Abstract
Selective gene silencing by RNA interference (RNAi) involves double-stranded small interfering RNA (ds siRNA) composed of single-stranded (ss) guide and passenger RNAs. siRNA is recognized and processed by Ago2 and C3PO, endonucleases of the RNA-induced silencing complex (RISC). RISC cleaves passenger RNA, exposing the guide RNA for base-pairing with its homologous mRNA target. Remarkably, the 3′ end of passenger RNA can accommodate a DNA extension of 19-nucleotides without loss of RNAi function. This construct is termed passenger-3′-DNA/ds siRNA and includes a 3′-nuclease-resistant mini-hairpin structure. To test this novel modification further, we have now compared the following constructs: (I) guide-3′-DNA/ds siRNA, (II) passenger-3′-DNA/ds siRNA, (III) guide-3′-DNA/ss siRNA, and (IV) passenger-3′-DNA/ss siRNA. The RNAi target was SIRT1, a cancer-specific survival factor. Constructs I–III each induced selective knock-down of SIRT1 mRNA and protein in both noncancer and cancer cells, accompanied by apoptotic cell death in the cancer cells. Construct IV, which lacks the SIRT1 guide strand, had no effect. Importantly, the 3′-DNA mini-hairpin conferred nuclease resistance to constructs I and II. Resistance required the double-stranded RNA structure since single-stranded guide-3′-DNA/ss siRNA (construct III) was susceptible to serum nucleases with associated loss of RNAi activity. The potential applications of 3′-DNA/siRNA constructs are discussed.
Keywords: Ago2, C3PO, nuclease-resistant DNA mini-hairpin, RNAi, SIRT1, therapy, 3′-DNA/siRNA constructs
Introduction
Natural posttranscriptional regulation of gene expression involves a variety of conserved mechanisms which are dependent upon double-stranded RNA (dsRNA) (see for example refs. 1 and 2). In the case of RNA interference (RNAi), gene silencing involves long precursor dsRNAs which are processed into ~21 nucleotide small interfering RNAs (siRNAs) by Dicer, a ribonuclease III (RNase III). Each ds siRNA consists of a siRNA guide strand (with antisense complementarity to its mRNA target) base-paired with its passenger (sense) strand. During RNAi, the nascent ds siRNA is loaded onto endoribonuclease Argonaute 2 (Ago2) of the RNA-induced silencing complex (RISC).3,4,5,6 Studies using reconstituted RISC components indicate that Ago2 directly binds ds siRNA and nicks the passenger strand of the siRNA. Subsequent removal of the nicked passenger strand from Ago2 requires the endonuclease activity of C3PO (component 3 promoter of RISC).7,8 Thus, the siRNA passenger strand is cleaved and removed from the RISC complex, leaving the ss siRNA guide strand free to direct Ago2-mediated cleavage of its target mRNA. The process is highly efficient and operates with exquisite selectivity.
The RNAi process can be recapitulated by introduction of synthetic siRNA into cells, or via engineered expression of short hairpin RNA (shRNA), an siRNA precursor.9 The ability of siRNA to target and inhibit gene expression offers a route for development of novel therapeutics where key disease-related genes have been identified.10,11 Synthetic siRNA can be delivered to cells and tissues in vivo by siRNA carriers, including biodegradable nanoparticles and lipids.10,11 Chemical modifications to improve synthetic siRNA stability and resistance to serum endonucleases are continually being sought in order to increase the efficacy of siRNA-based drugs.10,11 In our own work, we have previously developed a bifunctional siRNA construct in which the 3′ end of the siRNA passenger strand was extended with a 19nt DNA sequence. The 19 nt DNA extension was originally designed to function as a PCR primer to enable detection of the very low levels of siRNA effective for RNAi,12 and we have described a protocol applicable for quantification of different siRNA sequences via the 3′ 19nt DNA extension (which is common to each construct).12 We have previously demonstrated that the modified 3′ 19nt DNA/siRNA construct is recoverable following induction of RNAi, and is quantifiable down to a molar levels per cell.12 It has been proposed that this design may be useful for in vivo studies on the uptake, distribution and pharmacokinetics of siRNA and for the development of siRNA-based therapeutics.12 In addition, the 3′ end of the DNA primer sequence forms a stable hairpin structure and confers nuclease resistance at normal body temperature.13,14,15,16,17 This 3′-DNA/siRNA construct (initially termed “crook siRNA”) is here referred to as “passenger-3′-DNA/ds siRNA” to reflect the position of the 19nt DNA extension at the 3′ end of the siRNA passenger strand. The siRNA guide strand sequence of this 3′-DNA/ds siRNA construct was directed against a viral mRNA of the human papillomavirus (HPV), namely HPV E7 mRNA. Passenger-3′-DNA/ds E7 siRNA-induced selective knock-down of HPV E7 mRNA in human cervical cancer cells grown in culture (SiHa cells).12 HPV E6 and E7 are the oncogenic driving force in human cervical cancer and, specifically, HPV E7 alone is essential for cervical cancer cell viability.18 In line with this survival function of HPV E7, it was observed that targeting HPV E7 with passenger-3′-DNA/ds E7 siRNA-induced apoptosis of SiHa cervical cancer cells.12 Importantly, the passenger-3′-DNA/ds E7 siRNA had no apparent effect when transfected into HPV-negative human cells, either noncancerous or cancerous.12 This indicates that the synthetic 3′-DNA/ds siRNA construct does not, by itself, induce apoptosis or other form of cell killing. Induction of apoptosis in the SiHa cervical cancer cells was therefore attributed to the observed silencing of exogenous HPV E7 by passenger-3′-DNA/ds E7 siRNA.12
It seems remarkable that the precise molecular interactions involved in RNAi can accommodate such substantial modification of the initiating ds siRNA. Indeed, it was this consideration that originally led us to choose the siRNA passenger strand for 3′-DNA extension, the rationale being that the passenger strand might require less structural stringency than the guide strand, which determines the sequence-specificity of target mRNA cleavage. However, others have subsequently reported elegant studies indicating that both passenger and guide strands are mechanistically involved in RISC activation.7,8 In addition, the chemical structure of the siRNA passenger strand appears to be important for the overall process of RNAi. Chemical modifications that impair cleavage of the passenger strand also impact upon the efficiency of RISC activation and induction of RNAi.8 We therefore decided to probe RNAi induction by synthetic 3′-DNA/ds siRNA constructs in more detail (see Supplementary Figure S1). Here, we have compared unmodified ds siRNA with the following 3′-DNA/siRNA constructs: (I) guide-3′-DNA/ds siRNA, (II) passenger-3′-DNA/ds siRNA, (III) guide-3′-DNA/ss siRNA, and (IV) passenger-3′-DNA/ss siRNA (see Figure 1a for schematic).
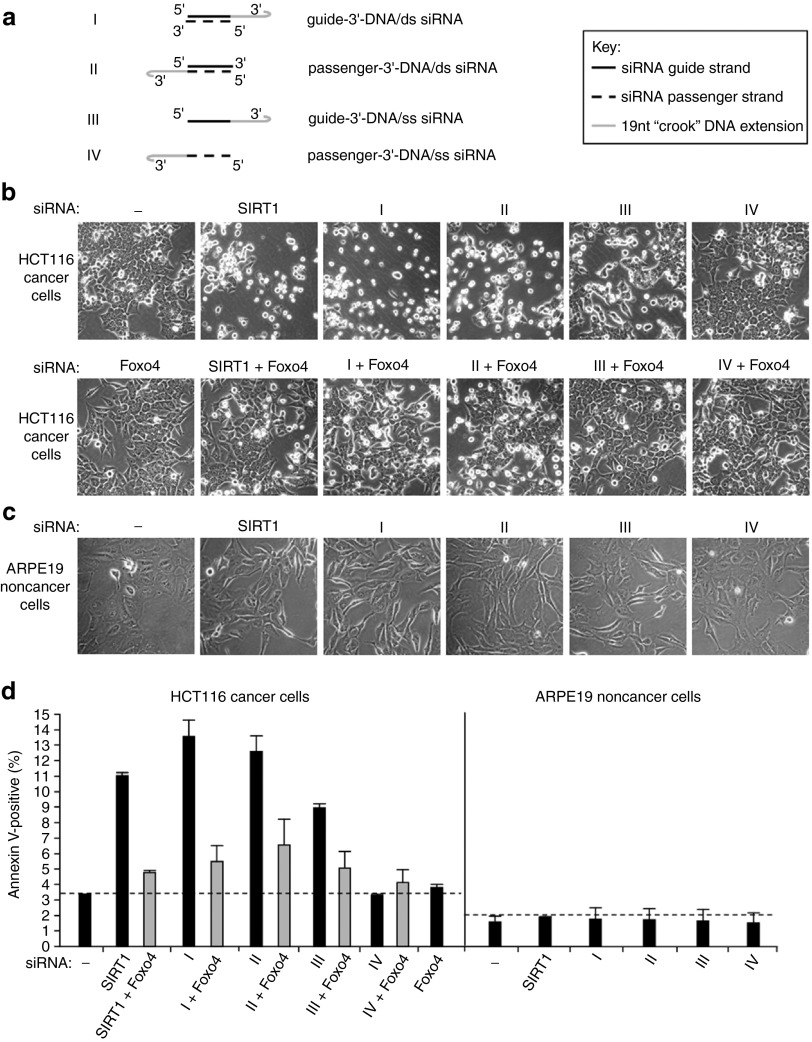
Figure 1.
