Abstract
There is a general opinion that penile skin lined neovagina of transsexual women is not able to support the growth of lactobacilli. This study was undertaken to prove if lactobacilli strains could survive in neovagina and to characterise the most dominant Lactobacillus species. Sixty three male-to-female transsexual women without abnormal vaginal discharge, clinical signs of infection were recruited on an ongoing basis from among transsexual outpatients in an academic research institution and tertiary care centre. Neovaginal smears were taken for molecular Lactobacillus spp. profiling by denaturing gradient gel electrophoresis (PCR–DGGE). Lactobacillus species were detected from 47/63 transsexual women (75%). The 279 Lactobacillus signals detected by PCR-DGGE technique belonged to 13 different species. Lactobacilli of the L. delbrueckii group (L. gasseri, L. crispatus, L. johnsonii, L. iners, L. jensenii) were predominant. More than 90% of women harboured a combination of two or more neovaginal Lactobacillus species. In this study we report the frequent occurrence of lactobacilli from neovagina of transsexual women. Both, frequency and composition were similar to the normal lactic acid bacterial microflora in both women of reproductive age and postmenopausal women.
Lactobacilli as bacterial symbionts constitute the predominant organisms of a normal vaginal microflora. They have the ability to adhere to vaginal epithelia, inhibit the adhesion and growth of pathogens, deplete nutrients otherwise available to pathogens, and modulate the host immune response and microenvironment1,2. Burton et al.3 and Reid et al.4 have shown Lactobacillus crispatus, L. jensenii, and L. iners to be the most common vaginal species. Vásquez et al.5 and Antonio et al.6 found L. jensenii, L. crispatus, and L. gasseri to be the predominant Lactobacillus species in the vagina. In our recent study7, L. casei, L. paracasei, L. jensenii, and L. iners were the most frequently occurring species in the vaginas of postmenopausal women, with the vaginal microflora dominated by either a single or a combination of two Lactobacillus species.
The neovagina of transsexual women is a relatively new microbial research field, and information on its characteristics is scarce8,9. The transsexual neovagina is a skin-lined cavity without mucosa that does not communicate with internal pelvic organs. Considering relatively constant neutral pH, cyclic shifts in flora as a response to monthly hormone cycles, would not be expected. The physiology and pathophysiology of the neovagina, while still lacking in evidence, is certainly different than that of a natal vagina. However, understanding the microfloral composition could be an important prerequisite for the follow-up of transsexual women. Based on Gram stain, the majority of smears revealed a mixed microflora of aerobe and anaerobe species usually found on the skin, in the intestinal microflora, or in bacterial vaginosis (BV) and contained various amounts of cocci as well as polymorphous Gram-negative and Gram-positive rods8,9. lactobacilli were found in only up to 4% of transsexual women8,9. PCR-DGGE (Denaturing gradient gel electrophoresis) detection of lactobacilli species in transsexual women has not yet been performed.
There is a general opinion that penile skin lined neovagina is not able to support the growth of lactobacilli. In this study we used PCR-DGGE, a technique capable of detecting even difficult or not culturable bacteria, to assess the prevalence and characterise Lactobacillus species colonizing the neovagina of transsexual women.
Results
Between April 2011 and December 2011, a total of 63 male-to-female transsexual women of Caucasian origin were included in this study. The mean age of the participants was 41.2±13.1 years, mean height was 176.5±6.4 cm, mean weight was 82.3±19.9 kg, and mean interval since sex reassignment surgery (SRS) was 5.2±4.8 years.
