The mammalian circadian clock requires the master transcription factors CLOCK and BMAL1 to drive rhythmic gene expression. Here, Menet et al. report that rhythmic binding of CLOCK:BMAL1 on DNA promotes rhythmic chromatin opening. Mechanisms include CLOCK:BMAL1 binding to nucleosomes and chromatin modifications such as incorporation of histone variant H2A.Z. The data indicate that clock regulation of transcription relies on rhythmic regulation of chromatin accessibility, thus extending the concept of pioneer factor function to acute gene regulation.
Keywords: circadian rhythms, regulation of transcription, nucleosome positioning, nucleosome occupancy, MNase-seq, chromatin modifications, H2A.Z
Abstract
The mammalian circadian clock relies on the master genes CLOCK and BMAL1 to drive rhythmic gene expression and regulate biological functions under circadian control. Here we show that rhythmic CLOCK:BMAL1 DNA binding promotes rhythmic chromatin opening. Mechanisms include CLOCK:BMAL1 binding to nucleosomes and rhythmic chromatin modification; e.g., incorporation of the histone variant H2A.Z. This rhythmic chromatin remodeling mediates the rhythmic binding of other transcription factors adjacent to CLOCK:BMAL1, suggesting that the activity of these other transcription factors contributes to the genome-wide CLOCK:BMAL1 heterogeneous transcriptional output. These data therefore indicate that the clock regulation of transcription relies on the rhythmic regulation of chromatin accessibility and suggest that the concept of pioneer function extends to acute gene regulation.
Circadian clocks drive the rhythmic expression of a large fraction of the transcriptome in many eukaryotic tissues and organisms to regulate biochemical, physiological, and behavioral functions. Circadian gene expression is generated by a set of core clock genes, which interact in transcriptional feedback loops. In mammals, they include the two master heterodimeric transcription factors CLOCK and BMAL1. This heterodimer rhythmically activates the expression of their transcriptional repressors, Period (Per1 and Per2) and Cryptochrome (Cry1 and Cry2) (for review, see Mohawk et al. 2012). Although these proteins and other clock components are well characterized, recent evidence suggest that the mechanisms by which they control genome-wide rhythmic gene expression are not well understood (Rey et al. 2011; Menet et al. 2012).
We recently characterized CLOCK:BMAL1 target genes and analyzed rhythmic transcription using Nascent-seq in mouse livers (Menet et al. 2012). The data revealed a surprising disconnect between the phase of CLOCK:BMAL1 DNA binding and the phase of target gene transcription, including the transcription of key core clock genes. CLOCK:BMAL1 binding occurs at the same phase of the cycle for all target genes, whereas the peaks of cycling transcription are heterogeneous, with little or no relationship to the singular phase of CLOCK:BMAL1 binding (Menet et al. 2012). This indicates that other transcription factors are involved in core clock gene transcription, for which there is experimental support (Ukai-Tadenuma et al. 2011).
There is evidence that CLOCK:BMAL1 function extends beyond transcriptional activation. For example, CLOCK is reported to have histone acetyl transferase activity (Doi et al. 2006), and genome-wide rhythmic modifications of CLOCK target gene chromatin have been recently described (Feng and Lazar 2012; Koike et al. 2012; Le Martelot et al. 2012; Vollmers et al. 2012; Aguilar-Arnal and Sassone-Corsi 2013). Importantly, ectopic expression of Drosophila CLOCK generates ectopic circadian clocks (Zhao et al. 2003; Kilman and Allada 2009), indicating that CLOCK can function to establish a circadian program analogous to the developmental programs generated by key factors like Pax6 (Osumi et al. 2008). Because some transcription factors such as the glucocorticoid receptor (Truss et al. 1995; Nagaich et al. 2004; Voss et al. 2011) establish their programs by remodeling chromatin and promoting nucleosome removal at lineage-specific target genes (Magnani et al. 2011; Zaret and Carroll 2011), we hypothesized that CLOCK:BMAL1 plays a similar role at its target gene chromatin.
In the present study, we report that the rhythmic binding of CLOCK:BMAL1 on DNA promotes the rhythmic removal of nucleosomes at its binding sites. Relevant mechanisms include CLOCK:BMAL1 binding to nucleosomes as well as rhythmic chromatin modifications such as incorporation of the histone variant H2A.Z. We also show that this rhythmic chromatin opening at CLOCK:BMAL1 DNA-binding sites is associated with rhythmic binding of another transcription factor (HNF6). This suggests that the activity of other transcription factors contributes to the heterogeneous transcriptional output of CLOCK:BMAL1 target genes and that the activity of these other factors relies on the rhythmic regulation of chromatin accessibility of CLOCK:BMAL1.