Phenotypic effects of 3′-DNA/SIRT1 siRNA constructs I–IV on HCT116 cancer cells and ARPE19 noncancer cells. (a) Schematical representation of the four different 3′-DNA/SIRT1 siRNA constructs that are used in this study and compared throughout to standard unmodified SIRT1 siRNA. I = double-stranded SIRT1 siRNA with a 19 nucleotide “crook” DNA extension on the 3′ end of the siRNA guide strand; II = double-stranded SIRT1 siRNA with the 19nt DNA extension on the 3′ end of the siRNA passenger strand; III = single-stranded SIRT1 siRNA (guide strand) with the 19nt DNA extension on the 3′ end of the siRNA guide strand; IV = single-stranded SIRT1 siRNA (passenger strand) with the DNA extension on the 3′ end of the siRNA passenger strand (b) Phase contrast micrograph images of HCT116 cells 48 hours after transfection with 3′-DNA/SIRT1 siRNA constructs or standard SIRT1 siRNA (upper panel) or following cotransfection of cells with Foxo4 siRNA (lower panel) as indicated. Untreated control = (−). (c) Phase contrast images of ARPE19 cells 72 hours after transfection with the indicated 3′-DNA/SIRT1 siRNA constructs or standard SIRT1 siRNA. (d) Quantification by annexin V-labeling of the apoptotic phenotype induced by 3′-DNA/SIRT1 siRNA constructs 48 hours after transfection of HCT116 cells and its rescue by Foxo4 cosilencing (left). Apoptotic levels in ARPE19 cells as determined by annexin V-labeling 72 hours after transfection of ARPE19 cells (right). Background levels of apoptosis are indicated by the dotted line.
The chosen cellular target was SIRT1 mRNA. SIRT1 is a highly conserved NAD+-dependent deacetylase. SIRT1 functions as an epigenetic regulator and is essential for normal embryonic development, differentiation and homeostasis (see ref. 19 for a recent review). The broad range of SIRT1 functions is achieved by targeted de-acetylation of histones, and also de-acetylation of regulatory proteins and transcription factors, including the tumor suppressor p53.19 Imbalance of SIRT1 is implicated in various disease processes, including neurodegeneration, diabetes and cancer.19 SIRT1 thus represents an important target for the development of novel therapeutic agents.
The ability of synthetic 3′-DNA/siRNA constructs to deplete SIRT1 expression in human cell lines was assessed at the levels of cellular mRNA, protein, and phenotypic outcome. Molecular complexing between RISC/SIRT1 mRNA was evidenced by coimmunoprecipitation with Ago2, eIF6, and C3PO, suggesting that silencing by 3′-DNA/siRNA constructs occurs via RNAi and activated RISC rather than a RNase H-driven mechanism. The 3′ DNA/siRNA constructs containing the guide strand for SIRT1 mRNA (constructs I–III; Figure 1a) induced efficient SIRT1 knock-down and cancer-specific apoptosis. As expected, passenger-3′-DNA/ss siRNA, which lacks the SIRT1 guide strand, had no effect. The 3′-DNA mini-hairpin structure formed by the DNA extension increased siRNA stability in serum and the advantages of 3′-DNA/RNA constructs for development of novel RNA-based therapeutics are considered.
Results
Synthetic siRNA constructs were transfected into human HCT116 cancer and ARPE19 noncancer cell lines grown in culture. A schematic of the four 3′-DNA/SIRT1 siRNA constructs used in this study is shown in Figure 1a. To facilitate initial screening for SIRT1 depletion, we exploited the cancer-specific survival function of SIRT1. It is established that RNAi-mediated silencing of SIRT1 induces apoptosis in human cancer cell lines, whereas in noncancer cell lines SIRT1 silencing has no apparent effect on cell viability.20,21
Induction of apoptosis in HCT116 cancer cells by 3′-DNA/SIRT1 siRNA constructs
Early passage human HCT116 cancer cells were monitored by phase contrast microscopy following 3′-DNA/siRNA transfections. Apoptosis was assessed by annexin V-labeling with FACS analysis (Materials and Methods). Visualization showed typical monolayer growth of HCT116 cells at subconfluence with a few floating apoptotic cells (Figure 1b, top left-hand panel). The remaining upper panels in Figure 1b show the cells 48 hours following transfection with unmodified SIRT1 siRNA or with 3′-DNA/SIRT1 siRNA constructs I–IV, as indicated. The unmodified SIRT1 siRNA-induced massive apoptosis of the HCT116 cancer cells within 48 hours of transfection (Figure 1b,d). This confirms previous observations on SIRT1 silencing in HCT116 cells.20 Passenger-3′-DNA/ds SIRT1 siRNA also induced apoptosis (construct II, Figure 1b,d). This is consistent with, and extends, previous results in which passenger-3′-DNA/ds HPV E7 siRNA was shown to induce apoptosis via selective knock-down of exogenous viral HPV E7 mRNA in HPV-positive human cancer cells.12
The novel 3′-DNA/SIRT1 siRNA constructs I and III also induced apoptosis when transfected into HCT116 cells (Figure 1b,d). Both these constructs, guide-3′-DNA/ds SIRT1 siRNA and guide-3′-DNA/ss SIRT1 siRNA, contain the siRNA guide strand for SIRT1 mRNA. Their ability to induce apoptosis is therefore consistent with SIRT1 silencing. In contrast, passenger-3′-DNA/ss SIRT1 siRNA (construct IV, Figure 1a), which lacks antisense homology for SIRT1 mRNA, had no apparent effect and failed to induce apoptosis (Figure 1b,d).
The failure of passenger-3′-DNA/ss SIRT1 siRNA to induce apoptosis in HCT116 cancer cells is important. This demonstrates that the 3′-DNA/siRNA construct is not intrinsically cytotoxic, nor does it induce apoptosis by any stress-related mechanism. It follows that the induction of apoptosis by passenger-3′-DNA/ds siRNA, by guide-3′-DNA/ds siRNA and by guide-3′-DNA/ss siRNA (constructs I–III; Figure 1b,d) is attributable to the sequence of the guide strand, which is targeted against SIRT1 mRNA. For subsequent experiments, passenger-3′-DNA/ss siRNA, which carries the 3′-DNA extension but lacks homology for the SIRT1 target mRNA, was included as a negative control for comparison with the three 3′-DNA/siRNA constructs containing the SIRT1 siRNA guide strand (Figure 1a; constructs I–III).
Cosilencing Foxo4 rescues HCT116 cells from apoptosis
The apoptotic pathway induced in HCT116 cells by SIRT1 silencing requires the transcription factor Foxo4.20 RNAi-mediated cosilencing of Foxo4 together with SIRT1 rescues the cells from apoptosis, which is otherwise induced by SIRT1 depletion alone.20 We next asked if Foxo4 is required for the proapoptotic effects induced by 3′-DNA/SIRT1 siRNA constructs in HCT116 cells. Cosilencing Foxo4 with the 3′-DNA/SIRT1 siRNA constructs I–III rescued HCT116 cells from apoptosis (Figure 1b, lower panel and Figure 1d). RNAi-induced silencing of Foxo4 alone, or Foxo4 cosilencing with passenger-3′-DNA/ss siRNA (construct IV), had no effect on HCT116 cell viability (Figure 1b, lower panel and Figure 1d). Thus, the apoptotic phenotype induced by constructs I–III was Foxo4 dependent and equivalent to that induced by unmodified SIRT1 siRNA (see above).
ARPE19 noncancer cells are refractory to 3′-DNA/SIRT1 siRNA constructs
SIRT1 is known to be required for HCT116 cell viability, but SIRT1 is not required for ARPE19 cell viability.20 We now demonstrate that in ARPE19 cells, none of the 3′-DNA/SIRT1 siRNA constructs I–IV induce apoptosis (Figure 1c,d). Indeed, no adverse effects on the cells were apparent over a period of 72 hours after transfection. This confirms that none of the 3′-DNA/SIRT1 siRNA constructs (I–IV; see Figure 1a) is intrinsically cytotoxic, as evidenced by transfection into human ARPE19 noncancer epithelial cells.
