Abstract
In situ reduction of selenite to elemental selenium (Se(0)), by microorganisms in sediments and soils is an important process and greatly affects the environmental distribution and the biological effects of selenium. However, the mechanism behind such a biological process remains unrevealed yet. Here we use Shewanella oneidensis MR-1, a widely-distributed dissimilatory metal-reducing bacterium with a powerful and diverse respiration capability, to evaluate the involvement of anaerobic respiration system in the microbial selenite reduction. With mutants analysis, we identify fumarate reductase FccA as the terminal reductase of selenite in periplasm. Moreover, we find that such a reduction is dependent on central respiration c-type cytochrome CymA. In contrast, nitrate reductase, nitrite reductase, and the Mtr electron transfer pathway do not work as selenite reductases. These findings reveal a previously unrecognized role of anaerobic respiration reductases of S. oneidensis MR-1 in selenite reduction and geochemical cycles of selenium in sediments and soils.
Selenium is an important element for life and exhibits a redox activity in the environment. Selenium is released to environments either from weathering of Se-rich rocks1 (e.g., black shales, carbonaceous, limestones, carbonaceous cherts, mudstones, and seleniferous coal) or from anthropogenic sources of industrial and agricultural activities2. As a valence-variable element, selenium can exist in environments in multiple organic and inorganic forms, including ionic selenate or selenite, solid-state Se(0), and selenocysteine/selenoproteins3. Among these, selenite is the most toxic inorganic selenium4,5,6. The lifetime of selenite in soils is closely associated with the microbial activity7,8. In particular, the process of selenite reduction to Se(0) is of great significance for its bioremediation and geochemical cycles5,9,10,11.
A wide variety of microorganisms can reduce selenite under appropriate redox conditions12,13,14,15,16. The intracellular selenite reduction is usually driven by reduced thiols, e.g., glutathione, in microorganisms17,18. Selenite reacts with glutathione to form selenodiglutathione (GS-Se-SG), which can be further reduced by NADPH to unstable selenopersulfide (GS-Se−) in the presence of glutathione reductase. Then, dismutation of GS-Se− will produce GSH and Se(0). In addition to the thiol groups, terminal reductases for anaerobic respiration in some microorganisms may also reduce selenite as they are redox-reactive in cells. It is reported that two nitrite reductases and an inducible sulfite reductase are able to conduct selenite reduction in cells19,20,21. However, the possible involvement of other various respiration reductases in selenite reduction, as well as the physiological and ecological influence of this process to cells, has not been reported.
S. oneidensis MR-1 is a well-known dissimilatory metal-reducing bacterium with a unique respiration pattern. It possesses modular electron transport pathways and a large number of terminal reductases to respire ferric oxides, manganese oxides, nitrate, fumarate, sulfur, sulfur oxyanions, dimethyl sulfoxide, and trimetlylamine oxide22,23. Analysis of the S. oneidensis MR-1 genome sequence suggests that there is a highly diverse electron-transport system consisting of 42 putative c-type cytochromes, pivotal in reducing chromate, cobalt (III), vanadium (V), and uranium (VI)24,25,26,27. Meanwhile, flavins, including flavin mononucleotide (FMN) and riboflavin, secreted by S. oneidensis MR-1 have also been demonstrated to accelerate the bioreduction of extracellular electron acceptors28,29. These features make S. oneidensis MR-1 a perfect target to study the roles of respiration reductases in selenite reduction.
Selenite reduction in S. oneidensis has drawn a special interest for nanoparticle synthesis or selenium sequestration30. Selenite was reduced to Se(0) and deposited differently under aerobic and anaerobic conditions31,32. Taratus et al. reported that S. oneidensis mutants deficient in selenite reduction showed an impaired ability of anaerobic respiration33, implying a possible role of the anaerobic respiration system in selenite reduction. Here, we experimentally demonstrated the ingenious involvement of the anaerobic respiratory system of S. oneidensis MR-1 in selenite reduction. Mutants deficient in synthesizing reductases were tested to reveal the mechanism of synergy between anaerobic respiration and selenite reduction. Results suggest that fumarate reductase FccA contributed greatly to the selenite reduction in S. oneidensis MR-1. The Mtr cluster proteins used for solid iron (hydro) oxides respiration, as well as the nitrate/nitrite reductase, were not preferred selenite reductases in S. oneidensis MR-1 for selenite reduction.
Results
Outer membrane and extracellular respiratory pathway
Extracellular respiratory system is the most striking feature of dissimilatory metal-reducing bacteria like S. oneidensis MR-1. This strain contains at least two electron corridors to transfer intracellular electrons to extracellular metallic oxides34. The first one, constructed with periplasmic decaheme c-type cytochrome MtrA, outer-membrane cytochromes MtrC and OmcA, plus the integral protein MtrB, is recognized as the most important molecular apparatus in anaerobic respiration and extracellular bioelectrochemistry of S. oneidensis MR-135. Another one is composed of cytochromes MtrD, MtrE, and MtrF36. In this work, we tested and compared the selenite reduction activities of the wide type strain and the mutants deficient in individual genes for encoding Mtr proteins.