Results and Discussion
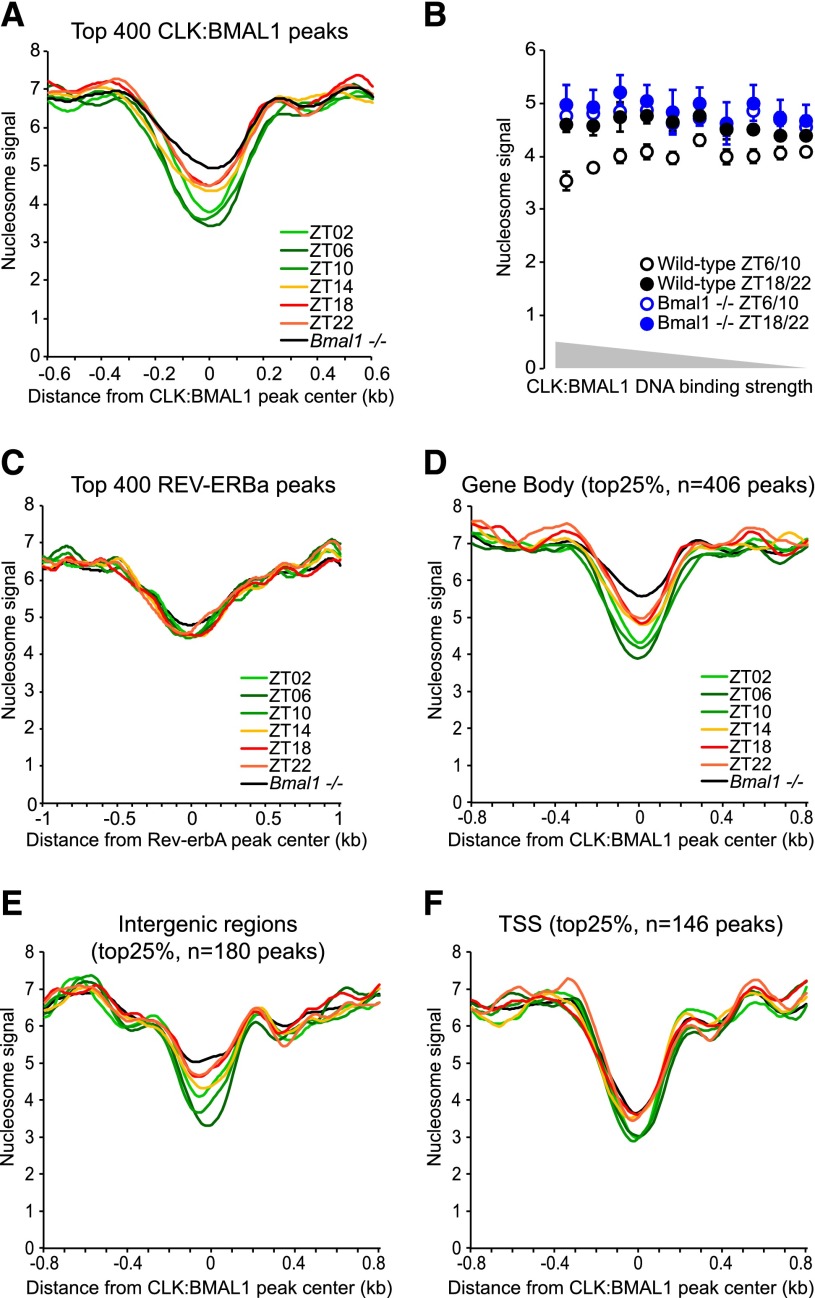
To test the hypothesis that CLOCK:BMAL1 remodels chromatin, we first performed a genome-wide nucleosome analysis using MNase-seq (digestion of chromatin with micrococcal nuclease and high-throughput sequencing of mononucleosomes) in mouse livers at six time points across the light:dark cycle. Four wild-type animals were used for each time point, and a minimum of 84 million nucleosomes was sequenced per time point. Analysis of CLOCK:BMAL1 DNA-binding sites (3217 peaks from Koike et al. 2012) showed that the nucleosome signal is rhythmic at these sites for all four individual rhythms and lower when CLOCK:BMAL1 binds to DNA during the light phase (e.g., Zeitgeber time 02 [ZT02] to ZT10) (Fig. 1A; Supplemental Fig. 1; Rey et al. 2011; Koike et al. 2012). In contrast, the identical MNase-seq assay in Bmal1−/− livers showed a more limited decrease in nucleosome signal and no rhythmicity, indicating that CLOCK:BMAL1 directly contributes to nucleosome removal (Fig. 1A; Supplemental Fig. 2). Consistent with this conclusion, the magnitude of the loss of nucleosome signal correlates with the strength of CLOCK:BMAL1 DNA binding (Fig. 1B).
Figure 1.
