Abstract
Prostate cancer is the most prevalent cancer in males in developed countries. Tumor suppressor candidate 3 (TUSC3) has been identified as a putative tumor suppressor gene in prostate cancer, though its function has not been characterized. TUSC3 shares homologies with the yeast oligosaccharyltransferase (OST) complex subunit Ost3p, suggesting a role in protein glycosylation. We provide evidence that TUSC3 is part of the OST complex and affects N-linked glycosylation in mammalian cells. Loss of TUSC3 expression in DU145 and PC3 prostate cancer cell lines leads to increased proliferation, migration and invasion as well as accelerated xenograft growth in a PTEN negative background. TUSC3 downregulation also affects endoplasmic reticulum (ER) structure and stress response, which results in increased Akt signaling. Together, our findings provide first mechanistic insight in TUSC3 function in prostate carcinogenesis in general and N-glycosylation in particular.
In developed countries, prostate cancer is the most prevalent cancer in males and the second most common cause of cancer related death1. Despite PSA screening and availability of multiple therapeutic options a number of patients experience relapse in the form of hormone-refractory or castration-resistant prostate cancer (CRPC). These patients constitute a heavily pretreated and highly heterogeneous population and despite current advances in treatment of CRPC patients, the 5-year survival rate remains less than 5%. Molecular factors for onset and development of prostate cancers are largely elusive, although their number and characterizations are growing rapidly. Most prostate cancers harbor mutations or deletions of tumor suppressor genes, such as PTEN, SMAD4, MAGI2, CADM2 or TP532. Recently, a genome wide expression, mutation and copy number alteration analysis identified genomic loss on chromosome 8p as the most common genetic aberration in prostate cancers3, confirming earlier observations of chromosomal loss and loss of heterozygosity (LOH) in this region4,5. These losses include large regions of this chromosomal arm encompassing a multitude of genes including prostate cancer tumor suppressors, such as the homeodomain-containing transcription factor NKX3-1. TUSC3, originally named N33, was identified as a potential tumor suppressor gene on the chromosomal band 8p22 in prostate cancer in the mid 1990's5,6,7,8,9. Our previous work described high frequency of LOH on the short arm of chromosome 8 in ovarian cancer10 and specifically defined the prognostic influence11,12 and carcinogenic effects13 of TUSC3 loss in this cancer entity.
TUSC3 has been described and identified as a homologue of the yeast Ost3p subunit of the oligosaccharyltransferase (OST) complex14,15. OST is an integral membrane protein complex that catalyzes N-linked glycosylation of proteins in the endoplasmic reticulum (ER)16. TUSC3 mutations have been found in families with non-syndromic autosomal recessive mental retardation17,18,19,20. In analogy to this observation, several congenital disorders of glycosylation present phenotypically with variable degrees of mental retardation. N-glycosylation is a ubiquitous posttranslational modification of eukaryotic proteins that modulates protein folding, protects them from degradation, and regulates their function as well as their immunogenicity21. In general, glycosylation is involved in biological processes such as intercellular or cell-matrix interactions, which play an important role in cancer initiation and progression22,23,24. In PTEN driven prostate cancer, increase in N-glycosylation results in increased tumorigenicity due to the activity of an endoplasmic reticulum UDPase ENTPD525. Changes in protein glycosylation patterns lead to accumulation of unfolded or misfolded proteins in the endoplasmic reticulum and induce the unfolded protein response (UPR)26. UPR then facilitates cellular adaptation to ER stress by several distinct mechanisms in order to modulate the crosstalk between autophagy and apoptosis, and its deregulation might thus further contribute to carcinogenesis27,28.
So far, function of TUSC3 in neither N-glycosylation nor ER stress has been well characterized. In our work we present the first evidence of TUSC3 involvement in protein N-glycosylation and demonstrate the effects of TUSC3 loss on ER stress response in prostate carcinogenesis.
Results
TUSC3 interacts with the STT3B subunit of the oligosaccharyltransferase complex and affects N-glycosylation
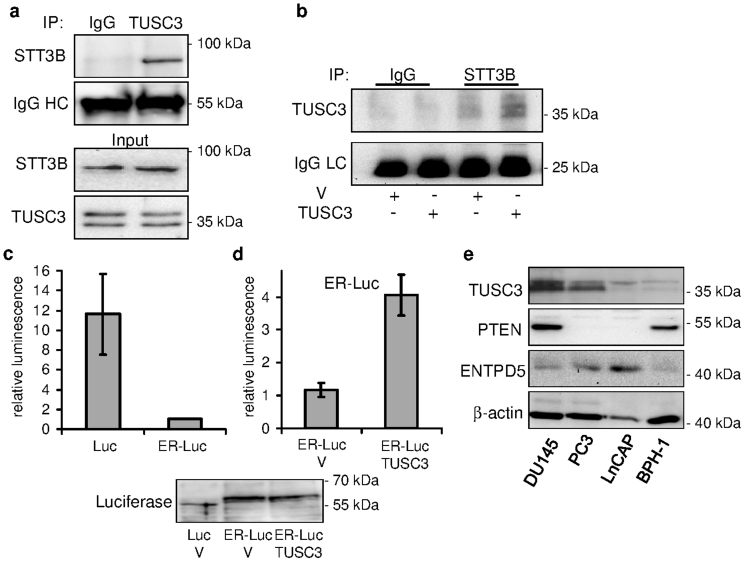
TUSC3 homologue Ost3p has been described as a subunit of the yeast OST complex responsible for OST substrate specificity and efficiency14,29. We could confirm the physical interaction between endogenous and exogenous human TUSC3, respectively, and the STT3B (Figures 1a and b), the core catalytic protein of the complex, by co-immunoprecipitation in HEK293T cells. In contrast, STT3A did not co-immunoprecipitate with TUSC3 in these cells (data not shown). To answer the question if and how TUSC3 regulates N-glycosylation within the OST complex, we employed a luciferase based assay described by Contessa et al30. In this assay, wild type firefly luciferase containing three N-glycosylation consensus sites is fused with the EGFR derived endoplasmic reticulum targeting sequence. N-glycosylation of the wild type firefly luciferase in HEK293T cells leads to a change in molecular weight in SDS-PAGE (Supplementary Figure S1a) and decreased enzymatic activity (Figure 1c). We used an overexpression approach to study the effects of TUSC3 on N-glycosylation of ER-luciferase (ER-Luc). Silencing would possibly lead to further inhibition of already decreased activity of the ER-Luciferase, making an evaluation difficult (Figure 1c). We overexpressed ER-Luc and wild-type TUSC3 in HEK293T cells and assessed the enzymatic activity of the ER-luciferase after 48 hours. We treated transfected HEK293T cells with 0.5 μM tunicamycin for 24 hours to show that deglycosylation results in a decrease in molecular weight of the ER-Luc (Supplementary Figure S1a). We observed an increase in luciferase activity in TUSC3 overexpressing cells compared to controls (Figure 1d), suggesting reduced N-glycosylation efficiency caused by TUSC3 overexpression, although not attaining the wild-type luciferase activity. TUSC3 overexpression in HEK293T cells also does not lead to a large band shift due to decreased glycosylation (Figure 1d), suggesting a small effect of TUSC3 on ER-luciferase N-glycosylation. This result might support the notion of a TUSC3 role in the regulation of N-glycosylation substrate specificity29.