Selective knock-down of SIRT1 mRNA and protein in both HCT116 and ARPE19 cells
The above results with HCT116 cells indicate selective knock-down of SIRT1 by 3′-DNA/siRNA constructs which contain the SIRT1 siRNA guide strand. To confirm this, we next assessed SIRT1 mRNA levels following transfection with the 3′-DNA/ SIRT1 siRNA constructs I–IV. For both HCT116 cells and ARPE19 cells, >80% SIRT1 mRNA knock-down was observed for each of the three 3′-DNA/siRNA constructs containing the SIRT1 siRNA guide strand (constructs I–III; Figure 2a,b respectively). In contrast, little if any SIRT1 mRNA depletion was observed following transfection with passenger-3′-DNA/ss siRNA, which lacks antisense homology for SIRT1 mRNA (construct IV; Figure 2a,b). These effects on SIRT1 mRNA levels were mirrored at the level of SIRT1 protein (Figure 2c, HCT116 cells; Figure 2d, ARPE19 cells). We conclude that 3′-DNA/SIRT1 siRNA constructs containing the SIRT1 siRNA guide strand induce SIRT1 mRNA depletion, with consequent depletion of SIRT1 protein. Importantly, SIRT1 knock-down is observed in both HCT116 cells and ARPE19 cells (Figure 2). Thus, the lack of apoptosis in ARPE19 cells (Figure 1b,d) cannot be explained by failure of SIRT1 knock-down in this particular cell line.
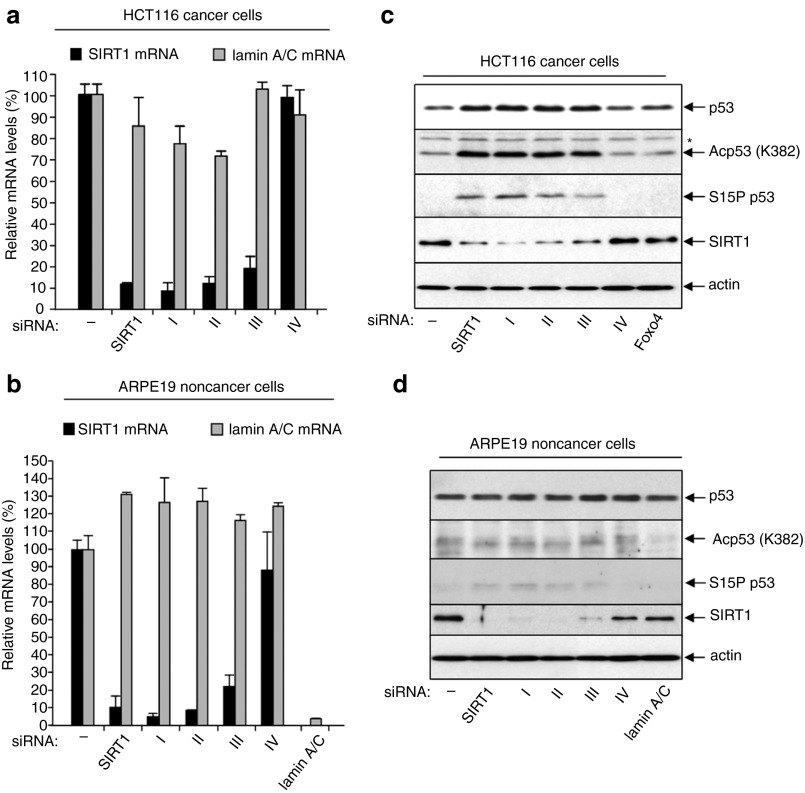
Figure 2.
Selective knock-down of SIRT1 mRNA and protein by 3′-DNA/SIRT1 siRNA constructs I–III in HCT116 and ARPE19 cells. Levels of SIRT1 mRNA and lamin A/C mRNA in (a) HCT116 cells and (b) ARPE19 cells 48 hours after transfection with the indicated 3′-DNA/SIRT1 siRNA constructs; quantitative RT-PCR data are shown, mean ± SD of four independent mRNA determinations. Immunoblots showing the effects of the 3′-DNA/SIRT1 siRNA constructs on SIRT1 protein levels in (c) HCT116 cells and (d) ARPE19 cells. Actin shows equivalent total protein loading. Effects on p53 protein and p53 acetylation and phosphorylation at specific sites is also shown (see text for details; *= nonspecific band).
Effects on p53 protein levels, phosphorylation, and acetylation
In nonstressed cells, p53 protein is present in its latent form with a very short half-life. In response to stress, p53 is stabilized by posttranslational modifications, including S15 phosphorylation and K382 acetylation. Different permutations of p53 modification help couple the p53 stress response with the nature of the stress suffered by the cell.22 Downregulation of activated p53 involves SIRT1-mediated de-acetylation, and this attenuates the p53 stress response.22
Importantly, RNAi can be induced experimentally without activating the p53 response.20,23,24,25 This is evident in the present study for siRNA-induced knock-down of Foxo4 in HCT116 cells (Figure 2c), and also for siRNA-induced knock-down of lamin A/C in ARPE19 cells (Figure 2d). In both cases, cellular p53 protein levels remain low, and no changes in phosphorylation at S15P nor acetylation at K382Ac are evident. In addition, there is no evidence of p53 activation in ARPE19 cells transfected with any of the 3′-DNA/SIRT1 siRNAs used in this study (constructs I–IV, Figure 2c,d). This demonstrates that the 3′-DNA/siRNA construct per se does not induce a p53 response.
In contrast to ARPE19 noncancer cells, transfection with 3′-DNA/siRNAs containing the SIRT1 siRNA guide strand (constructs I–III) induced apoptosis in the HCT116 cancer cells (see above, Figure 1b,d). This correlated with activation of the p53 protein, marked by S15 phosphorylation and K382 acetylation (Figure 2c). Since p53 is constitutively suppressed by SIRT1 in HCT116 cells,20 this result is consistent with targeted SIRT1 silencing by each of the 3′-DNA/siRNA constructs I–III. The negative control, passenger-3′-DNA/ss siRNA (construct IV), had no effect on p53 protein levels, nor upon p53 posttranslational modifications at S15 and K382 (Figure 2c). Thus, activation of p53 following transfection of HCT116 cells with 3′-DNA/siRNAs containing the SIRT1 siRNA guide strand is not attributable to a nonspecific response to the 3′-DNA/siRNA, but instead reflects selective silencing of SIRT1 via the SIRT1 siRNA guide strand.
Overall, the above results indicate that 3′-DNA/SIRT1 siRNA constructs that contain a siRNA guide strand can induce selective knock-down of the target SIRT1 mRNA, together with the predicted phenotypic consequences of SIRT1 silencing, the latter being dependent upon cellular context (i.e. HCT116 cancer cells versus ARPE19 noncancer cells). In contrast, passenger 3′-DNA/SIRT1 ssRNA, which lacks the SIRT1 guide strand, is ineffective for SIRT1 mRNA knock-down and also fails to induce any SIRT1-associated phenotypic changes established for human cells grown in culture.
Ago2 immunoprecipitation pulls down SIRT1 mRNA and RISC components C3PO and eIF6
The above results indicate that 3′-DNA/SIRT1 siRNA constructs containing the SIRT1 guide strand are competent for endogenous gene silencing via selective SIRT1 mRNA depletion in human cells. We next asked if activated RISC is involved in 3′-DNA/SIRT1 siRNA-mediated depletion of SIRT1 mRNA. For this purpose, we immunoprecipitated Ago2, a major component of RISC, and probed for coimmunoprecipitating SIRT1 mRNA (Materials and Methods). We reasoned that any association of SIRT1 mRNA with Ago2 would be indicative of activated RISC. The site of mRNA cleavage is determined by the siRNA guide strand sequence, in this case located in exon 8 of SIRT1 mRNA (nts 1550–1568, see Materials and Methods). The quantification of SIRT1 mRNA employed qPCR primers located on either side of the siRNA guide sequence (nts 1401–1421 and 1704-1684 respectively, see Materials and Methods) and therefore report on intact, uncleaved SIRT1 mRNA. Since complexing between Ago2 and target mRNA is a transient association, prior to mRNA cleavage and degradation, it was predicted that the recovery of full-length SIRT1 mRNA would be very low. Nonetheless, SIRT1 mRNA was detectable in complex with Ago2, and the levels were statistically significant (Figure 3a; P < 0.005). Moreover, the pull-down of full-length SIRT1 mRNA was six- to eightfold greater following transfection with constructs I–III (which induce SIRT1 mRNA degradation), compared with construct IV (inactive for induction of SIRT1 mRNA degradation) (Figures 2 and 4). These results are consistent with transient association of full-length SIRT1 mRNA with activated RISC in HCT116 cells transfected with 3′-DNA/SIRT1 siRNA constructs containing the SIRT1 guide sequence.