CLOCK:BMAL1 promotes the rhythmic removal of nucleosomes at its DNA-binding sites. (A) Average nucleosome signal at the top 400 CLOCK:BMAL1 DNA-binding sites (±0.6 kb) in mouse livers during the light phase (ZT2, ZT6, and ZT10; green) and dark phase (ZT14, ZT18, and ZT22; red/orange) of wild-type mice and in Bmal1−/− mice (average signal for six time points; black). (B) Effect of CLOCK:BMAL1 DNA-binding strength on the average nucleosome signal at CLOCK:BMAL1 DNA-binding sites (±75 bp), at times of high binding (ZT6 and ZT10; open circles) or low binding (ZT18 and ZT22; closed circles) to DNA in wild-type (black) and Bmal1−/− (blue) mice. The 3217 CLOCK:BMAL1 peaks were equally distributed in 10 bins based on the strength of CLOCK:BMAL1 DNA binding. (C) Average nucleosome signal at the top 400 Rev-erbA DNA-binding sites (±1 kb) in mouse livers. (D–F) Average nucleosome signal at the top 25% of CLOCK:BMAL1 DNA-binding sites (±0.8 kb) located in gene bodies (D), intergenic regions (E), and TSSs (F) in mouse livers.
We next examined nucleosome signal at REV-ERBα DNA-binding sites. This protein is a circadian transcription factor, and its gene is a direct CLOCK:BMAL1 target. Like CLOCK:BMAL1, REV-ERBα rhythmically binds to DNA, with higher binding at the end of the light phase (Feng et al. 2011; Bugge et al. 2012; Cho et al. 2012). In contrast to CLOCK:BMAL1-binding sites, however, REV-ERBα sites had indistinguishable nucleosome signals at all time points. Moreover, the signals were not affected in Bmal1−/− livers despite a blunting of Rev-erbα transcription and expression in this genetic background (Fig. 1C; Supplemental Fig. 3; Kornmann et al. 2007; Menet et al. 2012). These results show that not all rhythmic transcription factors promote nucleosome removal and suggest that CLOCK:BMAL1 transcriptional regulation is special.
Interestingly, CLOCK:BMAL1-dependent nucleosome removal is more pronounced within gene bodies and intergenic regions than at transcription start sites (TSSs) (Fig. 1D–F; Supplemental Figs. 4, 5). Consistent with this observation are the nucleosome signals from Bmal1−/− liver chromatin, which are strongly dependent on the genomic location of the CLOCK:BMAL1 peaks: TSSs are depleted of nucleosomes similar to wild-type chromatin, whereas nucleosome depletion in gene bodies and intergenic regions is much less pronounced in Bmal1−/− mice. The low TSS nucleosome signal even in the absence of CLOCK:BMAL1 binding (e.g., during the night at ZT18 and ZT22 as well as in Bmal1−/− mice) is consistent with the literature; i.e., TSSs are generally more nucleosome-depleted because of intrinsic DNA sequence bias and/or the presence of the transcription machinery (Hughes et al. 2012; Iyer 2012; Thurman et al. 2012). In contrast, nucleosome removal at enhancers (within genes or at intergenic regions) is more dependent on cis-regulatory mechanisms such as sequence-specific transcription factors (Calo and Wysocka 2013). The data show that CLOCK:BMAL1 promotes nucleosome removal and opens chromatin more potently at enhancers than at TSSs.
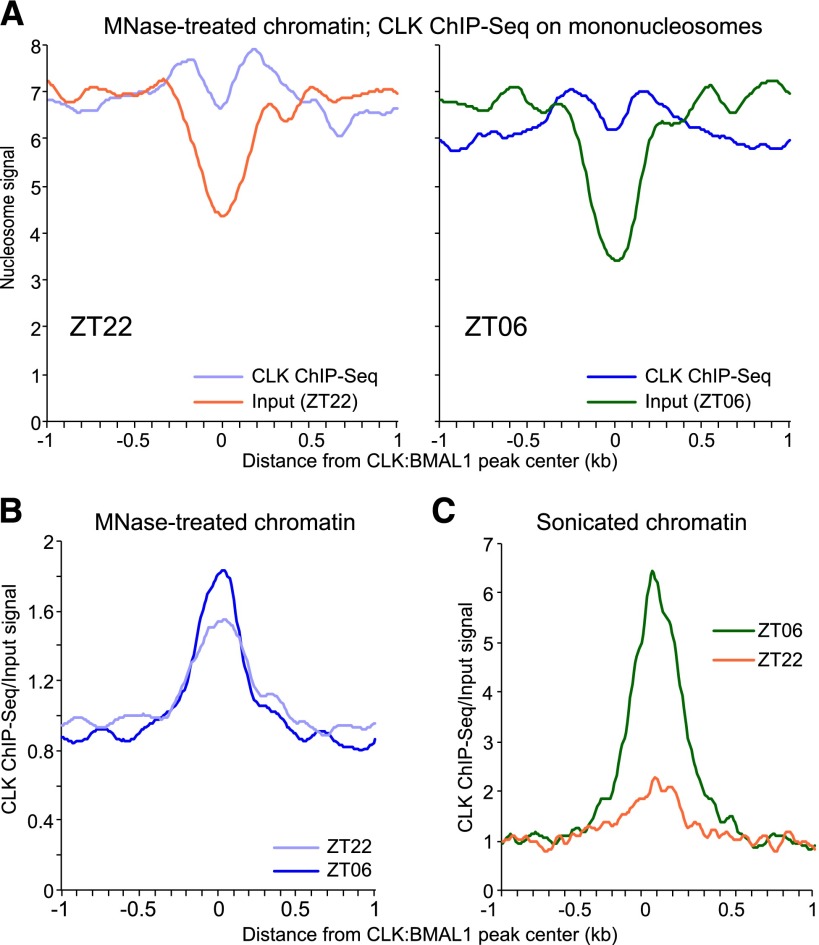
If a major role of CLOCK:BMAL1 is to open chromatin, how is this achieved? Nucleosomes present a physical barrier to most transcription factors and inhibit consensus sequence recognition (Magnani et al. 2011; Zaret and Carroll 2011; Dunham et al. 2012; Thurman et al. 2012). However, some developmental transcription factors, called pioneer factors, can bind to DNA within nucleosomes and promote histone repositioning and/or removal (Magnani et al. 2011; Zaret and Carroll 2011). To address whether CLOCK:BMAL1 has similar properties, we performed a CLOCK chromatin immunoprecipitation (ChIP) on mouse livers DNA digested by MNase at two different time points, ZT06 and ZT22 (Fig. 2; Supplemental Fig. 6). These time points are the times of maximal CLOCK:BMAL1 DNA binding and when CLOCK:BMAL1 initiates DNA binding, respectively (Rey et al. 2011; Koike et al. 2012); importantly, nucleosome signals at CLOCK:BMAL1 sites are still maximal at ZT22 (Fig. 1).
