Abstract
Background/Aims
To identify the predicting factors of present hepatitis C virus (HCV) infection among patients with positivity for antibodies to HCV (anti-HCV).
Methods
We analyzed patients who showed positive enzyme immunoassay (EIA) results and performed an HCV RNA test as a confirmatory test at Kyung Hee University Hospital at Gangdong from June 2006 to July 2012. The features distinguishing the groups with positive and negative HCV RNA results were reviewed.
Results
In total, 490 patients were included. The results of the HCV RNA test were positive and negative in 228 and 262 patients, respectively. The index value of anti-HCV, mean age, platelet counts, total bilirubin, prothrombin time international normalized ratio, albumin and alanine transaminase (ALT) levels differed significantly between the two groups. On multivariable analysis, an index value of anti-HCV >10 [odds ratio (OR)=397.27, P<0.001), ALT >40 IU/L (OR=3.64, P=0.001), and albumin <3.8 g/dL (OR=2.66, P=0.014) were related to present HCV infection.
Conclusions
Although EIA is not a quantitative test, considering the anti-HCV titer with ALT and albumin levels may be helpful in predicting present of HCV infection.
Keywords: Hepatitis C, Hepatitis C Antibodies, Diagnosis, Immunoassay
INTRODUCTION
The hepatitis C virus (HCV) infection is a major public health problem.1 It is estimated that approximately 130-210 million individuals are chronically infected with HCV worldwide.2 In Korea, the prevalence of antibodies to HCV (anti-HCV) is assumed to be about 0.4-2.1%.3-4 As the incidence is increasing year by year, there is a growing interest in hepatitis C.5
As for the diagnosis of HCV infection, current guidelines indicate that patients suspected of having acute or chronic HCV infection should first be tested for anti-HCV by enzyme immunoassays (EIAs), commonly available screening tests.1 In patients with a positive anti-HCV test or who are immunocompromised or suspected of having acute HCV infection, an HCV RNA test should be performed to confirm HCV infection.1 By directly detecting HCV RNA, the HCV RNA test has the advantage of distinguishing the presence of active HCV infection from past (but resolved) infection as well as verifying the presence of anti-HCV.6 A recombinant immunoblot assay (RIBA) is the supplemental anti-HCV test with high specificity. A negative RIBA result indicates a false positive antibody test in most individuals with the exceptions of individuals in the acute early phase of infection and immunocompromised populations, while a positive RIBA result indicates past or current infection.
Although the high sensitivity and specificity of current EIAs for anti-HCV were estimated particularly in high-risk patient groups with chronic liver disease, false-positive results are more likely to occur among populations with a low risk of HCV infection.6,7 In addition, the detection of anti-HCV may be seen during the recovery period of HCV infection as well as during a period of transient clearance of HCV RNA in acute HCV infection.1 Therefore even if the EIA result for anti-HCV is positive, it cannot be diagnosed as HCV infection without a confirmatory test such as a HCV RNA test or RIBA.6
Due to the development of HCV treatment, there is a growing interest of sociology of health about diagnostic and therapeutic approach to asymptomatic patients with HCV infection. As a result, a screening test for anti-HCV is getting a lot. By the way, when the positive anti-HCV result comes out at the time of screening test, many patients are embarrassed particularly if they do not have risk factors for HCV infection or biochemical evidences of chronic liver disease. And their clinicians want to know the factors that may be helpful to predict a correct diagnosis while waiting the confirmatory test result. Accordingly, we conducted an observational study to determine the factors that can predict present HCV infection among the patients positive for anti-HCV.