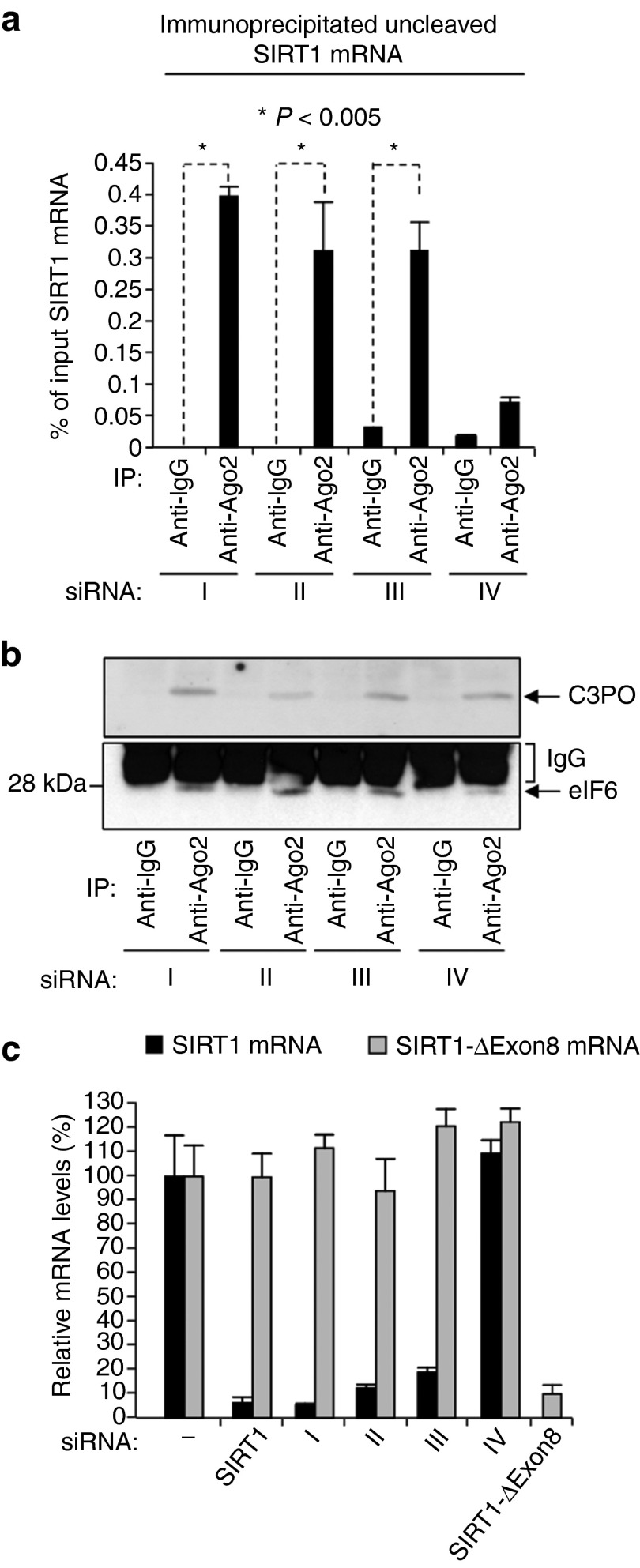
Figure 3.
Coimmunoprecipitation of endogenous SIRT1 mRNA with Ago2 protein in HCT116 cells transfected with 3′-DNA/SIRT1 siRNA constructs. (a) Levels of SIRT1 mRNA immunoprecipitated with Ago2 antibody or normal mouse IgG control antibody. Immunoprecipitated SIRT1 mRNA levels are expressed as a percentage of the input SIRT1 mRNA levels. HCT116 cell extracts for the immunoprecipitations were prepared 24 hours after transfection with the indicated 3′-DNA/SIRT1 siRNA constructs (see Materials and Methods). (b) Immunoblots showing coimmunoprecipitation of C3PO and eIF6 with Ago2 in cells transfected with 3′-DNA/SIRT1 siRNA constructs. eIF6 (arrowed) runs just ahead of IgG light chain (indicated). (c) Levels of full-length SIRT1 mRNA and splice variant SIRT1-ΔExon8 mRNA in HCT116 cells 48 hours after transfection with the indicated siRNAs or 3′-DNA/SIRT1 siRNA constructs; quantitative RT-PCR data are shown, mean ± SD of four independent mRNA determinations.
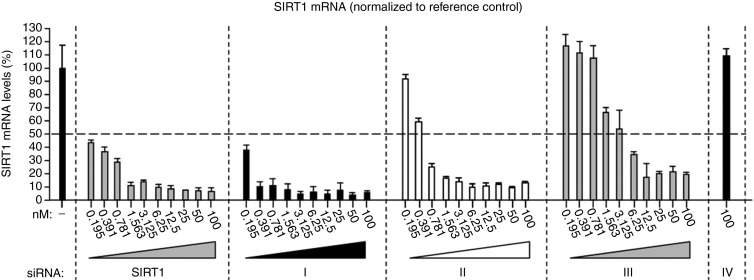
Figure 4.
Dose responses for SIRT1 mRNA knock-down by SIRT1 siRNA and 3′-DNA/SIRT1 siRNA constructs. SIRT1 mRNA levels in HCT116 cells 48 hours after transfection with indicated concentrations of the 3′-DNA/SIRT1 siRNA constructs (100 nmol/l to 195 pmol/l, twofold dilution series). SIRT1 mRNA knock-down by standard SIRT1 siRNA was also assessed for comparison of efficacy. mRNA levels determined by quantitative RT-PCR; mean ± SD of four independent mRNA determinations. The SIRT1 mRNA results were normalized to lamin A/C mRNA in each individual sample.
The observed association of SIRT1 mRNA with Ago2 indicates its recognition by 3′-DNA/SIRT1 siRNA-activated RISC. In further support of this, we also detected C3PO, another component of RISC (see Introduction) and eIF6 in the same samples (Figure 3b). The eIF6 protein is known to associate with activated RISC and is required for RNAi-mediated regulation of gene expression in human cells.26 Overall, the above results indicate that 3′-DNA/SIRT1 siRNA constructs I–III can enter the RISC complex via Ago2 and direct RNAi-mediated depletion of SIRT1 mRNA. Thus, despite their abnormal 3′-molecular structure, 3′-DNA/SIRT1 siRNA constructs appear able to induce RNAi via activating the RISC complex and selectively targeting SIRT1 mRNA via the siRNA guide strand and Ago2 complexes.
Evidence for siRNA-induced silencing versus RNase H-mediated silencing
In the above experiments, we cannot formally exclude the involvement of RNase H in the observed depletion of SIRT1 mRNA. In order to investigate this further, we compared the depletion of SIRT1 mRNA with the depletion of SIRT1-ΔExon8 mRNA, a newly discovered splice variant of SIRT1 which is expressed in HCT116 cells.27 The rationale underlying this experiment arises from the observation that the RNase H-driven antisense mechanism targets both pre-mRNA and mRNA.28 RNase H is thus predicted to silence all mRNA splice variants of a target gene via depletion of the precursor pre-mRNA. In contrast to RNase H, siRNA-induced silencing by RNA interference is specific for the mRNA target, and nuclear pre-mRNA remains intact.28 Thus, for siRNA-induced silencing the pre-mRNA and any mRNA splice variants which lack the siRNA target sequence should be unaffected. From this, we predicted that RNAi-induced silencing of SIRT1 should be selective for SIRT1 full-length mRNA over the SIRT1-ΔExon8 splice variant, since the siRNA target sequence lies within exon 8 of SIRT1 mRNA.27 The SIRT1-ΔExon8 splice variant lacks exon 8 and hence also lacks the siRNA target sequence.27
Using PCR primers that distinguish between SIRT1 full-length mRNA and SIRT1-ΔExon8 splice variant mRNA, we show selective depletion of SIRT1 full-length mRNA with little, if any effect on SIRT1-ΔExon8 splice variant mRNA (Figure 3c; 3′-DNA/siRNA constructs I–III). As expected construct IV, which lacks SIRT1 antisense sequence homology, had no effect on either mRNA species (Figure 3c). We conclude that 3′-DNA/SIRT1 siRNA constructs I–III selectively target full-length SIRT1 mRNA, without effect on SIRT1-ΔExon8 splice variant mRNA. The SIRT1-ΔExon8 splice variant mRNA is nonetheless susceptible to RNAi, and can be selectively depleted by an siRNA targeted against the junctional sequence of this splice variant27 (see also Figure 3c). Overall, these results argue against involvement of an RNase H-driven mechanism accounting for the observed phenotypes and depletion of SIRT1 mRNA in cells transfected with 3′-DNA/SIRT1 siRNA constructs I–III (Figures 1–5). Thus, we conclude that the observed depletion of SIRT1 mRNA (Figures 2–4) is most likely effected by siRNA-induced, RISC-mediated RNAi.
Figure 5.