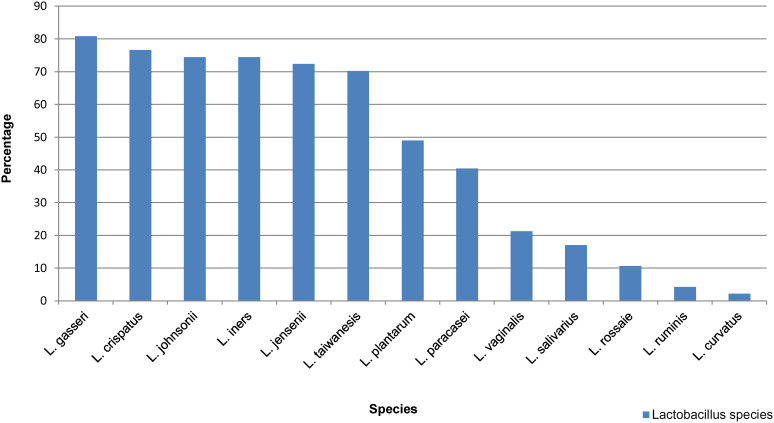
Using a PCR-DGGE based detection and identification strategy based on the primers Lac1-f and Lac-2r, it was possible to detect Lactobacillus species, which are difficult to detect or not detectable by cultural methods. In 75% (47 from 63) male to female transsexual a total of 279 Lactobacillus signals (DNA bands) presumptively corresponding to members of the Genus Lactobacillus were recognized. They belonged to 13 different species and the most frequently detected Lactobacillus species were L. gasseri (n = 38), L. crispatus (n = 36), L. johnsonii (n = 35), and L. iners (n = 35) (Figure 1). More than 90% of the 47 Lactobacillus - positive women harboured two or more species. Around 60% of transsexual women harboured the species L. gasseri, L. crispatus, L. johnsonii and L. iners at the same time.
Figure 1. Diversity and frequency of Lactobacillus species observed in neovagina of lactobacilli positive woman (n = 47).
Discussion
Previous studies have reported that the penile skin lined neovagina of transsexual women was dominated by a mixed microflora of aerobe and anaerobe species with very limited number of lactobacilli8,9. Our main research goal was to detect the lactobacilli from skin lined neovagina of transsexual women using PCR-DGGE, as a technique capable of detecting even difficult or not culturable bacteria and to assess the frequency of the most common neovaginal Lactobacillus strains in transsexual women. We verified that L. gasseri and L. crispatus were the most frequently detected neovaginal species, followed by L. johnsonii, L. iners, L. jensenii, and L. taiwanensis. The neovaginal microflora of the 47 participants in whom lactobacilli were detectable either contained a single or a combination of two or more Lactobacillus species.
In their study of the neovaginal microflora of transsexual women, Weyers et al. isolated lactobacilli in only 1 of 30 transsexual women after culture on 5 different media and through identification of the cultured isolates by tDNA-PCR8. The authors concluded that, although transsexual women have serum oestradiol levels comparable to those of substituted postmenopausal women, the neovaginal environment does not support the growth of lactobacilli. In another report, Weyers et al.9 used microscopic analysis of neovaginal cytology with papanicolaou (PAP) smear and found lactobacilli in 4% of transsexual women. In our study using a molecular detection technique (PCR–DGGE), we verified lactobacilli in 75% of transsexual women. The most important reason for these contradictory data is the different methodologies applied. The lacking reliability of cultural and phenotypic techniqes for the enumeration and species identification obviously constitutes a major problem in characterizing the vaginal Lactobacillus microbiota. Therefore molecular tools are necessary to overcome the drawbacks of the cultural and phenotypical techniques.
There are no previous reports about the normal Lactobacillus microflora of transsexual women. So far, data on what constitutes a normal Lactobacillus microbiota is derived from studies in pregnant, postmenopausal, or non-pregnant sexually active women of reproductive age3,13,14,15. Originally, the microbiota of healthy women of childbearing age had been believed to be dominated by L. acidophilus and L. fermentum, followed by L. brevis, L. jensenii, and L. casei with all subgroups14, and other species. More recently, molecular methods have shown L. crispatus, L. jensenii, and L. iners to be the most common isolates3,4. In our previous study13, L. crispatus and L. gasseri were the most frequent species in the vagina of pregnant women, followed by L. jensenii and L. rhamnosus.
To characterize the lactobacilli microbiota of transsexual women, we used the taxonomy proposed by Felis et al.15 Our results are in agreement with previous reports on the normal Lactobacillus microflora of women13,14,15, with almost the same lactobacilli species of the L. delbrueckii group (L. gasseri, L. crispatus, L. johnsonii, L. iners, L. jensenii) predominating the neovaginal microflora of transsexual women. Also, the current study applying PCR–DGGE showed that more than 90% of the transsexual women harboured two or more neovaginal Lactobacillus species. Surprisingly, about 25% of these women even had up to 8 different Lactobacillus species at the same time in neovagina. Referring to the applied analysis technique no quantification of the detected single species is possible in the sense of bacterial counts.