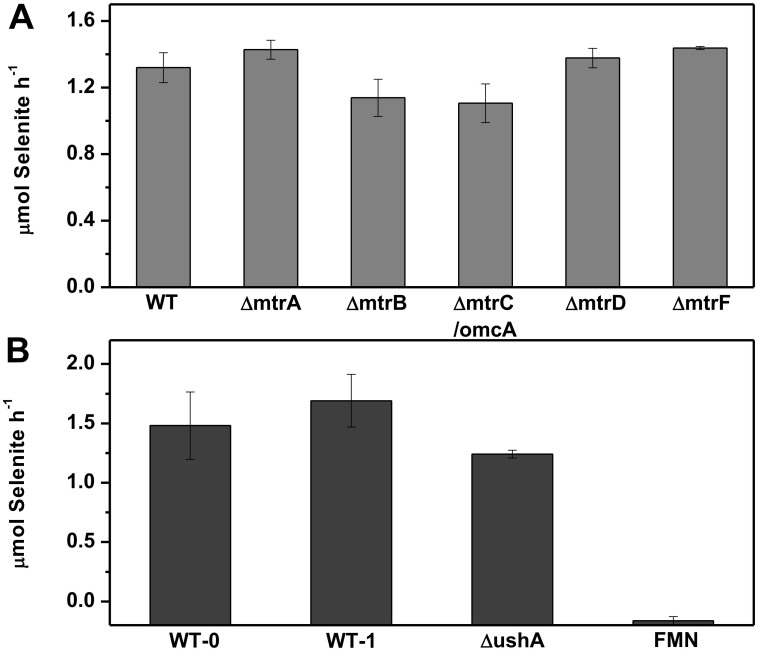
S. oneidensis MR-1 could easily reduce selenite and exhibited negligible adsorption to it (see Supplementary Fig. S1). More than 82% of selenite was reduced during 12 h. The reduced selenite fitted well with produced Se(0) in amount (Table 1). Thus, the decrease in selenite was used to quantify the selenite reduction. The selenite reduction rates of every strain were calculated over the initial linear portion of kinetics curve (12 h). As shown in Fig. 1A, the ΔmtrB and ΔmtrC/omcA exhibited slightly lower reduction rates than the wild-type. However, ΔmtrA and ΔmtrD showed a slightly enhanced selenite reduction. These results suggest that the extracellular respiratory proteins of S. oneidensis MR-1 might not be involved in the selenite reduction.
Table 1. Balance of elemental Se during S. oneidensis MR-1 wide type cells reducing selenite.
| 0 h | 24 h | 48 h | |||
|---|---|---|---|---|---|
| C0 (mM) | C24 (mM) | C24-C0 (mM) | C48 (mM) | C48-C0 (mM) | |
| Selenite | 0.5236 ± 0.0124 | 0.0612 ± 0.0131 | −0.4624 | 0.0295 ± 0.0014 | −0.4941 |
| Se(0) formed | 0.00 | 0.4248 ± 0.0112 | 0.4248 | 0.4999 ± 0.0128 | 0.4999 |
C0, C24, C48: the concentrations at 0 h, 24 h and 48 h. Values shown as mean value ± standard error.
Figure 1. Effect of extracellular respiratory system on selenite reduction by S. oneidensis MR-1.
(A) Selenite reduction rates of the wild-type strain and the extracellular respiratory proteins mutants. Both MtrA-MtrB-MtrC/OmcA and its functional analogue MtrD-MtrE-MtrF were examined for their contributions to selenite reduction. Periplasmic c-type cytochrome MtrA and MtrD were also studied here for integrity. Dual deletion of omcA and mtrC was constructed for their functional similarity in electron transfer. The reduction ability of all the mutants is expressed as the average rate (calculated over the linear portion of the kinetics curve, based on 4 time points) of selenite reduced in μmol h−1. The adjust (Adj.) R2 of each fitting were 0.9129, 0.8580, 0.9267, 0.9915, 0.9120, 0.8549. Error bars indicate the standard error of triplicate cultures; and (B) Effect of extracellular flavins to selenite reduction. FMN, instead of riboflavin, was dosed to the reduction culture of the wild-type and mutants, as MR-1 directly secreted FMN outside and riboflavin is in the form of hydrolyzate. The reduction ability is expressed as the average rate (calculated over the linear portion of the kinetics curve) of selenite reduced in μmol h−1.WT-0, wild-type without FMN dose; WT-1, wild-type with 1 μM FMN; ΔushA, ushA mutant without FMN dose; FMN, non-cells control with 10 μM FMN. The Adj. R2 of each fitting were 0.8640, 0.7816, 0.9978, 0.7962. Error bars indicate the standard error of triplicate cultures.
It is well known that S. oneidensis MR-1 secretes flavins, which drastically facilitate extracellular reduction of many chemicals, typically as metal oxides37. Secreted FMN, together with its hydrolyzed product, riboflavin, could transfer electrons from out-membrane c-type cytochromes to extracellular electron acceptors28. To explore the possible effect of extracellular flavins on selenite reduction, 1 μM FMN, a concentration close to the level naturally secreted by cells under anaerobic conditions28, was dosed to the culture for selenite reduction. It was observed that the dose of FMN only slightly increased the selenite reduction rate (Fig. 1B). The non-cell control with even 10 μM FMN showed no selenite reduction, excluding the direct reduction of selenite by FMN in the absence of cells. To further study the possible contribution of FMN to selenite reduction by S. oneidensis MR-1, a ushA mutant (ΔushA) with impaired ability to secrete FMN38 was used to test its ability to reduce selenite. As shown in Fig. 1B, ΔushA reduced selenite at a similar rate comparable to the wild-type strain. These results suggest that the extracellular FMN at the concentrations naturally produced by S. oneidensis MR-1 had no contribution to the selenite reduction.
