Abstract
Nonviral gene delivery systems are rapidly becoming a desirable and applicable method to overexpress genes in various types of cells. We have recently developed a piggyBac transposase-based, helper-independent and self-inactivating delivery system (pmGENIE-3) capable of high-efficiency transfection of mammalian cells including human cells. In the following study, we have assessed the potential of this delivery system to drive the expression of short hairpin RNAs to knock down genes in human cells. Two independent pmGENIE-3 vectors were developed to specifically target knockdown of an endogenous gene, telomerase reverse transcriptase (TERT), in telomerase-positive human immortalized cell lines. As compared with a transposase-deficient vector, pmGENIE-3 showed significantly improved short-term transfection efficiency (~4-fold enhancement, 48 hours posttransfection) and long-term integration efficiency (~5-fold enhancement) following antibiotic selection. We detected a significant reduction of both TERT expression and telomerase activity in both HEK293 and MCF-7 breast carcinoma cells transfected with two pmGENIE-3 construct targeting distinct regions of TERT. Importantly, this knockdown of expression was sufficient to abrogate telomerase function since telomeres were significantly shortened (3–4 Kb, P < 0.001) in both TERT-targeted cell lines following antibiotic selection of stable integrants. Together, these data show the capacity of the piggyBac nonviral delivery system to stably knockdown gene expression in mammalian cells and indicate the potential to develop novel tumor-targeting therapies.
Keywords: cell line, piggyBac transposase, pmGENIE-3, RNA interference, telomerase
Introduction
Transposon-based systems are simple and efficient transfection tools suitable for a variety of gene transfer applications.1 These plasmid-based gene delivery vehicles represent alternatives to popular integrating viral approaches. Advantages over viral vectors include tolerance by the immune system, decreased preference for integration into genes, and increased cargo capacity.2,3,4,5,6 In addition, viral approaches are burdened by expense in production and biosafety considerations.7 Initial transposition experiments in vertebrate cells used the Sleeping Beauty (SB) system. The SB transposon, originally reconstructed from inactivated sequences found in fish genomes, has proven to be a flexible and effective nonviral genetic tool.8,9,10 Recently, piggyBac (pB), isolated from the moth Trichoplusia Ni, has emerged as a highly efficient gene delivery vector for numerous in vitro and in vivo applications.11,12,13,14,15,16 The typical 2-plasmid system includes both a “helper” plasmid encoding the transposase enzyme and a “donor” plasmid containing the intended integration sequence, such as a gene of interest, flanked by the transposon terminal repeat elements (TREs). Upon entrance to the cell, the pB transposase recognizes the TREs and excises the transposon from the donor plasmid. pB subsequently integrates the transposon permanently into the genome at a TTAA tetranucleotide sequence. By introducing pB TREs into bacterial artificial chromosomes, it is possible to facilitate integration of large pB inserts greater than 100 kb.17 pB's unprecedented cargo capacity can allow for the integration of large genes, regulatory elements, and multiple reading frames. pB has been used to generate induced pluripotent stem cells due to its ability to precisely excise its transposon from the genome without leaving a DNA “footprint”.18 This feature allows for the removal of transgenes following complete reprogramming without leaving residual sequences.19 In addition, pB has been shown to be amenable to DNA-binding domain fusions, allowing for targeted transposition to chromosomal locations.20
We have recently designed improved self-inactivating vectors that contain all transpositional machinery on a single helper-independent plasmid, termed pmGENIE-3.16 pmGENIE-3 encodes a specialized pB transposase containing a TRE within an engineered intron. Upon transfection, pB recognizes the TREs and subsequently excises the transposon from the plasmid, thereby truncating the transposase gene. This design renders the transposase inactive after excision. Consequentially, potentially negative genotoxic effects that may develop by the persistence of an active PB gene are eliminated.
Over the past 20 years, RNA interference, using silencing RNA molecules or short hairpin RNA molecules (shRNA) has developed into a reliable method to target and reduce the expression of specific genes. Eukaryotic expression vectors have been shown to effectively drive expression of shRNA in mammalian cells, however the relative ability of transposase-based delivery systems, in particular the pB transposase systems, have not yet been thoroughly assessed. Telomerase is an enzymatic complex that is required for prolonged cancer cell proliferation and is present in ~90% of all human tumors.21 The activity of the telomerase complex is entirely dependent on expression of the catalytic component of telomerase, TERT.22,23 Therefore, in the present study, we sought to assess the potential of a novel pB-based vector to express shRNA to specifically target TERT, in human telomerase-positive cell lines.