Reports about the absence of glycogen-rich epithelial cells and Lactobacillus binding sites in the neovagina that are usually upregulated by oestrogen in the normal vaginal mucosa8 open up a new discussion about the origin of lactobacilli in the penile skin lined neovagina.
Some pathogenic bacteria infecting the female urogenital tract emerge from the woman's own intestinal microbiota ascending along the perineum to the vagina. We assume the same mechanism in transsexual women. Several bacterial species are known to colonise both the gastrointestinal and the reproductive tracts. The rectum may play an important role as a source of organisms that colonise the vagina16, and the gut has been suggested as a reservoir for vaginal colonisation by Lactobacillus spp., thereby contributing to the maintenance of a normal vaginal microbiota17,18. El AiIa et al.19 isolated the same Lactobacillus species from both the vagina and the rectum in 37% of pregnant women. In our previous study7, 80% of pregnant women and 40% of postmenopausal women had the same lactobacilli species in both the vagina and rectum.
In summary, we assessed the prevalence and diversity of the Lactobacillus species colonizing the neovagina of transsexual women by molecular methods (PCR-DGGE). This research was undertaken to prove if penile skin lined neovagina of transsexual women could support the growth of lactic acid bacteria and not to explore the clinical relevance of lactobacilli in transsexual women using mentioned diagnostic techniques. In contrast to previous reports on up to 4% of transsexual women harbouring neovaginal lactobacilli, we found lactobacilli in 75% of women. Further investigations into the possible recto-neovaginal connection are warranted to explain the origin of lactobacilli in transsexual women with a penile skin linked neovagina.
Methods
This observational study was performed after approval of the ethics committee of the Medical University of Vienna (EK 982/2010) in accordance with the Declaration of Helsinki and the guidelines for Good Clinical Practice. Informed consent was obtained from all study participants prior to enrolment.
We observed male-to-female transsexual women without abnormal vaginal discharge, clinical signs of infection, neoplasia, or bleeding who were recruited on an ongoing basis from among transsexual outpatients of the Medical University of Vienna during standard routine follow-up examinations. Transsexual women with diarrhoea, constipation, rectal pathologies, including haemorrhoids, or urinary tract infection and patients having received antibiotic therapy in the previous 4 weeks or patients using vaginal probiotics were excluded from the study. All participants had undergone sex reassignment surgery (SRS) using the inverted penile skin flap technique10 at least 1 year before enrolment. All transsexual women were treated according to the Standards of Care of the World Professional Association of Transgender Health (WPATH)11.
From each participant, a smear from both lateral walls of neovagina was taken, transferred to transport medium (Copan innovation Italy), and sent to the University of Natural Resources and Life Sciences, Vienna, for further processing. The smears were examined by molecular profiling of lactobacilli using PCR-DGGE technique. Bands with a migration pattern different from the marker bands were excised followed by re-amplification and sequencing.
Examination of vaginal swabs
The cells of each swab were suspended in 1 ml sterile phosphate buffered saline (PBS) for 1 minute and the suspension was directly used for the extraction of bacterial DNA.
Bacterial strains and growth conditions
The DNA of the following bacterial type strains was applied for the preparation of an marker ladder in the PCR-DGGE analysis: L. plantarum (ATCC 14917), L. johnsonii (ATCC 33200), L. gasseri (ATCC 33323), L. jensenii (ATCC 25258), L. crispatus (ATCC 33820), L. salivarius subsp. salivarius (ATCC 11741), L. paracasei subsp. paracasei (ATCC 25302), and L. vaginalis (ATCC 49540). Lactobacilli were grown anaerobically (80% N2, 10% CO2, 10% H2) in MRS broth (Merck, Darmstadt, Germany) at 37°C. L. iners (LMG 19913) was cultured on Columbia blood agar plates (PB0123, Oxoid, Wesel, Germany) under microaerophilic conditions (GENbag microaer*, Biomérieux, Marcy l'Etoile, France) and for 72 h at 37°C.