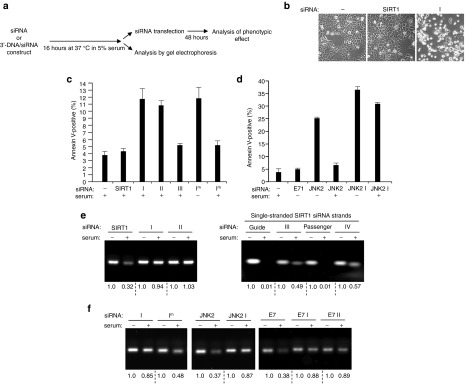
Comparison of the susceptibility of unmodified siRNAs and 3′-DNA/siRNA constructs to degradation by serum nucleases. (a) Schematic indicating the dual experimental approach used to assess the stability of unmodified siRNAs and 3′-DNA/siRNA constructs in serum. Analysis of the phenotypic effect 48 hours after transfection provides a readout of whether RNAi functionality is retained or lost. Visualization by gel electrophoresis allows levels of degradation of the siRNAs or constructs to be directly compared. (b) Phase contrast images of HCT116 cells 48 hours after transfection with serum-preincubated SIRT1 siRNA or serum-preincubated 3′-DNA/SIRT1 siRNA construct I. (c) Quantification by annexin V-labeling of apoptosis induced by SIRT1 siRNA and 3′-DNA/SIRT1 siRNA constructs I–III after 16 hours incubation in 5% serum prior to transfection. Apoptotic levels of HCT116 cells were determined 48 hours after transfection. Mean ± SD of three independent experiments. Induction of apoptosis by a novel 3′-DNA/SIRT1 siRNA construct that has a different 3′ 19 nucleotide DNA extension on the guide strand (Im) was also tested, both with no serum preincubation (−) and after serum preincubation (+). (d) Levels of apoptosis induced in HCT116 cells 48 hours after transfection with unmodified JNK2 siRNA or 3′-DNA/JNK2 siRNA construct I (JNK2 I; see Materials and Methods). The effects of no preincubation in serum (−) and 16 hours preincubation in 5% serum (+) are compared. The effects of transfection of 3′-DNA/E7 siRNA construct I (E7 I; see Materials and Methods) are also shown (no serum preincubation), which failed to induce apoptosis, as expected since HCT116 cells do not express HPV E7 mRNA (see also ref. 12). (e) Gel electrophoresis analysis of SIRT1 siRNA and 3′-DNA/SIRT1 siRNA constructs I–IV following incubation of equimolar amounts in no serum (−) or 5% serum (+) for 16 hours. (f) Effect of 16 hours incubation in no serum (−) or 5% serum (+) on levels of the indicated unmodified siRNAs or 3′-DNA/siRNA constructs. The stability of 3′-DNA/SIRT1 siRNA construct I and that of 3′-DNA/SIRT1 siRNA construct Im which differs only in the sequence of the 19 nucleotide DNA extension were compared. Unmodified JNK2 siRNA versus 3′-DNA/JNK2 siRNA construct I (JNK2 I); unmodified E7 siRNA versus 3′-DNA/E7 siRNA constructs I and II (E7 I; E7 II; see Materials and Methods).
Dose responses for SIRT1 mRNA knock-down by 3′-DNA/SIRT1 siRNA constructs
We next compared the efficacy of SIRT1 mRNA knock-down by each of the different 3′-DNA/SIRT1 siRNA constructs relative to each other, and also relative to unmodified ds SIRT1 siRNA. Twofold dilution series from 100 nmol/l to 195 pmol/l for each construct were performed in HCT116 cells with quantitative PCR at each dilution point to determine SIRT1 mRNA levels. Lamin A/C mRNA was quantitated as control. All the 3′-DNA/SIRT1 siRNA constructs containing the SIRT1 siRNA guide strand gave around 90% SIRT1 mRNA knock-down at 100 nmol/l (constructs I–III; Figure 4). The IC50s for unmodified SIRT1 siRNA and for guide-3′-DNA/ds SIRT1 siRNA (construct I) were both less than 0.195 nmol/l. Passenger-3′-DNA/ds SIRT1 siRNA (construct II) and guide-3′-DNA/ss SIRT1 siRNA (construct III) gave IC50s of 0.495 nmol/l and 3.2 nmol/l respectively. As expected, passenger-3′-DNA/ss siRNA (construct IV), which carries the 3′-DNA extension but lacks SIRT1 antisense homology, failed to deplete SIRT1 mRNA (Figure 4).
Enhanced stability in serum of 3′-DNA/ds siRNA constructs
The above results demonstrate that the efficacy of guide-3′-DNA/ds SIRT1 siRNA (construct I) is equivalent to unmodified SIRT1 siRNA for the targeted depletion of SIRT1 mRNA (Figure 4). For passenger-3′-DNA/ds SIRT1 siRNA (construct II) and guide-3′-DNA/ss SIRT1 siRNA (construct III), IC50s were in the low or subnanomolar range.
To ask if the 3′-DNA/SIRT1 siRNA constructs have any advantage over unmodified SIRT1 siRNA, we next compared susceptibility to nuclease attack. In HCT116 cancer cells, the selective depletion of SIRT1 by SIRT1 siRNA or by 3′-DNA/SIRT1 siRNA constructs I–III resulted in apoptotic cell death within 48 hours of siRNA transfection (Figures 1 and 2). This was exploited here by asking whether unmodified SIRT1 siRNA or the 3′-DNA/SIRT1 siRNA constructs retain their RNAi activity and are still able to induce apoptosis if preincubated in 5% serum for 16 hours prior to cell transfection (Figure 5a). Serum is rich in both DNA and RNA nucleases, and we anticipated that the proapoptotic activity of unmodified SIRT1 siRNA in HCT116 cells (Figure 1) may be partially or completely destroyed by preincubation in cell culture medium containing 5% serum. Indeed, this proved to be the case, with levels of apoptosis similar to those observed for the untreated controls (Figure 5b,c; see also Figure 1d). In contrast to unmodified SIRT1 siRNA, the guide-3′-DNA/ds SIRT1 siRNA construct (construct I) proved to be resistant to inactivation by serum, as indicated by its ability to induce apoptotic cell killing of HCT116 cancer cells at similar levels with and without serum preincubation (Figure 5b,c; see also Figure 1d). The passenger-3′-DNA/ds SIRT1 siRNA construct (construct II) also appeared to be resistant to serum inactivation as it also retained its ability to induce apoptosis in the HCT116 cancer cells (Figure 5c) following serum preincubation and at a similar level as without serum preincubation (see Figure 1d). In contrast, guide-3′-DNA/ss SIRT1 siRNA construct (construct III) that was preincubated in 5% serum had lost its ability to induce apoptosis in HCT116 cancer cells (Figure 5c, see also Figure 1d). This suggests that resistance to serum nucleases conferred by the 19nt DNA extension also requires the double-stranded RNA structure of the siRNA.
The 19nt DNA extension on these constructs forms a nuclease-resistant mini-hairpin structure at its 3′ end.13,14,15,16,17 To determine whether this mini-hairpin structure is important for the resistance conferred by the 19nt DNA extension, a different 19nt DNA extension was tested (see Materials and Methods) that is identical to the original except for the inversion of two bases which make it unable to form a mini-hairpin structure.17 This alternative 19 nt DNA extension was added to 3′ end of the guide strand of ds SIRT1 siRNA (guide-3′-DNA/ds SIRT1 siRNA construct Im). As expected, construct Im induced apoptosis in the HCT116 cancer cells, consistent with SIRT1 depletion, but induction of apoptosis was largely abolished if the construct was preincubated in serum (Figure 5c). This indicates that it is the mini-hairpin structure of the DNA extension that confers protection of the siRNA to nuclease attack and that any 19nt DNA extension will not suffice.
We assume that this DNA extension can provide similar nuclease protection to any siRNA. To test this assumption, we chose a second endogenous cellular target for depletion by RNAi. The second target chosen was the c-Jun N-terminal kinase JNK2 as we have previously shown that its depletion by RNAi also induces apoptosis in HCT116 cancer cells.25 This enabled the effects of serum upon the stability of the siRNA to be easily screened by quantification of levels of apoptosis induced as for SIRT1 siRNA. JNK2 siRNA induced higher levels of apoptosis in the HCT116 cancer cells than SIRT1 depletion consistent with previous studies25 (Figure 5d) but following preincubation in serum the ability of JNK2 siRNA to induce apoptosis was lost. In contrast, guide-3′-DNA/ds JNK2 siRNA (JNK2 construct I) induced similar levels of apoptosis with and without serum preincubation (Figure 5d, JNK2 I).
The susceptibility of the various 3′-DNA/siRNA constructs and unmodified siRNAs to degradation by serum nucleases was also analyzed by their direct visualization on agarose gels following incubation for 16 hours in either 5% serum or no serum (Figure 5a,e,f, see Materials and Methods). siRNAs targeted against SIRT1, JNK2, and HPV E7 were employed for this study. Incubation with 5% serum resulted in significant degradation of unmodified SIRT1 siRNA whereas 3′-DNA/ds SIRT1 siRNA constructs I and II appeared unaffected (Figure 5e). These results correlate with the apparent loss of RNAi activity by SIRT1 siRNA following serum preincubation and retention of activity by constructs I and II, as indicated by their abilities to still induce apoptosis in HCT116 cells (Figure 5b,c). Similar increased protection from degradation was also provided by the addition of the 19nt DNA extension to JNK2 siRNA or E7 siRNA (Figure 5f). The importance of the 3′ mini-hairpin structure formed by the DNA extension is indicated by the addition of a different 19nt DNA extension to SIRT1 siRNA which offered less protection from degradation (Figure 5f, compare construct I and Im). 3′-DNA/single stranded SIRT1 siRNA construct III was unable to induce apoptosis following serum incubation (Figure 5c), correlating with significant levels of degradation (Figure 5e). This indicates that the double-stranded RNA structure is required in addition to the 3′ DNA extension to protect the siRNA moiety from attack. Addition of the DNA extension to single-stranded siRNA did result in less degradation, however, than for unmodified single-stranded siRNA (Figure 5e).