Figure 2.
CLOCK binds to DNA wrapped around nucleosomes. (A) CLOCK ChIP-seq signal on mononucleosome (i.e., mouse liver chromatin digested by MNase) at ZT22 (light blue; left) and ZT06 (dark blue; right) for the top 400 CLOCK:BMAL1 DNA-binding sites. The signal corresponds to the average of three independent ChIP-seq experiments. Input MNase-seq signal at the same binding sites is displayed for both time points (ZT22 [dark orange] and ZT06 [green]). (B) CLOCK ChIP-seq over input signal ratio on MNase-treated chromatin at ZT22 (light blue) and ZT06 (dark blue) at the top 400 CLOCK:BMAL1 DNA-binding sites. (C) CLOCK ChIP-seq over input signal ratio on sonicated chromatin at ZT22 (orange) and ZT06 (green) at the top 400 CLOCK:BMAL1 DNA-binding sites.
We found that CLOCK could immunoprecipitate mononucleosomes (Illumina libraries were size-selected to ensure an insert length corresponding to only one nucleosome) (see the Materials and Methods for more details), and the signal strikingly resembles those usually seen for histone modifications; e.g., two peaks equidistant from the DNA-binding site and separated by ∼400 base pairs (bp) (Fig. 2A). Importantly, CLOCK was associated with mononucleosomes at both ZT22 and ZT06, with only slightly higher signal/input ratio at ZT06 (Fig. 2B; Supplemental Fig. 7). To validate this result, we also performed a control CLOCK ChIP on the same mouse liver nuclei but sonicated rather than MNase-treated. As previously described (Rey et al. 2011; Koike et al. 2012), there is much stronger CLOCK binding to sonicated DNA at ZT06 than at ZT22 (Fig. 2C). The data therefore indicate that CLOCK:BMAL1 first binds to its target sites within nucleosomes (e.g., ZT22) and then promotes nucleosome removal to effect subsequent binding to naked DNA (e.g., ZT06).
Nucleosome dynamics include histone post-translational modifications and histone variants. Modifications often affect positively charged lysines and weaken histone:DNA interactions (Cosgrove et al. 2004; Henikoff 2008), and the H2A variant H2A.Z similarly weakens histone:DNA interactions and aids nucleosome repositioning by chromatin remodelers (Henikoff 2008; Jin et al. 2009; Ku et al. 2012; Li et al. 2012; Hu et al. 2013). To determine whether CLOCK:BMAL1-mediated nucleosome remodeling involves H2A.Z, we performed a H2A.Z ChIP from mouse liver chromatin across a light:dark cycle.