DNA extraction
The peqGOLD Bacterial DNA Kit (peqlab, Erlangen, Germany) kit was used for the extraction of bacterial DNA. According to the instructions for Gram-positive bacteria, the DNA was extracted from the overnight cultures of lactobacilli and the swab suspensions described above.
Denaturing gradient gel electrophoresis (DGGE)
Within the isolated DNA, a specific region of the 16S rRNA gene was amplified by applying the primer combination Lac1-f: 5′-AGCAGTAGGGAATCTTCCA-3′ and Lac2r: 5′-ATTYCACCGCTACACATG-3′12. A GC-clamp (5′-CGC CCG GGG CGC GCC CCG GGC GGC CCG GGG GCA CCG GGG G -3′) was attached to the 5′-end of the reverse primer for the separation by denaturing gradient gel electrophoresis. The PCR reactions were carried out in a Mastercycler (Eppendorf, Hamburg, Germany) in a 0.2 ml reaction tubes format. Each PCR reaction mixture consisted of 10 μl DNA template, 1 μl of primer Lac1-f and Lac2r (10 pmol/μl for Lac1-f, 12.5 pmol/μl for Lac2r), (Eurofins MWG Operon, Ebersberg, Germany) 2.5 μl 10x PCR buffer (Finnzymes, Espoo, Finland), 0.5 μl dNTPs (10 mM, Carl Roth, Karlsruhe, Germany), 0.5 μl Dynazyme DNA polymerase (2 U/μl; Finnzymes, Espoo, Finland), and 9.5 μl sterile water. The amplification program was 95°C for 4 min, 35 cycles of 95°C for 30 s, 61°C for 40 s, 72°C for 1 min, and a final elongation step at 72°C for 5 min. The DCode™ Universal Mutation Detection System (BioRad, Munich, Germany) was applied for the DGGE according to the manufacturer's instructions. For the electrophoresis, an 8% polyacrylamide gel (37.5:1 acrylamide-bisacrylamide, Serva, Heidelberg, Germany) with a denaturing gradient of 35%–55% formed by urea and formamide was used. The gels were run in 1x TAE buffer at 60°C and 70 V for 16 hours. The separated bands were visualized by ethidium bromide staining, identified by comparing the banding pattern to a ladder consisting of bands originating from pure Lactobacillus type strains, and documented by digital photography (BioRad Gel Doc XR+).
DNA sequence analysis
DGGE bands that differed from the DGGE-marker applied for the identification were cut from the DGGE gel using a clean scalpel and incubated overnight in 1x PCR buffer at 4°C. The following amplification was carried out using the primers Lac1-f and Lac-2r (without GC-clamp)12. The PCR products obtained were purified using the PCRExtract Mini Kit (5 Prime GmbH, Hamburg, Germany) and sequenced (Eurofins MWG Operon, Ebersberg, Germany). The sequences were analysed with the BLASTn tool (http://blast.ncbi.nlm.nih.gov). A minimum sequence identity of 98% was chosen as criterion for species identification.
Statistics
Because this is the first study on lactobacilli isolation in transsexual women, no historical estimates for sample size calculations were available. We included all patients matching the inclusion criteria during one year. Demographic and background information is summarized and displayed using descriptive statistical techniques.
Main outcome measure
The main outcome measure was the molecularly detected diversity of Lactobacillus species in the neovagina of male-to-female transsexual women, including difficult to non-cultivable species.
Author Contributions
All authors fulfilled all conditions required for authorship. L.P. as a first author was responsible for organisation of the research and writing the paper. U.K. prepared the research, obtained clinical support and conducted the patient service. K.D. was microbiology researcher and writing the paper. M.K. provided microbiology research J.M. provided clinical support. W.K. was senior microbiology research supervisor and responsible for writing the paper. H.K. was senior author, responsible for co-organisation of the research and writing the paper.
Acknowledgments
The authors thank Gabriele Berghammer, the text clinic, for medical writing services.