In principle, this 19 nt DNA extension with its mini-hairpin structure can be added to any double-stranded siRNA to provide increased protection from nucleases, as indicated here for SIRT1 siRNA, JNK2 siRNA and HPV E7 siRNA. Future studies will address this further and, in particular, ask whether this 3′ 19nt DNA extension with its mini-hairpin structure is able to stabilize therapeutic siRNAs in vivo.
Discussion
In this work, we have employed SIRT1 mRNA as an endogenous mRNA target for testing selective gene silencing by synthetic 3′-DNA/siRNA constructs transfected into human cells in vitro. SIRT1 is an NAD+-dependent deacetylase.19 Following cellular stress, SIRT1 downregulates the p53 stress response by de-acetylating the p53 protein at K382.22 In addition, SIRT1 can act as a cancer-specific survival factor and is essential for the viability of many human cancer cell lines when grown in culture.20 Different 3′-DNA/SIRT1 siRNA constructs were assessed as follows: (i) for the selective depletion of SIRT1 mRNA and SIRT1 protein following transfection into noncancer (ARPE19) and cancer (HCT116) cells; (ii) for the selective killing of cancer versus noncancer cells; and (iii) for knock-on effects of SIRT1 depletion at the level of p53 acetylation in HCT116 cancer cells, which require SIRT1 in order to maintain constitutive de-acetylation of p53 under normal conditions of cell culture.20 The results indicate highly selective targeting and silencing of SIRT1 by the 3′-DNA/SIRT1 siRNA constructs which contain the SIRT1 guide strand (constructs I–III; Figures 1–3; and Results section). In contrast, passenger-3′-DNA/ss siRNA, which carries the 3′-DNA extension but lacks SIRT1 antisense homology, had no effect at any level. Overall these biochemical and phenotypic observations are consistent with the induction of RNA interference via synthetic 3′-DNA/SIRT1 siRNAs containing the SIRT1 guide strand.
Central to the process of RNA interference is RISC, a multi-protein complex within which RNA processing occurs. The conversion of pre-RISC (containing Ago2 loaded with ds siRNA) to activated RISC requires the slicer activity of Ago2 to cleave the siRNA passenger strand. This is followed by further cleavage and clearance of the passenger strand by endonuclease C3PO (Introduction). C3PO is itself composed of two TRAX and six translin subunits. These assemble into an asymmetric structure with specific residues positioned at the atomic level (i) for binding ssRNA and (ii) for nuclease activity (see ref. 8 for details). The interaction between Ago2 and siRNA is believed to be highly dynamic with large conformational shifts in both components.29,30,31,32 This dynamic interplay is likely to include C3PO subunits as well. Such considerations may explain why 2′-O-methylation of siRNA can interfere with passenger strand cleavage by C3PO, and hence with activation of RISC.8 They may also explain the aberrant mRNA cleavage observed when cells are treated with ssRNA (guide strand) modified with 2′F-ribose coupled with 5′-end phosphorylation.33
Given the molecular precision and interactions which operate during RISC activation and target mRNA degradation (outlined above), it seems remarkable that the initiating siRNA can accommodate a 3′ 19 nt DNA extension containing an intrinsic hairpin structure (see above). In theory, this novel 3′-DNA/siRNA construct may operate via a RISC-independent mechanism, possibly via direct mRNA association or perturbation of other RNA processing machinery. However, our present results indicate that 3′-DNA/SIRT1 siRNA constructs containing the SIRT1 siRNA guide strand are able (i) to induce targeted mRNA knock-down and (ii) to associate with Ago2 and other components of the RISC complex, such as C3PO and eIF6. These results support the notion that 3′-DNA/siRNA constructs are competent for selective gene silencing via RNAi-mediated mRNA depletion. Our results argue against a RNase H-driven antisense mechanism as this targets both pre-mRNA and mRNA,28 and the 3′-DNA/SIRT1 siRNA constructs had no effect on mRNA expression of the SIRT1-ΔExon8 splice variant (Figure 3c). RNAi-induced silencing of SIRT1 is selective for SIRT1 full-length mRNA over the SIRT1-ExonΔ8 splice variant since the siRNA target sequence lies within exon 8 of SIRT1 mRNA.27 The SIRT1-ExonΔ8 splice variant lacks exon 8 and hence also lacks the siRNA target sequence.27
We believe that 3′-DNA/ds siRNA constructs may prove useful for studying the molecular mechanism of RISC activation in mammalian cells. Our work shows that the 3′-DNA extension can be employed as a differential marker on either passenger or guide RNA strands, with little detrimental effect on RISC activation. Thus, it may be possible to employ differential labeling of each siRNA strand for detailed analysis of the functional interplay between RISC components Ago2, C3PO, and eIF6 during the initiation and process of RNA interference.
We show that both guide and passenger strands of the siRNA can accommodate the 3′-DNA extension with remarkably little reduction in RNAi efficacy (Figure 4). Even single-stranded guide-3′-DNA/SIRT1 siRNA gave good SIRT1 mRNA knock-down, with an IC50 of 3.2 nmol/l. We further show that this 3′ 19 nt DNA extension helps to protect the siRNA from degradation by serum nucleases (Figure 5). This was dependent upon the 3′ mini-hairpin structure of the DNA extension since a similar length DNA extension unable to form a hairpin offered little protection. In principle, this DNA extension with its mini-hairpin structure can be added to any double-stranded siRNA and provide protection from degradation by serum nucleases, as shown here for SIRT1 siRNA, for JNK2 siRNA and for HPV E7 siRNA (Figure 5). Overall, these observations identify the 19 nt DNA sequence with its 3′-mini-hairpin structure employed in this study as a novel siRNA modification with promise for overcoming problems associated synthetic siRNA stability in vivo and which may thereby aid the development of siRNA therapeutics.
From a manufacturing viewpoint, we predict that simply extending the 3′-end of synthetic siRNA with a defined DNA sequence (as employed in this work) will be more cost-effective compared with the production of chemically modified RNA for therapeutic use. The former simply requires 3′-DNA/siRNA synthesis, followed by purification to the level required for therapeutic application. In contrast, chemically modified siRNA involves siRNA synthesis, followed by purification, followed by chemical modification and finally followed by repurification to the level of purity required for therapeutic purposes.
This is a tightly controlled multi-parameter study with (i) molecular, biochemical and cellular readouts, (ii) comparison of human cancer versus noncancer cells, and (iii) an experimental model operated under basal conditions (i.e., without activating intrinsic cellular stress responses as occurs with shRNA and other silencing agents). We have performed in-depth testing of a series of 3′DNA/siRNA constructs which, although derivative, are novel. These novel constructs carry advantages for (i) their recovery and quantification from cells and tissues; (ii) efficacy under basal conditions, with no apparent adverse effects on normal cells; (iii) in the case of SIRT1 and JNK2 siRNA constructs the cancer-specific induction of apoptosis; and (iv) resistance to serum nucleases. The novel constructs may also serve as useful tools for further experimentation aimed at unravelling the mechanism of RNAi in mammalian cells. In conclusion, we propose that 3′-DNA/siRNA constructs offer an exciting new approach for (i) the therapeutic development of novel siRNAs with ease of manufacture and quantification, coupled with (ii) benefits of nuclease resistance and (iii) efficient targeted knock-down of disease-related targets.
Materials and methods
3′-DNA/siRNA constructs and siRNAs. In order to selectively knock-down full-length SIRT1 mRNA and not that of its splice variants, it is necessary for the siRNA to target a nucleotide sequence within exon 8 of SIRT1 mRNA.20,27,34 We have designed two independent SIRT1 siRNAs targeted against discrete RNA sequences within SIRT1-Exon 8. Both siRNAs give equivalent and selective knock-down of human SIRT1 mRNA.20 For the purposes of the present study, we selected one of these well-validated siRNAs, which is targeted against nts 1550–1568 in exon 8 of human SIRT1 (NM_012238): SIRT1 siRNA guide strand sequence 5′-UACAGGGUUACAGCAAAGU(dTdT)-3′; SIRT1 siRNA passenger strand sequence 5′-ACUUUGCUGUAACCCUGUA(dTdT)-3′.20 The 3′-DNA/SIRT1 siRNA constructs have the same RNA sequence as the SIRT1 siRNA but differ in the 3′ addition of a 19 nucleotide DNA extension in place of a 3′-dTdT overhang. This DNA extension (5′-tcacctcatcccgcgaagc-3′) forms a 3′-nuclease resistant DNA hairpin and can function as a DNA primer for PCR amplification enabling quantification of the 3′-DNA/siRNA constructs.12 The 3′-DNA/SIRT1 siRNA constructs were chemically synthesized 3′ to 5′, HPLC-purified and annealed as appropriate (Biospring, Frankfurt, Germany). The organization of each construct is outlined schematically in Figure 1a. Unmodified SIRT1 siRNA was included as a control throughout all experiments.
