Summary
This review provides a practical introduction to superresolution microscopy from the perspective of microbiological research. Because of the small sizes of bacterial cells, superresolution methods are particularly powerful and suitable for revealing details of cellular structures that are not resolvable under conventional fluorescence light microscopy. Here we describe the methodological concepts behind three major categories of super-resolution light microscopy: photoactivated localization microscopy (PALM) and stochastic optical reconstruction microscopy (STORM), structured illumination microscopy (SIM) and stimulated emission-depletion (STED) microscopy. We then present recent applications of each of these techniques to microbial systems, which have revealed novel conformations of cellular structures and described new properties of in vivo protein function and interactions. Finally, we discuss the unique issues related to implementing each of these superresolution techniques with bacterial specimens and suggest avenues for future development. The goal of this review is to provide the necessary technical background for interested microbiologists to choose the appropriate super-resolution method for their biological systems, and to introduce the practical considerations required for designing and analysing superresolution imaging experiments.
Introduction
Light microscopy is a powerful and widely used research tool in biology that employs visible light to resolve small cellular objects and dynamic processes in biological samples. The biological questions that can be investigated with light microscopy are limited by the imaging resolution, which defines the minimum distance between two distinguishable features. For conventional optical microscopes, the resolution is bounded by the diffraction of light, which causes the signal from a point source to spread as it travels to the detector. The shape of the resulting signal is termed the ‘point spread function (PSF)’ and can be well approximated by a Gaussian distribution. The full width at half maximum (FWHM) of this Gaussian determines the ‘diffraction limit’ of resolution, which is approximately half the wavelength of detected light (> 250 nm for most biocompatible fluorophores) (Abbe, 1873; Rayleigh, 1896).
Electron microscopes (EM) can achieve superior resolution because the wavelength of an electron is subnanometre. However, it is difficult to specifically label proteins to allow their unambiguous identification under EM. Additionally, fixation or vitrification of samples, which is required for EM, is not compatible for live-cell imaging.
Recently, a variety of superresolution fluorescence microscopy techniques have been developed to take advantage of the specific labelling and live-cell compatibility afforded by fluorescence light miscopy, while achieving resolutions (10–50 nm) approaching that of electron microscopy. These techniques can be divided into three categories: first, methods based on single-molecule localization such as photoactivated localization microscopy (PALM) (Betzig et al., 2006) and stochastic optical reconstruction microscopy (STORM) (Rust et al., 2006; Bates et al., 2007); second, derivations of structured illumination microscopy (SIM) (Heintzmann and Cremer, 1999; Frohn et al., 2000; Gustafsson, 2000; 2005); and third, point-scanning methods such as stimulated emission-depletion (STED) microscopy (Hell and Wichmann, 1994; Klar et al., 2000).
The technical details and history of these methods, as well as their application to eukaryotic cytoskeletons, organelles and membrane receptors, have been excellently reviewed elsewhere (Hell, 2007; 2009; Huang et al., 2009; Heilemann, 2010; Schermelleh et al., 2010; van de Linde et al., 2012). In this review, we focus on the use of these methods for microbiology and the unique issues associated with microbial specimens. We first provide conceptual introductions to typical techniques in the three categories, review recent applications of superresolution imaging for microbiological questions, and discuss issues related to applying these techniques to microbial systems.
