diminish but not obviate the dependency of c-MYC-driven mouse lymphomas on MCL-1. Importantly, targeting of MCL-1 killed c-MYC-driven human Burkitt lymphoma cells, even those bearing mutations in p53. Given that loss of one allele of Mcl-1 is well tolerated in healthy tissues, our results suggest that therapeutic targeting of MCL-1 would be an attractive therapeutic strategy for MYC-driven cancers.
The c-MYC transcription factor regulates ∼10,000 genes, including many that control cell growth and division (Dang 1999). Deregulated c-MYC expression is detected in up to 70% of human cancers, including lymphomas and leukemias (Boxer and Dang 2001; Sanchez-Beato et al. 2003). Much of what we know about c-MYC-driven tumorigenesis has emerged from studies using Eμ-Myc transgenic mice, in which the c-Myc transgene is subjugated to the immunoglobulin (Ig) heavy chain gene enhancer (Eμ), mimicking the consequences of the c-MYC/IGH or c-MYC/IGL chromosomal translocations that drive human Burkitt lymphoma (Adams et al. 1985). The Eμ-Myc mice exhibit a preleukemic expansion of pre-B cells that precedes the outgrowth of clonal sIg− pre-B-cell or sIg+ B-cell lymphomas arising with a median latency of ∼110 d (Adams et al. 1985; Langdon et al. 1986).
Evasion from cell death is a hallmark of cancer, particularly those driven by c-MYC (Hanahan and Weinberg 2011; Kelly and Strasser 2011). Deregulated c-MYC expression enhances cell death under unfavorable growth conditions, such as limited availability of growth factors, through the intrinsic apoptotic pathway that is regulated by interactions of proteins belonging to three functionally distinct subgroups of the BCL-2 family (Youle and Strasser 2008). Following apoptotic stimuli, the proapoptotic BH3-only proteins (e.g., BIM and PUMA) become transcriptionally and/or post-transcriptionally up-regulated. For example, in response to DNA damage or oncogenic stress, the tumor suppressor p53 directly transcriptionally activates PUMA and NOXA to cause cell death (Jeffers et al. 2003; Villunger et al. 2003). The BH3-only proteins directly or indirectly activate BAX/BAK, the executioners of apoptosis, which permeabilize the outer mitochondrial membrane and thereby unleash the downstream caspase cascade for cellular demolition (Merino et al. 2009; Llambi et al. 2011).
Apoptotic blocks, such as overexpression of prosurvival BCL-2-like proteins (Strasser et al. 1990; Swanson et al. 2004) or loss of proapoptotic BIM or PUMA (Egle et al. 2004; Hemann et al. 2004; Michalak et al. 2009), accelerate c-MYC-driven lymphomagenesis. Interestingly, mutations that deregulate the intrinsic apoptotic pathway, including amplifications of the genomic regions encoding MCL-1 and BCL-X (Beroukhim et al. 2010), are detected in human tumors and are often associated with poor chemotherapy responses (Khaw et al. 2011). Burkitt lymphoma is associated with frequent loss of BIM and/or PUMA expression, in part due to gene hypermethylation (Mestre-Escorihuela et al. 2007; Garrison et al. 2008; Richter-Larrea et al. 2010; Giulino-Roth et al. 2012). Moreover, many Burkitt lymphomas contain defects in the p53 tumor suppressor pathway, particularly mutations in p53 itself (Farrell et al. 1991; Bhatia et al. 1992) but also overexpression of the ubiquitin ligase HDM2 (called MDM2 in mice) or loss of the p14/ARF locus (Lindstrom et al. 2001).
Although overexpression of prosurvival BCL-2 family members accelerates c-MYC-induced lymphomagenesis, it is unclear whether the sustained survival and growth of c-MYC-driven lymphomas depends on the expression of these proteins under endogenous regulation. This remains an important question because knowledge of which prosurvival protein is essential for the sustained growth of a particular cancer will pinpoint the family member that should be targeted by the emerging BH3 mimetic drugs (Lessene et al. 2008). Using innovative gene targeting and drug-mimicking tools, we show for the first time that MCL-1 targeting kills c-MYC-driven mouse and human lymphoma cells even when p53 is mutated. This is remarkable given that we found that p53 does affect the dependency of these lymphoma cells on MCL-1 and suggest that therapeutic targeting of MCL-1 may be a promising strategy for cancer therapy.
Results
Experimental strategy to determine which anti-apoptotic BCL-2 family member is essential for the sustained survival and expansion of MYC-driven lymphomas
To examine the roles of BCL-XL and MCL-1 expression under endogenous control in the sustained growth of Eμ-Myc lymphomas, we generated mice in which we could genetically delete Mcl-1 or Bcl-x at will exclusively within c-MYC-driven lymphoma cells. Specifically, we produced Eμ-Myc transgenic mice that express a tamoxifen-regulated Cre recombinase-estrogen receptor fusion protein from the ubiquitously expressed Rosa26 locus (Rosa26-CreERT2 mice) (Seibler et al. 2003) and bear loxP targeted (floxed, denoted fl) alleles of the prosurvival gene Bcl-x (Wagner et al. 2000) or Mcl-1 (Vikstrom et al. 2010) (Supplemental Fig. 1). As expected, the progeny with all three genetic alterations developed lymphomas with the same latency as standard Eμ-Myc mice (Supplemental Fig. 2A) because they expressed normal levels of BCL-XL or MCL-1. The lymphoma cells were transplanted into C57BL/6-Ly5.1+ recipients, which were treated with tamoxifen to activate the Cre-ERT2 recombinase, leading to deletion of Bcl-xfl or Mcl-1fl alleles exclusively within the malignant cells (Supplemental Fig. 1). Selected primary lymphomas were infected with an Ub-GFP-Luciferase lentivirus to facilitate monitoring lymphoma progression/regression following Bcl-x or Mcl-1 deletion by imaging for bioluminescence in C57BL/6-albino recipient mice (Supplemental Fig. 1). In mice transplanted with Eμ-Myc;CreERT2 lymphoma cells lacking any floxed allele (for simplicity, hereafter termed “control”), tumor expansion did not differ significantly between untreated and tamoxifen-treated recipients (P = 0.06) (Supplemental Fig. 2B). This shows that CreERT2 activation per se does not impair the growth of Eμ-Myc lymphomas.
Homozygous loss of Bcl-x only slightly impairs the sustained growth of Eμ-Myc lymphomas
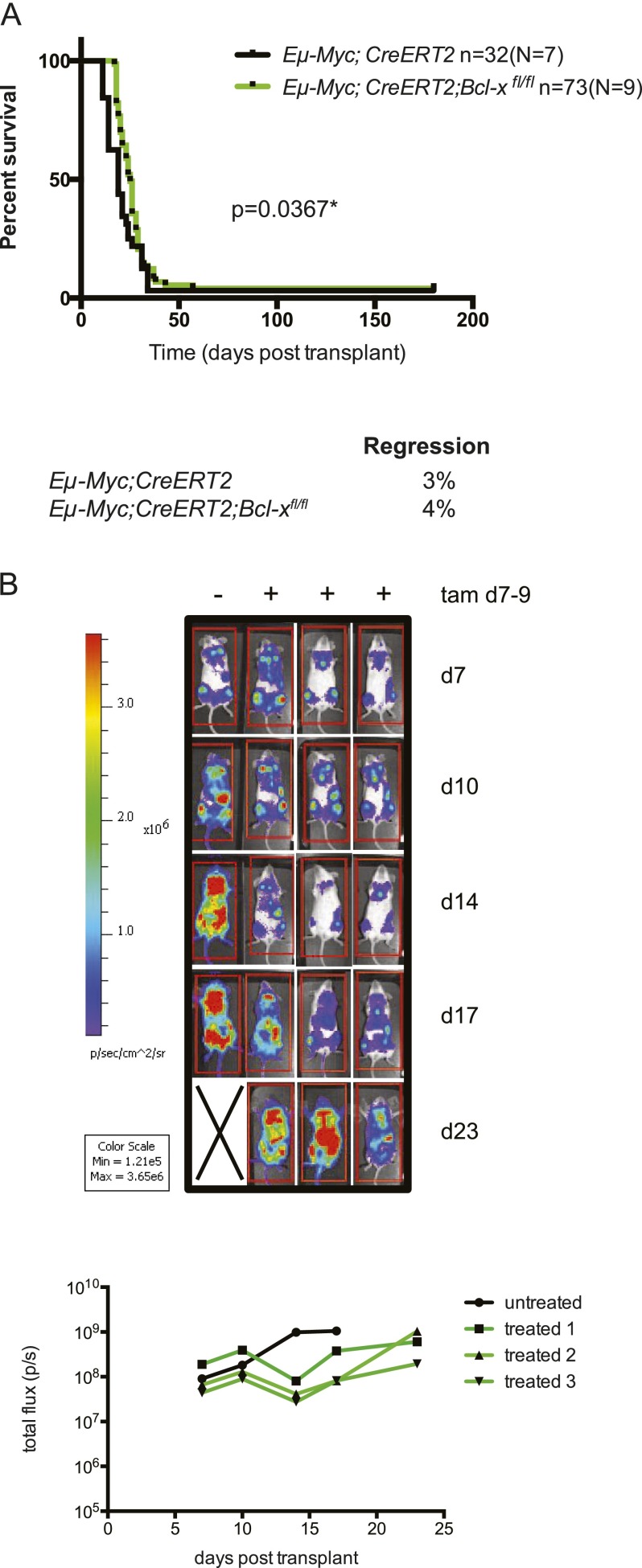
Since endogenous BCL-XL is necessary for Eμ-Myc-induced lymphoma development (Kelly et al. 2011), we predicted that it might also be essential for the sustained expansion of such malignant lymphomas. To examine this, primary lymphomas from nine Eμ-Myc;CreERT2;Bcl-xfl/fl and seven control mice were transplanted into C57BL/6-Ly5.1+ recipient mice and cohorts treated with tamoxifen. Homozygous Bcl-x loss resulted in only a modest delay in tumor expansion and slightly prolonged survival of the mice (median survival 25 d for Eμ-Myc;CreERT2;Bcl-xfl/fl vs. 19 d for the controls; [*] P = 0.0367) (Fig. 1A). Following homozygous Bcl-x deletion, only 4% of the recipient mice (three from 73) showed complete lymphoma regression, which is comparable with the 3% of the tamoxifen-treated mice (one from 32) bearing control tumors.