Results
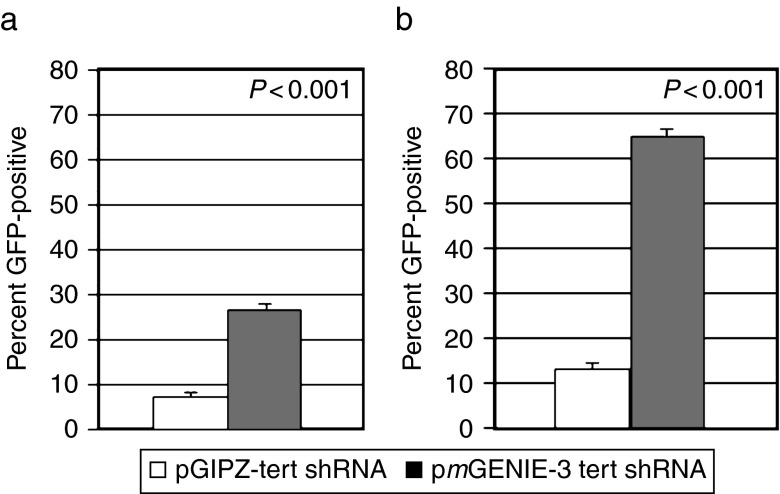
We have previously shown that the pmGENIE-3 transposase-based nonviral delivery system can efficiently express genes in mammalian cells both in vitro and in vivo in transgenic mice.16,24 To assess the capacity of this system to knock down target genes using RNA interference, we developed two vectors, pmGENIE-3-Trt1 and pmGENIE-3-Trt2, to express shRNA-targeting TERT (Figure 1). We initially compared transfection efficiency of our actively integrating pmGENIE-3 to a passive, transposase-deficient pGIPZ shRNAmir vector expressing the same TERT-targeting shRNA. We found the transient transfection levels (72 hours posttransfection) to be enhanced ~4-fold using the pmGENIE-3 vector (Figure 2a). Furthermore, following selection with antibiotic, we observed ~5-fold enhancement of the frequency of stably transfected cells receiving pmGENIE-3 based on green fluorescent protein expression (Figure 2b).
Figure 1.
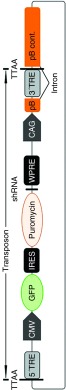
Map of the pmGENIE-3 pB transposase-based vector. Included in the vector design are ubiquitously expressed green fluorescent protein to facilitate detection of transfectants, internal ribosome entry site-Puro to allow stable selection of integrants, and a site to allow insertion of short hairpin RNA sequences.
Figure 2.
Comparison of the short-term and long-term transfection efficiency of pmGENIE-3 to a transposase-deficient vector. (a) HEK293 cells were transiently transfected (FuGENE6) in triplicate with equal amounts of either pmGENIE-3 or pGIPZ shRNAmir plasmid and the fraction of green fluorescent protein (GFP)-positive cells (the ratio of GFP+ to GFP- cells) was assessed by FACS analysis at 72 hours posttransfection. (b) HEK293 cells were transfected as in a, followed by 4-week selection with puromycin. Values represent the number of total GFP+ cells relative to a control reference sample transfected with an empty pmGENIE-3 vector grown without selection. Standard error bars and P value (Student's t test) are shown.