PATIENTS AND METHODS
Study population
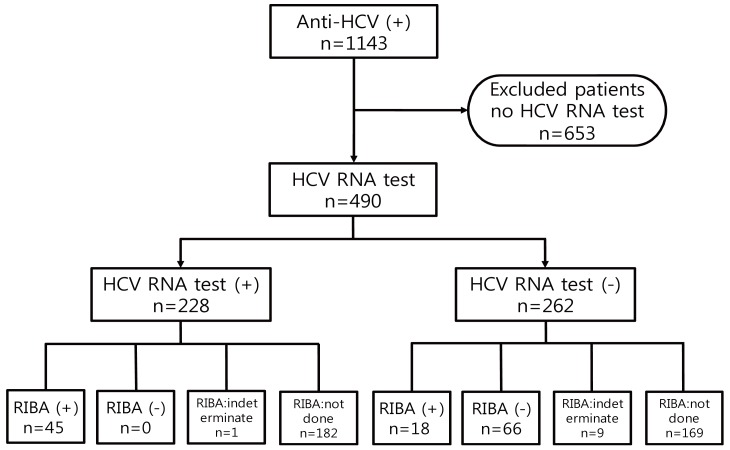
The subjects who attended the Kyung Hee University Hospital at Gangdong from June 2006 to July 2012 were investigated. We evaluated all patients who showed positive results of EIA for anti-HCV (index value of anti-HCV >1.00) and performed the HCV RNA test as confirmatory test. A total of 1,143 subjects were positive for anti-HCV EIA, 653 subjects who did not complete confirmatory test, including the patients already diagnosed with hepatitis C were excluded. Accordingly, 490 patients were included. We divided the patients into the positive HCV RNA group and the negative HCV RNA group (Fig. 1).
Figure 1.
Flow diagram of the patients. RIBA, recombinant immunoblot assay.
Methods
Enzyme immunoassay
The presence of anti-HCV was determined by an Advia Centaur HCV immunoassay (Siemens Healthcare Diagnostics, Tarrytown, NY, USA), the third generation version of EIA, using an indirect two wash sandwich immunoassay.8 It uses two recombinant HCV-encoded (c200 and NS5) antigens and one synthetic HCV-encoded core (c22) peptide. The c200 protein is derived from both the NS3 and NS4 sequences. The relative light units are used to calculate the index value from the master curve. Samples with an index value of <0.80, ≥0.80 but <1.00, and ≥1.00 were considered nonreactive, equivocal, and reactive for anti-HCV immunoglobulin G (IgG), respectively.
Recombinant immunoblot assay
The third generation immunoblot assay (LG HCD Confirm, LG Life Sciences, Seoul, South Korea) was performed for detection of anti-HCV using five recombinant antigens consisting of Core 14 (core), Core 518 (core/NS3), E1E2NS4 (E1,E2 membrane/NS4), KHCV897 (NS3), and NS5 1.2 (NS5).
HCV RNA test
HCV RNA was detected by the COBAS TaqMan HCV quantitative assay and the COBAS Amplicor HCV 2.0 qualitative assay (both from Roche Molecular Systems, Branchburg, NJ, USA).9,10 The lower detection limits of the each assay are 15 IU/mL and 50 IU/mL, respectively. All assays were performed according to the manufacturer's recommendations.
Clinical parameters and laboratory data
Age, gender, past medical history (including diabetes mellitus (DM), hemodialysis treatment, hemophilia, and HIV infection), index values of anti-HCV IgG, and laboratory test results (including platelet count, hemoglobin, total bilirubin, prothrombin time international normalized ratio (PT INR), albumin, alanine transaminase (ALT), and alkaline phosphatase (ALP) were reviewed retrospectively.
Statistical analysis
Chi-square test and Fisher's exact test were used to compare categorical data while continuous data were compared using a t-test. Platelet count, hemoglobin, albumin levels were divided by the lower limit of the normal range, and total bilirubin, PT INR, ALT, ALP levels were divided by the upper limit of the normal range for our laboratory. The cutting point for the index value of anti-HCV IgG was defined as 10 considering the reports of other researchers.11 A logistic regression test was applied to the variables with a P-value less than 0.10 in the univariate analysis. A two-sided P-value less than 0.05 was considered significant. Statistical analyses were performed using PASW Statistics for Windows, Version 18.0 (SPSS Inc. Chicago, IL, USA).
RESULTS
Clinical characteristics of study subjects
Among the 490 subjects positive for anti-HCV EIA, present infection was confirmed among 228 (46.5%) of the patients who showed positive HCV RNA test results and 262 (53.5%) of the patients revealed negative HCV RNA results. Sixty-six patients who showed negative RIBA results revealed false positive results of anti-HCV EIA. One hundred seventy-eight patients with negative HCV RNA results did not have present HCV infection but false positivity of the anti-HCV EIA test or past infection was not concluded because they revealed an indeterminate RIBA result or did not perform a RIBA. Among the 63 RIBA positive patients, HCV RNA was detected in 45 patients (71.4%), and 18 patients (28.6%) showed negative HCV RNA test results. These results mean that the former is present HCV infection, while the latter is past infection (Fig. 1).