Figure 1. TUSC3 associates with oligosaccharyltransferase subunit STT3B.
(a) TUSC3 co-immunoprecipitates with STT3B in HEK293T cells. Lysates from HEK293T cells were immunoprecipitated with TUSC3 antibody or control IgG and detected with STT3B antibody on a western blot. IgG heavy chain (IgG HC) represents the loading control. Endogenous input STT3B and TUSC3 levels are shown. (b) HEK293T cells were transfected with TUSC3 or an empty control pcDNA 3.1 vector (V). Lysates from transfected cells were immunoprecipitated using STT3B antibody or control IgG and TUSC3 was detected using western blot. IgG light chain (IgG LC) serves as loading control. TUSC3 overexpression affects ER-Luciferase N-glycosylation in HEK293T cells. (c) HEK293T cells were transfected with wild type firefly luciferase (Luc) or luciferase fused to an endoplasmic reticulum targeting sequence (ER-Luc) and seeded in 96-well plates. Luminesence was measured 32–48 hours after transfection and normalized to the cell count. (d) HEK293T cells were co-transfected with ER-Luc and empty pcDNA 3.1 vector (V) or full length TUSC3. Luminesence was measured and normalized to the cell count. The luciferase experiments are representative of several biological replicates and were performed in triplicates (depicted as means and standard deviations). Electrophoretic gel shift of the fusion construct (ER-Luc) in contrast to wild type luciferase (Luc) is shown. TUSC3 overexpression does not have an effect on the molecular weight of the ER-Luciferase. (e) A panel of cell lines derived from human prostate cancers (DU145, PC3, LNCaP) and benign prostatic hyperplasia (BPH-1) were assessed for expression of TUSC3, PTEN and ENTPD5. Cropped blots are depicted for clarity reasons and full-length blots will be provided upon request.
We also analyzed the expression of TUSC3 in three cell lines derived from human prostate cancer (DU145, PC3, LNCaP) and one cell line from benign prostatic hyperplasia (BPH-1). We can demonstrate varied levels of TUSC3 expression in these cell lines, with LNCaP and BPH-1 cells having lowest levels of TUSC3 expression. The ER UDPase ENTPD5 has been recently implicated in the pathogenesis of PTEN negative prostate cancer through ER stress modulation25, and for this reason we also assessed its expression. In contrast to the PTEN negative LNCaP cells, DU145 and PC3 cells display higher TUSC3 expression levels while their basal expression of ENTPD5 is relatively low (Figure 1e).
Knockdown of TUSC3 leads to increased proliferation and invasion of prostate cancer cells in vitro
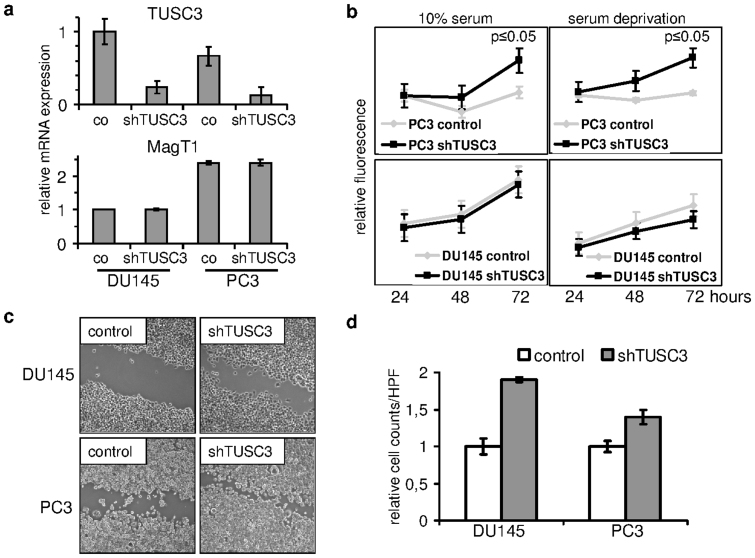
We were curious about the phenotypic effects of TUSC3 loss in a prostate cancer cell line model. We knocked down TUSC3 expression in two well characterized androgen resistant prostate cancer cell lines, PC3 and DU145, and analyzed them in vitro. Silencing efficiency of 5 short hairpin RNAs against TUSC3 was first tested in the HEK293T cell line (Supplementary Figure S1c). PC3 and DU145 cell lines were lentivirally transduced to achieve a considerable reduction of TUSC3 expression (Figure 2a). mRNA levels of the closest TUSC3 homologue, MagT1 remained unchanged, thus validating knockdown specificity (Figure 2a). Cell lines with silenced TUSC3 expression (shTUSC3) and cell lines transduced with scrambled shRNA (control) were assessed for proliferation using a resazurin based cell viability assay. TUSC3 silenced PC3 prostate cancer cell lines demonstrate a significant growth advantage over control cells, which is retained under conditions of cellular stress, such as serum deprivation. Interestingly, knockdown of TUSC3 in DU145 cells did not demonstrate similar effects (Figure 2b). Next, we tested if shTUSC3 cells were displaying increased migratory and invasive properties consistent with increased tumorigenicity. In a wound healing assay, 18 hours after wounding, both silenced cell lines show accelerated wound closure (Figure 2c). To assess invasion through extracellular matrix, we used a Matrigel transwell invasion assay using medium with 20% FCS as chemoattractant. In comparison to control cells, and corrected for migration through the 8 μm pore membrane, shTUSC3 cells demonstrate increased invasion through extracellular matrix (Figure 2d). These results suggest that TUSC3 loss might accelerate tumorigenesis in prostate cancer cells under specific circumstances.
Figure 2. TUSC3 loss promotes carcinogenesis.
(a) Prostate cancer cell lines DU145 and PC3 demonstrated shRNA mediated downregulation of TUSC3 mRNA (shTUSC3) versus scrambled shRNA (co) and unchanged expression of its closest homologue MagT1. (b) TUSC3 silenced cells proliferate in serum and under serum deprivation in vitro. Cells were plated in 96-well plates and allowed to adhere overnight, followed by addition of serum free or serum complemented medium. Cell viability for up to 72 hours was assessed in triplicates. (c) Cell motility was compared 18 hours after wounding of a DU145 and PC3 monolayer under conditions of serum deprivation. Representative images are shown in 200× magnification. (d) Invasion of prostate cancer cell lines through Matrigel was assessed in a transwell migration assay towards cell culture medium with 20% FCS as chemoattractant. Crystal violet stained cells from triplicate experiments were counted in 3 high power fields (HPF, 400× magnification) per well and normalized to migration without Matrigel. Results are depicted as means and standard deviations.