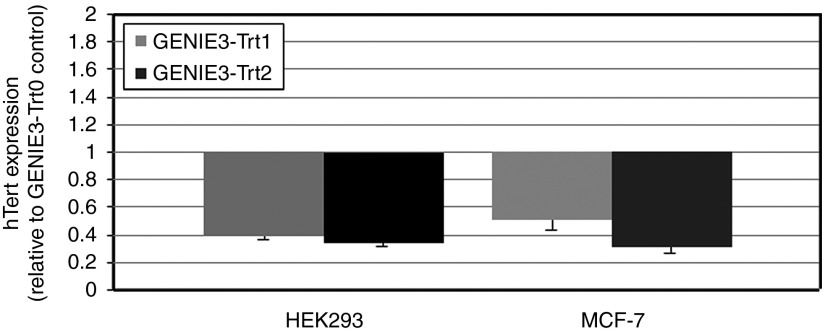
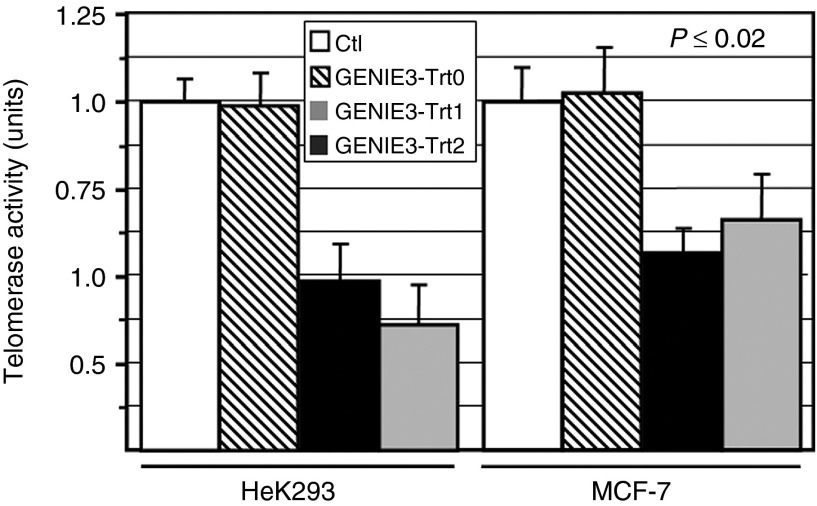
To assess the ability of pmGENIE-3-Trt1 and pmGENIE-3-Trt2 to knock down TERT, we stably transfected both HEK293 cells and MCF-7 cells with these constructs. We also generated control cell lines transfected with a pmGENIE-3 construct without shRNA (pmGENIE-3-Trt0). At ~2 months postselection, we observed significant reduction in relative levels of TERT mRNA for both constructs in HEK293 and MCF-7 (P ≤ 0.001) cells (Figure 3). To determine whether this level of knockdown was sufficient to have an effect on the telomerase enzymatic complex, we also measured telomerase activity in these cells. We observed significantly (P ≤ 0.02) reduced levels of telomerase activity with both constructs in HEK293 and MCF-7 cells, in agreement with the reduced levels of TERT mRNA (Figure 4). These observations confirm that the nonviral pmGENIE-3-TERT delivery system is capable of knocking down target genes via stable expression of shRNA.
Figure 3.
Analysis of the potential of pmGENIE-3 to perform RNAi-mediated knockdown of gene expression in human cells. Human cell lines (HEK293 and MCF-7) were transfected with pmGENIE-3 vectors expressing short hairpin RNA targeting telomerase reverse transcriptase. Following Puromycin selection, the relative expression of telomerase reverse transcriptase was assessed using real- time reverse transcription polymerase chain reaction. Expression values, normalized to GAPDH are shown relative to corresponding control cells (transfected with Trt0). All analyses were performed in triplicate; standard error bars are shown.
Figure 4.
Analysis of pmGENIE-3 mediated telomerase reverse transcriptase knockdown of telomerase activity in human cell lines. Telomerase activity was measured using the TRAP assay in the same cells as assessed in Figure 3. Telomerase activity levels for an equivalent number of untreated cells are also shown. All analyses were performed in triplicate; standard error bars are shown.