References
- Erickson K. L. & Hubbard N. E. Probiotic immunomodulation in health and disease. J. Nutr. 130, 403–409 (2000). [DOI] [PubMed] [Google Scholar]
- Reid G., Cook R. L. & Bruce A. W. Examination of strains of lactobacilli for properties that may influence bacterial interference in the urinary tract. J. Urol. 138, 330–5 (1987). [DOI] [PubMed] [Google Scholar]
- Burton J. P., Cadieux P. A. & Reid G. Improved understanding of the bacterial vaginal microbiota of women before and after probiotic instillation. Appl Environ Microbiol. 69, 97–101 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid G., McGroarty J. A., Tomeczek L. & Bruce A. W. Identification and plasmide profiles of Lactobacillus species from the vagina of 100 healthy women. FEMS Immunol Med Microbiol. 15, 23–26 (1996). [DOI] [PubMed] [Google Scholar]
- Vasquez A., Jakobsson T., Ahrne S., Forsum U. & Molin G. Vaginal Lactobacillus flora of healthy Swedish women. J. Clin Microbiol. 40, 2746–2749 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonio M. A. D., Hawes S. E. & Hillier S. L. The Identification of Vaginal Lactobacillus Species and the Demographic and Microbiologic Characteristics of Women Colonized by These Species. J. Infect Dis. 180, 1950–1956 (1999). [DOI] [PubMed] [Google Scholar]
- Petricevic L. et al. Characterisation of the oral, vaginal and rectal Lactobacillus flora in healthy pregnant and postmenopausal women. Eur J. Obstet Gynecol Reprod Biol. 160, 93–9 (2012). [DOI] [PubMed] [Google Scholar]
- Weyers S. et al. Microflora of the penile skin-lined neovagina of transsexual women. BMC Microbiol. 9, 102 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyers S. et al. Cytology of the ‘penile' neovagina in transsexual women. Cytopathology. 21, 111–115 (2010). [DOI] [PubMed] [Google Scholar]
- Sohn M. & Bosinski H. A. Gender identity disorders: diagnostic and surgical aspects. J. Sex Med. 4, 1193–207 (2007). [DOI] [PubMed] [Google Scholar]
- Coleman E. et al. Standards of Care for the Health of Transsexual, Transgender and Gender-Nonconforming People, Version 7. Int J. Transgend. 13, 165–232 (2011). [Google Scholar]
- Walter J. et al. Detection of Lactobacillus, Pediococcus, Leuconostoc and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl Environ Microbiol. 67, 2578–85 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss H. et al. Vaginal Lactobacillus microbiota of healthy women in the late first trimester of pregnancy. BJOG. 114, 1402–1407 (2007). [DOI] [PubMed] [Google Scholar]
- Kandler O. & Weiss N. [Genus Lactobacillus.] Bergey's Manual of Systematic Bacteriology vol. 2, 9th edition. [Sneath, P.H. A., Mair, N. S., Sharpe, M. E. & Holt, J. G. (eds.)] [1063–1065] (Williams and Wilkins, Baltimore, 1986).
- Felis G. E. & Dellaglio F. Taxonomy of Lactobacilli and Bifidobacteria. Curr Issues Intest Microbiol. 8, 44–61 (2007). [PubMed] [Google Scholar]
- Antonio M. A., Rabe L. K. & Hillier S. L. Colonization of the rectum by Lactobacillus species and decreased risk of bacterial vaginosis. J. Infect Dis. 192, 394–398 (2005). [DOI] [PubMed] [Google Scholar]
- Reid G. et al. Oral probiotics can resolve urogenital infections. FEMS Immunol Med Microbiol. 30, 49–52 (2001). [DOI] [PubMed] [Google Scholar]
- Hilton E., Isenberg H. D., Alperstein P., France K. & Borenstein M. T. Ingestion of yogurt containing Lactobacillus acidophilus as prophylaxis for candidal vaginitis. Ann Intern Med. 116, 353–357 (1992). [DOI] [PubMed] [Google Scholar]
- El Aila N. A. et al. Identification and genotyping of bacteria from paired vaginal and rectal samples from pregnant women indicates similarity between vaginal and rectal microflora. BMC Infect Dis. 14, 167 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]