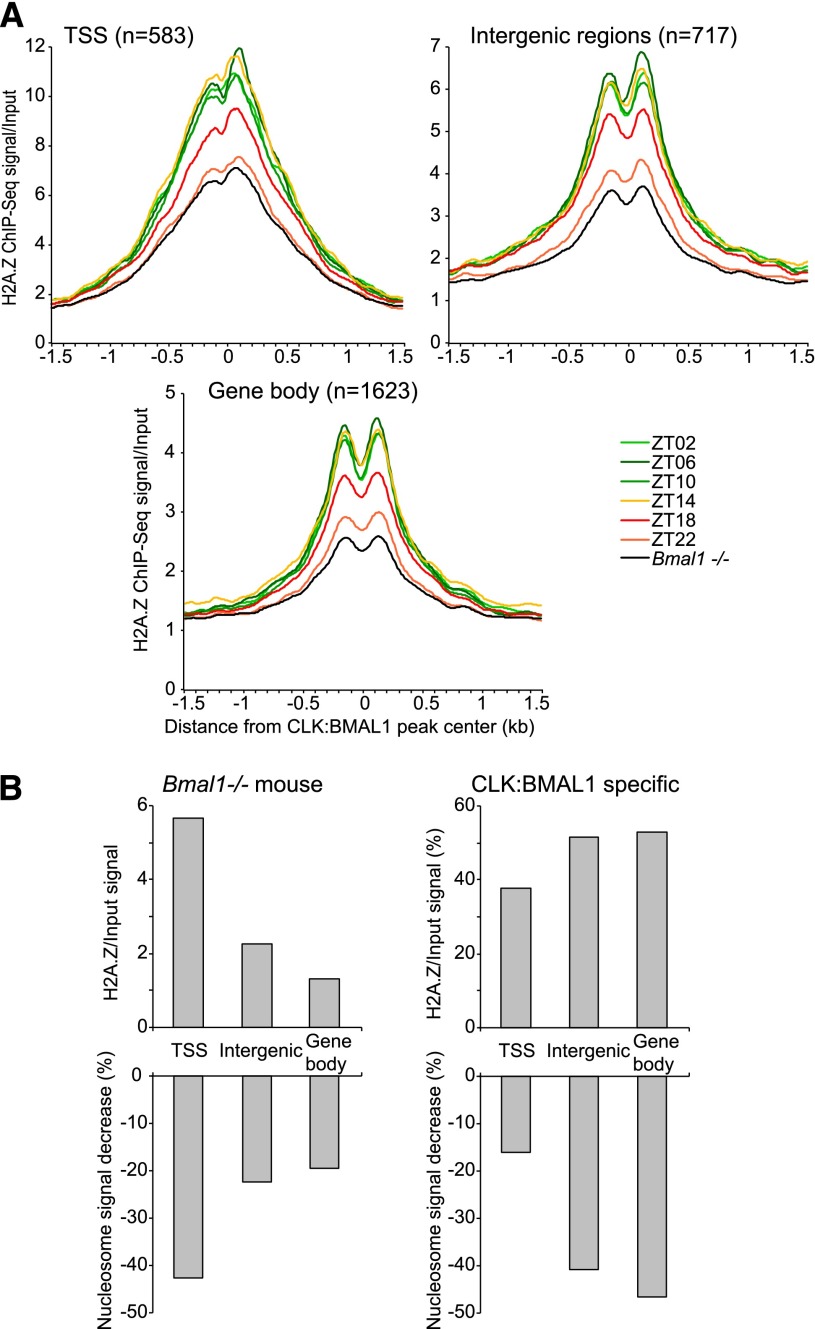
The H2A.Z signal is strongly rhythmic at CLOCK:BMAL1 DNA-binding sites: high from ZT02 to ZT14 and then decreasing during the night to reach a trough at ZT22 (Fig. 3A). Importantly, incorporation of H2A.Z is severely compromised in Bmal1−/− mice, and levels do not exceed the trough levels observed in wild-type mice at ZT22 (Fig. 3A; Supplemental Fig. 8). Furthermore, there is a striking correlation between H2A.Z signal and CLOCK:BMAL1-mediated decrease in nucleosome signal (Fig. 3B). In Bmal1−/− mice, a higher H2A.Z signal at TSSs is associated with a stronger decrease in nucleosome signal compared with intergenic regions or gene bodies (Figs. 1F, 3B). Conversely, a higher amplitude of H2A.Z signal occurs in wild-type mice within intergenic regions and gene bodies, which is associated with a stronger effect of CLOCK:BMAL1 on nucleosome removal (Figs. 1D,E, 3B). This is similar to what has been shown recently for FoxA2 during differentiation of embryonic stem cells into endoderm/hepatic progenitors (Li et al. 2012); namely, CLOCK:BMAL1 binding to nucleosomes promotes use of the histone variant H2A.Z and nucleosome removal.
Figure 3.
Rhythmic CLOCK binding on DNA is associated with rhythmic H2A.Z signal at CLOCK:BMAL1 DNA-binding sites. (A) H2A.Z ChIP-seq over input signal ratio on MNase-treated chromatin in wild-type mice during the light phase (green) and dark phase (orange/red) and in Bmal1−/− mice (average of six time points; black). Signal ratio is displayed at CLOCK:BMAL1 peaks located within gene bodies, intergenic regions, or TSSs. (B) Average H2A.Z ChIP-seq/input signal (top) and percentage of nucleosome signal decrease (bottom) at TSSs, intergenic regions, and gene bodies. (Left) Values in Bmal1−/− mice. (Right) CLOCK:BMAL1-specific contribution (e.g., values above those observed in Bmal1−/− mice).
Nucleosomes at CLOCK:BMAL1 DNA-binding sites have been recently shown to be rhythmically acetylated and methylated (H3K9ac, H3H27ac, H3K4me1, and H3K4me3) (Koike et al. 2012). Interestingly, the phases of H3K27ac and H3K4me3 rhythms are influenced by the genomic location of CLOCK:BMAL1 sites (Supplemental Fig. 9) and better match the phase of CLOCK:BMAL1 DNA binding at gene bodies and intergenic regions (from ZT0.4 to ZT12.5) than the later phase of DNA binding at TSSs (H3K27ac phase: ZT16.6; H3K4me3 phase: ZT14.9). As the gene bodies and intergenic sites are also the most potent sites of nucleosome removal (Fig. 1D–F), the data suggest that these rhythmic histone modifications partner with H2A.Z incorporation to help promote nucleosome removal, as proposed in other systems (Ku et al. 2012; Hu et al. 2013).