Univariate analysis between the positive HCV RNA group and the negative HCV RNA group
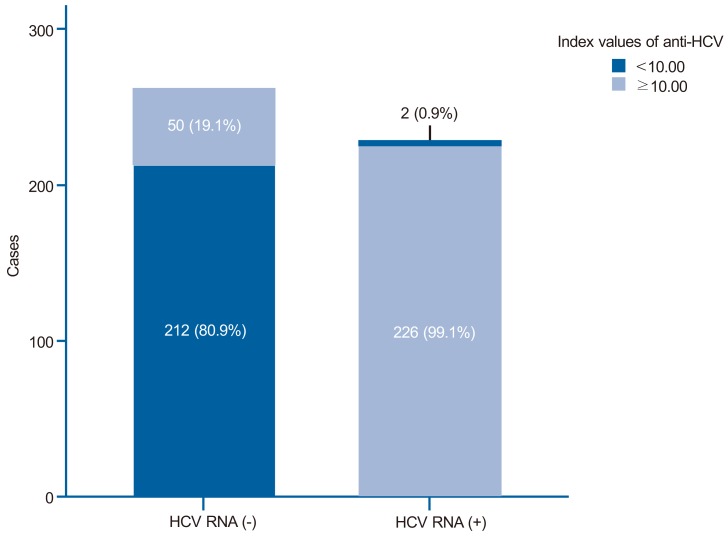
In the positive HCV RNA group, the mean age (59.4±14.5 vs. 53.6±20.9) was higher than in the negative HCV RNA group. Diabetes mellitus was more common in the positive HCV RNA group (20.6% vs. 14.9%), but statistically there was no significant difference between the two groups (P=0.096). Other comorbidities such as renal disease on hemodialysis, hemophilia, and HIV infection showed no differences between the two groups. Most of the positive HCV RNA group showed index values of anti-HCV IgG >10 (99.1%), while only 19.1% of the negative HCV RNA group showed index values of anti-HCV IgG >10 (Fig. 2). Low levels of platelet counts, albumin and high levels of total bilirubin, PT INR and ALT were frequently discovered in the positive HCV RNA group (P<0.05) (Table 1).
Figure 2.
Index values of anti-HCV immunoglobulin G in the positive and negative HCV RNA test groups. The index value of anti-HCV was >10 in 99.1% of those in the positive HCV RNA group but only 19.1% of those in the negative HCV RNA group.