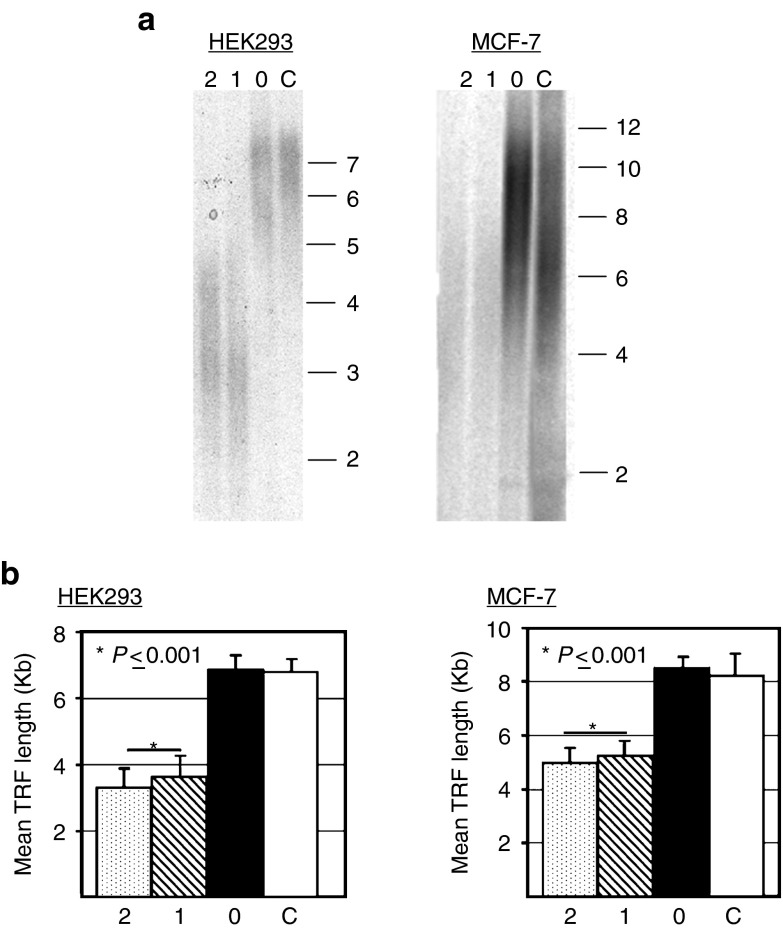
We next sought to determine if the reduced level of telomerase had a physiological effect in these cell lines. Specifically, we measured telomere length by southern analysis of terminal restriction fragments at ~2 months posttransfection (Figure 5). For both HEK293 and MCF-7 cells transfected with either pmGENIE-3-Tert1 or pmGENIE-3-Tert2, we observed a significant reduction in telomere length. For the HEK293 cell line, which has relatively short initial telomere length, we noted that telomeres were approaching critical length required to maintain cell viability (~3 Kb).25,26 An analysis of proliferation rate in cells transfected with the TERT targeting vectors revealed a marked (>2-fold) reduction in growth by ~2 months posttransfection in cells (Supplementary Figure S1).
Figure 5.
Analysis of the physiological effect of pmGENIE-3-mediated gene knockdown in human cells. Following puromycin selection, telomere length analysis was performed on the same cells that were transfected with the telomerase reverse transcriptase targeting pmGENIE-3 in Figure 3. (a) Sample blots of southern analysis of TRF length for HEK293 and MCF-7. C, no treatment control; 0, pmGENIE-3-Trt0; 1, pmGENIE-3-Trt1; 2, pmGENIE-3-Trt2. (b) Quantitative analysis of mean TRF length for all samples. All analyses were performed in triplicate; standard error bars are shown. P value represents Student's t test.
Discussion
Transposition-based gene delivery systems are appealing tools which bypass the use of viral vectors for transgene delivery. Transposase/transposon vectors have been shown to have less of an affinity for transcriptional start sites, transcribed regions, and protooncogenes than lenti- or retroviruses and their ease of production at low expense make them ideal candidates to evaluate human applications of gene therapy.27 We have modified the pB transposon system to consist of a single self-inactivating vector for highly efficient generation of stable transformants in mammalian cells in vitro as well as in transgenic animals.16,24 Due to our previous success at utilizing helper-independent pB constructs to stably transfect cell lines and animals with marker genes such as enhanced green fluorescent protein, we speculated that transgenes that affect the biological function of cells would have the same successful delivery and integration.
In the present study, we have assessed the ability of our single plasmid pB vector, pmGENIE-3, to drive expression of shRNA and affect the target gene expression in human cell lines. The practical utility of silent interfering RNA to suppress cancer has been intensively studied over the past 10 years,28 however, truly effective RNAi-based therapies have yet to be developed. Telomerase, specifically TERT, has been studied as a target for RNAi technology.29 Indeed, a TERT-targeting RNAi therapeutic could potentially complement other antisense targeting methods, such as using stable antisense oligonucleotides that target the telomerase RNA component30 to dramatically reduce telomerase levels and induce cancer cell senescence or apoptosis. We show in both HEK293 and MCF-7 cells that pmGENIE-3 vectors designed to target the TERT gene transcript, effectively knockdown TERT expression and reduce telomerase activity levels. Furthermore, this knockdown of telomerase was sufficient to effect the physiological function of telomerase, since in both cell lines, it was accompanied by significant telomere shortening (P < 0.001). These results suggest that the pB transposase-based delivery system is an effective technique for expressing shRNA and knocking down target genes in human cell lines, including tumor cell lines.