TUSC3 loss under serum deprivation promotes Akt activity
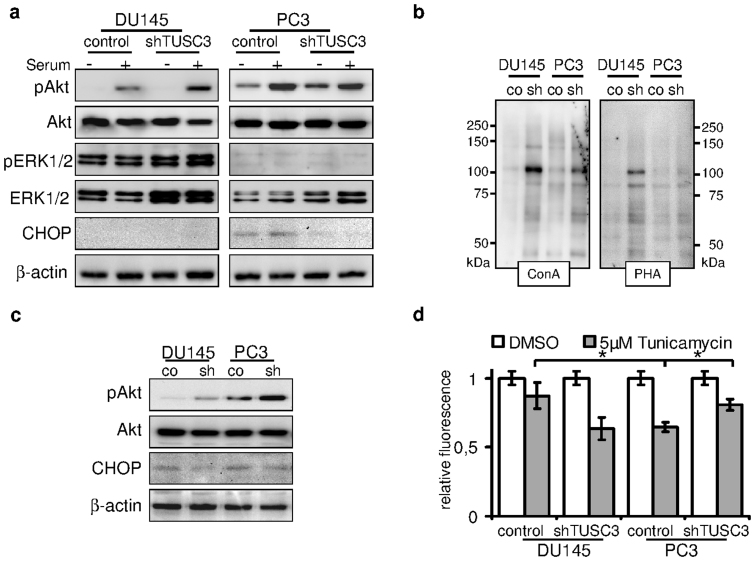
Growing cancer cells often encounter lack of nutrients leading to cellular stress response and adaptation, including induction of ER stress and autophagy. Interestingly, both cell lines continue to proliferate without serum for up to 72 hours (Figure 2b) and the difference between TUSC3 silenced cells and controls remains unaffected. Next, we looked at the downstream activation of MAPK and PI3K/Akt pathways as major drivers of cellular proliferation and survival in the two prostate cancer cell lines. For this purpose, we serum starved the prostate cells for 36 hours and stimulated growth factor activation with FCS for 30 minutes before lysis. We observe differential phosphorylation of ERK1/2 and Akt (Ser473) between the PC3 and DU145 cell lines as a result of their PTEN mutational status (Figure 3a). PTEN levels remain unaffected by TUSC3 status (Supplementary Figure S1d). MAPK pathway seems to be the predominant signaling pathway in the DU145 cell line as evidenced by ERK1/2 phosphorylation. In PTEN negative PC3 cells however, loss of TUSC3 further induces basal Akt activity in serum starved cells (Figure 3c).
Figure 3. TUSC3 loss leads to increased viability, N-glycosylation and Akt signaling.
(a) shTUSC3 and control cells were serum starved for 36 hours (−) and serum was added for 30 minutes before lysis (+). PI3K/Akt and MAPK signaling pathway were evaluated by immunoblotting. Increased downstream activation of Akt can be observed in serum starved TUSC3 silenced PC3 cells as well as in DU145 cells after stimulation with serum. ER stress and CHOP are induced by prolonged serum starvation (lane 5 and 6) in PC3 cells. Loss of TUSC3 decreases CHOP levels in PC3 cells (lane 7 and 8). (b) Increased N-glycosylation in shTUSC3 cells. Lectin blotting using Concanavalin A and Phytohaemagglutinin-L lectins on isolated cell surface proteins was performed in cell membrane fractions of PC3 and DU145 cells following 72 h serum starvation. Control for protein loading was performed by amido black staining (Supplementary Figure S2C). (c) DU145 and PC3 prostate cancer cell lines were serum starved for 72 hours before lysis. Silencing of TUSC3 (sh) leads to sustained phosphorylation of Akt and decreased expression of CHOP in both cell lines. (d) Viability of TUSC3 silenced (shTUSC3) and control (scrambled shRNA) prostate cancer cells was assessed with the CellTiter-Blue® Assay after treatment with 5 μM tunicamycin or DMSO for 72 hours in full medium. Experiments were performed in triplicates and results are representative of several independent experiments. * p = 0.01.
TUSC3 function within the OST complex may define distinct OST isoforms and confer target specificity16,29. In order to assess the level of global protein glycosylation in shTUSC3 cells, we employed lectin based assays using Concanavalin A and Phytohaemagglutinin-L. There was no difference in binding of these two lectins to glycosylated residues of isolated whole cell protein lysates or whole cells (Supplementary Figures S2a and b). To specifically analyze the N-glycosylation of cell surface proteins, we used cell surface biotinylation and purification. Surprisingly, within the isolated cell membrane fraction we could detect increased N-glycosylation of unspecified cell surface proteins in both cell lines (Figure 3b).
Given the possible alterations in N-glycosylation and ER stress induction through loss of TUSC3, we looked at a downstream target of UPR, the pro-apoptotic transcription factor CHOP (GADD 153). When we evaluated induction of CHOP in serum starved PC3 and DU145 cells at 36 h, we can see an increase of CHOP primarily in PC3 cells while we simultaneously observe a reduction of CHOP levels in PC3 shTUSC3 cells (Figure 3a). To induce physiological and long term ER stress induction in prostate cancer cells, we deprived them of serum for up to 72 hours. Under these conditions, sustained Akt phosphorylation in both shTUSC3 cell lines is present, although a stronger signal is observable in PC3 cells (Figure 3b). At this time point, we also observe CHOP induction in both cell lines. Repeatedly, this pro-apoptotic response is partly abolished in shTUSC3 cells. We were curious how loss of TUSC3 in prostate cancer cells may affect the ER stress induced cell death in general. We treated shTUSC3 and control cell lines with tunicamycin at concentrations ranging from 0.5 μM to 10 μM and evaluated their viability for up to 72 hours after induction of ER stress. IWe observed that at 5 μM tunicamycin, PC3 cell were generally more sensitive to ER stress than DU145 cells (Figure 3d) and that knockdown of TUSC3 significantly increased their resistance to tunicamycin (Figure 3d), while no difference was observed in DU145 cells. Taken together, we speculate that TUSC3 loss may control and increase Akt activity in serum deprived tumor cells especially under ER stress conditions.
TUSC3 loss alleviates endoplasmic reticulum stress in prostate cancer cells
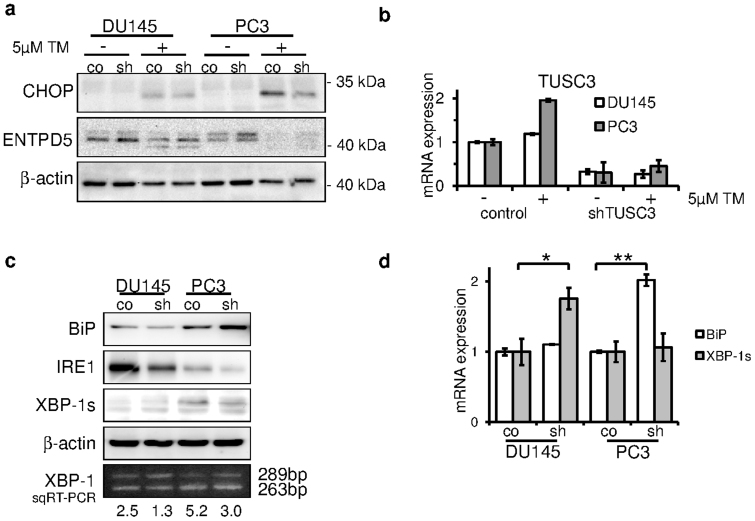
Disruption of the OST complex and N-glycosylation leads to accumulation of misfolded proteins in the ER lumen, which triggers the unfolded protein response and activation of pro-apoptotic PERK/CHOP as well the adaptive IRE-1/XBP-1 pathways. In resting cells, the ER stress receptors are inactivated through their association with the ER chaperone BiP(GRP78). We wondered, if TUSC3 loss may lessen the pro-apoptotic ER stress caused by tunicamycin treatment. Induction of CHOP following tunicamycin is markedly reduced in shTUSC3 PC3 cells (Figure 4a). Previously, induction of N-glycosylation by ENTPD5 expression was shown to alleviate ER stress in prostate cancer cells31. ENTPD5 expression is slightly increased in TUSC3 knockdown cells, consistent with adaptation to increased glycosylation. However, its expression is abolished upon tunicamycin treatment in PC3 cells, thus it is not sufficient to explain ER stress reduction in these cells.