Table 1.
Comparison of characteristics in the positive and negative HCV RNA groups among the enzyme immunoassay positive patients
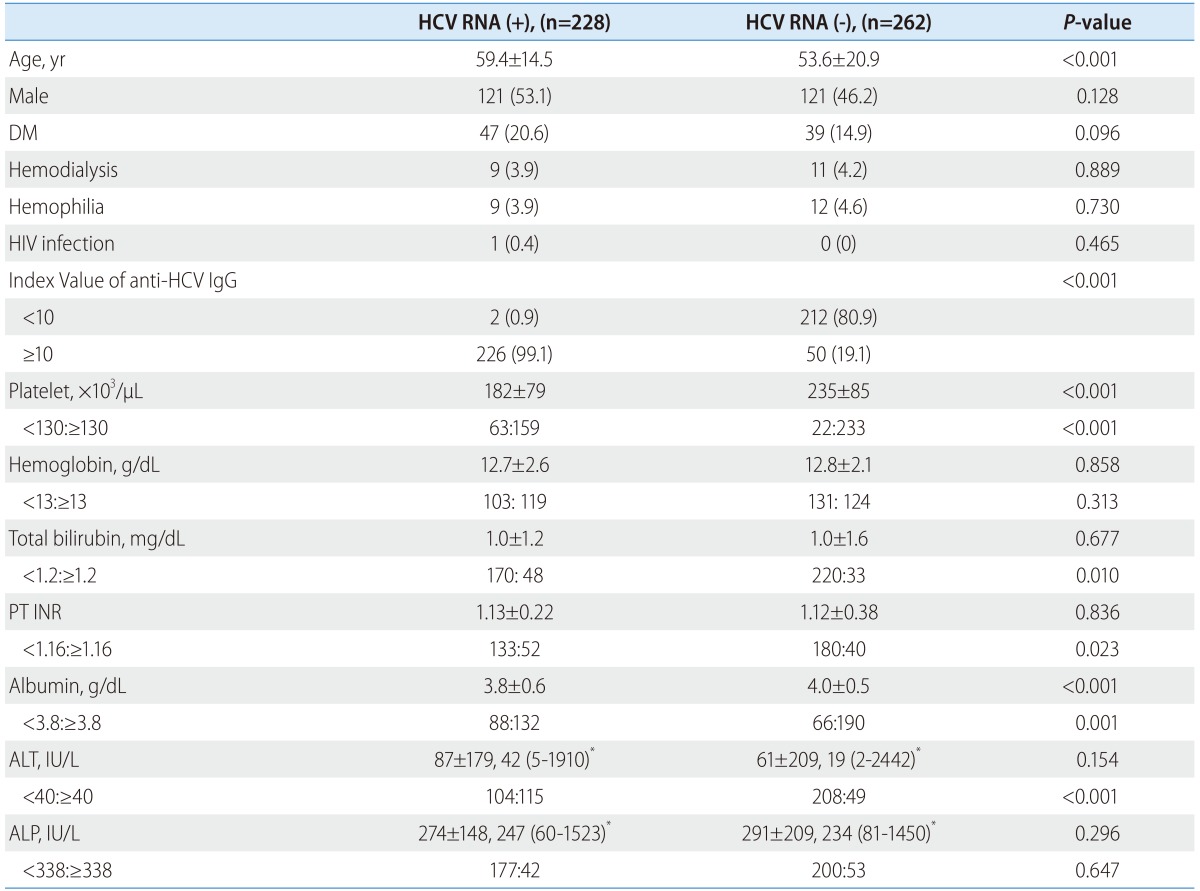
Data are presented as number (%).
IgG, immunoglobulin G; PT, prothrombin time; INR, international normalized ratio; ALT, alanine transaminase; ALP, alkaline phosphatase.
*Mean±SD and median (range).
Multivariable analysis of factors for predicting present HCV infection
According to a multivariable logistic regression analysis, a positive HCV RNA test result was independently associated with index values of anti-HCV IgG >10 (odds ratio: 397.27, P<0.001), ALT levels >40 IU/L (odds ratio: 3.64, P=0.001), and albumin levels <3.8 g/dL (odds ratio: 2.66, P=0.014) (Table 2).
Table 2.
Results of multivariable analysis of the predictive factors for positive HCV RNA test results
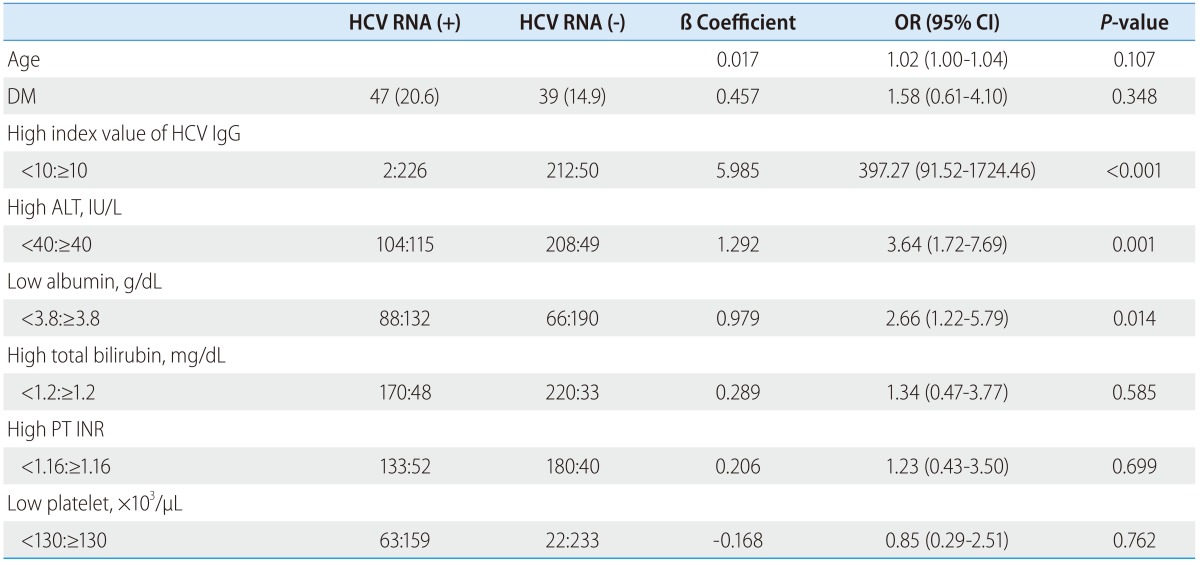
Data are presented as number (%).