Involvement of the terminal reductases in periplasm
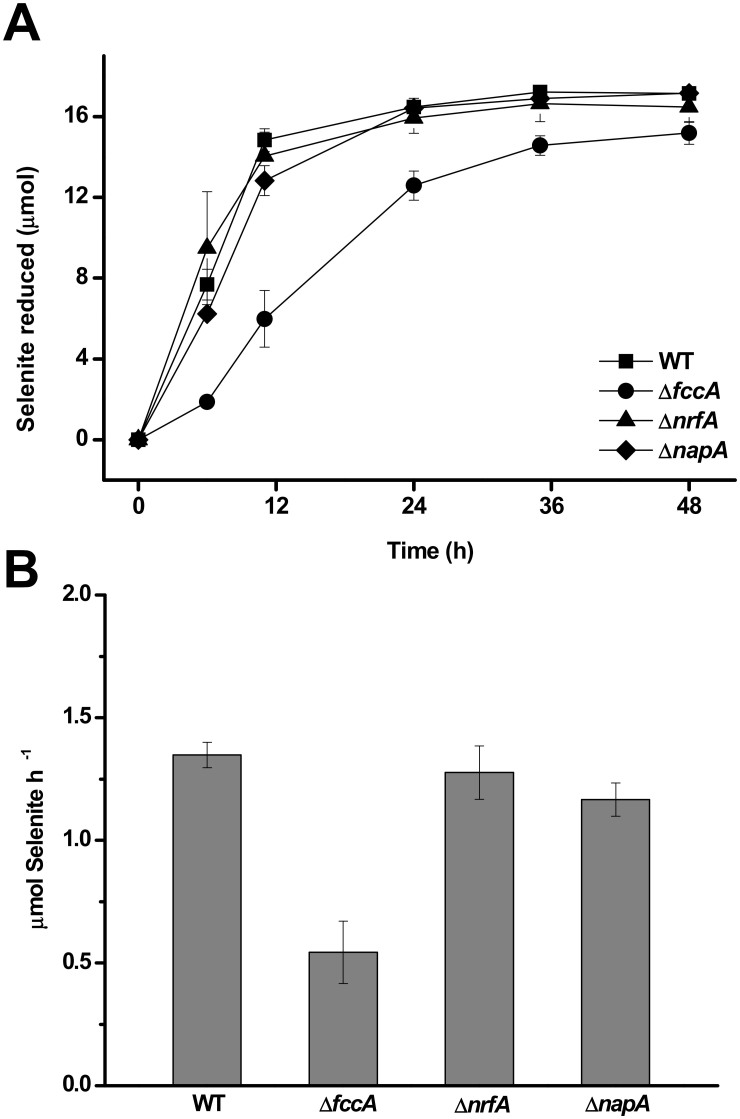
S. oneidensis MR-1 synthesizes several periplasmic terminal reductases, e.g., fumarate reductase FccA, nitrate reductase NapA, nitrite reductase NrfA, and two other periplasmic mediators for anaerobic respiration, i.e., MtrA and DmsE22,39,40,41,42. These c-type cytochromes catalyze terminal reduction in periplasm, or serve as an electronic relay to the downstream c-type cytochromes. The selenite reduction performance of the mutants deficient in fccA, napA or nrfA was examined. The results show that ΔnrfA and ΔnapA reduced selenite at nearly the same rate as the wild-type strain (Fig. 2A and B), indicating that neither nitrate reductase nor nitrite reductase was involved in selenite reduction. Conversely, selenite reduction by ΔfccA was severely suppressed, with a 60% decrease in the reduction rate within the initial 12 h (0.54 ± 0.13 μmol Se h−1 for ΔfccA vs. 1.35 ± 0.05 μmol Se h−1 for the wild-type strain). Such a result indicates that fccA played an important role in selenite reduction by S. oneidensis MR-1. fccA encodes a terminal reductase FccA, which is the key enzyme for fumarate reduction in S. oneidensis MR-1.
Figure 2. Effect of periplasmic reductases on selenite reduction by S. oneidensis MR-1.
(A) Selenite reduction kinetics of the wild-type strain and fumarate reductase mutant (ΔfccA), nitrate reductase mutant (ΔnapA), nitrite reductase mutant (ΔnrfA). The reduction ability is expressed as the total quantities of reduced selenite in μmol. Error bars indicate the standard error of triplicate cultures; and (B) Selenite reduction rates of the wild-type, ΔfccA, ΔnapA, and ΔnrfA. The linear portion of the kinetics curves (in the initial 12 h) in Fig. 2A was used to calculate the rates. The Adj. R2 of each fitting were 0.9980, 0.8624, 0.9567, 0.9906. Error bars indicate the standard error of triplicate cultures.
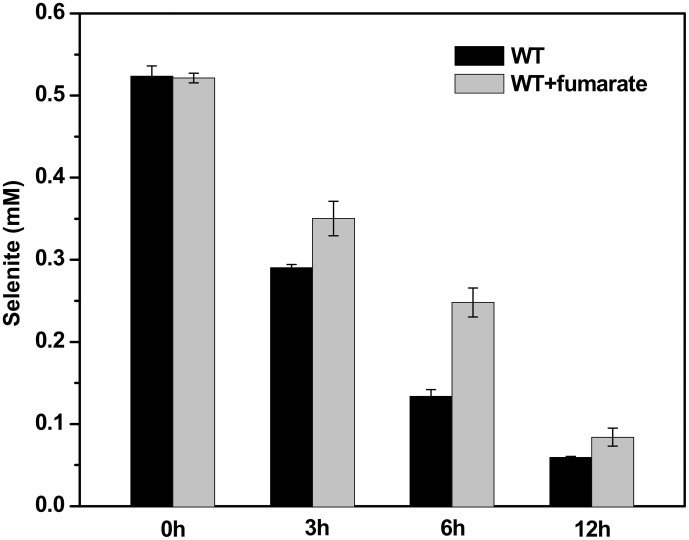
In anaerobic respiration with fumarate as electron acceptor, S. oneidensis MR-1 reduces it to succinate by FccA. Dosing of fumarate to the wild-type strain led to a significantly weakened selenite reduction, suggesting a competition between fumarate and selenite for FccA (Fig. 3). After6 h, there was 47.6% and 25.5% detectable selenite remained in the cultures with and without dosing fumarate, respectively. It should be noted that S. oneidensis MR-1 didn't grow under our cultivation conditions even in the presence of fumarate. The biomass represented by the total proteins gradually decreased during the reduction process (see Supplementary Fig. S2), which might result from the toxicity of selenite.
Figure 3. Competitive inhibition of selenite reduction by fumarate.
Comparative selenite concentrations of the initial 12 h in the culture of S. oneidensis MR-1 wild type strain with and without fumarate (20 mM) are shown.