Figure 4. TUSC3 alleviates ER stress in prostate cancer cell lines.
(a) CHOP and ENTPD5 expression was measured after 24 hours of 5 μM tunicamycin (TM) or DMSO treatment. Decreased CHOP induction in shTUSC3 PC3 cells indicates reduced ER stress response (lane 7 vs. 8). (b) Cells were treated with 5 μM tunicamycin (TM) or DMSO for 24 hours. TUSC3 mRNA expression was assessed using qRT-PCR. (c) UPR was assessed by expression levels of BiP, IRE-1 and XBP-1s in serum starved (72 h) prostate cancer cells. XBP-1 mRNA splicing was also measured by semiquantitative RT-PCR. Intensity of the bands was quantified and ratio of spliced to unspliced isoform was calculated. Increased ratio indicates elevated levels of ER stress. Cropped blots are depicted for clarity reasons and full-length blots will be provided upon request. (d) qRT-PCR was used to evaluate the mRNA expression of BiP and spliced XBP-1 isoform. Silencing of TUSC3 leads to an increase in BiP expression in PC3 cells and conversely to an increase in spliced XBP-1 in DU145 cells, suggesting opposing effects on UPR signaling.* p = 0.03 and ** p = 0.04.
To further examine the role of TUSC3 in ER stress response, we looked at the transcriptional regulation of TUSC3 expression under ER stress. Following a 24 h treatment with 5 μM tunicamycin, we measured a twofold induction of TUSC3 mRNA in PC3 cells (Figure 4b).
Next, in order dissect the responsible ER stress pathways and addressing adaptive as well as pro-apoptotic branches, we analyzed BiP, IRE-1 and XBP-1 expression in TUSC3 silenced cell lines after 72 h of serum starvation. Interestingly, BiP (GRP78) was recently identified as an essential factor for Akt activation in PTEN negative prostate cancer32. Interestingly, levels of BiP were elevated in shTUSC3 PC3 cells (Figure 4c) whereas the induction of IRE-1 and spliced XBP-1 was repressed. The effect was more pronounced in PC3 cells, although the general expression of IRE-1 was higher in DU145. This effect was confirmed using different methods. We performed a semi-quantitative and quantitative PCR of the XBP-1 splice variants as well as BiP. We find that serum starvation induces BiP mRNA expression predominantly in the PC3 shTUSC3 cell line (Figure 4d). In addition, a shift from spliced to unspliced, inactive isoform of XBP-1 can be detected upon downregulation of TUSC3, which is more pronounced in PC3 cells, indicating less ER stress induction (Figure 4c and d).
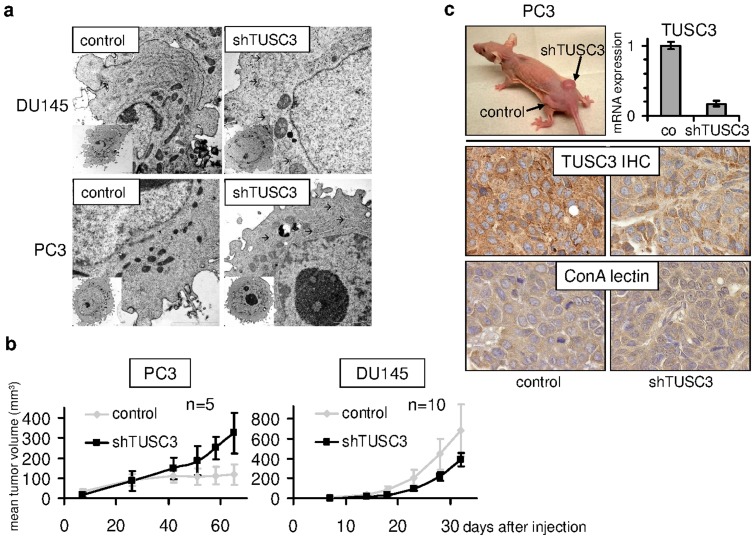
These observations prompted us to examine the ultrastructural changes of endoplasmic reticulum in TUSC3 silenced prostate cancer cell lines. Both, PC3 and DU145 cells deficient for TUSC3 displayed ER rearrangement consistent with cross-linked and slightly dilated ER cisternae when compared to controls. These effects on the endoplasmic reticulum may represent morphological correlates of disturbed ER function under ER stress conditions. In our analyses, only cells with clearly visible nucleus, nucleolus, mitochondria, Golgi apparatus and rough endoplasmic reticulum were evaluated and representative images are shown (Figure 5a).
Figure 5. TUSC3 silencing is associated with ultrastructural changes in endoplasmic reticulum.
(a) shTUSC3 and control (scrambled shRNA) PC3 and DU145 cells, respectively, were evaluated using transmission electron microscopy in ultrathin sections. Only cells with intact plasma membrane, nucleus, nucleolus, mitochondria, ER and Golgi apparatus were included into the analysis. We examined 20 cells of each cell type. Arrows indicate branching or dilatation of ER cisternae. Scale bars correspond to 2 μm. TUSC3 silencing promotes prostate cancer xenograft growth. (b) shTUSC3 and control (scrambled shRNA) PC3 and DU145 cells, respectively, were injected into the flanks of 6–8-week old male nude mice. Tumor growth was monitored and is shown as mean tumor size (+/− SEM). (c) Representative tumor formed by the TUSC3 silenced PC3 xenograft in the right flank of a Foxn1nu/nu mouse (top left). Sustained downregulation of TUSC3 in mouse tumors was analyzed by qRT-PCR (top right) and immunohistochemistry (TUSC3 IHC, middle graphs). Major differences in N-glycosylation in mouse tumors were not identified by Concanavalin A lectin histochemistry (ConA lectin, bottom graphs).
TUSC3 loss leads to increased tumor formation in vivo
To assess the influence of TUSC3 on in vivo tumorigenesis, we used a nude mouse xenograft model. Relatively small differences in proliferation, migration and invasion, which we observed in vitro, might have a summative effect in an in vivo model. We injected the modified shTUSC3 cell lines into the dorsal flanks of male nude athymic (Foxn1nu/nu) mice. The contralateral flanks were inoculated with control (scrambled shRNA) cells of the respective cell line. In vivo growth of TUSC3 silenced DU145 and PC3 cell lines did rather resemble the in vitro proliferation pattern, disregarding the TUSC3 mediated differences in extracellular matrix invasion and migration of the DU145 cell line. Tumor formation of shTUSC3 DU145 cells did not differ from their control counterparts while shTUSC3 PC3 xenografts developed a growth advantage over control cells after 6–7 weeks in vivo (Figure 5b). TUSC3 silencing was maintained in isolated mouse tumors, as evidenced by mRNA levels and histochemistry (Figure 5c). Not unexpectedly, pronounced changes in protein glycoslyation were not apparent by ConA lectin histochemistry of mouse tumors as suggested by the in vitro data (Figure 5c).