A simple model to explain the disconnect between the uniform phase of nucleosome removal at CLOCK:BMAL1 sites and the heterogeneous phases of transcription is that other transcription factors bind to the open chromatin at these sites. To address this possibility, we first analyzed 31 previously published ChIP-seq mouse liver data sets to identify transcription factors that bind close to CLOCK:BMAL1 sites (see the Materials and Methods). There is a significant coassociation of most transcription factors; i.e., transcription factor DNA-binding sites cluster in DNase I-hypersensitive sites (Fig. 4A; as described in Dunham et al. 2012; Thurman et al. 2012). This analysis also confirmed that nuclear receptors associate better with the circadian repressors Cry1 and Cry2 than with other clock components (Lamia et al. 2011; Koike et al. 2012). To extend this analysis, we also assayed the percentage of base-pair overlap between these 31 mouse liver ChIP-seq data sets. This analysis revealed that many liver transcription factors bind adjacent to CLOCK:BMAL1 DNA-binding sites. Some factors even overlap with >70% of CLOCK:BMAL1 sites; e.g., HNF4A, Bcl6, STAT5, CEBPA, or HNF6 (Fig. 4B).
Figure 4.
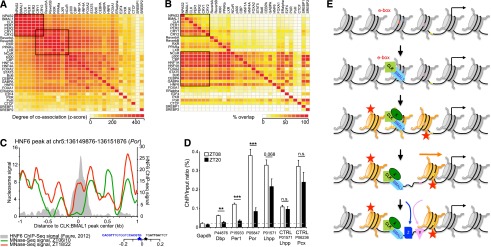
CLOCK:BMAL1-mediated rhythmic nucleosome removal promotes the rhythmic binding of transcription factors to DNA. (A) Coassociation between transcription factors in mouse livers. Thirty-one publicly available mouse liver ChIP-seq data sets were analyzed by pairs using the Genome Structure Correction statistic as previously described (Dunham et al. 2012). Black rectangles denote core clock genes and nuclear receptors (see the text for more details). (B) Percentage of overlap between 31 publicly available mouse liver ChIP-seq data sets. Black rectangles denote transcription factors that exhibit an overlap superior to 40% with core clock genes (see the text for more details). (C) Rhythmic nucleosome signal at a CLOCK:BMAL1 DNA-binding site located near HNF6 DNA-binding sites. Nucleosome signal is displayed for wild-type mice at time of high (average ZT6 and ZT10; green) or low (average ZT18 and ZT22; red) CLOCK:BMAL1 DNA binding. The HNF6 ChIP-seq signal from Faure et al. (2012) is shown in gray. Genomic locations of CLOCK:BMAL1 (blue) and HNF6 (black) consensus sequences are also displayed. (D) HNF6 ChIP-seq signal in mouse livers at ZT08 (white) and ZT20 (black) at several specific DNA-binding sites (n = 4 mice per time point). Values represent the average ± SEM. (**) P < 0.05; (***) P < 0.01. (E) Model illustrating a new mechanism by which CLOCK:BMAL1 regulates the expression of its target genes.
To compare transcription factor-binding dynamics adjacent to CLOCK:BMAL1 sites with nucleosome occupancy rhythms, we performed HNF6 ChIP assays at two opposite time points. Consistent with the model, HNF6 rhythmically associates with DNA-binding sites located close to a CLOCK:BMAL1 DNA-binding site (Fig. 4C). The amplitude of binding was surprisingly high (greater than fourfold for many sites), suggesting that HNF6 affects CLOCK:BMAL1 transcriptional output. Importantly rhythmic binding does not occur at control sites; e.g., HNF6 sites without nearby CLOCK:BMAL1-binding sites (Fig. 4D). Assuming that the activity of other transcription factors is similarly potentiated, their nature (e.g., activator or repressor) as well as their level of expression must contribute to the heterogeneity of CLOCK:BMAL1 target gene transcription quantitatively and qualitatively (Fig. 4E).
Taken together, our data indicate that CLOCK:BMAL1 functions like pioneer transcription factors and regulates the DNA accessibility of other transcription factors. Contrary to the permanent chromatin opening associated with lineage commitment, however, CLOCK:BMAL1-mediated chromatin opening is dynamic and occurs every day. This may reflect differences between the downstream physiologically relevant transcription factors like HNF6 and the downstream developmentally relevant factors that follow pioneer proteins.