Figure 1.
Homozygous loss of Bcl-x has only a minor impact on the growth of Eμ-Myc lymphomas. (A) Survival curves of C57BL/6-Ly5.1+ recipient mice transplanted with Eμ-Myc;CreERT2;Bcl-xfl/fl (green line) or control (Eμ-Myc;CreERT2; black line) lymphoma cells and treated with tamoxifen to inactivate Bcl-x where applicable. (n) Total number of recipient mice analyzed; (N) number of independent lymphomas tested. Homozygous deletion of Bcl-x resulted in a small but significant delay in tumor growth. (*) P = 0.0367. For the mice transplanted with the control Eμ-Myc;CreERT2 lymphomas, 3% regressed, and the overall median survival was 19 d. For the mice transplanted with the Eμ-Myc;CreERT2;Bcl-xfl/fl lymphomas, 4% regressed, and the overall median survival was 25 d. (B) Bioluminescence imaging of the tumor burden in C57BL/6-albino recipient mice injected with primary Eμ-Myc;CreERT2;Bcl-xfl/fl lymphoma cells that had been transduced with a lentiviral vector coexpressing GFP and luciferase. At 7 d post-transplant, a cohort of these mice was treated with tamoxifen. Mice were subsequently imaged for bioluminescence to monitor lymphoma burden every 3–4 d by measuring the total photon flux per second emitted from a region of interest (ROI) drawn around the whole mouse. See also Supplemental Figure 3.
This result was confirmed by in vivo lymphoma bioluminescence imaging (Fig. 1B). As early as 7 d after tumor transplant, the recipients displayed a significant lymphoma burden (≥1 × 107 photon flux per second). These lymphomas continued to grow following tamoxifen treatment to delete both Bcl-x alleles and overwhelmed the recipients only shortly after the untreated mice had succumbed to the same lymphomas that retained Bcl-x. Efficient deletion of the Bcl-x alleles and loss of the BCL-XL protein were confirmed by quantitative RT–PCR (qRT–PCR) and Western blot analyses in paired tamoxifen-treated and untreated lymphomas taken from mice transplanted with Eμ-Myc;CreERT2;Bcl-xfl/fl lymphomas (Supplemental Fig. 3). These data reveal that malignant Eμ-Myc lymphomas can continue to grow without endogenous BCL-XL expression.
Loss of Mcl-1 substantially impairs the sustained growth of Eμ-Myc lymphomas
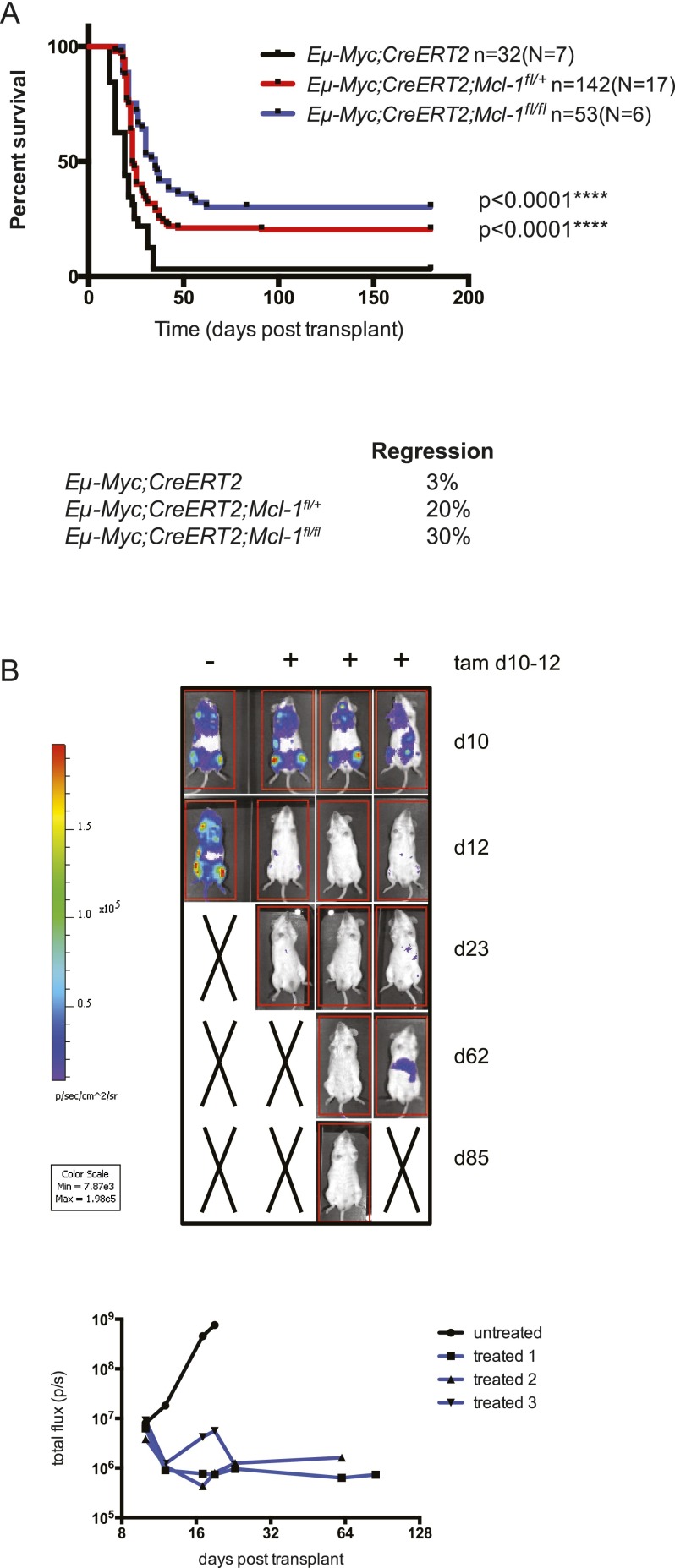
In parallel experiments, we investigated whether MCL-1 was essential for the sustained growth of Eμ-Myc lymphomas. Cohorts of C57BL/6-Ly5.1+ mice were transplanted with independent primary lymphomas from six Eμ-Myc;CreERT2;Mcl-1fl/fl and seven control mice, and their survival was compared following tamoxifen treatment (Fig. 2). Strikingly, in 30% of the recipients, homozygous deletion of Mcl-1 provoked complete lymphoma regression and extended survival (>180 d post-transplant) (Fig. 2A). The dramatic tumor regression was confirmed by bioluminescence imaging of lymphomas transplanted into C57BL/6-albino recipient mice (Fig. 2B). Importantly, even the tamoxifen-treated recipients that relapsed exhibited substantially extended survival compared with those bearing control lymphomas (median survival 35 d for mice bearing Eμ-Myc;CreERT2;Mcl-1fl/fl lymphomas vs. 19 d for those bearing control lymphomas; [****] P < 0.0001).
Figure 2.
Loss of Mcl-1, even loss of a single allele, greatly impairs the sustained growth of Eμ-Myc lymphomas within the whole animal. (A) Survival curves of C57BL/6-Ly5.1+ recipient mice transplanted with Eμ-Myc;CreERT2;Mcl-1fl/+ (red line), Eμ-Myc;CreERT2;Mcl-1fl/fl (blue line), or control (Eμ-Myc;CreERT2; black line) lymphoma cells and treated with tamoxifen to inactivate Mcl-1 where applicable. (n) Total number of recipient mice analyzed; (N) number of independent lymphomas tested. Heterozygous and homozygous deletion of Mcl-1 significantly delayed lymphoma growth. (****) P < 0.0001. For the mice transplanted with the control Eμ-Myc;CreERT2 lymphomas, 3% regressed, and the overall median survival was 19 d. For the mice transplanted with the Eμ-Myc;CreERT2;Mcl-1fl/+ lymphomas, 20% regressed, and the overall median survival was 23 d. For the mice transplanted with the Eμ-Myc;CreERT2;Mcl-1fl/fl lymphomas, 30% regressed, and the overall median survival was 35 d. (B) Bioluminescence imaging of the tumor burden in C57BL/6-albino recipient mice injected with primary Eμ-Myc;CreERT2;Mcl-1fl/fl lymphoma cells. At 10 d post-transplant, a cohort of these mice was treated with tamoxifen. These mice were subsequently imaged for bioluminescence.