IgG, immunoglobulin G; PT, prothrombin time; INR, international normalized ratio; ALT, alanine transaminase; OR, odds ratio; CI, confidence interval.
DISCUSSION
This study was conducted to identify the predictive factors associated with positive results of an HCV RNA test in the case of positive screening results of anti-HCV EIA. As a result, high index values of anti-HCV, high ALT and low albumin levels were related to the present HCV infection. These findings are consistent with the Center for Disease Control and Prevention's guidelines describing a good correlation between high signal cut-off (S/CO) ratios of anti-HCV and HCV infection.6 In several studies on the diagnostic efficacy of cut-off ratios of anti-HCV, it was suggested that the S/CO ratios of screening tests could be used to determine need for reflex supplemental testing.6, 11-13 Therefore if anti-HCV titer is low, the positive EIA result requires careful interpretation in order to diagnose present HCV infection. However, the diagnosis should not be made solely based on the S/CO ratios because negative HCV RNA test results occasionally can occur despite the high S/CO ratios of anti-HCV. Additionally, high ALT and low albumin levels were associated with the present HCV infection. It appears that the subjects with liver disease such as hepatitis or liver cirrhosis were included as part of the study population. Accordingly, in the case of a positive result of anti-HCV, considering the above findings comprehensively may be helpful to anticipate confirmative results during the screening stage.
In this study, the diagnostic accuracy of the EIA test cannot be dealt with because EIA negative patients were not included. But the HCV RNA positive rate among the subjects with positive results of EIA for anti-HCV was relatively low at about 46.5% (228/490). This seems to be due to the circumstance that this study was not performed focused on the high-risk patient population, and included the subjects with a low pretest probability of infection in a general hospital where anti-HCV test is routinely performed at admission, preoperational workup and so on.
The DM prevalence of 20.6% in the positive HCV RNA group was higher than that of the 14.9% in the negative HCV RNA group. It is correlated with the findings that HCV is associated with insulin resistance and DM through the complex interplay between host and viral factors such as the direct effects of HCV in modulating insulin signaling via core proteins and NS5A.14
As a retrospective observational study, this study has several limitations. First, in the case of positive anti-HCV EIA results and negative HCV RNA test results, a single negative HCV RNA result does not rule out active infection. During the period of low-level viremia in acute HCV infection, HCV RNA may be undetected. Though there is a possibility of the presence of acute HCV infection among the patients showing negative HCV RNA test results, we did not perform a follow-up HCV RNA test in all patients. Second, patients with negative anti-HCV and positive HCV RNA were not enrolled in the study because acute HCV infection could not be distinguished from chronic HCV infection within the clinical context. Also because anti-HCV negative patients were not included in this study, diagnostic value of anti-HCV titer >10 could not be evaluated. Third, in some patients RIBA was not performed supplementally due to various clinical situations, detailed diagnostic information could not be given in all patients.
In conclusion, although EIA is not a quantitative test, it is suggested that considering index values of anti-HCV with ALT and albumin levels may be helpful to predict present HCV infection.
Abbreviations
- ALP
alkaline phosphatase
- ALT
alanine transaminase
- anti-HCV
antibodies to HCV
- EIA
enzyme immunoassay
- HCV
hepatitis C virus
- IgG
immunoglobulin G
- INR
international normalized ratio
- PT
prothrombin time
- RIBA
Recombinant Immunoblot Assay
- RNA
ribonucleic acid
Footnotes
The authors have no conflicts to disclose.