For the experiments shown in Figure 5, additional 3′-DNA/siRNA constructs directed against other mRNA targets were tested and compared with unmodified siRNA. mRNA targets chosen were JNK2, silencing of which has also been shown to induce apoptosis in HCT116 cancer cells,25 and HPV16 E7. Unmodified JNK2 siRNA and HPV16 E7 siRNA were as previously published.12,18,25 For JNK2 siRNA, the effect of the 19 nucleotide DNA extension on the 3′ end of the siRNA guide strand was tested. This construct is designated 3′-DNA/JNK2 siRNA construct I (JNK2 I) as it is equivalent to 3′-DNA/SIRT1 siRNA construct I except for the siRNA sequence. For E7, the effect of the 19 nucleotide DNA extension on the 3′ end of either the guide or the passenger siRNA strand was tested (3′-DNA/E7 siRNA constructs I and II respectively (E7 I and E7 II)). For SIRT1 siRNA, the effects of a different 19 nucleotide DNA extension (5′-tcacctcatccggccaagc-3′) that is unable to form a mini-hairpin structure17 was tested on the 3′ end of the guide strand; this is designated 3′-DNA/SIRT1 siRNA construct Im (altered bases indicated in red). Additional unmodified siRNAs were Foxo4 (guide sequence 5′-GGUCCACAUAUCGGCUUCU(dTdT)-3′; passenger sequence 5′-AGAAGCCGAUAUGUGGACC (dTdT)-3′),20 SIRT1-ΔExon8 (guide sequence 5′-UACUGAUUACUUGGAAUUA(dTdT)-3′; passenger sequence 5′-UAAUUCCAAGUAAUCAGUA(dTdT)-3′),27 lamin A/C (guide sequence 5′-UGUUCUUCUGGAAGUCCAG(dTdT)-3′; passenger sequence 5′-CUGGACUUCCAGAAGAACA(dTdT)-3′).20 All unmodified siRNAs have previously been extensively verified in human cells in vitro.20,27
Cell lines and culture. Cell lines were maintained at low passage and cultured in antibiotic-free media. HCT116 human colorectal epithelial cancer cells35 were cultured in Dulbecco's modified Eagle medium (DMEM) with 10% fetal calf serum. ARPE19 cells are a human noncancer retinal epithelial cell line36 that senesce after prolonged passage (>8 passages). They can be induced to undergo neuronal transdifferentiation by the retinoic acid analogue fenretinide.37 ARPE19 cells were cultured in DMEM-F12 with 10% fetal calf serum.
Transfection of 3′-DNA/siRNA constructs. Twenty-four hours prior to transfection, cells were seeded in six-well plates at a cell density of 6 × 104 cells (ARPE19) or 1.2 × 105 cells (HCT116) per well. Cells were transfected with 3′-DNA/siRNA constructs or control siRNAs formulated into liposomes (Oligofectamine, Invitrogen, Carlsbad, CA) as described.12,23,24,25 All siRNAs and 3′-DNA/siRNA constructs were initially used at a concentration of 100 nmol/l except for the DNA/siRNA construct dilution series (Figure 4; 10 point 2-fold dilution series; 100 nmol/l to 195 pmol/l and for Figure 5 siRNA and 3′-DNA/siRNA starting concentration in serum of 20 nmol/l). The phenotypic effects shown in Figures 1–3 are for transfections at 100 nmol/l. Please note that similar phenotypic effects were observed down to 3 nmol/l for guide-3′-DNA/ds SIRT1 siRNA; down to 6 nmol/l for passenger-3′-DNA/ds SIRT1 siRNA; and down to 12.5 nmol/l for guide-3′-DNA/ss SIRT1 siRNA (data not shown).
Quantification of mRNA knock-down. Total RNA was isolated from cells using an RNeasy kit (Qiagen) and quantitated by UV spectroscopy (GeneSpecV), as previously described.24 For quantitative real-time RT-PCR, 50 ng total RNA was used per reaction. Reactions were run in quadruplicate using Quantitect SYBRGreen RT-PCR kit (Qiagen) on a DNA Engine Opticon2 system. For quantification of full-length SIRT1 mRNA, primers 5′-CTAATTCCAAGTTCCATACCC-3′ (NM_012238, nts 1401–1421, exons 7/8 boundary) and 5′-CTGAAGAATCTGGTGGTGAAG-3′ (NM_012238, nts 1704-1684, exon 8) were used. These primers are specific for full-length SIRT1 mRNA and do not amplify splice variant SIRT1-ΔExon8 mRNA due to their location within exon 8.27 Cycling conditions were 50 °C for 30 minutes, 94 °C for 15 minutes, followed by 36 cycles of 94 °C for 30 seconds, 55 °C for 30 seconds, 72 °C for 30 seconds. SIRT1-ΔExon8 specific primers were 5′-ACTGTGAAGCTGTACGAGGAG-3′ (NM_012238, nts 1189–1209, exon 6) and 5′-AACAGATACTGATTACTTGGA-3′ (NM_012238, nts 1984-1969 and 1410-1406, exons 7/9 boundary).27 Cycling conditions were as for full-length SIRT1 except that the annealing temperature was 53 °C. For lamin A/C mRNA quantification, primers 5′-AAGCAGCGTGAGTTTGAGAGC-3′ and 5′-AGGGTGAACTTTGGTGGGAAC-3′ were used in the thermal cycle: 50 °C for 30 minutes, 94 °C for 15 minutes, followed by 36 cycles of 94 °C for 45 seconds, 58 °C for 45 seconds, and 72 °C for 1 minutes.
RNA immunoprecipitation. RNA immunoprecipitations were performed as described38 using 10 six-well plates per 3′-DNA/siRNA construct. Twenty-four hours after transfection, equivalent numbers of transfected cells were lysed using polysome lysis buffer (100 mmol/l KCl, 5 mmol/l MgCl2, 10 mmol/l Hepes pH 7, 0.5% NP40, 1 mmol/l DTT, 100 U/ml RNase inhibitor, 2 mmol/l vanadyl ribonucleoside complexes solution, 25 μl/ml protease inhibitor cocktail (Sigma, St Louis, MO)). Following centrifugation at 16000g for 15 minutes at 4 °C, an aliquot of supernatant was removed for determination of the starting amount of SIRT1 mRNA in each immunoprecipitation reaction. The residual supernatant was divided equally into two, and incubated overnight at 4 °C with 5 µg of anti-Ago2 antibody (clone 9E8.2 RIPAb+, Millipore) or 5 µg of normal mouse IgG (Millipore). Protein-RNA complexes were collected on protein A/G agarose beads for 3 hours at 4 °C, followed by eight washes with polysome lysis buffer. One-tenth of the slurry of washed beads was added to 2xLaemmli's SDS buffer for immunoblotting. The remaining beads were resuspended in polysome lysis buffer containing 0.5% SDS and 30 µg proteinase K and incubated at 50 °C for 30 minutes to elute any immunoprecipitated RNA. RNA was extracted once with phenol-chloroform-isoamyl alcohol (25:24:1) and once with chloroform-isoamyl alcohol (24:1), and then precipitated with 2.5 vol ice-cold 100% ethanol in the presence of 1 µl glycoblue (Ambion). Purified RNA from the immunoprecipitation reactions, and inputs were analyzed for SIRT1 mRNA levels by quantitative real-time RT-PCR. For quantification of intact, uncleaved full-length SIRT1 mRNA, primers 5′-CTAATTCCAAGTTCCATACCC-3′ (NM_012238, nts 1401–1421, exons 7/8 boundary) and 5′-CTGAAGAATCTGGTGGTGAAG-3′ (NM_012238, nts 1704-1684, exon 8) were used.
Statistical analysis and calculation of IC50 values. Statistical analysis for Figure 3 was carried out using a two-tailed Student's t-test calculated with Excel. A P value of <0.05 was considered statistically significant. IC50 values representing the concentration of 3′-DNA/SIRT1 siRNA construct or unmodified SIRT1 siRNA that gives 50% knock-down of SIRT1 mRNA were calculated from the titration data of Figure 4 (qRT-PCR measurements for SIRT1 mRNA in quadruplicate normalized to lamin A/C mRNA for each concentration of 3′-DNA/siRNA (10 point 2-fold dilution series; 100 nmol/l to 195 pmol/l)). The formula used to calculate the IC50 values is, IC50 = ((a/b) × concentration at a) + concentration at a. a = % of SIRT1 mRNA obtained at a particular concentration of 3′-DNA/siRNA that is nearest to and just above 50% – 50; b= % of SIRT1 mRNA obtained at a particular concentration of 3′-DNA/siRNA that is nearest to and just above 50% to % of SIRT1 mRNA obtained at a particular concentration of 3′-DNA/siRNA that is nearest to and just below 50%.