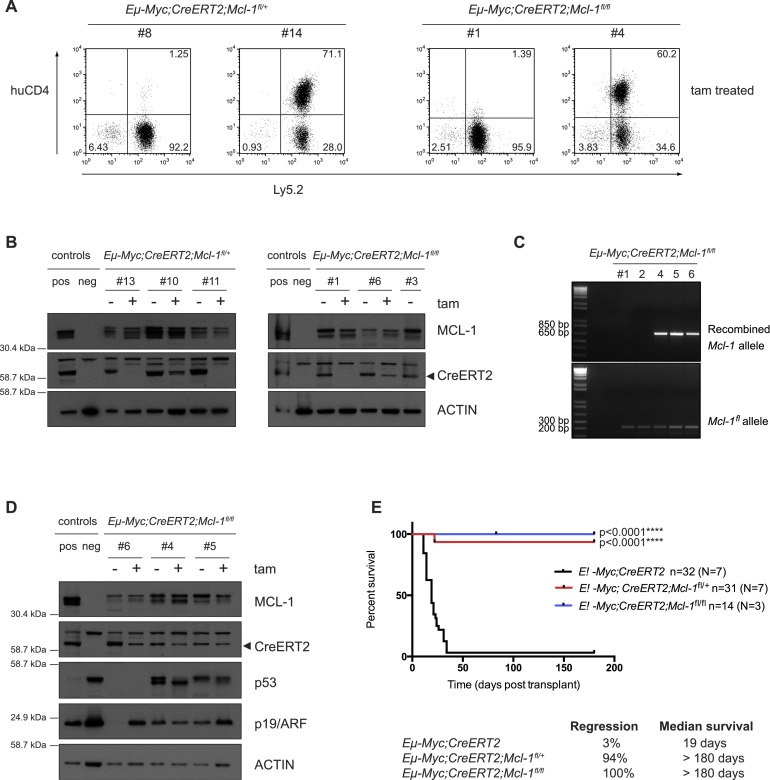
Remarkably, even heterozygous Mcl-1fl loss substantially impaired sustained lymphoma growth: 20% of recipient mice bearing Eμ-Myc;CreERT2;Mcl-1fl/+ lymphomas showed prolonged survival after tamoxifen treatment (median survival was 23 d for relapsing lymphomas vs. 19 d for control mice; [****] P < 0.0001) (Fig. 2A). These results demonstrate that the level of MCL-1 is highly critical for the sustained survival and expansion of MYC-driven lymphomas. This conclusion was strengthened by further analysis of the relapsing tamoxifen-treated lymphomas. The conditional allele of Mcl-1 carries a human (hu) CD4 reporter gene, which is expressed following Mcl-1fl recombination, thus permitting single-cell analysis of the recombination efficiency (Vikstrom et al. 2010; Glaser et al. 2012). Strikingly, ∼60% of Eμ-Myc;CreERT2;Mcl-1fl/+ and 40% of Eμ-Myc;CreERT2;Mcl-1fl/fl lymphomas that relapsed following tamoxifen treatment had escaped deletion of their conditional Mcl-1 alleles mostly because they had undergone selection for loss of CreERT2 expression (Fig. 3A,B). Western blot analysis of these tamoxifen-treated tumors confirmed that MCL-1 protein expression was not reduced (Fig. 3B). In addition, the minority of Eμ-Myc;CreERT2;Mcl-1fl/fl lymphomas that did relapse as huCD4-positive following tamoxifen treatment had only recombined one allele of Mcl-1 and continued to express MCL-1 protein (Fig. 3C,D). By inference, the survival curves described earlier grossly underestimate the role that MCL-1 plays in the sustained growth of Eμ-Myc lymphomas. Indeed when animals that succumbed to relapsed lymphomas that had selected against loss of Mcl-1fl alleles were removed from the analysis (and those lymphomas that had mutated p53 alleles were also removed from the survival curves) (Fig. 5A; see below), nearly 100% of animals survived lymphoma-free long-term (>180 d post-transplant) (Fig. 3E). These findings reveal that even heterozygous Mcl-1 loss is highly detrimental for the sustained in vivo growth of malignant Eμ-Myc lymphomas, indicating that relatively weak targeting of MCL-1 might have therapeutic benefits in MYC-driven cancers.
Figure 3.
Most Eμ-Myc lymphomas that relapse following tamoxifen treatment have escaped Mcl-1fl allele recombination. (A) FACS analysis to detect expression of huCD4 (reporter for Mcl-1fl recombination) on the surface of two representative Eμ-Myc;CreERT2;Mcl-1fl/+ and Eμ-Myc;CreERT2;Mcl-1fl/fl lymphomas that had relapsed in recipient mice after tamoxifen treatment (transplanted lymphoma cells stained positive for Ly5.2). Approximately 60% of the relapsed Eμ-Myc;CreERT2;Mcl-1fl/+ and 40% of the relapsed Eμ-Myc;CreERT2;Mcl-1fl/f lymphoma cells had escaped Mcl-1fl allele recombination and, as such, were huCD4-negative (like the sample at the left of each pair), whereas ∼40% of the relapsed Eμ-Myc;CreERT2;Mcl-1fl/+ and 60% of the relapsed Eμ-Myc;CreERT2;Mcl-1fl/fl lymphomas had efficiently recombined at least one Mcl-1fl allele, as reflected by staining positive for huCD4 (like the sample at the right of each pair). (B) Immunoblotting to detect MCL-1 and CreERT2 protein expression (probing for Actin was used as a loading control) in extracts from the spleens, lymph nodes, or thymuses of sick mice that had been transplanted with Eμ-Myc;CreERT2;Mcl-1fl/+ or Eμ-Myc;CreERT2;Mcl-1fl/fl lymphomas. Three independent, paired control, and tamoxifen-treated tumors of each genotype were analyzed (note that one of the Eμ-Myc;CreERT2;Mcl-1fl/fl tumors [#3] examined never relapsed after tamoxifen treatment). Mcl-1 knockout (KO) mouse embryonic fibroblasts (MEFs) were used as a control to confirm the specificity of the MCL-1 antibody (neg control). (C) DNA PCR analysis of recombined Mcl-1fl and unrecombined (intact) floxed alleles in Eμ-Myc;CreERT2;Mcl-1fl/fl lymphomas that relapsed following tamoxifen treatment. (D) Immunoblotting to detect MCL-1, CreERT2, p53, p19/ARF and Actin (loading control) protein expression in extracts from the spleens, lymph nodes, or thymuses of sick mice transplanted with Eμ-Myc;CreERT2;Mcl-1fl/fl lymphomas that relapsed as huCD4-positive following tamoxifen treatment. Three independent, paired control, and tamoxifen-treated tumors were analyzed. Mcl-1 knockout (KO) MEFs were used as a control (neg control) to confirm the specificity of the MCL-1 antibody. (E) Survival curves of C57BL/6-Ly5.1+ recipient mice transplanted with Eμ-Myc;CreERT2;Mcl-1fl/+ (red line) or Eμ-Myc;CreERT2;Mcl-1fl/fl (blue line) lymphoma cells that had wild-type p53 genes following tamoxifen treatment and excluding those lymphomas that escaped Cre-mediated deletion of one or both Mcl-1fl alleles. C57BL/6-Ly5.1+ recipient mice transplanted with control (Eμ-Myc;CreERT2; black line) lymphoma cells and treated with tamoxifen are shown for comparison. (n) Total number of recipient mice analyzed; (N) number of independent lymphomas tested. Efficient heterozygous and homozygous deletion of Mcl-1 significantly delayed lymphoma growth. (****) P < 0.0001. For the mice transplanted with the control Eμ-Myc;CreERT2 lymphomas, 3% regressed, and the overall median survival was 19 d. For the mice transplanted with the Eμ-Myc;CreERT2;Mcl-1fl/+ lymphomas, 94% regressed, and the overall median survival was >180 d. For the mice transplanted with the Eμ-Myc;CreERT2;Mcl-1fl/fl lymphomas, 100% regressed, and the overall median survival was >180 d.
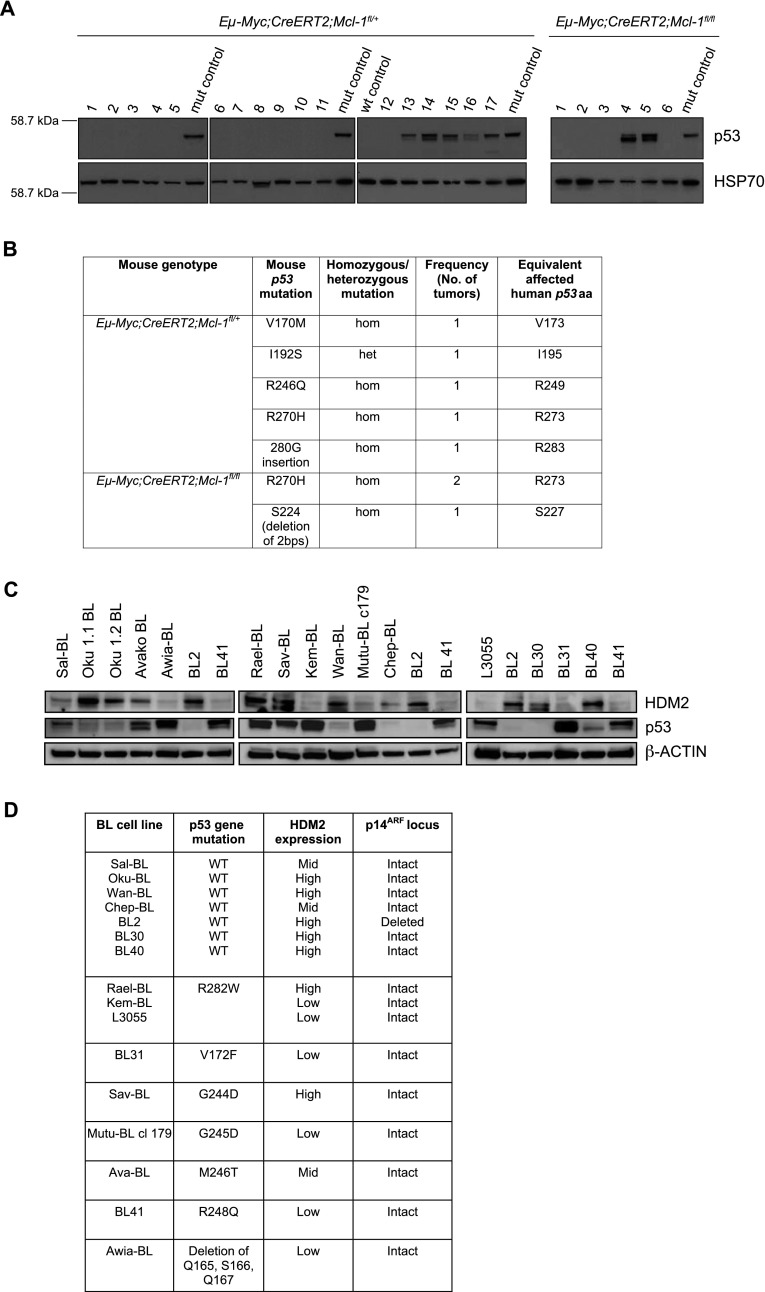
Figure 5.