The ability of the pmGENIE-3 vector to efficiently knockdown TERT and reduce telomerase levels, in conjunction with the high level of long-term expression, indicative of genomic integration associated with this vector, suggest that pB transposase-based delivery systems may provide a new method to suppress tumor progression. We are presently undertaking experiments to assess the potential and effect of using pmGENIE-3 to knockdown TERT in tumor cells in an in vivo murine model.
The results from this study have further implications for the utility of pmGENIE-3 vector beyond cancer therapy, for example, to study cell aging. The use of pmGENIE-3 vector to target TERT and/or other genes in rodents and other laboratory animals could be a powerful way to assess the combinatorial affects of altered expression of these genes on cell aging in vitro and in vivo. The ability to express multiple genes or shRNAs, from a single construct is another advantage of the pB transposase-based delivery system, as we (data to be published elsewhere) and others31 have recently demonstrated by our use of pmGENIE-3 vector to efficiently derive induced pluripotent stem cellsiPSCs by expression of the Yamanaka factors (Oct4, Sox2, Klf4, and c-Myc).
In summary, we have shown that the pB transposase-based delivery system is effective at shRNA-driven targeted gene knockdown in mammalian (human) cells. The ability to affect both telomerase and telomeres in a tumor cell line implies a potential utility for the pB vector in cancer therapy.
Materials and methods
Vector design. The pmGENIE-3 transposase-based delivery system has been described previously.16 pmGENIE-3 is compatible with Gateway recombineering cloning (Life Technologies) for simple addition of target genes into the transposon portion of the construct. The two shRNA sequences were modeled after human miR-30 and were designed to target distinct regions of the TERT mRNA, specifically, 5′-AGCAAGTTGCAAAGCAT-3′ (Tert1 shRNA) and 5′-CGAGCTGCTCAGGTCTTTCTT-3′ (Tert2 shRNA). The shRNAs were located 3′ to a bicistronic TurboGFP and puromycin-coding region driven by the cytomegalovirus promoter. Each cassette including shRNA was excised from lentiviral pGIPZ shRNAmir plasmids (catalog #s RHS4430-101132965 and RHS4430-99161517, Thermo Scientific, Rockford, IL) via XbaI and BglII (New England Biolabs, Ipswich, MA) and cloned into an empty pENTR1A (Life Technologies) vector in preparation for Gateway recombineering into pmGENIE-3.
Cell lines and transfection. The HEK293 and MCF-7 cell lines were grown in standard culture conditions (37 °C, 5% CO2) in Dulbecco's modified Eagle medium for HEK293 and Dulbecco's modified Eagle medium F12 for MCF-7, both in high glucose, complete media with 10% fetal bovine serum. Cells were transfected using FuGENE 6, (Roche, Indianapolis, IN) per manufacturers' instructions. Cells were selected with puromycin (HEK293 cells with 500 ng/ml, MCF-7 cells with 300 ng/ml).