Single-molecule localization methods
Concept
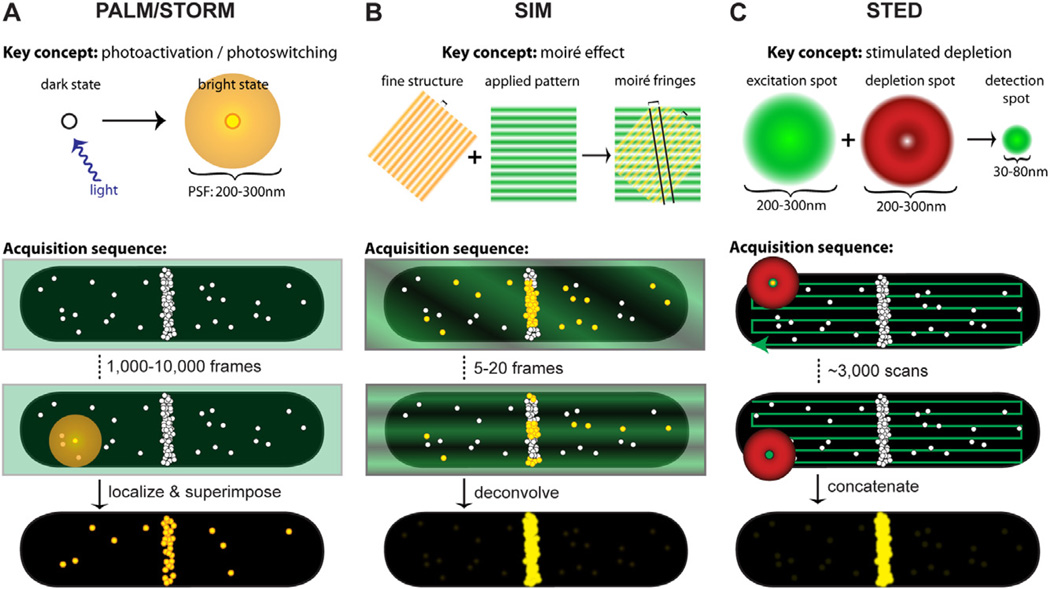
Photoactivatable and photoswitchable fluorophores are the key to implementing localization-based superresolution techniques. These fluorophores can transition stochastically between bright and dark emission states by exposure to specific wavelengths of light. During super-resolution imaging, activation light is kept at a low level such that within a diffraction-limited area only a single fluorophore is fluorescing at a time (Fig. 1A). The positions of these single molecules are then localized with nanometre accuracy by fitting their intensity profiles with a Gaussian function that approximates the microscope’s PSF. Positions collected from thousands of frames are then overlaid to reconstruct a superresolution image. During this long acquisition time, fiducial beads are often added to the sample to track and calibrate stage drift.
Fig. 1.
Key concepts and acquisition schematics for each superresolution technique. In each schematic, molecule positions are shown as white circles, excitation light is represented in green, depletion light is represented in red, and fluorescing molecules are highlighted in yellow. In acquisition schematics, molecule positions mimic cellular distributions of the FtsZ protein, which is the only bacterial protein that has been analysed by all three methods.
A. Single-molecule localization-based techniques such as PALM and STORM apply low levels of activation light (violet arrow) so that single molecules are stochastically activated and localized. An activated molecule produces a diffraction-limited spot (diffuse yellow circle), which is fit with a Gaussian function to localize the molecule’s position with nanometre precision. After hundreds to thousands of molecules have been localized, their positions are superimposed to create the superresolution image (bottom).
B. SIM utilizes the moiré effect, which results when an illumination pattern (green stripes) is applied to a specimen with fine structures that are smaller than the diffraction limit (e.g. closely spaced yellow stripes or molecule positions). Interference between the illumination pattern and the sample produces moiré fringes (two are shown as diagonal black lines) that are spaced further apart than the underlying sample spacing, thus visualizing sub-diffraction-limited features. Several illumination patterns are applied to the sample, then spatial information extracted from the Fourier transforms of each image is combined to generate the superresolution image (bottom). Although the emission from fluorescing molecules (yellow circles) is diffraction-limited, the diffraction-limited profiles are omitted for clarity.
C. For STED imaging, concentric excitation and depletion beams (green circle and red donut respectively) are projected onto a sample. Although fluorophores are excited throughout the diffraction-limited excitation spot (large green circle), the depletion beam (red donut) stimulates molecules outside the central 30–80 nm region back to the ground state before they fluoresce, generating a superresolution PSF (small green circle). These beams are scanned across the specimen to collect the superresolution image (bottom).