Mutations in p53 reduce but do not ablate the dependency of c-MYC-driven mouse and human lymphomas on MCL-1. (A) Immunoblotting to detect stabilized p53 proteins, indicative of a mutant p53 protein, in extracts from 17 Eμ-Myc;CreERT2;Mcl-1fl/+ and six Eμ-Myc;CreERT2;Mcl-1fl/fl lymphomas. All but two of the samples were extracted from transplanted and tamoxifen-treated lymphomas. The two exceptions were Eμ-Myc;CreERT2;Mcl-1fl/+ #9, which was a primary lymphoma, and Eμ-Myc;CreERT2;Mcl-1fl/fl #3, which was a transplanted but not tamoxifen-treated lymphoma (this particular lymphoma has the deletion of two bases that introduces a premature stop codon in the p53 gene). Probing for HSP70 served as a loading control. (B) Summary of the p53 mutations detected in Eμ-Myc;CreERT2;Mcl-1fl/+ and Eμ-Myc;CreERT2;Mcl-1fl/fl lymphomas as detected by DNA sequence analysis. The mutations are listed as wild-type (wt) amino acid (aa) affected, amino acid position, and mutant amino acid. The frequencies at which the mutations were detected in the lymphomas and the human equivalents of the mutations are listed. The sequence alignment for murine p53 was performed using Ensembl transcript number ENSMUST00000108658 as the reference transcript. (C) Immunoblotting to detect the expression of HDM2, p53, and β-Actin (loading control) in human Burkitt lymphoma cell lines. (D) Summary of the p53 pathway aberrations detected in the human Burkitt lymphoma cell lines. The p53 mutations present in each cell line is listed as wild-type (wt) amino acid (aa) affected, amino acid position, and mutant amino acid, and the number of alleles affected is also detailed.
The sustained growth of human Burkitt lymphoma cells depends on the expression of MCL-1
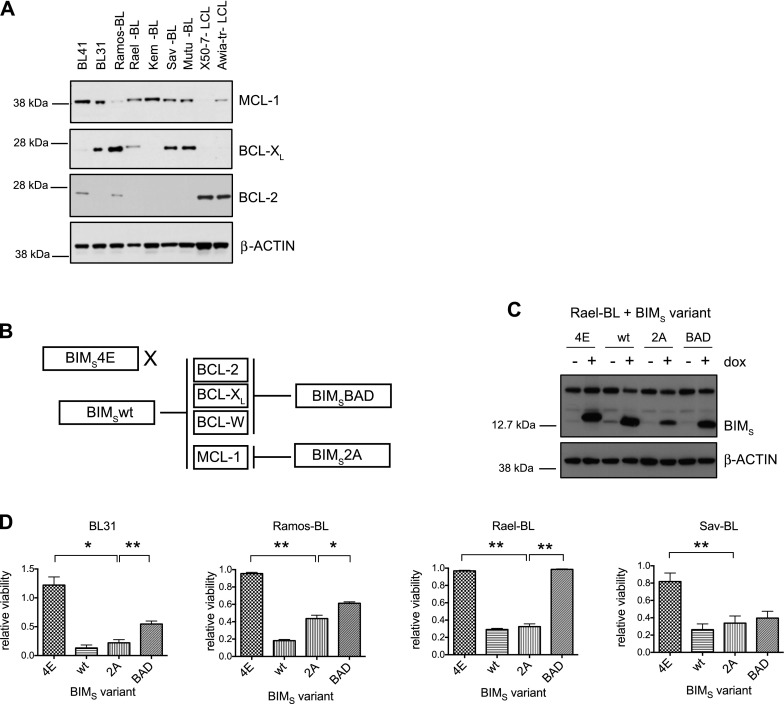
We sought to translate these findings into the human disease setting. Western blot analysis of Burkitt lymphoma cell lines revealed that all expressed MCL-1 with heterogeneous expression of BCL-XL and, as previously reported (Henderson et al. 1991) little to no BCL-2 (Fig. 4A). As a control for the BCL-2 antibody, we showed that the X50-7 and Awia-tr lymphoblastoid cell lines expressed high levels of this prosurvival protein (Fig. 4A), as reported (Henderson et al. 1991).
Figure 4.
MCL-1 blockade kills human Burkitt lymphoma cells, but some also show minor dependency on BCL-XL. (A) Immunoblotting to detect MCL-1, BCL-XL, BCL-2, and β-Actin (loading control) in extracts from Burkitt lymphoma cell lines and, as controls for BCL-2 expression, the X50-7 and Awia lymphoblastoid cell lines. (B) Schematic showing the binding specificities of the BIMS variants to the BCL-2 prosurvival proteins. (C) Immunoblotting to detect the inducible expression of the BIMS variants before and after 24 h of dox treatment in the Rael-BL cell lines carrying the different lentiviral-inducible BIMS expression constructs. Cells were maintained in medium supplemented with the pan-caspase inhibitor qVD-OPh to prevent protein degradation due to apoptosis. Note that the endogenous BIMEL, BIML, and BIMS proteins can also be detected but at a lower level. Probing for β-Actin served as a loading control. (D) Viability of BL cell lines stably infected with lentiviral constructs carrying vectors for dox-inducible expression of BIMS variants was determined 72 h after the addition of dox to the medium by staining the cells with propidium iodide (PI) followed by FACS analysis for PI and GFP fluorescence (GFP is expressed from the lentivirus encoding the BIMs variants). The PI-negative/GFP-positive viable cells were recorded. The percentage of viable/GFP-positive untreated cells was assigned an arbitrary value of 1, and the percentage of viable/GFP-positive dox-treated cells was expressed as a proportion of this. Each line was assayed in triplicate, and data are presented as the mean and standard error of the mean of three independent experiments. Statistical analysis using a paired two-tailed t-test showed that BIMs2A induced significantly more death in all of the Burkitt lymphoma cells examined compared with the negative control BIMs4E (BL31, [*] P = 0.0179; Ramos-BL, [**] P = 0.0025; Rael-BL, [**] P = 0.0017; Sav-BL, [**] P = 0.0044) and that BIMs2A induced significantly more death than BIMsBAD in BL31 ([**] P = 0.0056), Ramos-BL ([*] P = 0.0421), and Rael-BL ([**] P = 0.0023) cells but not in the Sav-BL cells ([ns] P = 0.1348). See also Supplemental Figure 4.
To examine the impact of targeting BCL-2 prosurvival proteins on Burkitt lymphoma growth, we developed a novel assay based on doxycycline (dox)-inducible expression (from a lentiviral vector) of genetically engineered variants of the BH3-only protein BIMS with select binding specificities for distinct BCL-2 prosurvival proteins (Fig. 4B; Chen et al. 2005; Lee et al. 2008). These polypeptides have a mechanism of action similar to that of BH3 mimetic drugs; i.e., they antagonize BCL-2 prosurvival protein function by engaging a hydrophobic ligand-binding groove on their surface (Lee et al. 2007, 2008; Souers et al. 2013). The wild-type BIMS, which binds with high affinity to all BCL-2 prosurvival proteins, served as a positive control for the integrity of the intrinsic apoptotic pathway. Mutant BIMS4E, which does not bind to any BCL-2 prosurvival family member, thus served as a negative control. L62A/F69A mutant BIMS2A preferentially binds MCL-1, and BIMSBAD binds to BCL-2, BCL-XL, and BCL-W. Western blotting verified inducible expression of all BIMS variants in Rael-BL (Fig. 4C) and other Burkitt lymphoma cell lines (Supplemental Fig. 4A). All Burkitt lymphoma cell lines tested were highly sensitive to the expression of wild-type BIMS, demonstrating that they had an intact intrinsic apoptotic pathway. Conversely, as expected, these cells were not affected by BIMS4E (representative FACS plots are shown in Supplemental Fig. 4B). Notably, all Burkitt lymphoma cells were sensitive to BIMS2A, indicating a dependency on MCL-1 (Fig. 4D; Supplemental Fig. 4D). Ramos-BL, BL31, and Sav-BL were also sensitive to BIMSBAD (and, accordingly, also to treatment with ABT-737, which targets BCL-XL, BCL-2, and BCL-W), albeit for Ramos-BL and BL31, significantly less than to BIMS2A, indicating only a partial dependency on BCL-XL (Fig. 4D; Supplemental Fig. 4C,D). Kinetic binding analyses using the Biacore confirmed that the more potent killing activity of BIMS2A versus BIMSBAD on most cell lines reflected the dependence of these cells on MCL-1 rather than a greater capacity of BIMS2A to engage MCL-1 compared with the ability of BIMSBAD to bind BCL-XL. Indeed these Biacore binding assays showed that the affinity of the BAD-BH3 is actually ∼20-fold higher for BCL-XL (similar to that of wild-type BIMS BH3) than the affinity of BIMS2A for MCL-1 due to a significantly faster dissociation rate in the MCL-1 interaction (Supplemental Fig. 4E). Collectively, these results show that the sustained survival and growth of Burkitt lymphoma cells is dependent on MCL-1 either alone or with a contribution by BCL-XL.