Immunoblotting. Total cell extracts were prepared39 and equivalent protein amounts were resolved by SDS-PAGE and electroblotted onto nitrocellulose for immunoblotting. Primary antibodies were anti-SIRT1 (H300; Santa Cruz, Santa Cruz, CA), anti-p53 (DO1; Santa Cruz), anti-K382 acetylated p53 (Cell Signalling Technology), anti-S15 phosphorylated p53 (Cell Signalling Technology), anti-eIF6 (Cell Signalling Technology), anti-actin (Chemicon), and C3PO antibody.7 Visualization of bound antibodies was by ECL.
Apoptosis. Apoptotic cells were identified and quantified by flow cytometry and annexin V/propidium iodide labeling using annexin-V-Fluos kit (Roche, Basel, Switzerland) following the manufacturer's protocol.24
Gel electrophoresis analysis of 3′-DNA/siRNA constructs. For the experiments shown in Figure 5, equivalent amounts of 3′-DNA/siRNA constructs or unmodified siRNAs were preincubated in 5% serum or no serum for 16 hours at 37 °C before then either being used for transfection (starting concentration assuming no degradation of 20 nmol/l) or analyzed by gel electrophoresis. For gel electrophoresis, samples were run on a 2% agarose-1xTAE gel at 90 V for 30 minutes and visualized either by ethidium bromide staining or, for the single-stranded siRNAs and constructs, by SYBR Gold staining. Quantification of band intensities by densitometry was performed using Quantity One software (Biorad).
SUPPLEMENTARY MATERIAL Figure S1 siRNA/DNA oligonucleotide sequences of constructs.
Acknowledgments
This work was supported in part by C2D2, Funder grant number: 097829/Z/11/A, York grant number: A0119732, and in part by a Yorkshire Cancer Research core research funding award to J.M. C3PO antibody was a generous gift from Professor Qinghua Liu (Southwestern Medical Centre, University of Texas). The authors declare no conflict of interest.
Supplementary material
References
- Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, et al. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- Kim K, Lee YS, Carthew RW. Conversion of pre-RISC to holo-RISC by Ago2 during assembly of RNAi complexes. RNA. 2007;13:22–29. doi: 10.1261/rna.283207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Ye X, Jiang F, Liang C, Chen D, Peng J, et al. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science. 2009;325:750–753. doi: 10.1126/science.1176325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Huang N, Liu Y, Paroo Z, Huerta C, Li P, et al. Structure of C3PO and mechanism of human RISC activation. Nat Struct Mol Biol. 2011;18:650–657. doi: 10.1038/nsmb.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao DD, Vorhies JS, Senzer N, Nemunaitis J. siRNA vs. shRNA: similarities and differences. Adv Drug Deliv Rev. 2009;61:746–759. doi: 10.1016/j.addr.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Castanotto D, Rossi JJ. The promises and pitfalls of RNA-interference-based therapeutics. Nature. 2009;457:426–433. doi: 10.1038/nature07758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett JC, Rossi JJ, Tiemann K. Current progress of siRNA/shRNA therapeutics in clinical trials. Biotechnol J. 2011;6:1130–1146. doi: 10.1002/biot.201100054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Arzumanov AA, Gait MJ, Milner J. A bi-functional siRNA construct induces RNA interference and also primes PCR amplification for its own quantification. Nucleic Acids Res. 2005;33:e151. doi: 10.1093/nar/gni144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirao I, Nishimura Y, Tagawa Y, Watanabe K, Miura K. Extraordinarily stable mini-hairpins: electrophoretical and thermal properties of the various sequence variants of d(GCGAAAGC) and their effect on DNA sequencing. Nucleic Acids Res. 1992;20:3891–3896. doi: 10.1093/nar/20.15.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan IM, Coulson JM. A novel method to stabilise antisense oligonucleotides against exonuclease degradation. Nucleic Acids Res. 1993;21:2957–2958. doi: 10.1093/nar/21.12.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa S, Ueda T, Ishido Y, Miura K, Watanabe K, Hirao I. Nuclease resistance of an extraordinarily thermostable mini-hairpin DNA fragment, d(GCGAAGC) and its application to in vitro protein synthesis. Nucleic Acids Res. 1994;22:2217–2221. doi: 10.1093/nar/22.12.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Réfrégiers M, Laigle A, Jollès B, Chinsky L. Fluorescence resonance energy transfer analysis of the degradation of an oligonucleotide protected by a very stable hairpin. J Biomol Struct Dyn. 1996;14:365–371. doi: 10.1080/07391102.1996.10508131. [DOI] [PubMed] [Google Scholar]
- Jollès B, Réfrégiers M, Laigle A. Opening of the extraordinarily stable mini-hairpin d(GCGAAGC) Nucleic Acids Res. 1997;25:4608–4613. doi: 10.1093/nar/25.22.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Milner J. Selective silencing of viral gene expression in HPV-positive human cervical carcinoma cells treated with siRNA, a primer of RNA interference. Oncogene. 2002;21:6041–6048. doi: 10.1038/sj.onc.1205878. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford J, Jiang M, Milner J. Cancer-specific functions of SIRT1 enable human epithelial cancer cell growth and survival. Cancer Res. 2005;65:10457–10463. doi: 10.1158/0008-5472.CAN-05-1923. [DOI] [PubMed] [Google Scholar]
- Knight JR, Milner J. SIRT1, metabolism and cancer. Curr Opin Oncol. 2012;24:68–75. doi: 10.1097/CCO.0b013e32834d813b. [DOI] [PubMed] [Google Scholar]
- Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harb Perspect Biol. 2010;2:a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Milner J. Bcl-2 constitutively suppresses p53-dependent apoptosis in colorectal cancer cells. Genes Dev. 2003;17:832–837. doi: 10.1101/gad.252603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison SJ, Milner J. SIRT3 is pro-apoptotic and participates in distinct basal apoptotic pathways. Cell Cycle. 2007;6:2669–2677. doi: 10.4161/cc.6.21.4866. [DOI] [PubMed] [Google Scholar]
- Ahmed SU, Milner J. Basal cancer cell survival involves JNK2 suppression of a novel JNK1/c-Jun/Bcl-3 apoptotic network. PLoS ONE. 2009;4:e7305. doi: 10.1371/journal.pone.0007305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, et al. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- Lynch CJ, Shah ZH, Allison SJ, Ahmed SU, Ford J, Warnock LJ, et al. SIRT1 undergoes alternative splicing in a novel auto-regulatory loop with p53. PLoS ONE. 2010;5:e13502. doi: 10.1371/journal.pone.0013502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers TA, Koo S, Bennett CF, Crooke ST, Dean NM, Baker BF. Efficient reduction of target RNAs by small interfering RNA and RNase H-dependent antisense agents. A comparative analysis. J Biol Chem. 2003;278:7108–7118. doi: 10.1074/jbc.M210326200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, et al. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS. How to slice: snapshots of Argonaute in action. Silence. 2010;1:3. doi: 10.1186/1758-907X-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haringsma HJ, Li JJ, Soriano F, Kenski DM, Flanagan WM, Willingham AT. mRNA knockdown by single strand RNA is improved by chemical modifications. Nucleic Acids Res. 2012;40:4125–4136. doi: 10.1093/nar/gkr1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah ZH, Ahmed SU, Ford JR, Allison SJ, Knight JR, Milner J. A deacetylase-deficient SIRT1 variant opposes full-length SIRT1 in regulating tumor suppressor p53 and governs expression of cancer-related genes. Mol Cell Biol. 2012;32:704–716. doi: 10.1128/MCB.06448-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- Dunn KC, Aotaki-Keen AE, Putkey FR, Hjelmeland LM. ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Exp Eye Res. 1996;62:155–169. doi: 10.1006/exer.1996.0020. [DOI] [PubMed] [Google Scholar]
- Chen S, Samuel W, Fariss RN, Duncan T, Kutty RK, Wiggert B. Differentiation of human retinal pigment epithelial cells into neuronal phenotype by N-(4-hydroxyphenyl)retinamide. J Neurochem. 2003;84:972–981. doi: 10.1046/j.1471-4159.2003.01608.x. [DOI] [PubMed] [Google Scholar]
- Peritz T, Zeng F, Kannanayakal TJ, Kilk K, Eiríksdóttir E, Langel U, et al. Immunoprecipitation of mRNA-protein complexes. Nat Protoc. 2006;1:577–580. doi: 10.1038/nprot.2006.82. [DOI] [PubMed] [Google Scholar]
- Ford J, Ahmed S, Allison S, Jiang M, Milner J. JNK2-dependent regulation of SIRT1 protein stability. Cell Cycle. 2008;7:3091–3097. doi: 10.4161/cc.7.19.6799. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.