TUSC3 is downregulated in prostate cancer patients
We set out to identify and characterize a putative tumor suppressor gene in prostate cancer. Although we identified loss of TUSC3 expression through promoter hypermethylation in ovarian cancer11,12, TUSC3 has never been evaluated in a large prostate cancer patient cohort. In order to assess TUSC3 protein expression in clinical samples, we analyzed 143 prostate cancer patients using a tissue microarray. We observed strong cytoplasmic staining of TUSC3 in prostate cancer as well as normal tissues, consistent with its subcellular localization in the endoplasmic reticulum (representative stainings are shown in Supplementary Figure S3a). We found that 13.3% of tissue samples demonstrated starkly reduced protein expression of TUSC3 (assessed as absent and weak staining in more than 30% of cells, Supplementary Figure S3b). We could not detect significant differences in TUSC3 expression between the clinicopathological subgroups (Supplementary Table 1). The follow-up of this cohort was not sufficient to assess the influence of TUSC3 on progression-free or overall survival (Supplementary Table 1). Additionally, we performed a comprehensive in silico evaluation of TUSC3 gene expression, promoter methylation and gene copy number using publicly available datasets of The Cancer Genome Atlas (TCGA). We could define a large number of homozygous (11.8%) and heterozygous (42.9%) deletions of TUSC3, confirming a high percentage of 8p deletions in prostate cancer3 (Supplementary Figure S3b). Consistent with the data in ovarian cancer11,12, TUSC3 expression was significantly correlated with promoter hypermethylation as well as downregulated in higher stage tumors (Supplementary Figure S3c). This data further supports the notion of TUSC3 acting as a tumor suppressor in prostate cancer.
Discussion
Functional knowledge about the origins of altered glycosylation of tumor cell surface proteins is limited33,34. Deregulated enzymatic activities of proteins directly involved in N-glycosylation or the availability of potential glycosylation sites determined by the branching of N-glycans are considered to be crucial for these effects25,35. TUSC3 is the human homologue to S. cerevisiae Ost3p, a non-catalytic subunit of the OST complex15,29. Analyses of the Ost3p and its yeast paralogue Ost6p (human MagT1/IAP) demonstrated their function in regulating glycosylation substrate specificity29, glycosylation efficiency36 as well as their recently uncovered role in magnesium transport37. We find that mammalian TUSC3 binds the STT3B isoform of the catalytic subunit of the OST complex, but not STT3A. It is conceivable that TUSC3 modulates the function of STT3B, which has been described to have a salvage role in N-glycosylation, while the STT3A based complex is primarily responsible for the bulk of co-translational N-glycosylation15,38,39. This distinct role of STT3B complex might explain the relatively modest effect of TUSC3 overexpression on ER-Luc glycosylation and no global changes in lectin binding upon TUSC3 knockdown. In contrast to ENTPD5, which has been described as an oncogenic factor in PTEN negative prostate cancer25, TUSC3 might have a selective role in regulating N-glycosylation, affecting target specificity and OST efficiency. Moreover, ENTPD5 acts downstream of the OST complex, hydrolyzing UDP in the endoplasmic reticulum to promote further glycosylation and protein folding following OST mediated transfer of the core glycan on the nascent protein. Therefore it is conceivable that TUSC3 downregulation and ENTPD5 overexpression in prostate cancer may confer an additive effect on tumor growth.
Our finding of increased N-glycosylation of cell surface proteins being mediated by TUSC3 loss is somehow unexpected, as we previously described an opposite role for TUSC3 in ovarian cancer13. These differences might be attributable to a number of factors, such as different cancer model (prostate vs. ovarian), different expression system (knockdown vs. overexpression) as well as different lectins used (ConA and PHA vs. SNA). We believe that TUSC3 mediated N-glycosylation might exert several different functions in tumorigenesis depending on cancer type and genetic background. Integrins as well as growth factor receptors are likely candidates for N-glycosylated target proteins involved in carcinogenesis40,41,42, an our further efforts are aimed at the successful identification of a TUSC3 target protein in prostate and ovarian cancers. In our study, however, we cannot prove a direct effect of increased N-glycosylation caused by TUSC3 loss to prostate carcinogenesis.
We observe that ER structure is altered depending on TUSC3 status in prostate cancer cells and we detect an effect of TUSC3 on the unfolded protein response. UPR is triggered by endoplasmic reticulum stress caused by accumulation of misfolded proteins, which causes simultaneous activation of both adaptive and pro-apoptotic pathways. ER stress receptors are activated by binding of unfolded proteins to the BiP(GRP78) and its dissociation from IRE-1, ATF6 and PERK27. The pro-apoptotic response consists of the PERK mediated activation of the transcription factor CHOP. The adaptive pathway is primarily regulated by by IRE-1, a conserved UPR stress sensor, which splices XBP-1 into its active form26. We observe ER stress induction in both prostate cancer cell lines studied, although serum starved PTEN negative cells demonstrate generally higher levels of XBP-1 splicing and CHOP induction. This response is blunted in TUSC3 silenced cell lines and particularly in PTEN negative cells, which may result in a better adaptation to ER stress and growth advantage in vivo. We are aware that there might be further differences between the two prostate cancer cell lines than their PTEN status, but we have reason to believe that the PTEN/Akt pathway disruption is the main driver for increased ER stress in prostate cancer31. The importance of a crosstalk between ER stress response through BiP/GRP78 and Akt activation in prostate cancer pathogenesis has been demonstrated recently32. We speculate that TUSC3 might be responsible for regulation of N-glycosylation and UPR under ER stress conditions of serum starvation, leading to alteration of Akt signaling. The possibly deleterious effects of increased ER stress are ameliorated in PTEN negative cells, leading to a survival advantage.
In our previous work, we characterized the prognostic relevance of TUSC3 promoter hypermethylation in ovarian cancer11,12. TUSC3 promoter methylation has been described in prostatic intraepithelial neoplasia as well as prostate adenocarcinoma43. Here we offer further evidence for a loss of TUSC3 expression in two large cohorts of prostate cancer patients as well as indications for its epigenetic regulation through promoter hypermethylation.
We hypothesize that loss of TUSC3 in prostate epithelium may facilitate cancer initiation and progression under certain circumstances, such as PTEN loss. In conclusion, we propose that loss of TUSC3 in PTEN negative prostate cancer cells further enhances their carcinogenic potential by increased N-glycosylation of yet unknown target proteins and alleviation of ER stress and unfolded protein response.
Methods
Cell culture conditions and treatments
The cancer cell lines PC3, DU145, BPH-1, LNCaP, HEK293T were obtained from European Collection of Cell Cultures (ECACC) or American Type Culture Collection (ATCC). Cells were cultivated at 37°C in a humidified atmosphere with 5% CO2 in growth medium (HEK293T: DMEM; DU145, PC3: RPMI; all enriched with 10% FCS (fetal calf serum), 50 units ml−1 penicillin G, and 50 μg ml−1 streptomycin sulfate; LNCaP, BPH-1: RPMI enriched with 20% FCS (fetal calf serum), 50 units ml−1 penicillin G, and 50 μg ml−1 streptomycin sulfate. In addition, BPH-1 cells received 5 mg/ml transferrin, 5 ng/ml sodium selenite, 5 mg/ml insulin (Invitrogen, Carlsbad, CA, USA). Tunicamycin (T7765) was obtained from Sigma Aldrich (St. Louis, MO, USA) and reconstituted in DMSO.