Mutations in p53 reduce but do not ablate the dependency of c-MYC-driven mouse and human lymphomas on MCL-1
Although most transplanted Eμ-Myc mouse lymphomas (14 from 23 analyzed) could not continue to grow following heterozygous recombination of Mcl-1fl, a minority of relapsing lymphomas (nine from 23 analyzed) did display heterozygous Mcl-1fl allele deletion, as demonstrated by huCD4 immunostaining (Fig. 3A). We were interested to understand why these lymphoma cells were less dependent on MCL-1 for their growth, since such insights will be critical for predicting the therapeutic response of c-MYC-driven lymphomas and other cancers to MCL-1 antagonists. Interestingly, we noticed that most lymphomas capable of growth following transplantation and heterozygous Mcl-1fl loss (seven of nine identified) displayed dramatically increased expression of the tumor suppressor p53 (Figs. 3D, 5A), a hallmark of p53 gene mutations. DNA sequence analysis confirmed the presence of mutations affecting known “hot spot” residues (Freed-Pastor and Prives 2012) within the DNA-binding domain of p53 in the primary lymphomas capable of growth following transplantation and heterozygous deletion of Mcl-1 (Figs. 3D, 5B). This suggests that the presence of existing mutations in the p53 gene decreased the dependency of these tumor cells on MCL-1. An additional relapsed lymphoma of genotype Eμ-Myc;CreERT2;Mcl-1fl/fl (#6) showed evidence of p53 pathway mutation (increased p19/ARF protein expression) upon tamoxifen treatment (increasing the frequency to eight of nine lymphomas showing perturbation of the p53 pathway) (Fig. 3D). As a control, we sequenced the p53 gene in seven of the 14 lymphomas that expressed physiological (i.e., very low/barely detectable in the absence of genotoxic stress) levels of p53 (indicative of wild-type nonmutated p53 genes) and were not capable of growing following heterozygous loss of Mcl-1 and confirmed that their p53 genes were wild type. This highlights a novel and highly specific interplay between malignant cell survival dependency on MCL-1 and the p53 tumor suppressor pathway. Importantly, a lymphoma of genotype Eμ-Myc;CreERT2;Mcl-1fl/fl (#3) carrying the p53 mutation that introduced a stop codon almost always regressed following tamoxifen treatment. These results suggest that mutations in the p53 pathway and complete loss of wild-type p53 were not sufficient to allow sustained expansion of Eμ-Myc lymphomas with homozygous Mcl-1 loss.
Therefore, and since mutations in p53 are a common feature of many human cancers (Vousden and Lane 2007), we investigated whether p53 pathway defects could render Burkitt lymphoma cells resistant to MCL-1 targeting. Mutations of the p53 gene, clustering around “hot spot” residues in the DNA-binding domain that are known to abrogate p53 function (Cho et al. 1994; Petitjean et al. 2007; Freed-Pastor and Prives 2012), were detected in Rael-BL, Sav-BL, and BL-31 and globally in ∼60% of the 16 Burkitt lymphoma cell lines tested (note that many lines were in early passage) (Fig. 5C,D). Importantly, all Burkitt lymphoma cells that retained wild-type p53 genes displayed other abnormalities in the p53 tumor suppressor pathway, including HDM2 overexpression (Fig. 5C) or p14ARF locus deletion (BL2) (Fig. 5D). Thus, functional inactivation of the p53 tumor suppressor pathway appears to constitute a critical step in the pathogenesis of c-MYC-driven Burkitt lymphoma but is insufficient to overcome the exquisite growth dependency of these malignant cells on MCL-1. Therefore, therapeutic targeting of MCL-1 would still remain an effective treatment regime for Burkitt lymphoma and possibly other c-MYC-driven human cancers.
Discussion
Up to 70% of human cancers display deregulated c-MYC overexpression (Boxer and Dang 2001; Sanchez-Beato et al. 2003). Identifying the cellular factors critical for the growth of c-MYC-driven cancers therefore remains an important objective for designing novel treatment strategies.
We investigated this by using two novel, innovative approaches: conditional deletion of Mcl-1 or Bcl-x genes at will in Eμ-Myc lymphomas within the whole mouse or inducible expression of polypeptides that inhibit specific BCL-2 prosurvival proteins (mimicking the action of drugs that target these proteins; e.g., ABT-737 or ABT-263/navitoclax) in human Burkitt lymphoma cells. We found that although BCL-XL is essential for Eμ-Myc-induced lymphoma development (Kelly et al. 2011), the loss of BCL-XL had only a minimal impact on sustained tumor growth. Consistent with this, treatment of Eμ-Myc lymphomas with the BH3 mimetic ABT-737, which targets BCL-2, BCL-XL, and BCL-W (Oltersdorf et al. 2005; van Delft et al. 2006), does not cause tumor regression (Mason et al. 2008), suggesting that these three prosurvival proteins are dispensable for sustained Eμ-Myc lymphoma growth. In contrast, sustained growth of Eμ-Myc lymphomas was exquisitely dependent on MCL-1. Remarkably, even heterozygous deletion of Mcl-1 was sufficient to result in complete regression of ∼20% of tumors, allowing long-term survival of formerly lymphoma-burdened mice. Strikingly, analysis of the lymphomas that did relapse revealed that ∼60% had escaped Mcl-1 deletion (mostly due to loss of the Cre recombinase). Indeed, when the animals bearing lymphomas that escaped Mcl-1fl allele deletion (and had not acquired mutations in p53) were removed from the analysis, nearly 100% survived lymphoma-free long-term (>180 d post-transplant) (Fig. 3E). Importantly, this translated into the human disease setting, where we found that MCL-1 blockade efficiently killed Burkitt lymphoma cells, with a lesser impact of BCL-XL targeting observed in some lines.
An MCL-1 dependency was also detected in AML mouse models and human AML-derived cell lines (Xiang et al. 2010; Glaser et al. 2012). This finding and the observation that ∼10% of diverse tumors contain somatically acquired amplifications of the genomic region harboring MCL-1 (Beroukhim et al. 2010) highlight the importance of developing BH3 mimetic drugs that selectively target MCL-1 for treating human cancers. A concern is that MCL-1 is required for the survival of hematopoietic stem/progenitor cells (Opferman et al. 2005) and several other critical cell types, including cardiomyocytes (Opferman et al. 2003; Vikstrom et al. 2010; Thomas et al. 2013; Wang et al. 2013). Importantly, our discovery that MYC-driven lymphomas cannot tolerate even heterozygous loss of Mcl-1 (which should mimic 50% drug-mediated inhibition of the protein), whereas its heterozygous loss is well tolerated in normal tissues (Opferman et al. 2003, 2005; Vikstrom et al. 2010), indicates that it should be possible to establish a therapeutic window for MCL-1 inhibitory drugs.
A novel and highly important finding from this study is the observation that mutations in “hot spot” residues in the DNA-binding domain of the p53 gene can reduce but not abrogate the lymphoma cells' dependency on MCL-1. Consistent with a recent study that found that p53 mutations are the fourth most commonly detected genetic change in a large panel of Burkitt lymphoma biopsies (Love et al. 2012), we found that ∼60% of Burkitt lymphoma lines had acquired mutations in the DNA-binding domain of p53 (Petitjean et al. 2007). Remarkably, all of the remaining Burkitt lymphoma lines examined displayed some other abnormalities in components of the p53 pathway, such as HDM2 overexpression or p14/ARF loss. Importantly, MCL-1 antagonism was able to kill Burkitt lymphomas with p53 pathway defects, suggesting that MCL-1 inhibitory drugs could be efficacious in the treatment of c-MYC-driven cancers bearing p53 mutations.
The network of processes that p53 activates to suppress tumorigenesis are still emerging (Brady et al. 2011; Li et al. 2012; Valente et al. 2013). The observation that mutation and loss of p53 constitute critical steps in c-MYC-driven lymphomagenesis (Vousden and Lane 2007; Michalak et al. 2009) and that they can facilitate the sustained growth of malignant Eμ-Myc lymphomas with reduced MCL-1 expression highlights the connection between the p53 tumor suppressor pathway and the dependency of malignant cells on prosurvival MCL-1.
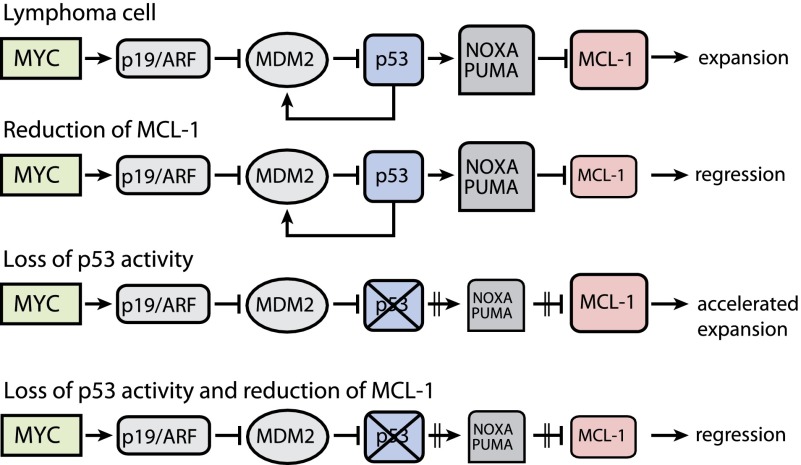
Our results are consistent with a model (Fig. 6) in which c-MYC-driven lymphomas are highly dependent on MCL-1 for survival. A reduction of MCL-1 expression through either genetic means or, in the future, therapeutics could induce cell death by disturbing the balance between the proapoptotic p53 targets PUMA/NOXA and prosurvival MCL-1. However, when designing therapeutic strategies for such lymphomas, we need to be mindful that mutations in p53 would prevent oncogenic stress-induced up-regulation of PUMA and NOXA and may thereby render lymphoma cells less dependent on MCL-1. Therefore, for such (p53 mutated) c-MYC-driven lymphomas, it may be beneficial to activate the intrinsic apoptotic pathway more potently than is needed for tumors that carry wild-type p53.
Figure 6.
A proposed model in which c-MYC-driven lymphoma cells that are highly dependent on MCL-1 for their sustained expansion can no longer survive once MCL-1 expression is reduced and the balance between this prosurvival protein and the p53 proapoptotic targets PUMA/NOXA is disturbed. The c-MYC-driven lymphoma cells that have acquired p53 mutations display an accelerated lymphoma progression. These lymphomas express less PUMA/NOXA, and, consequently, less MCL-1 expression is required to maintain the lymphoma progression. Despite this, we showed that targeting of MCL-1 in lymphomas with p53 mutations was sufficient to result in tumor regression. Note that the mouse protein nomenclature is used.
In conclusion, our findings suggest that MCL-1 targeting may be a viable strategy for cancer therapy, particularly in tumors where deregulated c-MYC expression instills an exquisite dependence (much more so than in nontransformed cells) on this prosurvival protein.