This close parallel between CLOCK:BMAL1 and pioneer transcription factors can explain why ectopic expression of CLOCK in Drosophila leads to the development of functional ectopic clocks (Zhao et al. 2003; Kilman and Allada 2009): The chromatin of key core clock genes is opened like the chromatin of key developmental target genes. It is notable that a large number of additional CLOCK:BMAL1 target genes are probably not core clock genes but output genes. Many of them also undergo comparable nucleosome changes but are only marginally expressed. CLOCK:BMAL1 may therefore promote a rhythmically permissive state to accommodate circumstances not yet encountered, also reflecting the absence of one or more transcription factors under these conditions. Last, the suggested pioneer mechanism for CLOCK:BMAL1 provides a different perspective on the core circadian clock: Circadian feedback may directly control the temporal regulation of key target gene chromatin and only indirectly impact the transcriptional activation of many key core clock genes.
Materials and methods
Animals
Adult wild-type and Bmal1−/− mice (Bunger et al. 2000) entrained on a 12 h-light:12 h-dark schedule were used. All experiments were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the Brandeis Institutional Animal Care and Use Committee (IACUC; protocols no. 0809-03 and no. 12013).
Generation of MNase-seq libraries
Formaldehyde-cross-linked mouse liver nuclei, collected as previously described (Menet et al. 2012), were incubated with 1000 U/mL micrococcal nuclease for 15 min at 37°C before the reaction was stopped by addition of EDTA. These conditions resulted in ∼70%–80% mononucleosomes and ∼20%–30% dinucleosomes. Sequencing libraries were generated using 50 ng of DNA purified from the MNase-digested chromatin (Illumina TruSeq DNA sample prep kit) and size-selected to ensure an insert size of a mononucleosome. Four MNase-seq libraries per time point (except for Bmal1−/− ZT22, for which n = 3) were sequenced. Each library corresponds to one mouse.
ChIP
MNase-digested chromatin was immunoprecipitated using H2A.Z (Active Motif, 39113) or CLOCK antibody (Santa Cruz Biotechnology, sc-6927X). The number of mice used was as follows: for H2A.Z, four wild-type and two Bmal1−/− mice per time point (six time points), and for CLOCK, three wild-type mice per time point (ZT06 and ZT22)
Sonicated chromatin (Diagenode Bioruptor Plus) was immunoprecipitated using CLOCK or HNF6 (Santa Cruz Biotechnology, sc-13050X) antibody. A detailed protocol is provided as Supplemental Material.
Generation of ChIP-seq libraries
Sequencing libraries were generated from ∼10 ng of immunoprecipitated DNA using the Illumina TruSeq DNA sample preparation protocol with some changes: (1) Illumina TruSeq-indexed adapters were diluted to a 3:1 adapter:insert ratio in each ligation reaction, (2) ligation products were size-selected (inserts of ∼130–280 bp) prior to PCR amplification, and (3) libraries generated from MNase-digested chromatin were size-selected after the amplification (inserts of a mononucleosome).
Computational analysis
Analysis details are available as Supplemental Material. Sequences were mapped to the mouse genome (version mm9) using bowtie (Langmead et al. 2009). Only those that mapped uniquely to the mouse genome were used for further analysis. A summary of the alignment results is provided in Supplemental Table 2. Analysis of rhythmic expression was performed as previously described (Menet et al. 2012).
MNase-treated chromatin data sets
Sequences were expanded to 147 nucleotides (nt). Nucleosome signal was then retrieved at genomic locations of interest and normalized to the sequencing depth. These locations include CLOCK:BMAL1 binding (Koike et al. 2012), REV-ERBα binding (Feng et al. 2011; Cho et al. 2012), and DNase I-hypersensitive sites (Ling et al. 2010). The signal obtained for each library was averaged by time points.
Sonicated chromatin data sets
The CLOCK ChIP-seq signal was retrieved at genomic locations of interest, normalized to sequencing depth, and averaged for each library.
Coassociation analysis
The list of the publicly available mouse liver ChIP-seq data sets used in the coassociation analysis is provided as Supplemental Table 3 (see also the Supplemental Material). Coassociation analysis of transcription factor DNA-binding sites was performed using the algorithm described in the original ENCODE project paper (Dunham et al. 2012). The percentage of overlap (Fig. 4B), defined as the base-pair overlap ratio between two data sets, was calculated using the BEDTools suite (Quinlan and Hall 2010).
Additional information
All data sets are publicly available on Gene Expression Omnibus database at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=vreftasemesumjw&acc=GSE47145.
Acknowledgments
We thank Christopher Bradfield for kindly providing the Bmal1−/− mouse, Akhilesh Reddy for sharing unpublished information at an early stage of this project, Kevin Yip for helping with the analysis of coassociation of transcription factor DNA-binding sites, and Gung-Wei Chirn, Joseph Rodriguez, and Shuai Zhan for helping with the bioinformatics analysis. We are also grateful to Michael Marr, Nelson Lau, Sebastian Kadener, Christine Merlin, and Kate Abruzzi and other members of the Rosbash laboratory for helpful comments and discussions. This research was supported by the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.228536.113.