Reduction dependence on quinone dehydrogenases
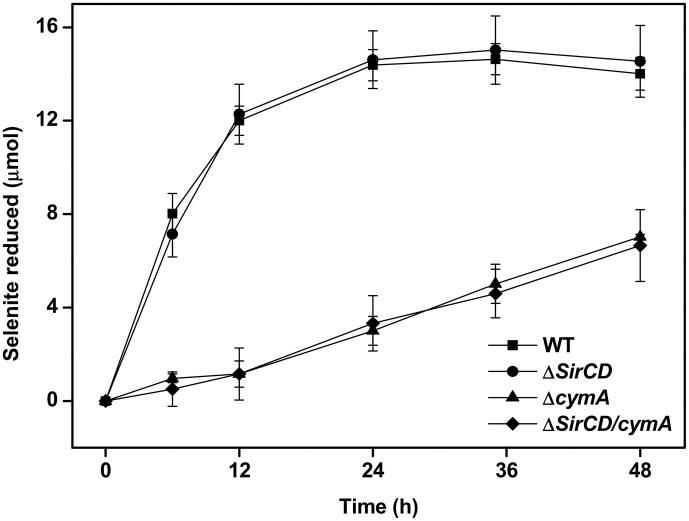
The cytoplasmic membrane-anchored cytochrome CymA has been recognized as a key component of S. oneidensis MR-1 to bridge electron transfer from quinone pool to several respiratory reductases in the anaerobic respiration process, including the fumarate reductase FccA22. Since ΔfccA shows a considerable defect to reduce selenite, CymA may also be involved in selenite reduction. To test this and further unveil the mechanism of selenite reduction by S. oneidensis MR-1, the ΔcymA mutant was constructed and its selenite reduction kinetics by ΔcymA was investigated. As expected, deletion of cymA suppressed selenite reduction severely (Fig. 4). The selenite reduction rate of ΔcymA was only 9.6% of that of the wild-type strain (0.096 μmol h−1vs. 1.000 μmol h−1). The transmission electronic microscope images in Supplementary Fig. S3 also illustrate fewer Se(0) accumulation in the ΔcymA cells than in the wild-type cells. All these results confirm that CymA was essential for selenite reduction by S. oneidensis MR-1.
Figure 4. Selenite reduction by S. oneidensis MR-1 wild-type strain and the mutants deficient in quinone dehydrogenases.
The reduction ability is expressed with the total quantities of reduced selenite in μmol. Error bars indicate the standard error of triplicate cultures.
As a substitute of CymA, the protein complex SirCD can also transfer electrons from quinone pool to fumarate or DMSO reductase43. SirCD can be induced in ΔcymA and restore the respiration ability of S. oneidensis MR-1 using fumarate, DMSO, or Fe(III) as electrons acceptors. Hence, it is possible that SirCD might offer another bridge to pass electrons to FccA. sirCD was deleted from both the wild-type strain and ΔcymA to completely suppress its possible expression. Fig. 4 shows that the ΔsirCD reduced selenite at nearly the same rate as the wild-type strain. Moreover, there was no difference between the selenite reduction kinetics of ΔsirCD/cymA and ΔcymA, suggesting that SirCD was not involved in the electron transfer for selenite reduction.
Accumulation of Se(0) nanoparticles
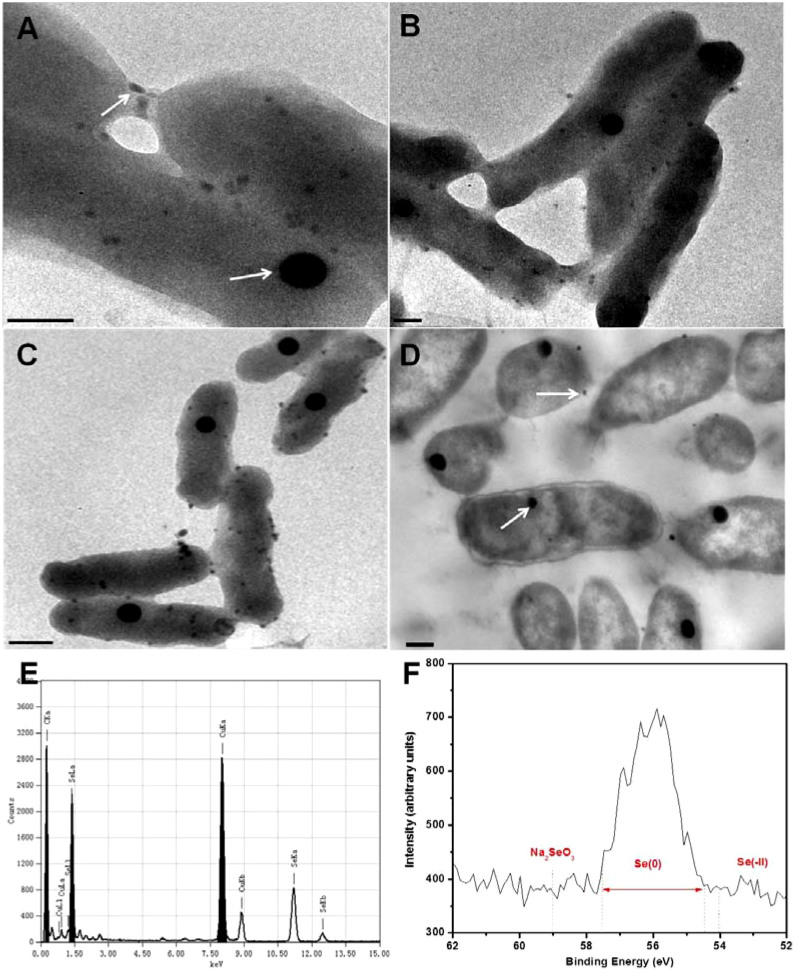
The selenite reduction process in S. oneidensis MR-1 was tracked by electronic microscope observations. The culture of S. oneidensis MR-1with 0.5 mM selenite rapidly turned red after selenite was reduced. The TEM images clearly show the formation of Se(0) particles (Fig. 5). Two different-shaped Se(0) nanoparticles were observed after 6 h. Spherical particles with approximately 100-nm diameter quickly accumulated in cells, and the size remained almost unchanged during the last reduction period (Fig. 5A–C). The formation of this type of Se(0) might represent a stress response of S. oneidensis MR-1 to selenite44. In addition, many smaller particles with a diameter of about 20 nm were gradually observed on the cell surface, possibly bound with extracellular polymeric substances. The thin section image (Fig. 5D) shows that Se(0) assembly was principally an intracellular process, further validating our hypothesis of periplasmic reductase-based selenite reduction. With the energy dispersive X-ray analysis, the element of the particles in the TEM images was confirmed to be selenium (Fig. 5E). High-resolution X-ray photoelectron spectroscopy (XPS) of the sample after selenite reduction shows clearly the 3d spectral peak of Se(0) (Fig. 5F).