Materials and methods
Mice
Experiments with mice were conducted according to the guidelines of The Walter and Eliza Hall Institute Animal Ethics Committee. Eμ-Myc transgenic (Adams et al. 1985), Bcl-xfl/fl (Wagner et al. 2000), Mcl-1fl/fl (Vikstrom et al. 2010), Rosa26-CreERT2 (Seibler et al. 2003), and p53−/− (Jacks et al. 1994) gene targeted mice have all been described previously. All mouse strains were on a C57BL/6-Ly5.2+ genetic background generated on either this background (Mcl-1fl/fl and Rosa26-CreERT2) using C57BL/6-derived embryonic stem cells, a mixed C57BL/6x129SV background (Bcl-xfl/fl) using 129SV-derived embryonic stem cells, or a mixed C57BL/6xSJL background (Eμ-Myc) using microinjection of C57BL/6xSJL-derived oocytes and then backcrossed for 10 to >20 generations with C57BL/6 mice.
Single-cell suspensions of 3 × 106 Eμ-Myc lymphoma cells in PBS were injected into C57BL/6-Ly5.1+ or C57BL/6J-Tyrc-2J (referred to as C57BL/6-albino; The Jackson Laboratory) recipient mice by intravenous (i.v.) tail vein injection. Mice were administered by oral gavage with 200 mg/kg tamoxifen (Sigma-Aldrich) in peanut oil/10% ethanol per day (Anastassiadis et al. 2010) for two consecutive days on either days 5 and 6 or days 10 and 11 post-injection of the tumor cells. Transplants in which the matched control untreated mice did not become sick by 28 d post-tumor cell injection were excluded from the analysis.
Statistical analysis
Graphpad Prism software was used for generating Kaplan-Meier plots and performing statistical analysis (using a log-rank test) to compare the survival of mice injected with lymphoma cells of different genotypes. Graphpad Prism was also used to carry out paired two-tailed t-tests on the death induced in the Burkitt lymphoma cells following dox-inducible expression of the BIMs variants. Specifically, the significance of the extent of cell death induced by BIMs2A (to target MCL-1) expression compared with either inducible expression of BIMs4E (negative control) or BIMsBAD (to target BCL-2, BCL-XL, and BCL-W) was calculated for each of the Burkitt lymphoma cell lines.
Cell culture
Eμ-Myc mouse lymphoma cells were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Gibco), 50 μM β-mercaptoethanol (Sigma-Aldrich), 100 μM asparagine (Sigma-Aldrich), 100 U/mL penicillin, and 100 mg/mL streptomycin (Gibco) at 37°C and 10% CO2. OP9 cells were cultured in αMEM (Gibco) supplemented with 20% heat-inactivated FBS, 1 mM glutamine (Gibco), 10 mM Hepes (Gibco), 1 mM sodium pyruvate (Gibco), 50 μM β-mercaptoethanol, 100 U/mL penicillin, and 100 μg/mL streptomycin. Human cell lines were cultured in a humidified incubator at 37°C and 5% CO2. Virus-producing 293T cells were maintained in DMEM supplemented with 10% FBS. Approximately 6 h prior to transfection, 293T cells were cultured in DME glutamax (Gibco) supplemented with 10% FBS and 25 mM Hepes. Rael-BL, Ramos-BL, Sav-BL, and BL-31 human Burkitt lymphoma-derived cell lines were cultured in RPMI 1640 supplemented with 10% FBS, 1 mM glutamine (Gibco), 1 mM sodium pyruvate (Gibco), 50 μM α-thioglycerol (Sigma-Aldrich), and 20 nM bathocuproine disulfonic acid (BCS) (Sigma-Aldrich). X50-7 and Awia lymphoblastoid cell lines were cultured in RPMI 1640 supplemented with 10% FBS and 1 mM glutamine.
Lentiviral plasmids and virus production
The dox-responsive lentiviral vector pFTRE3G_pGK3G_GFP was generated by digesting PacI/AscI and cloning the TRE3G_pGK3G_GFP cassette. The TRE3G_pGK3G GFP cassette was amplified by PCR as pTRE3G_pGK3G_GFP (Yamamoto et al. 2012). The PCR products of the BIMS variants (Lee et al. 2008) were cloned into pFTRE3G_pGK3G_GFP. The lentiviral imaging construct was generated by inserting Luciferase2 linked via a T2A peptide to eGFP downstream from the human ubiquitin promoter of the FUGW vector (Lois et al. 2002). Lentivirus-containing supernatants were produced by transiently transfecting 293T cells with the expression constructs of interest alongside the packaging plasmids, VSV-G, MDL, and RSV-Rev using the standard CaPO4 method (Herold et al. 2008). Supernatants containing infectious virus particles were harvested 2–3 d post-transfection.
Lentiviral transduction of Eμ-Myc lymphoma cells and Burkitt lymphoma cell lines
For infection, aliquots of 0.5 × 106 to 1 × 106 primary Eμ-Myc lymphoma cells or 1 × 105 Burkitt lymphoma cells were suspended in 4 mL of virus-containing supernatant containing 10 ng/μL polybrene, incubated for 30 min at 37°C and 5% or 10% CO2, and then centrifuged at 2200 rpm for 2.5 h at 32°C. The supernatant was subsequently discarded, and the cells were resuspended in fresh medium for culture.
Inducible expression of BIMS variants in human Burkitt lymphoma cells
To induce expression of the BIMS variants, cells were cultured in medium supplemented with 1 μg/mL dox. The viability of the GFP-positive cells was determined by propidium iodide (PI) staining and FACS analysis in an LSR1 machine. The PI-negative/GFP-positive cells were considered as live cells. The cell death following inducible expression of the BIMs variant was expressed relative to the untreated Burkitt lymphoma cells, which were assigned a value of 1. For the analysis of the BIMS variant expression by immunoblotting, the pan-caspase inhibitor qVD-OPh (25 μM; MP Biomedicals) was added to the dox-treated cells for 24 h to inhibit cell killing.
Direct binding assays to determine the binding parameters of BH3 ligands for BCL-XL and MCL-1
Direct binding assays were performed at room temperature using a Biacore S51 biosensor exactly as described previously (Lee et al. 2007). The peptides used were BIMs BH3, DMRPEIWIAQELRRIGDEFNAYYARR; BIMS 2A BH3, DMRPEIWIAQEARRIGDEANAYYARR; and BADS BH3, NLWAAQRYGRELRRMSDEFVDSFKKG.
Immunofluorescent staining, flow cytometric analysis, and cell sorting
To immunophenotype Eμ-Myc lymphomas, single-cell suspensions were stained as described (Strasser et al. 1991) with surface marker-specific antibodies and analyzed using an LSR1 machine (Becton Dickinson). The following fluorochrome-conjugated antibodies were used: CD19 (clone ID3), B220 (RA3-6B2), IgM (clone 5.1), IgD (clone 11-26C), CD4 (clone H129), CD8 (clone YTS.169), Thy1 (clone T3.24.1), Mac1 (M1/70), and GR1 (clone RB6-8C5). To discriminate host-derived (Ly5.1+) from donor-derived (Ly5.2+) cells, monoclonal antibodies to Ly5.1 (clone A201.1) and Ly5.2 (clone 5.450.15.2 or AL14A7) were used. To verify recombination of the loxP-flanked allele of Mcl-1, cells were stained with antibodies to human CD4 (clone RPA-T4), since Mcl-1fl recombination subjugates a human CD4 reporter cassette to the Mcl-1 promoter/enhancer (Vikstrom et al. 2010). Antibodies were produced in our laboratory and conjugated to FITC, R-phycoerythrin (R-PE), or allophycocyanin (APC) according to the manufacturers' instructions. Where required, Burkitt lymphoma cells were sorted for GFP expression using an AriaW cell sorter (Becton Dickinson).
Immunoblotting
Total protein extracts were prepared from cells (primary Eμ-Myc lymphomas and Burkitt lymphoma cell lines) by lysis in onyx buffer, NP40 buffer, or urea buffer containing protease inhibitors (Complete protease inhibitor cocktail, Roche). Protein content was quantified using the Bio-Rad Bradford assay. Total protein extracts (20 μg) were separated on the basis of molecular weight by SDS-PAGE and Western-blotted onto nitrocellulose membranes. The membranes were blocked in 5% skim milk in PBS and 0.1% Tween20 (blocking buffer) before incubation with antibodies. Polyclonal antibodies were used to detect mouse MCL-1 (Rockland Antibodies and Assays), BIM (Enzo Life Sciences), BCL-x (BD Biosciences), and human MCL-1 (s-19, Santa Cruz Biotechnology); monoclonal antibodies were used to detect mouse p53 (clone IMX25, Novocastra), p19/ARF (clone 5.C3.1, Rockland Antibodies and Assays), ERα (to detect the CreERT2 protein) (clone HC-20, Santa Cruz Biotechnology), Actin (clone AC40, Sigma-Aldrich), and β-Actin (clone AC74, Sigma-Aldrich); and monoclonal antibodies were used to detect human p53 (clone FL393), HDM2 (clone N20), BCL-2 (clone C-2), BCL-XL (clone H5) (all from Santa Cruz Biotechnology), β-Actin (clone AC15, Sigma-Aldrich), and HSP70 (mouse monoclonal antibody clone N6; detects both mouse and human protein) (a gift from Dr R Anderson, Peter MacCallum Cancer Research Institute, Melbourne, Australia). All antibodies were diluted in blocking buffer.
qRT–PCR analysis
Total RNA was extracted from paired untreated and tamoxifen-treated Eμ-Myc lymphoma cells using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. The RNA was treated with DNase to remove contaminating DNA using the RNase-free DNase Qiagen kit according to the manufacturer's instructions. RNA quality and quantity were determined using the NanoDrop assay (Thermo Fisher Scientific). Aliquots of 1 μg of RNA were reverse-transcribed into cDNA using the SuperScript III first strand synthesis Supermix kit (Invitrogen) in a 20-μL reaction volume according to the manufacturer's instructions. The cDNA was diluted 10-fold in H2O, and PCR amplifications of 1 μL of cDNA were performed with an Applied Biosystems 7900HT thermal cycler using TaqMan Universal PCR Mastermix and 0.5 μL of TaqMan primer/probes (both Applied Biosystems) in a 10-μL reaction volume. Assays specific for Bcl-x (Mm00437783) and (as an endogenous control for RNA quality/input) HMBS (Mm01143545) transcripts were performed. Three replicates of each reaction were performed. All qPCR data were analyzed using the 2−ΔΔCT method and expressed relative to the untreated sample of each pair.