References
- 1.Ghany MG, Strader DB, Thomas DL, Seeff LB. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology. 2009;49:1335–1374. doi: 10.1002/hep.22759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.European Association for the Study of the Liver. EASL Clinical Practice Guidelines: management of hepatitis C virus infection. J Hepatol. 2011;55:245–264. doi: 10.1016/j.jhep.2011.02.023. [DOI] [PubMed] [Google Scholar]
- 3.Park KS, Lee YS, Lee SG, Hwang JY, Chung WJ, Cho KB, et al. A study on markers of viral hepatitis in adults living in daegu and Gyungbuk area. Korean J Gastroenterol. 2003;41:473–479. [Google Scholar]
- 4.Na HY, Park MH, Park KS, Sohn YH, Joo YE, Kim SJ. Geographic Characteristics of Positivity of Anti - HCV and Chonnam Province: Survey Data of 6, 790 Health Screenees. Korean J Gastroenterol. 2001;38:177–184. [Google Scholar]
- 5.Kim SJ. Viral hepatitis surveillance system and statue of C hepatitis sentinel surveillance in Korea. Public Health Wkly Rep. 2012;5:214–219. [Google Scholar]
- 6.Alter MJ, Kuhnert WL, Finelli L. Guidelines for laboratory testing and result reporting of antibody to hepatitis C virus. Centers for Disease Control and Prevention. MMWR Recomm Rep. 2003;52:1–13. [PubMed] [Google Scholar]
- 7.Colin C, Lanoir D, Touzet S, Meyaud-Kraemer L, Bailly F, Trepo C. Sensitivity and specificity of third-generation hepatitis C virus antibody detection assays: an analysis of the literature. J Viral Hepat. 2001;8:87–95. doi: 10.1046/j.1365-2893.2001.00280.x. [DOI] [PubMed] [Google Scholar]
- 8.Denoyel G, van Helden J, Bauer R, Preisel-Simmons B. Performance of a new hepatitis C assay on the Bayer ADVIA centaur immunoassay system. Clin lab. 2004;50:75–82. [PubMed] [Google Scholar]
- 9.Vermehren J, Kau A, Gartner BC, Gobel R, Zeuzem S, Sarrazin C. Differences between two real-time PCR-based hepatitis C virus (HCV) assays (RealTime HCV and Cobas AmpliPrep/Cobas TaqMan) and one signal amplification assay (Versant HCV RNA 3.0) for RNA detection and quantification. J Clin Microbiol. 2008;46:3880–3891. doi: 10.1128/JCM.00755-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nolte FS, Fried MW, Shiffman ML, Ferreira-Gonzalez A, Garrett CT, Schiff ER, et al. Prospective multicenter clinical evaluation of AMPLICOR and COBAS AMPLICOR hepatitis C virus tests. J Clin Microbiol. 2001;39:4005–4012. doi: 10.1128/JCM.39.11.4005-4012.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oh EJ, Chang J, Yang JY, Kim Y, Park YJ, Han K. Different signal-to-cut-off ratios from three automated anti-hepatitis C virus chemiluminescence immunoassays in relation to results of recombinant immunoblot assays and nucleic acid testing. Blood Transfus. 2013;11:471–473. doi: 10.2450/2012.0085-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moretti M, Pieretti B, Masucci A, Sisti D, Rocchi M, Delprete E. Role of signal-to-cutoff ratios in hepatitis C virus antibody detection. Clin Vaccine Immunol. 2012;19:1329–1331. doi: 10.1128/CVI.00175-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stramer SL, Dodd RY, Brodsky JP. The value of screening signal-to-cutoff ratios for hepatitis C virus antibody confirmation. Transfusion. 2013;53:1497–1500. doi: 10.1111/j.1537-2995.2012.03955.x. [DOI] [PubMed] [Google Scholar]
- 14.Eslam M, Khattab MA, Harrison SA. Insulin resistance and hepatitis C: an evolving story. Gut. 2011;60:1139–1151. doi: 10.1136/gut.2010.228262. [DOI] [PubMed] [Google Scholar]