Figure 5. Observation of the synthesized Se(0).
TEM images of the unstained S. oneidensis MR-1 cells showing the subcellular localization of Se deposition in the cells. Selenite-reducing cells at 6 h (A); 12 h (B); 24 h (C); and thin sections of cells at 12 h (D) are given. Arrows denote Se(0) particles. Scale bar: 200 nm; (E) Typical EDX analysis of the particles in (A). Both types of nanoparticles at different points were analyzed and the results were consistent; and (F) High-resolution Se 3d XPS of cells after Se(0) deposited (48 h). The energy positions/ranges of Na2SeO3, Se(0), as well as typical inorganic selenides were marked.
Discussion
Selenite reduction occurs widely in many eukaryotes and prokaryotes, but is found to be driven by reduced thiols in the cytoplasm of these microorganisms17,46,47. This reduction process can lead to the formation of superoxide anions (O2−), which is toxic to microorganisms and should be eliminated with superoxide dismutase and catalase3. S. oneidensis MR-1 possesses multiple respiration reductases and can reduce a wide range of electron acceptors. The diverse anaerobic respiration modules greatly benefit for its survival in nature by enabling flexible respiration adjustment and high toxicity resistance45. Although the respective functions of these redox complexes in anaerobic respiration of S. oneidensis MR-1 have been well defined, their involvements in the reduction of selenite have not been well elucidated. Taratus et al. reported that deficiency in selenite reduction suppressed anaerobic growth on other terminal electron acceptors by S. oneidensis MR-133. Besides, S. oneidensis MR-1 was found to reduce selenite faster than Veillonella atypica, a strain lacking of the multiple respiration redoxases31. In the present work, we systematically studied the possible contributions of multiple respiration reductases to selenite reduction with premeditated mutation of gene synthesizing these reductases. By comparing the selenite reduction rates of different mutants, we identified that FccA, which catalyzes the reduction of fumarate to succinate in periplasm, might serve as the executor for selenite reduction in S. oneidensis MR-1. We also found that CymA might work as a relay to continuously supply electrons to FccA, just as the case in respiring with fumarate in S. oneidensis MR-1. Our results suggest a new possible mechanism of anaerobic selenite reduction by respiratory reductases in microorganisms.
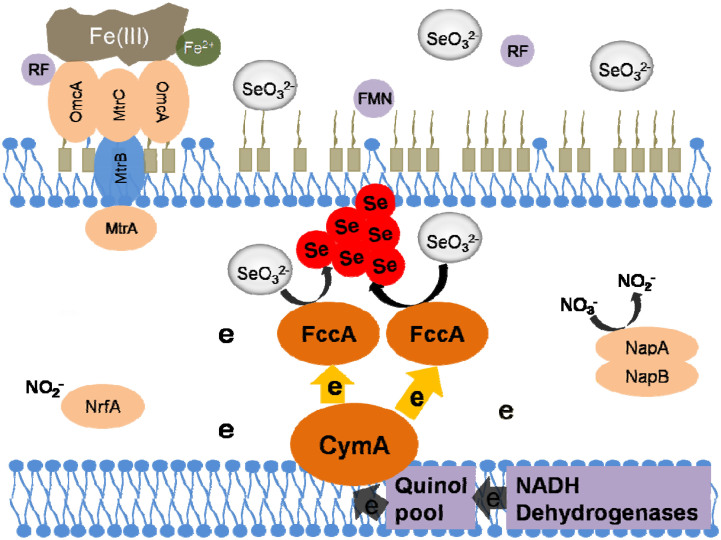
The most typical terminal electron acceptors in natural environment for S. oneidensis MR-1 are Fe(III) oxides and nitrate45. However, S. oneidensis MR-1 also reduces many ‘harmful' acceptors, such as metavanadate, U(VI), Co(III)-EDTA, and pertechnetate. Previous studies have proven that these electron acceptors, although most of which are soluble and may penetrate across the outer membrane into the periplasm and cytoplasm, are reduced extracellularly and the electron transfer to them is dependent on the outer membrane c-type cytochromes and hydrogenases24,26,27,48. Interestingly, these respiratory redoxases showed a higher difference and specificity when reducing selenite. Our tests with ΔnapB and ΔnrfA mutants showed that neither nitrate reductase nor nitrite reductase in S. oneidensis MR-1 contributed to selenite reduction. This finding is different from the previous reports that nitrite reductase in Thauera selenatis and in Rhizobium sullae contributes to selenite reduction13,20. Likewise, the Mtr proteins and extracellular flavins also exhibited little contribution to the reduction. This neutralism of reductases/molecules to selenite reduction precludes the competition for electrons from natural Fe(III)/nitrate respiration. Hence, it is possible for S. oneidensis MR-1 to conduct anaerobic respiration using Fe(III)/nitrate as an electron acceptor, while synchronously reducing selenite. Energy produced in respiration may facilitate cells' survival and resistance to toxicity of selenite, while a less common respiratory reductase, FccA, is used to remove the selenite already entered into the periplastic space. Thus, it is likely that synergetic anaerobic respiration and embezzlement of respiratory reductases act as the primary apparatus for selenite reduction by S. oneidensis MR-1 (Fig. 6). In addition, this ingenious selenite reducing ability conferred by the anaerobic respiration system in S. oneidensis MR-1 prevents selenite from entering the cytoplasm, thereby reducing the generation of free radicals due to selenite reduction by glutathione3,17. The proposed glutathione-mediated selenite reduction is consistent with our results that the slow reduction occurred in ΔcymA and ΔfccA.