Measurement of TERT expression. Relative levels of TERT mRNA were assessed by real-time polymerase chain reaction (PCR) using a Bio-Rad IQ cycler. Total RNA was isolated from cells using TRIzol reagent (Invitrogen, Grand Island, NY). RNA cleanup and DNase treatment was done using the RNeasy Mini Kit (Qiagen, Germantown, MD). One microgram of total RNA was reverse transcribed using the iScript Advanced cDNA Synthesis Kit for real-time quantitative PCR (Bio-Rad). The real-time PCR assays were performed in 25 μl reactions using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA), 50 ng cDNA, and 0.4 μmol/l of forward and reverse primers. Analysis was done using a MyiQ single-color real-time PCR detection system (Bio-Rad). Thermal cycling conditions were the following: 2 minutes at 50 °C and 10 minutes at 95 °C for initial denaturation, followed by 40 cycles at 95 °C for 15 seconds and 54 °C for 1 minute, and a dissociation step at 95 °C for 15 seconds, 54 °C for 30 seconds and 95 °C for 15 seconds. Gene relative quantification of TERT was calculated as described by Livak and Schmittgen32 and normalized with the housekeeping gene hypoxanthine phosphoribosyltransferase. All samples were analyzed in triplicate along with no RT controls. Primers used for amplification were as follows: TERT F, 5′-CGTCGAGCTGCTCAGGTCTT-3′ and R, 5′-AGTGCTGTCTGATTCCAATGCTT-3′:33 hHPRT F, 5′-TGACACTGGCAAAACAATGCA-3′ and R, 5′-GGTCCTTTTCACCAGCAAGCT-3′.34
Detection of telomerase. Telomerase activity was measured using the TRAP assay (TRAPeze detection kit; Millipore, Billerica, MA) per manufacturers' protocol. Briefly, 1 × 106 cells per sample were lysed on ice in CHAPS buffer (30 minutes) followed by centrifugation (12,000g, 30 minutes). The supernatant was then used directly in TRAP reactions (PCR cycling conditions: 27 cycles of 94 °C/30 seconds, 60 °C/30 seconds, 72 °C/60 seconds).
Telomere length analysis. Telomere length was assessed by Southern analysis of terminal restriction fragment length as previously described.26,35
Statistics. All measurements are shown as the mean of three or more independent measurements. Error bars represent standard error. P values represent Student's t test.
SUPPLEMENTARY MATERIAL Figure S1. Effect of TERT-targeting shRNA expression on growth rate of HEK293 cells.
Acknowledgments
The authors have no conflicts of interest to declare. This work was supported by Castle Foundation and Tilker Foundation awards to RA and National Institutes of Health Grants (5P20RR024206, R01GM083158-01A1) to SM.
Supplementary material
References
- Katter K, Geurts AM, Hoffmann O, Mátés L, Landa V, Hiripi L, et al. Transposon-mediated transgenesis, transgenic rescue, and tissue-specific gene expression in rodents and rabbits. FASEB J. 2013;27:930–941. doi: 10.1096/fj.12-205526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel R, Smith JA. Integration site selection by retroviral vectors: molecular mechanism and clinical consequences. Hum Gene Ther. 2008;19:557–568. doi: 10.1089/hum.2007.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Matteo M, Belay E, Chuah MK, Vandendriessche T. Recent developments in transposon-mediated gene therapy. Expert Opin Biol Ther. 2012;12:841–858. doi: 10.1517/14712598.2012.684875. [DOI] [PubMed] [Google Scholar]
- Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:E234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muruve DA, Barnes MJ, Stillman IE, Libermann TA. Adenoviral gene therapy leads to rapid induction of multiple chemokines and acute neutrophil-dependent hepatic injury in vivo. Hum Gene Ther. 1999;10:965–976. doi: 10.1089/10430349950018364. [DOI] [PubMed] [Google Scholar]
- Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Kay MA, Glorioso JC, Naldini L. Viral vectors for gene therapy: the art of turning infectious agents into vehicles of therapeutics. Nat Med. 2001;7:33–40. doi: 10.1038/83324. [DOI] [PubMed] [Google Scholar]
- Ivics Z, Hackett PB, Plasterk RH, Izsvák Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- Ivics Z, Izsvák Z. Nonviral gene delivery with the sleeping beauty transposon system. Hum Gene Ther. 2011;22:1043–1051. doi: 10.1089/hum.2011.143. [DOI] [PubMed] [Google Scholar]