IVIS imaging
Single-cell suspensions of lymphoma cells taken from the lymph nodes, spleens, or thymuses of sick Eμ-Myc;CreERT2,Bcl-xfl/fl and Eμ-Myc;CreERT2;Mcl-1fl/fl mice were transduced with a GFP-luc lentiviral vector as described above. Transduced cells were cultured for three passages on an OP9 cell feeder layer (Zúñiga-Pflücker et al. 1994) and then washed in PBS, and 4 × 105 to 1 × 106 live lymphoma cells were injected i.v. into C57BL/6-albino recipient mice. To visualize the tumor burden, 200 μL of 15 mg/mL D-luciferin potassium salt (Caliper Life Sciences) in PBS was administered to the mice by intraperitoneal (i.p.) injection. Mice were then anaesthetized with isoflurane inhalant and imaged using the IVIS live-imaging system (Perkin Elmer) to detect luciferase bioluminescence exactly 15 min after administration of the luciferin substrate. The tumor burden was quantified by measuring the total photon flux per second emitted from a region of interest (ROI) drawn around the whole mouse. When the lymphoma burden was sufficiently high (photon flux per second of >1 × 107), some mice from each cohort were administered tamoxifen by oral gavage once per day for three consecutive days, and the tumor regression/progression was monitored by further bioluminescence imaging at indicated time points.
Human p53 cDNA sequence analysis
The human p53 gene status was determined by sequencing of cDNA (Lindstrom et al. 2001). Total RNA was extracted from cells using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. The cDNA was made from total RNA using the Advantage one-step RT–PCR kit according to the manufacturer's instructions (Clontech). p53 was sequenced from pooled PCR reactions using the following primers: p53F1 (5′-GAAGGATCCGAGGAGCCGCAGT-3′), p53F781 (5′-CTGAGGTTGGCTCTGACTGTACCACCATCC-3′), p53R789 (5′-AACAAGCTTATTACCACTGGAGTCTTC-3′), and p53R1179 (5′-AGGGAATTCAGTCTGAGTCAGGC-3′).
DNA extraction, PCR, and DNA sequence analysis
DNA was extracted from cells using the Qiagen DNEasy tissue kit (Qiagen) according to the manufacturer's instructions, and the DNA concentration was quantified using the Nanodrop assay. Exons 4, 5–6, 7, 8–9, and 10 of the mouse p53 gene were PCR-amplified from 100 ng of DNA using GoTaq green master mix (M712, Promega) and 3.3 μM primers using 32 cycles of 1 min at 94°C, 1 min at 52°C, and 1 min at 72°C followed by a 5-min extension at 72°C. DNA sequence analysis was performed by Micromon Monash. The following primer combinations were used to PCR-amplify and sequence mouse p53: Ex4F (5′-GGTTCTTCTTTGTCCCATCC-3′) with Ex4R (5′-GAGGCATTGAAAGGTCACAC-3′), Ex5F 5′-TTAGTTCCCCACCTTGACAC-3′) with Ex6R (5′-AGGCTGGAGTCAACTGTCTC-3′), Ex7F (5′-TAGTGAGGTAGGGAGCGACTTC-3′) with Ex7R (5′-CCAAGAGGAAACAGAGGAGG-3′), Ex8F (5′-CTTCTCGGGGTTCCTGTAAC-3′) with Ex9R (5′-CCTGGCAACCTGCTAATAAC-3′), and Ex10F (5′-AAACCTGTAAGTGGAGCCAG-3′) with Ex10R (5′-AGTCAGTTCTCGTAGGGTGC-3′). The sequence alignment for murine p53 was performed using Ensembl transcript number ENSMUST00000108658 as the reference transcript.
DNA PCR to detect Mcl-1fl, Mcl-1wt, and recombined Mcl-1 alleles
The wild-type, Mcl-1fl, and recombined Mcl-1fl alleles were PCR-amplified from 100 ng of DNA using GoTaq green master mix (M712, Promega) and 0.5 μM primers using 30 cycles of 40 sec at 94°C, 30 sec at 55°C, and 1 min at 72°C followed by a 5-min extension at 72°C. The following primers were used to detect Mcl-1fl and Mcl-1wt alleles: 5′-GCACAATCCGTCCGCGAGCCAA-3′ and reverse 5′-GCCGCAGTACAGGTTCAAG-3′. The wild-type PCR product was smaller than the floxed PCR product. The following primers were used to detect recombined Mcl-1fl alleles: 5′-CGACACAGATCAGCAGGCGTTC-3′ with 5′-GAGTCAGCGCGATCATTCAGCT-3′.
DNA PCR to detect homozygous deletion of the CDKN2A (INK4A/ARF) locus
The following PCR primers were used: exon 1β, (F, 5′-TGCAGTTAAGGGGGCAGGAG-3′; R, 5′-TTATCTCCTCCTCCTCCTAGCCTG-3′) and exon 2 (551R, 5′-TCTGAGCTTTGGAAGCTCT-3′; 42F, 5′-GGAAATTGGAAACTGGAAGC-3′). A GAPDH exon 8 fragment served as control for integrity of DNA.
Acknowledgments
We thank G. Siciliano and his team for help with animal husbandry, C. McLean for editorial assistance, Dr. J.M. Adams and Dr. S. Cory for advice and discussions, and Dr. Kelly Rogers for help with imaging of mice. This work is supported by a Kay Kendall Leukemia Fund Intermediate Fellowship (KKL331) and an EMBO short-term fellowship (both awarded to G.L.K.); the National Health and Medical Research Council, Australia (program grant 1016701 and fellowship 1020363 to A.S., project grant APP1049720 to M.J.H., project grant APP1041936 to W.D.F., and career development fellowship APP1024620 to E.F.L.); Cancer Research UK (program grant awarded to L.F., M. Rowe, and A.B.R.); the Leukemia Foundation; the Leukemia and Lymphoma Society (specialized center of research 7001-13); and a German Research Council Fellowship (He 5740/1-1 awarded to M.J.H.). This work was made possible through Victorian State Government Operational Infrastructure Support and Australian Government National Health and Medical Research Council Independent Research Institutes Infrastructure Support Scheme.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.232009.113.
References
- Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL 1985. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 318: 533–538 [DOI] [PubMed] [Google Scholar]
- Anastassiadis K, Glaser S, Kranz A, Berhardt K, Stewart AF 2010. A practical summary of site-specific recombination, conditional mutagenesis, and tamoxifen induction of CreERT2. Methods Enzymol 477: 109–123 [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Mermel C, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm J, Dobson J, Urashima M, et al. 2010. The landscape of somatic copy-number alteration across human cancers. Nature 463: 899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia KG, Gutierrez MI, Huppi K, Siwarski D, Magrath IT 1992. The pattern of p53 mutations in Burkitt's lymphoma differs from that of solid tumors. Cancer Res 52: 4273–4276 [PubMed] [Google Scholar]
- Boxer LM, Dang CV 2001. Translocations involving c-myc and c-myc function. Oncogene 20: 5595–5610 [DOI] [PubMed] [Google Scholar]
- Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM, Broz DK, Basak S, Park EJ, McLaughlin ME, et al. 2011. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell 145: 571–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, Colman PM, Day CL, Adams JM, Huang DCS 2005. Differential targeting of pro-survival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell 17: 393–403 [DOI] [PubMed] [Google Scholar]
- Cho Y, Gorina S, Jeffrey PD, Pavletich NP 1994. Crystal structure of a p53 tumor suppressor-DNA complex: Understanding tumorigenic mutations. Science 265: 346–355 [DOI] [PubMed] [Google Scholar]
- Dang CV 1999. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol 19: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egle A, Harris AW, Bouillet P, Cory S 2004. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc Natl Acad Sci 101: 6164–6169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell PJ, Allan GJ, Shanahan F, Vousden KH, Crook T 1991. p53 is frequently mutated in Burkitt's lymphoma cell lines. EMBO J 10: 2879–2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed-Pastor WA, Prives C 2012. Mutant p53: One name, many proteins. Genes Dev 26: 1268–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison SP, Jeffers JR, Yang C, Nilsson JA, Hall MA, Rehg JE, Yue W, Yu J, Zhang L, Onciu M, et al. 2008. Selection against PUMA gene expression in Myc-driven B-cell lymphomagenesis. Mol Cell Biol 28: 5391–5402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giulino-Roth L, Wang K, MacDonald TY, Mathew S, Tam Y, Cronin MT, Palmer G, Lucena-Silva N, Pedrosa F, Pedrosa M, et al. 2012. Targeted genomic sequencing of pediatric Burkitt lymphoma identifies recurrent alterations in antiapoptotic and chromatin-remodeling genes. Blood 120: 5181–5184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser S, Lee EF, Trounson E, Bouillet P, Wei A, Fairlie WD, Izon DJ, Zuber J, Rappaport AR, Herold MJ, et al. 2012. Anti-apoptotic Mcl-1 is essential for the development and sustained growth of acute myeloid leukemia. Genes Dev 26: 120–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA 2011. Hallmarks of cancer: The next generation. Cell 144: 646–674 [DOI] [PubMed] [Google Scholar]
- Hemann MT, Zilfou JT, Zhao Z, Burgess DJ, Hannon GJ, Lowe SW 2004. Suppression of tumorigenesis by the p53 target PUMA. Proc Natl Acad Sci 101: 9333–9338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S, Rowe M, Gregory C, Croom Carter D, Wang F, Longnecker R, Kieff E, Rickinson A 1991. Induction of bcl-2 expression by Epstein-Barr virus latent membrane protein 1 protects infected B cells from programmed cell death. Cell 65: 1107–1115 [DOI] [PubMed] [Google Scholar]