This principle was first demonstrated in 2006 using photoactivatable fluorescent proteins (PALM) (Betzig et al., 2006) and cyanine dye pairs (STORM) (Rust et al., 2006), and resolutions of 10 nm and 20 nm were achieved respectively. Later, this principle was extended to conventional organic dyes, which have been shown to photoswitch robustly under proper buffer conditions (Folling et al., 2008; Heilemann et al., 2008; Burnette et al., 2011). The general principle of isolating single fluorophores can be further extended to non-photon-driven switching. For example, PAINT (Point Accumulation for Imaging in Nanoscale Topography) utilizes fluorophores that can only be detected when their fluorescence is enhanced ∼ 1000-fold by binding reversibly to a lipid membrane (Sharonov and Hochstrasser, 2006).
New observations and insights
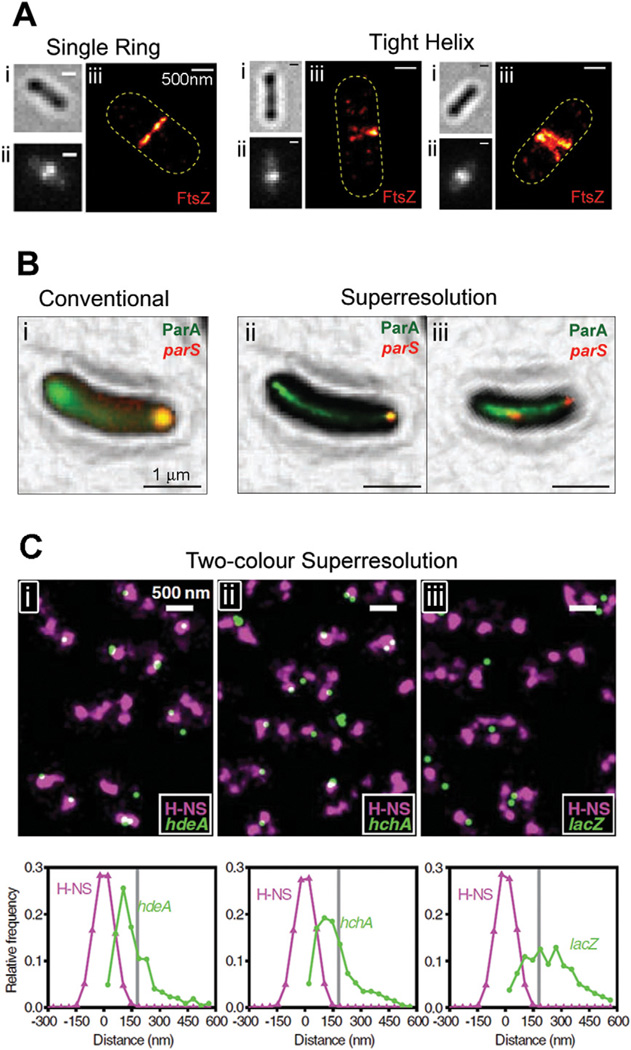
Single-molecule localization-based superresolution imaging techniques are the simplest to implement instrumentally, and have consequently been most quickly adopted by microbiologists. Using these methods, fine details of a variety of bacterial superstructures that are unresolvable under conventional light microscopy have been revealed. For example, the actin homologue MreB, which is necessary for cell shape maintenance, was shown to adopt helix-like structures in early stages of the Caulobacter crescentus cell cycle and ring-like structures in late stages (Biteen et al., 2008). The tubulin homologue FtsZ, which is essential for cytokinesis, was found to exist in either a compact helix or ring conformation in non-constricting Escherichia coli cells (Fig. 2A) (Fu et al., 2010). SpoIIIE, the translocase that sequesters DNA into nascent spores in Bacillus subtilis, was found to be confined to the leading edges of constricting spore septa (Fleming et al., 2010), providing unprecedented structural insight into the protein’s function.
Fig. 2.
Single-molecule localization-based superresolution images.
A. Superstructures formed by FtsZ-mEos2 in E. coli. For each cell, the brightfield (i), conventional fluorescence (ii) and superresolution (iii) images are shown. Although each conventional fluorescence image (ii) shows a single midcell band, the two right panels show that these structures are sometimes resolved into tight helix conformations by PALM imaging (iii). Bars, 500 nm.