Figure 6. Schematic diagram of the proposed pathways of selenite reduction and anaerobic respiration in S. oneidensis MR-1.
The oxidation of lactate provides electrons in the form of NADH, which further deliver the electrons to CymA through NADH dehydrogenases and the quinol pool. Shunt of electrons from CymA to various reductases enables execution of usual anaerobic respiration and selenite reduction.
The involvement of anaerobic respiration system in selenite reduction by S. oneidensis MR-1 playes a role in the geochemical cycles of selenium and other elements such as iron49. Selenite is exceptionally absorbable to iron (hydro)oxides in sediments and soils50, showing possible concomitance between selenite and iron (hydro)oxides in econiche of S. oneidensis. The possible selenite reduction along with bio-dissolution of iron minerals implies complexity of biogeochemical transformation of selenium in the environment. However, this warrants further investigations. Furthermore, the selenite reduction and detoxification by respiratory reductases support the physiological significance of diverse anaerobic respiration in dissimilatory metal-reducing bacteria as well as the understanding about the bioremediation of selenite-polluted environments.
Methods
Growth conditions and reduction medium of strains
S. oneidensis MR-1 and mutant strains were routinely grown in Luria-Bertani (LB) medium under 30°C. For selenite reduction tests, anaerobic mineral medium was used. The medium contains (per liter) NaCl 5.85 g, sodium 4-(2-hydroxyethyl)-1-piperazineethanesulphonic acid (HEPES) 11.91 g, NaOH 0.3 g, NH4Cl 1.498 g, KCl 0.097 g, NaH2PO4·2H2O 0.67 g, and 1 mL of trace elements solution. The trace minerals solution contains (in per liter): Nitrilotriacetic acid 1.5 g, MgSO4·7H2O 30 g, MnSO4·H2O 5 g, NaCl 10 g, FeSO4·7H2O 1 g, CaCl2·2H2O 1 g, CoCl2·6H2O 1 g, ZnCl21.3 g, CuSO4·5H2O 0.1 g, AlK(SO4)2·12H2O 0.1 g, H3BO3 0.1 g, Na2MoO4·2H2O 0.25 g, NiCl2·6H2O 0.25 g, Na2WO4·2H2O 0.25 g. No vitamins or amino acids were added. The medium was deoxygenated by boiling, packed into bottles, sealed with rubber stoppers under N2 atmosphere and then autoclaved. Before use, selenite and lactate were added to a final concentration of 0.5 mmol/L and 20 mmol/L, respectively.
Construction of mutants
Mutants with in-frame deletion of desired genes were constructed as reported elsewhere51. Briefly, chimerical DNA fragments with flanking regions of target genes were amplified and ligated by polymerase chain reaction (PCR), ligated with pRE112 and subsequently transformed into E. coli WM3064. The resulting plasmids were introduced into S. oneidensis MR-1 through conjugation with E. coli WM3064. After two rounds of selection, the mutant with specific gene deleted was validated by PCR using primer pairs up and down the deletion's location.
Selenite reduction assay
Strains cultured in LB medium were collected by centrifugation, and washed twice with minerals medium. In the selenite reduction tests, cells were added to 40 mL anaerobic mineral medium with a final OD600 of 1.0. The reduction tests were carried out at 30°C and the reduction efficiency was expressed by the decrease in selenite concentration. 20 mM fumarate was added when needed. To quantify selenite concentration, subsample at each time point was centrifugated to remove cells and Se(0) particles. Supernatant of 600 μL was mixed with 300 μL of hydrochloric acid (4 mol/L), and then with 600 μL of ascorbic acid (1 mol/L) with vortex. Selenite concentration was measured spectrophotometrically at 500 nm after 10 min23. In the flavins tests, plate reader was used to record the absorbance. To deduct the background absorption of flavins, samples were replaced with distilled water for comparison.
For the selenium balance tests, 1 mL sample was taken from each reduction cultures at 0, 24 and 48 h. Cells and extracellular Se(0) were collected by centrifugation at 8,000 × g for 8 min and re-dispersed in 500 μL NaOH solution (1 M). Then, the samples were placed in boiling water bath for 10 min, and cooled down in cold water. Both the cells and Se(0) were dissolved in this way. As a result, Se(0) would be disproportionated to selenide and selenite53,52. Neutralization the excess NaOH with H2SO4, and dose of 0.5 M trichloroacetic acid were used to remove the proteins coming from the cells. The remaining samples were digested with HNO3/HClO4 (4:1) to oxidize selenide to selenite, which was determine with ICP-AES.
Characterization of Se(0) particles
For TEM observation, cells were fixed with 5% glutaraldehyde for 12 h prior to being adsorbed onto copper grids for imaging and elemental analysis54. For XPS analysis, cells were collected from the reduction cultures at 48 h and centrifuged and washed twice with the deoxygenized PBS (50 mM) in an anaerobic glove chamber. Then, the cells were lyophilized to form red powder, which was analyzed using a monochromatized Al K alpha source. Survey spectra were recorded using a fixed pass energy of 20 eV. The spectrometer was calibrated in energy to the 1 s electronic level of carbon (284.9 eV). The estimated precision of bend energy values is 0.1 eV.
Author Contributions
D.B.L., C.W. and Y.Y.C. designed the experiments; C.W. and Y.Y.C. constructed strains and plasmids and conducted other molecular manipulations; D.B.L., C.W., N.L. and Z.C.Y. conducted selenite reduction tests. Y.Y.C., Z.H.T. and H.Q.Y. contributed to the planning and coordination of the project; D.B.L., Y.Y.C., W.W.L. and H.Q.Y. wrote and edited the manuscript. All authors contributed to discussion about the results and the manuscript.
Supplementary Material
SUPPLEMENTARY INFO
Acknowledgments
This work is partially supported by the National Natural Science Foundation of China (51129803, 21107105 and 51278479) and the Program for Changjiang Scholars and Innovative Research Team in University, China. The authors wish to thank Prof. K.H. Nealson from University of Southern California, USA for providing the bacteria used in this work.