Semiquantitative and quantitative reverse transcription-polymerase chain reaction
Total RNA was isolated from a confluent 6-well plate using RNAzol® RT (Molecular Research Center, Inc.). RNA concentration was measured on a Nanodrop 8000 and cDNAs were generated by reverse transcription of isolated mRNAs using SuperScript II RNase H-Reverse Transcriptase (Invitrogen) according to the manufacturer's instructions. Quantitative real-time PCR was performed using TaqMan® Gene Expression Assays by Applied Biosystems (Carlsbad, CA, USA) for TUSC3 (Hs00185147_m1), MagT1 (Hs00259564_m1), huHPRT (Hs99999909_m1) or SYBR® Green based assays for CHOP, BiP and XBP-1 using published primers and conditions44,45. For the semiquantitative determination of XBP-1 splicing, primers and conditions as described previously were used46.
Proliferation and cytotoxicity assays
Cells were plated at a density of 5 × 103 in 100 μl of culture medium in a 96-well plate and were left overnight to adhere. On the next day medium was aspirated, cells were washed once with 1× PBS, and appropriate medium containing 5 μM tunicamycin (Sigma Aldrich) or DMSO was added. Cells were then incubated for 24 to 72 hours. Cell viability was measured by CellTiter-Blue® Assay (Promega, Madison, WI, USA) according to the manufacturer's instructions.
Matrigel assay
50,000 cells were seeded in triplicates onto a 24-well plate with modified Boyden chambers (BD Biosciences, San Jose, CA, USA). The lower chamber contained 20% FCS/culture medium as a chemoattractant. Cells were incubated for 18 hours, stained with crystal violet and counted under a microscope.
Lentiviral transduction and transfection
Downregulation of TUSC3 in prostate cancer cell lines DU145 and PC3 was performed by lentiviral delivery using pLKO.1 vector containing TUSC3 shRNA (Open Biosystems, Thermo Fisher Scientific, Lafayette, CO, USA) and HEK293T packaging cell line. Transduced cells were selected and maintained in medium containing puromycin (3 μg/ml). ER-Luc, Luc and TUSC3 constructs were cloned into the pcDNA 3.1+ vector according to Contessa et al.30 and transfected into HEK293T cells using the CalPhos™ Mammalian Transfection Kit (Clontech, Saint-Germain-en-Laye, France).
Luciferase assay
Luciferase activity was measured using Promega's Luciferase Assay System according to the manufacturer's protocol. Measurements were performed on a Berthold TriStar LB 941 Multimode Microplate Reader (Berthold Technologies, Bad Wildbad, Germany). In experiments using tunicamycin at cytotoxic concentrations, the final values of bioluminescence were normalized to cell counts of treated wells.
Immunoblotting and co-immunoprecipitation
Cells were lysed with RIPA buffer supplemented with cOmplete Protease Inhibitor Cocktail Tablets (Roche Diagnostics, Mannheim, Germany) and PhosSTOP Phosphatase Inhibitor Cocktail Tablets (Roche Diagnostics). After incubation for 10 minutes on ice, cell lysates were cleared by centrifugation at 15,000 rpm for 10 minutes at 4°C and protein concentration was determined by Bradford absorbance assay (Sigma Aldrich). Equal amounts of protein lysates (40 μg) were separated by SDS-PAGE, blotted on PVDF membranes (GE Healthcare, Chalfont St. Giles, UK), incubated with the appropriate primary antibody and horseradish peroxidase (HRP)-conjugated secondary antibodies and detected with enhanced chemilumescence detection system (Pierce ECL Western Blotting Substrate, Thermo Scientific, Rockford, IL, USA). Following antibodies and dilutions were used: luciferase (1:500, AB3256, Millipore, Billerica, MA, USA), TUSC3 (1:500, ab65213, Abcam), actin (1:500, sc-1616, Santa Cruz Biotechnology, Santa Cruz, CA), phospho-ERK 1/2 (Thr202/Tyr204, 1:1000, #9101, Cell Signaling Technology, Danvers, MA, USA), ERK (1:1000, sc-93, Santa Cruz), phospho-Akt (Ser473, 1:1000, Cell Signaling #4060), Akt (1:1000, Cell Signaling #9272), PTEN (1:500, Cell Signaling #9552), ENTPD5 (1:500, HPA002927, Sigma Aldrich), CHOP (1:500, MA1-250, Thermo Scientific), IRE1α (14C10) (Cell Signaling #3294), BiP (C50B12) (Cell Signaling #3177), STT3B (1:500, 15323-1-AP, Proteintech, Chicago, IL, USA) and XBP-1s (D2C1F) (Cell Signaling #12782).
For co-immunoprecipitations, cells were lysed in PBS with 0.75% NP40 and cOmplete Protease Inhibitor as well as PhosSTOP Phosphatase Inhibitor. After incubation for 10 minutes on ice, cell lysates were cleared by centrifugation at 15,000 rpm for 10 minutes at 4°C. 1000 μg of the protein lysate were incubated with Protein A/G Sepharose beads for 1 hour at 4°C. Precleared lysates were then incubated overnight at 4°C with the respective antibody (TUSC3, ab65213, Abcam; STT3B, 15323-1-AP, Proteintech; normal rabbit IgG, sc-2027, Santa Cruz), followed by precipitation with protein A Sepharose (GE Healthcare) at 4°C for 1 hour. Beads were washed three times in lysis buffer, bound proteins separated with SDS/PAGE and visualized by western blotting.
Cell Surface Protein isolation
Selective isolation of the cell membrane proteins was performed using the Pierce Cell Surface Protein Isolation Kit according to the manufacturer's instructions (Thermo Scientific).
Immunohistochemistry
5 μm thick tissue sections were deparaffinized by heating at 60°C and subsequently rehydrated in xylene and graded alcohols. Antigen retrieval was performed with DEPP-9 epitope retrieval solution (EB-depp9-250, eubio, Vienna, Austria), followed by treatment with 0.3% H2O2 in PBS (pH 7.4) to quench endogenous peroxidase activity. After blocking with 10% secondary antibody host serum for 10 minutes, the sections were incubated in primary antibody (rabbit polyclonal to TUSC3, dilution 1:300, ab65213, Abcam, Cambridge, UK) for 1 hour at room temperature. Primary antibody dilutions were made in 10% secondary antibody host serum. The sections, after 2xPBS washes, were incubated in respective biotinylated secondary antibodies [biotinylated anti-rabbit IgG (BA-1000), Vector Laboratories, Burlingame, CA, USA], diluted 1:200 in 10% serum for 30 minutes at room temperature, followed by 45 minute incubation in StreptABComplex/HRP (K0377 Dako, Glostrup, Denmark). The sections were again washed twice with PBS and incubated in Dako Liquid DAB + Substrate-Chromogen System (K3468) until the development of brown color. This was followed by counterstaining with Meyer's hematoxylin, dehydration, and mounting using Eukitt medium.
Three tissue microarrays of prostate cancer from 143 patients were used to assess the TUSC3 protein expression. Each tissue microarray consisted of 3 biopsies of tumor material and 2 biopsies of adjacent benign tissue from each patient. Stained cells per 5 high-power fields were counted and grouped according to percentage of positive cells as < 10%, 10%–30%, and > 30%. The intensity of staining was determined on a scale of 0 to 3, with 0 for absent, 1 for weak, 2 for moderate, and 3 for strong. The microscopic assessment and quantification of the staining was performed independently by three investigators (R.H., M.A. and P.H.).