- Izsvák Z, Ivics Z. Sleeping beauty transposition: biology and applications for molecular therapy. Mol Ther. 2004;9:147–156. doi: 10.1016/j.ymthe.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Shinohara ET, Kaminski JM, Segal DJ, Pelczar P, Kolhe R, Ryan T, et al. Active integration: new strategies for transgenesis. Transgenic Res. 2007;16:333–339. doi: 10.1007/s11248-007-9077-z. [DOI] [PubMed] [Google Scholar]
- Wu SC, Meir YJ, Coates CJ, Handler AM, Pelczar P, Moisyadi S, et al. piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. Proc Natl Acad Sci USA. 2006;103:15008–15013. doi: 10.1073/pnas.0606979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- Feschotte C. The piggyBac transposon holds promise for human gene therapy. Proc Natl Acad Sci USA. 2006;103:14981–14982. doi: 10.1073/pnas.0607282103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saridey SK, Liu L, Doherty JE, Kaja A, Galvan DL, Fletcher BS, et al. PiggyBac transposon-based inducible gene expression in vivo after somatic cell gene transfer. Mol Ther. 2009;17:2115–2120. doi: 10.1038/mt.2009.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urschitz J, Kawasumi M, Owens J, Morozumi K, Yamashiro H, Stoytchev I, et al. Helper-independent piggyBac plasmids for gene delivery approaches: strategies for avoiding potential genotoxic effects. Proc Natl Acad Sci USA. 2010;107:8117–8122. doi: 10.1073/pnas.1003674107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MA, Turner DJ, Ning Z, Yusa K, Liang Q, Eckert S, et al. Mobilization of giant piggyBac transposons in the mouse genome. Nucleic Acids Res. 2011;39:e148. doi: 10.1093/nar/gkr764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser MJ, Ciszczon T, Elick T, Bauser C. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Mol Biol. 1996;5:141–151. doi: 10.1111/j.1365-2583.1996.tb00048.x. [DOI] [PubMed] [Google Scholar]
- Yusa K, Rad R, Takeda J, Bradley A. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat Methods. 2009;6:363–369. doi: 10.1038/nmeth.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens JB, Urschitz J, Stoytchev I, Dang NC, Stoytcheva Z, Belcaid M, et al. Chimeric piggyBac transposases for genomic targeting in human cells. Nucleic Acids Res. 2012;40:6978–6991. doi: 10.1093/nar/gks309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, et al. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- Bodnar AG, Ouellette M, Frolkis M, Holt SE, Chiu CP, Morin GB, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- Marh J, Stoytcheva Z, Urschitz J, Sugawara A, Yamashiro H, Owens JB, et al. Hyperactive self-inactivating piggyBac for transposase-enhanced pronuclear microinjection transgenesis. Proc Natl Acad Sci USA. 2012;109:19184–19189. doi: 10.1073/pnas.1216473109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- Allsopp RC, Harley CB. Evidence for a critical telomere length in senescent human fibroblasts. Exp Cell Res. 1995;219:130–136. doi: 10.1006/excr.1995.1213. [DOI] [PubMed] [Google Scholar]
- Galvan DL, Nakazawa Y, Kaja A, Kettlun C, Cooper LJ, Rooney CM, et al. Genome-wide mapping of PiggyBac transposon integrations in primary human T cells. J Immunother. 2009;32:837–844. doi: 10.1097/CJI.0b013e3181b2914c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau RL, Davidson BL. Generation of hairpin-based RNAi vectors for biological and therapeutic application. Meth Enzymol. 2012;507:275–296. doi: 10.1016/B978-0-12-386509-0.00014-4. [DOI] [PubMed] [Google Scholar]
- Zaffaroni N, Pennati M, Folini M. Validation of telomerase and survivin as anticancer therapeutic targets using ribozymes and small-interfering RNAs. Methods Mol Biol. 2007;361:239–263. doi: 10.1385/1-59745-208-4:239. [DOI] [PubMed] [Google Scholar]
- Dikmen ZG, Gellert GC, Jackson S, Gryaznov S, Tressler R, Dogan P, et al. In vivo inhibition of lung cancer by GRN163L: a novel human telomerase inhibitor. Cancer Res. 2005;65:7866–7873. doi: 10.1158/0008-5472.CAN-05-1215. [DOI] [PubMed] [Google Scholar]
- Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hämäläinen R, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Zhu CQ, Cutz JC, Liu N, Lau D, Shepherd FA, Squire JA, et al. Amplification of telomerase (hTERT) gene is a poor prognostic marker in non-small-cell lung cancer. Br J Cancer. 2006;94:1452–1459. doi: 10.1038/sj.bjc.6603110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Schächter F, Uchida I, Wei L, Zhu X, Effros R, et al. Loss of telomeric DNA during aging of normal and trisomy 21 human lymphocytes. Am J Hum Genet. 1993;52:661–667. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.