References
- Winkel L. H. E. et al. Environmental selenium research: from microscopic processes to global understanding. Environ. Sci. Technol. 46, 571–579 (2011). [DOI] [PubMed] [Google Scholar]
- Fernández-Martínez A. & Charlet L. Selenium environmental cycling and bioavailability: a structural chemist point of view. Rev. Environ. Sci. Biotechnol. 8, 81–110 (2009). [Google Scholar]
- Zannoni D., Borsetti F., Harrison J. J. & Turner R. J. The Bacterial Response to the Chalcogen Metalloids Se and Te. Adv. Microb. Physiol. 53, 71 (2008). [DOI] [PubMed] [Google Scholar]
- Frankenberger Jr W. T. & Engberg R. A. (eds.) Environmental Chemistry of Selenium. (Marcel Dekker, Inc., New York, NY.; 1998).
- Jayaweera G. R. & Biggar J. W. Role of redox potential in chemical transformations of selenium in soils. Soil Sci. Soc. Am. J. 60, 1056–1063 (1996). [Google Scholar]
- Stolz J. F. & Oremland R. S. Bacterial respiration of arsenic and selenium. FEMS Microbiol. Rev. 23, 615–626 (1999). [DOI] [PubMed] [Google Scholar]
- Frankenberger W. & Arshad M. Bioremediation of selenium-contaminated sediments and water. Biofactors 14, 241–254 (2001). [DOI] [PubMed] [Google Scholar]
- Losi M. E. & Frankenberger W. T. J. Bioremediation of selenium in soil and water. Soil Sci. 162, 692–702 (1997). [Google Scholar]
- Ma J., Kobayashi D. Y. & Yee N. Chemical kinetic and molecular genetic study of selenium oxyanion reduction by Enterobacter cloacae SLD1a-1. Environ. Sci. Technol. 41, 7795–7801 (2007). [DOI] [PubMed] [Google Scholar]
- Oremland R. S. et al. Selenate reduction to elemental selenium by anaerobic bacteria in sediments and culture: biogeochemical significance of a novel, sulfate-independent respiration. Appl. Environ. Microbiol. 55, 2333–2343 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weres O., Jaouni A. R. & Tsao L. The distribution, speciation and geochemical cycling of selenium in a sedimentary environment, Kesterson Reservoir, California, U.S.A. Appl. Geochem. 4, 543–563 (1989). [Google Scholar]
- Hunter W. J. & Manter D. K. Bio-reduction of selenite to elemental red selenium by Tetrathiobacter kashmirensis. Curr. Microbiol. 57, 83–88 (2008). [DOI] [PubMed] [Google Scholar]
- Hunter W. J. & Kuykendall L. D. Reduction of selenite to elemental red selenium by Rhizobium sp strain B1. Curr. Microbiol. 55, 344–349 (2007). [DOI] [PubMed] [Google Scholar]
- Mishra R. R. et al. Reduction of selenite to red elemental selenium by moderately halotolerant Bacillus megaterium strains isolated from Bhitarkanika mangrove soil and characterization of reduced product. Chemosphere 84, 1231–1237 (2011). [DOI] [PubMed] [Google Scholar]
- Blum J. S., Bindi A. B., Buzzelli J., Stolz J. F. & Oremland R. S. Bacillus arsenicoselenatis, sp nov, and Bacillus selenitireducens, sp nov: two Haloalkaliphiles from Mono Lake, California that respire oxyanions of selenium and arsenic. Arch. Microbiol. 171, 19–30 (1998). [DOI] [PubMed] [Google Scholar]
- Lortie L., Gould W. D., Rajan S., McCready R. G. & Cheng K. J. Reduction of selenate and selenite to elemental selenium by a Pseudomonas stutzeri isolate. Appl. Environ. Microbiol. 58, 4042–4044 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessi J. & Hanselmann K. W. Similarities between the abiotic reduction of selenite with glutathione and the dissimilatory reaction mediated by Rhodospirillum rubrum and Escherichia coli. J. Biol. Chem. 279, 50662–50669 (2004). [DOI] [PubMed] [Google Scholar]
- Kessi J. Enzymic systems proposed to be involved in the dissimilatory reduction of selenite in the purple non-sulfur bacteria Rhodospirillum rubrum and Rhodobacter capsulatus. Microbiology 152, 13 (2006). [DOI] [PubMed] [Google Scholar]
- Basaglia M. et al. Selenite-reducing capacity of the copper-containing nitrite reductase of Rhizobiumsullae. FEMS Microbiol. Lett. 269, 124–130 (2007). [DOI] [PubMed] [Google Scholar]
- DeMoll-Decker H. & Macy J. M. The periplasmic nitrite reductase of Thauera selenatis may catalyze the reduction of selenite to elemental selenium. Arch. Microbiol. 160, 241–247 (1993). [Google Scholar]
- Harrison G., Curie C. & Laishley E. J. Purification and characterization of an inducible dissimilatory type sulfite reductase from Clostridium pasteurianum. Archive of Microblology 138, 7 (1984). [DOI] [PubMed] [Google Scholar]
- Fredrickson J. K. et al. Towards environmental systems biology of Shewanella. Nat. Rev. Microbiol. 6, 592–603 (2008). [DOI] [PubMed] [Google Scholar]
- Heidelberg J. F. et al. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20, 1118–1123 (2002). [DOI] [PubMed] [Google Scholar]
- Marshall M. J. et al. c-Type cytochrome-dependent formation of U(IV) nanoparticles by Shewanella oneidensis. PLoS Biol. 4, 1324–1333 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belchik S. M. et al. Extracellular reduction of hexavalent chromium by cytochromes MtrC and OmcA of Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 77, 4035–4041 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hau H. H., Gilbert A., Coursolle D. & Gralnick J. A. Mechanism and consequences of anaerobic respiration of cobalt by Shewanella oneidensis strain MR-1. Appl. Environ. Microbiol. 74, 6880–6886 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers J. M., Antholine W. E. & Myers C. R. Vanadium(V) reduction by Shewanella oneidensis MR-1 requires menaquinone and cytochromes from the cytoplasmic and outer membranes. Appl. Environ. Microbiol. 70, 1405–1412 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canstein H. v., Ogawa J., Shimizu S. & Lloyd J. R. Secretion of flavins by Shewanella species and their role in extracellular electron transfer. Appl. Environ. Microbiol. 74, 615–623 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsili E. et al. Shewanella secretes flavins that mediate extracellular electron transfer. Proc. Natl. Acad. Sci. USA 105, 3968–3973 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam K. et al. Growth mechanism of amorphous selenium nanoparticles synthesized by Shewanella sp HN-41. Biosci., Biotechnol., Biochem. 74, 696–700 (2010). [DOI] [PubMed] [Google Scholar]