Lectin histochemistry and lectin blotting
5 μm thick paraffin embedded tissue sections and cell lines grown on glass chamber slides (Nunc, Langenselbold, Germany) were used. Lectin histochemistry and cytochemistry was performed using Concanavalin A (ConA) HRP conjugate in a concentration of 1:40 in PBS (USBiological, Swampscott, MA, USA) overnight at 4°C, followed by 2xPBS washes and visualized with Dako Liquid DAB + Substrate-Chromogen System (K3468). This was followed by counterstaining with haematoxylin, dehydration and mounting using Eukitt medium (O. Kindler GmbH, Freiburg, Germany).
Lectin blotting was performed after cell lysis in lysis buffer (1% Triton X-100, 20 mM Tris-HCl (pH 7.4), 150 mM NaCl, and 5 mM EDTA) containing protease and phosphatase inhibitors. Protein lysates were separated by SDS-PAGE, blotted on PVDF membranes (GE Healthcare) and blocked with PBST + 2% gelatin for 1 hour. Membranes were incubated 4 hours at 4°C with 2 μg/ml Concanavalin A (ConA) or Phytohaemagglutinin-L (PHA) HRP lectin conjugates and washed three times in PBST. Bound lectins were visualized with enhanced chemilunescence detection system (Pierce ECL Western Blotting Substrate).
Transmission electron microscopy
For electron microscopy analysis, cell lines were harvested from cell culture plates using trypsin-EDTA and suspended in RPMI medium. Cells were fixed in 3% glutaraldehyde with 0.2% tannin in 0.1 M cacodylate buffer for 1 hour and post-fixed in 1% OsO4 in the same buffer for 50 minutes. Cells were then washed three times in cacodylate buffer and then embedded in small blocks of 1% agar of 1 mm3 in size. Those blocks were then dehydrated in ascending series of ethanol (50%, 70%, 96%, and 100%), treated 2 × 10 minutes with 100% acetone and embedded in Durcupan resin (Sigma Aldrich). Ultrathin sections were prepared on LKB 8802A ultramicrotome, stained with OsO4 and examined on Morgagni 286(D) (FEI, Hillsboro, OR, USA) transmission electron microscope.
Mouse xenograft experiments
We treated 8-week old male, athymic Foxn1nu/nu mice, which were maintained under specific pathogen-free conditions. shTUSC3 DU145 (107 cells) and shTUSC3 PC3 (5 × 106 cells) prostate cancer cell lines as well as the same number of scrambled shRNA control cells were subcutaneously injected into the right and left dorsal flanks of nu/nu mice, respectively. Matrigel (BD Biosciences, San Jose, CA, USA) was used for the engraftment of PC3 cells and mixed with shTUSC3 and control cells in a 1:1 volume ratio. Tumor size was measured three times weekly in two perpendicular axes using a caliper. Tumor volume was calculated using the formula: volume = ½(length × width2). Animal experiments were carried out in adherence to protocols approved by the Austrian Federal Ministry for Science and Research and the Animal Ethics Committee of the Medical University of Vienna.
Statistical and in silico analysis
All statistical computations were performed using the SPSS software version 20 (SPSS Inc., Chicago, IL, USA). Comparison of means in normally distributed data was performed using Student's t-test, otherwise the nonparametric Mann-Whitney U test was applied. Pearson's correlation coefficient (r) was used to examine significant correlations. P-values of equal or less than 0.05 were considered statistically significant. All bar graphs are depicted using means and standard deviations as error bars, unless stated otherwise. TUSC3 gene expression, promoter methylation and gene copy number were analyzed using the Caleydo visualization software47 and publicly available TCGA (http://cancergenome.nih.gov/) datasets for prostate adenocarcinoma after obtaining a written permission from the TCGA Research Network.
Author Contributions
P.H., P.V. and M.K. designed the study and the experiments; P.H., E.T., P.V., K.K., M.A., M.M., C.E.L., M.G. and D.P. collected and assembled data in figures 1–4 as well as supplementary figures; P.H., K.K. and M.A. collected and assembled data in figure 5; P.H., E.T., P.V., K.K., M.A., M.M. and C.E.L. analyzed and interpreted the data; P.H. and E.T. wrote the manuscript; R.H. provided pathological support and review; M.S. supervised the animal experiments; M.K. provided financial support; all authors reviewed the manuscript.
Supplementary Material
Supplementary data
Acknowledgments
This work was supported by the Austrian Science Fund (FWF) [grant number P17891]; Funds of the Oesterreichische Nationalbank [project number 14109]; Initiative Krebsforschung; Centre for International Cooperation & Mobility of the Austrian Agency for International Cooperation in Education and Research [project number CZ 04/2012]; Ministry of Education, Youth and Sports of Czech Republic [project number 7AMB12AT019]. We thank Miroslava Sedláčková for cytological assessment and ER analysis, Dobromila Klemová, Andrea Schanzer, Maria König and Martina Hammer for expert technical support. We are grateful to Christopher J. Sweeney, William K. Oh, Massimo Loda and Rosina Lis for providing the prostate cancer tissue microarray.