- Herold MJ, van den Brandt J, Seibler J, Reichardt HM 2008. Inducible and reversible gene silencing by stable integration of an shRNA-encoding lentivirus in transgenic rats. Proc Natl Acad Sci 105: 18507–18512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA 1994. Tumor spectrum analysis in p53-mutant mice. Curr Biol 4: 1–7 [DOI] [PubMed] [Google Scholar]
- Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, MacLean KH, Han J, Chittenden T, Ihle JN, et al. 2003. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell 4: 321–328 [DOI] [PubMed] [Google Scholar]
- Kelly GL, Strasser A 2011. The essential role of evasion from cell death in cancer. Adv Cancer Res 111: 39–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly PN, Grabow S, Delbridge ARD, Strasser A, Adams JM 2011. Endogenous Bcl-xL is essential for Myc-driven lymphomagenesis in mice. Blood 118: 6380–6386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaw SL, Huang DC, Roberts AW 2011. Overcoming blocks in apoptosis with BH3-mimetic therapy in haematological malignancies. Pathology 43: 525–535 [DOI] [PubMed] [Google Scholar]
- Langdon WY, Harris AW, Cory S, Adams JM 1986. The c-myc oncogene perturbs B lymphocyte development in Eμ-myc transgenic mice. Cell 47: 11–18 [DOI] [PubMed] [Google Scholar]
- Lee EF, Czabotar PE, Smith BJ, Deshayes K, Zobel K, Colman PM, Fairlie WD 2007. Crystal structure of ABT-737 complexed with Bcl-x(L): Implications for selectivity of antagonists of the Bcl-2 family. Cell Death Differ 14: 1711–1713 [DOI] [PubMed] [Google Scholar]
- Lee EF, Czabotar PE, van Delft MF, Michalak E, Boyle M, Willis SN, Puthalakath H, Bouillet P, Colman PM, Huang DCS, et al. 2008. A novel BH3 ligand that selectively targets Mcl-1 reveals that apoptosis can proceed without Mcl-1 degradation. J Cell Biol 180: 341–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessene G, Czabotar PE, Colman PM 2008. BCL-2 family antagonists for cancer therapy. Nat Rev Drug Discov 7: 989–1000 [DOI] [PubMed] [Google Scholar]
- Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, Baer R, Gu W 2012. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 149: 1269–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom MS, Klangby U, Wiman KG 2001. p14ARF homozygous deletion or MDM2 overexpression in Burkitt lymphoma lines carrying wild type p53. Oncogene 20: 2171–2177 [DOI] [PubMed] [Google Scholar]
- Llambi F, Moldoveanu T, Tait SW, Bouchier-Hayes L, Temirov J, McCormick LL, Dillon CP, Green DR 2011. A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol Cell 44: 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D 2002. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295: 868–872 [DOI] [PubMed] [Google Scholar]
- Love C, Sun Z, Jima D, Li G, Zhang J, Miles R, Richards KL, Dunphy CH, Choi WW, Srivastava G, et al. 2012. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet 44: 1321–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason KD, Vandenberg CJ, Scott CL, Wei AH, Cory S, Huang DC, Roberts AW 2008. In vivo efficacy of the Bcl-2 antagonist ABT-737 against aggressive Myc-driven lymphomas. Proc Natl Acad Sci 105: 17961–17966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino D, Giam M, Hughes PD, Siggs OM, Heger K, O'Reilly LA, Adams JM, Strasser A, Lee EF, Fairlie WD, et al. 2009. The role of BH3-only protein Bim extends beyond inhibiting Bcl-2-like prosurvival proteins. J Cell Biol 186: 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestre-Escorihuela C, Rubio-Moscardo F, Richter JA, Siebert R, Climent J, Fresquet V, Beltran E, Agirre X, Marugan I, Marin M, et al. 2007. Homozygous deletions localize novel tumor suppressor genes in B-cell lymphomas. Blood 109: 271–280 [DOI] [PubMed] [Google Scholar]
- Michalak EM, Jansen ES, Happo L, Cragg MS, Tai L, Smyth GK, Strasser A, Adams JM, Scott CL 2009. Puma and to a lesser extent Noxa are suppressors of Myc-induced lymphomagenesis. Cell Death Differ 16: 684–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, et al. 2005. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 435: 677–681 [DOI] [PubMed] [Google Scholar]
- Opferman JT, Letai A, Beard C, Sorcinelli MD, Ong CC, Korsmeyer SJ 2003. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature 426: 671–676 [DOI] [PubMed] [Google Scholar]
- Opferman J, Iwasaki H, Ong CC, Suh H, Mizuno S, Akashi K, Korsmeyer SJ 2005. Obligate role of anti-apoptotic MCL-1 in the survival of hematopoietic stem cells. Science 307: 1101–1104 [DOI] [PubMed] [Google Scholar]
- Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M 2007. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: Lessons from recent developments in the IARC TP53 database. Hum Mutat 28: 622–629 [DOI] [PubMed] [Google Scholar]
- Richter-Larrea JA, Robles EF, Fresquet V, Beltran E, Rullan AJ, Agirre X, Calasanz MJ, Panizo C, Richter JA, Hernandez JM, et al. 2010. Reversion of epigenetically mediated BIM silencing overcomes chemoresistance in Burkitt lymphoma. Blood 116: 2531–2542 [DOI] [PubMed] [Google Scholar]
- Sanchez-Beato M, Sanchez-Aguilera A, Piris MA 2003. Cell cycle deregulation in B-cell lymphomas. Blood 101: 1220–1235 [DOI] [PubMed] [Google Scholar]
- Seibler J, Zevnik B, Kuter-Luks B, Andreas S, Kern H, Hennek T, Rode A, Heimann C, Faust N, Kauselmann G, et al. 2003. Rapid generation of inducible mouse mutants. Nucleic Acids Res 31: e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, Dayton BD, Ding H, Enschede SH, Fairbrother WJ, et al. 2013. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med 19: 202–208 [DOI] [PubMed] [Google Scholar]
- Strasser A, Harris AW, Bath ML, Cory S 1990. Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature 348: 331–333 [DOI] [PubMed] [Google Scholar]
- Strasser A, Harris AW, Cory S 1991. Bcl-2 transgene inhibits T cell death and perturbs thymic self-censorship. Cell 67: 889–899 [DOI] [PubMed] [Google Scholar]
- Swanson PJ, Kuslak SL, Fang W, Tze L, Gaffney P, Selby S, Hippen KL, Nunez G, Sidman CL, Behrens TW 2004. Fatal acute lymphoblastic leukemia in mice transgenic for B cell-restricted bcl-xL and c-myc. J Immunol 172: 6684–6691 [DOI] [PubMed] [Google Scholar]
- Thomas RL, Roberts DJ, Kubli DA, Lee Y, Quinsay MN, Owens JB, Fischer KM, Sussman MA, Miyamoto S, Gustafsson AB 2013. Loss of MCL-1 leads to impaired autophagy and rapid development of heart failure. Genes Dev 27: 1365–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente LJ, Gray DHD, Michalak EM, Pinon-Hofbauer J, Egle A, Scott CL, Janic A, Strasser A 2013. p53 efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, Puma and Noxa. Cell Rep 3: 1339–1345 [DOI] [PubMed] [Google Scholar]
- van Delft MF, Wei AH, Mason KD, Vandenberg CJ, Chen L, Czabotar PE, Willis SN, Scott CL, Day CL, Cory S, et al. 2006. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell 10: 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikstrom I, Carotta S, Luethje K, Peperzak V, Jost PJ, Glaser S, Busslinger M, Bouillet P, Strasser A, Nutt SL, et al. 2010. Mcl-1 is essential for germinal center formation and B cell memory. Science 330: 1095–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ, Adams JM, Strasser A 2003. p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 302: 1036–1038 [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lane DP 2007. p53 in health and disease. Nat Rev Mol Cell Biol 8: 275–283 [DOI] [PubMed] [Google Scholar]
- Wagner KU, Claudio E, Rucker EB 3rd, Riedlinger G, Broussard C, Schwartzberg PL, Siebenlist U, Hennighausen L 2000. Conditional deletion of the Bcl-x gene from erythroid cells results in hemolytic anemia and profound splenomegaly. Development 127: 4949–4958 [DOI] [PubMed] [Google Scholar]
- Wang X, Bathina M, Lynch J, Koss B, Calabrese C, Frase S, Schuetz JD, Rehg JE, Opferman JT 2013. Deletion of MCL-1 causes lethal cardiac failure and mitochondrial dysfunction. Genes Dev 27: 1351–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Z, Luo H, Payton JE, Cain J, Ley TJ, Opferman JT, Tomasson MH 2010. Mcl1 haploinsufficiency protects mice from Myc-induced acute myeloid leukemia. J Clin Invest 120: 2109–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Okuyama M, Ma JS, Kimura T, Kamiyama N, Saiga H, Ohshima J, Sasai M, Kayama H, Okamoto T, et al. 2012. A cluster of interferon-γ-inducible p65 GTPases plays a critical role in host defense against Toxoplasma gondii. Immunity 37: 302–313 [DOI] [PubMed] [Google Scholar]
- Youle RJ, Strasser A 2008. The BCL-2 protein family: Opposing activities that mediate cell death. Nat Rev Mol Cell Biol 9: 47–59 [DOI] [PubMed] [Google Scholar]
- Zúñiga-Pflücker JC, Jiang D, Schwartzberg PL, Lenardo MJ 1994. Sublethal γ-radiation induces differentiation of CD4−/CD8− into CD4+/CD8+ thymocytes without T cell receptor β rearrangement in recombinase activation gene 2−/− mice. J Exp Med 180: 1517–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]