- Pearce C. I. et al. Investigating different mechanisms for biogenic selenite transformations: Geobacter sulfurreducens, Shewanella oneidensis and Veillonella atypica. Environ. Technol. 30, 1313–1326 (2009). [DOI] [PubMed] [Google Scholar]
- Klonowska A., Heulin T. & Vermeglio A. Selenite and tellurite reduction by Shewanella oneidensis. Appl. Environ. Microbiol. 71, 5607–5609 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taratus E. M., Eubanks S. G. & Dichristina T. J. Design and application of a rapid screening technique for isolation of selenite reduction of deficient mutants of Shewanella putrefaciens. Microbiol. Res. 155, 79–85 (2000). [DOI] [PubMed] [Google Scholar]
- Coursolle D. & Gralnick J. A. Modularity of the Mtr respiratory pathway of Shewanella oneidensis strain MR-1. Mol. Microbiol. 77, 995–1018 (2010). [DOI] [PubMed] [Google Scholar]
- Bretschger O. et al. Current production and metal oxide reduction by Shewanella oneidensis MR-1 wild type and mutants. Appl. Environ. Microbiol. 73, 7003–7012 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke T. A. et al. Structure of a bacterial cell surface decaheme electron conduit. Proc. Natl. Acad. Sci. USA 108, 9384–9389 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutinel E. D. & Gralnick J. A. Shuttling happens: soluble flavin mediators of extracellular electron transfer in Shewanella. Appl. Microbiol. Biotechnol. 93, 41–47 (2012). [DOI] [PubMed] [Google Scholar]
- Covington E. D., Gelbmann C. B., Kotloski N. J. & Gralnick J. A. An essential role for UshA in processing of extracellular flavin electron shuttles by Shewanella oneidensis. Mol. Microbiol. 78, 519–532 (2010). [DOI] [PubMed] [Google Scholar]
- Chen Y., Wang F. P., Xu J., Mehmood M. A. & Xiao X. Physiological and evolutionary studies of NAP systems in Shewanella piezotolerans WP3. ISME Journal 5, 843–855 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H. C. et al. Reduction of nitrate in Shewanella oneidensis depends on atypical NAP and NRF systems with NapB as a preferred electron transport protein from CymA to NapA. ISME Journal 3, 966–976 (2009). [DOI] [PubMed] [Google Scholar]
- Myers C. R. & Myers J. M. Ferric reductase is associated with the membranes of anaerobically grown Shewanella putrefaciens MR-1. FEMS Microbiol. Lett. 108, 15–22 (1993). [Google Scholar]
- Schwalb C., Chapman S. K. & Reid G. A. The membrane-bound tetrahaem c-type cytochrome CymA interacts directly with the soluble fumarate reductase in Shewanella. Biochem. Soc. Trans. 30, 658–662 (2002). [DOI] [PubMed] [Google Scholar]
- Cordova C. D., Schicklberger M. F. R., Yu Y. & Spormann A. M. Partial functional replacement of CymA by SirCD in Shewanella oneidensis MR-1. J. Bacteriol. 193, 2312–2321 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debieux C. M. et al. A bacterial process for selenium nanosphere assembly. Proc. Natl. Acad. Sci. USA 108, 13480–13485 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hau H. H. & Gralnick J. A. Ecology and biotechnology of the genus Shewanella. Annu. Rev. Microbiol. 61, 237–258 (2007). [DOI] [PubMed] [Google Scholar]
- Garbisu C., Ishii T., Leighton T. & Buchanan B. B. Bacterial reduction of selenite to elemental selenium. Chem. Geol. 132, 199–205 (1996). [Google Scholar]
- Garbi C. et al. Morphological and biochemical responses of Bacillus subtilis to selenite stress. BioFactors 10, 311–319 (1999). [DOI] [PubMed] [Google Scholar]
- Marshall M. J. et al. Hydrogenase- and outer membrane c-type cytochrome-facilitated reduction of technetium(VII) by Shewanella oneidensis MR-1. Environ. Microbiol. 10, 125–136 (2008). [DOI] [PubMed] [Google Scholar]
- Lovley D. R. Dissimilatory metal reduction. Annu. Rev. Microbiol. 47, 263–290 (1993). [DOI] [PubMed] [Google Scholar]
- Martin A. J. et al. Biogeochemical mechanisms of selenium exchange between water and sediments in two contrasting lentic environments. Environ. Sci. Technol. 45, 2605–2612 (2011). [DOI] [PubMed] [Google Scholar]
- Wang F., Xiao X., Ou H. Y., Gai Y. B. & Wang F. P. Role and regulation of fatty acid biosynthesis in the response of Shewanella piezotolerans WP3 to different temperatures and pressures. J. Bacteriol. 191, 2574–2584 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. X. et al. A hydrothermal synthesis of orthorhombic nanocrystalline cobalt diselenide CoSe2. Mater. Res. Bull. 35, 2403–2408 (2000). [Google Scholar]
- Hoffmann J. E. & King M. G. Selenium and Selenium Compounds. Kirk-Othmer Encyclopedia of Chemical Technology (John Wiley & Sons, Inc., 2010).
- Picard A. et al. Monitoring microbial redox transformations of metal and metalloid elements under high pressure using in situ X-ray absorption spectroscopy. Geobiology 9, 196–204 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY INFO