References
- Jemal A. et al. Global cancer statistics. CA Cancer J. Clin. 61, 69–90 (2008). [DOI] [PubMed] [Google Scholar]
- Berger M. F. et al. The genomic complexity of primary human prostate cancer. Nature 470, 214–220 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor B. S. et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergerheim U. S., Kunimi K., Collins V. P. & Ekman P. Deletion mapping of chromosomes 8, 10, and 16 in human prostatic carcinoma. Gene. Chromosome. Canc. 3, 215–220 (1991). [DOI] [PubMed] [Google Scholar]
- Bova G. S. et al. Physical mapping of chromosome 8p22 markers and their homozygous deletion in a metastatic prostate cancer. Genomics 35, 46–54 (1996). [DOI] [PubMed] [Google Scholar]
- MacGrogan D., Levy A., Bova G. S., Isaacs W. B. & Bookstein R. Structure and methylation-associated silencing of a gene within a homozygously deleted region of human chromosome band 8p22. Genomics 35, 55–65 (1996). [DOI] [PubMed] [Google Scholar]
- Levy A., Dang U. C. & Bookstein R. High-density screen of human tumor cell lines for homozygous deletions of loci on chromosome arm 8p. Gene. Chromosome. Canc. 24, 42–47 (1999). [DOI] [PubMed] [Google Scholar]
- Arbieva Z. H. et al. High-resolution physical map and transcript identification of a prostate cancer deletion interval on 8p22. Genome Res. 10, 244–257 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke S. L. et al. High-resolution array CGH clarifies events occurring on 8p in carcinogenesis. BMC Cancer 8, 288 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribill I. et al. High frequency of allelic imbalance at regions of chromosome arm 8p in ovarian carcinoma. Cancer Genet. Cytogenet. 129, 23–29 (2001). [DOI] [PubMed] [Google Scholar]
- Pils D. et al. Five genes from chromosomal band 8p22 are significantly down-regulated in ovarian carcinoma: N33 and EFA6R have a potential impact on overall survival. Cancer 104, 2417–2429 (2005). [DOI] [PubMed] [Google Scholar]
- Pils D. et al. Methylation status of TUSC3 is a prognostic factor in ovarian cancer. Cancer 119, 946–954 (2013). [DOI] [PubMed] [Google Scholar]
- Vanhara P. et al. Loss of the oligosaccharyl transferase subunit TUSC3 promotes proliferation and migration of ovarian cancer cells. Int. J. Oncol. 42, 1383–1389 (2013). [DOI] [PubMed] [Google Scholar]
- Kelleher D. J. & Gilmore R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16, 47R–62R (2006). [DOI] [PubMed] [Google Scholar]
- Kelleher D. J., Karaoglu D., Mandon E. C. & Gilmore R. Oligosaccharyltransferase isoforms that contain different catalytic STT3 subunits have distinct enzymatic properties. Mol. Cell 12, 101–111 (2003). [DOI] [PubMed] [Google Scholar]
- Mohorko E., Glockshuber R. & Aebi M. Oligosaccharyltransferase: the central enzyme of N-linked protein glycosylation. J. Inherit. Metab. Dis. 34, 869–878 (2011). [DOI] [PubMed] [Google Scholar]
- Garshasbi M. et al. A defect in the TUSC3 gene is associated with autosomal recessive mental retardation. Am. J. Hum. Genet. 82, 1158–1164 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M. A. et al. A novel deletion mutation in the TUSC3 gene in a consanguineous Pakistani family with autosomal recessive nonsyndromic intellectual disability. BMC Med. Genet. 12, 56 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinari F. et al. Oligosaccharyltransferase-subunit mutations in nonsyndromic mental retardation. Am. J. Hum. Genet. 82, 1150–1157 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garshasbi M. et al. A novel nonsense mutation in TUSC3 is responsible for non-syndromic autosomal recessive mental retardation in a consanguineous Iranian family. Am. J. Med. Genet. A 155A, 1976–1980 (2011). [DOI] [PubMed] [Google Scholar]
- Ohtsubo K. & Marth J. D. Glycosylation in cellular mechanisms of health and disease. Cell 126, 855–867 (2006). [DOI] [PubMed] [Google Scholar]
- Helenius A. & Aebi M. Intracellular functions of N-linked glycans. Science 291, 2364–2369 (2001). [DOI] [PubMed] [Google Scholar]
- Freeze H. H. Genetic defects in the human glycome. Nat. Rev. Genet. 7, 537–551 (2006). [DOI] [PubMed] [Google Scholar]
- Hakomori S. Glycosylation defining cancer malignancy: new wine in an old bottle. Proc. Natl. Acad. Sci. U S A 99, 10231–10233 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M. et al. The ER UDPase ENTPD5 promotes protein N-glycosylation, the Warburg effect, and proliferation in the PTEN pathway. Cell 143, 711–724 (2011). [DOI] [PubMed] [Google Scholar]
- Hetz C., Martinon F., Rodriguez D. & Glimcher L. H. The unfolded protein response: integrating stress signals through the stress sensor IRE1alpha. Physiol. Rev. 91, 1219–1243 (2011). [DOI] [PubMed] [Google Scholar]
- Ozcan L. & Tabas I. Role of endoplasmic reticulum stress in metabolic disease and other disorders. Annu. Rev. Med. 63, 317–328 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H. P., Calfon M., Urano F., Novoa I. & Ron D. Transcriptional and translational control in the Mammalian unfolded protein response. Annu. Rev. Cell. Dev. Biol. 18, 575–599 (2002). [DOI] [PubMed] [Google Scholar]
- Schulz B. L. et al. Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency. Proc. Natl. Acad. Sci. U S A 106, 11061–11066 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contessa J. N. et al. Molecular imaging of N-linked glycosylation suggests glycan biosynthesis is a novel target for cancer therapy. Clin. Cancer Res. 16, 3205–3214 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Huang S., Fang M. & Wang X. ENTPD5, an endoplasmic reticulum UDPase, alleviates ER stress induced by protein overloading in AKT-activated cancer cells. Cold Spring Harb. Symp. Quant. Biol. 76, 217–223 (2011). [DOI] [PubMed] [Google Scholar]
- Fu Y. et al. Pten null prostate tumorigenesis and AKT activation are blocked by targeted knockout of ER chaperone GRP78/BiP in prostate epithelium. Proc. Natl. Acad. Sci. U S A 105, 19444–19449 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis J. W., Granovsky M. & Warren C. E. Glycoprotein glycosylation and cancer progression. Biochim. Biophys. Acta 1473, 21–34 (1999). [DOI] [PubMed] [Google Scholar]
- Fukuda M. Possible roles of tumor-associated carbohydrate antigens. Cancer Res. 56, 2237–2244 (1996). [PubMed] [Google Scholar]
- Lau K. S. et al. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell 129, 123–134 (2007). [DOI] [PubMed] [Google Scholar]
- Karaoglu D., Kelleher D. J. & Gilmore R. Functional characterization of Ost3p. Loss of the 34-kD subunit of the Saccharomyces cerevisiae oligosaccharyltransferase results in biased underglycosylation of acceptor substrates. J. Cell Biol. 130, 567–577 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H. & Clapham D. E. Mammalian MagT1 and TUSC3 are required for cellular magnesium uptake and vertebrate embryonic development. Proc. Natl. Acad. Sci. U S A 106, 15750–15755 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Canada C., Kelleher D. J. & Gilmore R. Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell 136, 272–283 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T. et al. STT3B-dependent posttranslational N-glycosylation as a surveillance system for secretory protein. Mol. Cell 47, 99–110 (2012). [DOI] [PubMed] [Google Scholar]
- Goel H. L., Li J., Kogan S. & Languino L. R. Integrins in prostate cancer progression. Endocr. Relat. Cancer 15, 657–664 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kariya Y. & Gu J. N-glycosylation of ss4 integrin controls the adhesion and motility of keratinocytes. PLoS One 6, e27084 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabius H. J. M. V. D. W., Andre S. & Villalobo A. Down-regulation of the Epidermal Growth Factor Receptor by Altering N-Glycosylation: Emerging Role of beta1,4-Galactosyltransferases. Anticancer Res. 32, 1565–1572 (2012). [PubMed] [Google Scholar]
- Kekeeva T. V. et al. [Abberant methylation of p16, HIC1, N33 and GSTP1 genes in tumor epitelium and tumor-associated stromal cells of prostate cancer]. Mol. Biol. (Mosk) 41, 79–85 (2007). [PubMed] [Google Scholar]
- Hirota M., Kitagaki M., Itagaki H. & Aiba S. Quantitative measurement of spliced XBP1 mRNA as an indicator of endoplasmic reticulum stress. J. Toxicol. Sci. 31, 149–156 (2006). [DOI] [PubMed] [Google Scholar]
- van Schadewijk A., van't Wout E. F., Stolk J. & Hiemstra P. S. A quantitative method for detection of spliced X-box binding protein-1 (XBP1) mRNA as a measure of endoplasmic reticulum (ER) stress. Cell Stress Chaperones 17, 275–279 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samali A., Fitzgerald U., Deegan S. & Gupta S. Methods for monitoring endoplasmic reticulum stress and the unfolded protein response. Int. J. Cell Biol. 2010, 830307 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit M., Lex A., Kalkusch M., Zatloukal K. & Schmalstieg D. Caleydo: connecting pathways and gene expression. Bioinformatics 25, 2760–2761 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data