References
- Aguilar-Arnal L, Sassone-Corsi P 2013. The circadian epigenome: How metabolism talks to chromatin remodeling. Curr Opin Cell Biol 25: 170–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, Jager J, Lazar MA 2012. Rev-erbα and Rev-erbβ coordinately protect the circadian clock and normal metabolic function. Genes Dev 26: 657–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA 2000. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103: 1009–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, Wysocka J 2013. Modification of enhancer chromatin: What, how, and why? Mol Cell 49: 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, et al. 2012. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 485: 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove MS, Boeke JD, Wolberger C 2004. Regulated nucleosome mobility and the histone code. Nat Struct Mol Biol 11: 1037–1043 [DOI] [PubMed] [Google Scholar]
- Doi M, Hirayama J, Sassone-Corsi P 2006. Circadian regulator CLOCK is a histone acetyltransferase. Cell 125: 497–508 [DOI] [PubMed] [Google Scholar]
- Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, et al. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure AJ, Schmidt D, Watt S, Schwalie PC, Wilson MD, Xu H, Ramsay RG, Odom DT, Flicek P 2012. Cohesin regulates tissue-specific expression by stabilizing highly occupied cis-regulatory modules. Genome Res 22: 2163–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Lazar MA 2012. Clocks, metabolism, and the epigenome. Mol Cell 47: 158–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA 2011. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331: 1315–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S 2008. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet 9: 15–26 [DOI] [PubMed] [Google Scholar]
- Hu G, Cui K, Northrup D, Liu C, Wang C, Tang Q, Ge K, Levens D, Crane-Robinson C, Zhao K 2013. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell 12: 180–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Jin Y, Rando OJ, Struhl K 2012. A functional evolutionary approach to identify determinants of nucleosome positioning: A unifying model for establishing the genome-wide pattern. Mol Cell 48: 5–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VR 2012. Nucleosome positioning: Bringing order to the eukaryotic genome. Trends Cell Biol 22: 250–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G 2009. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet 41: 941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilman VL, Allada R 2009. Genetic analysis of ectopic circadian clock induction in Drosophila. J Biol Rhythms 24: 368–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS 2012. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338: 349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U 2007. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol 5: e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Jaffe JD, Koche RP, Rheinbay E, Endoh M, Koseki H, Carr SA, Bernstein BE 2012. H2A.Z landscapes and dual modifications in pluripotent and multipotent stem cells underlie complex genome regulatory functions. Genome Biol 13: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Papp SJ, Yu RT, Barish GD, Uhlenhaut NH, Jonker JW, Downes M, Evans RM 2011. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 480: 552–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Martelot G, Canella D, Symul L, Migliavacca E, Gilardi F, Liechti R, Martin O, Harshman K, Delorenzi M, Desvergne B, et al. 2012. Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles. PLoS Biol 10: e1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Gadue P, Chen K, Jiao Y, Tuteja G, Schug J, Li W, Kaestner KH 2012. Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell 151: 1608–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling G, Sugathan A, Mazor T, Fraenkel E, Waxman DJ 2010. Unbiased, genome-wide in vivo mapping of transcriptional regulatory elements reveals sex differences in chromatin structure associated with sex-specific liver gene expression. Mol Cell Biol 30: 5531–5544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani L, Eeckhoute J, Lupien M 2011. Pioneer factors: Directing transcriptional regulators within the chromatin environment. Trends Genet 27: 465–474 [DOI] [PubMed] [Google Scholar]
- Menet JS, Rodriguez J, Abruzzi KC, Rosbash M 2012. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. eLife 1: e00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohawk JA, Green CB, Takahashi JS 2012. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci 35: 445–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaich AK, Walker DA, Wolford R, Hager GL 2004. Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol Cell 14: 163–174 [DOI] [PubMed] [Google Scholar]
- Osumi N, Shinohara H, Numayama-Tsuruta K, Maekawa M 2008. Concise review: Pax6 transcription factor contributes to both embryonic and adult neurogenesis as a multifunctional regulator. Stem Cells 26: 1663–1672 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM 2010. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F 2011. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol 9: e1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. 2012. The accessible chromatin landscape of the human genome. Nature 489: 75–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truss M, Bartsch J, Schelbert A, Hache RJ, Beato M 1995. Hormone induces binding of receptors and transcription factors to a rearranged nucleosome on the MMTV promoter in vivo. EMBO J 14: 1737–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukai-Tadenuma M, Yamada RG, Xu H, Ripperger JA, Liu AC, Ueda HR 2011. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell 144: 268–281 [DOI] [PubMed] [Google Scholar]
- Vollmers C, Schmitz RJ, Nathanson J, Yeo G, Ecker JR, Panda S 2012. Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell Metab 16: 833–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss TC, Schiltz RL, Sung MH, Yen PM, Stamatoyannopoulos JA, Biddie SC, Johnson TA, Miranda TB, John S, Hager GL 2011. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell 146: 544–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS 2011. Pioneer transcription factors: Establishing competence for gene expression. Genes & Dev 25: 2227–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Kilman VL, Keegan KP, Peng Y, Emery P, Rosbash M, Allada R 2003. Drosophila clock can generate ectopic circadian clocks. Cell 113: 755–766 [DOI] [PubMed] [Google Scholar]