B. Colocalization between ParA-eYFP (green) and mCherry-ParB (red), which binds to the parS locus on the C. crescentus chromosome. The diffuse eYFP fluorescence in the conventional image (i) is resolved into tight linear bundles in the superresolution images (ii and iii). Both mCherry-labelled loci are at the same pole in a young cell (ii), but are clearly segregated in a late-stage cell (iii). Bars, 1 µm.
C. Colocalization between mEos2-labelled H-NS (purple) and eYFP-labelled gene loci (green) in live E. coli cells. Each field (i, ii, iii) shows several cells with two to four clusters of H-NS per cell. Histograms underneath each field describe distance distributions between gene loci and their nearest H-NS clusters for hdeA (i) and hchA (ii), which are organized by H-NS, and lacZ (iii), which is not organized by H-NS. Bars, 500 nm.
Images are reproduced with permission from Fu et al. (2010) (A), Ptacin et al. (2010) (B) and Wang et al. (2011) (C).
In addition to revealing fine details of cellular superstructures, localization-based superresolution techniques have also been used to describe the dynamics and activities of various proteins. By mapping the diffusion and distribution of ribosomes and RNA polymerases in live E. coli cells using PALM, Bakshi et al. determined that the most translation occurs on free mRNA and that co-transcriptional insertion of proteins into the membrane, or ‘transertion’, may exert an expansion force on the nucleoid in the lateral direction (Bakshi et al., 2012). In support of a chromosome-organizing role for the E. coli nucleoid-associated protein, H-NS, Wang et al. observed distinct clusters of the H-NS in live E. coli cells while other nucleoid-associated proteins (HU, Fis and IHF) were observed to be more scattered (Wang et al., 2011). Finally, English et al. probed the activity cycle of RelA, a stringent response factor, in E. coli by tracking the diffusion of RelA molecules before and during starvation (English et al., 2011). They observed that RelA diffuses as a ribosome-bound complex during normal conditions, but becomes cytoplasmic during starvation, indicating that RelA activity occurs off the ribosome. These experiments highlight the ability of superresolution techniques to answer fundamental questions of bacterial cell biology.
Quantitative measurements
Quantitative measurements such as structural dimensions can also be made from superresolution images. Feature dimensions are often measured as the FWHM that results from fitting feature profiles to Gaussian distributions. From superresolution studies we now know the width of the FtsZ ring in E. coli (110 nm) (Fu et al., 2010) and C. crescentus (67 nm prior to division; 92 nm during division) (Biteen et al., 2012), the width of linear ParA bundles that span C. crescentus cells to facilitate chromosome segregation (40 nm) (Ptacin et al., 2010), and the diameter of crescentin fibres that give C. crescentus its characteristic crescent shape (92 nm, converted to FWHM from reported σ-value) (Lew et al., 2011). When combined with average protein copy numbers, these measurements can provide valuable information about the arrangement of molecules within structures. For example, because the E. coli FtsZ ring width would be threefold smaller if all FtsZ molecules were packed next to each other, Fu et al. deduced that the FtsZ ring is a loose structure with unoccupied regions that can be occupied by other associated proteins (Fu et al., 2010).
Single-molecule localization-based superresolution imaging also allows molecule density measurements (number of molecules per unit area), which can be analysed for non-uniformity to provide information about molecular interactions. By measuring the distribution of molecule counts within chemotaxis clusters, Greenfield et al. were able to confirm the proposed stochastic assembly mechanism of chemotaxis clusters in E. coli (Greenfield et al., 2009). By comparison to simulated random distributions, Lee et al. determined that the distribution of HU, a nucleoid-associated protein, is non-uniform, perhaps because HU localization is enhanced in highly transcribed regions (Lee et al., 2011). Furthermore, using molecule density measurements, three-dimensional information can be extracted from two-dimensional projections. For example, Fu et al. determined that the number of molecules detected per pixel within E. coli FtsZ rings is consistent with a multilayer arrangement of FtsZ protofilaments (Fu et al., 2010). One caveat (discussed in the section Practical considerations) to molecule counting, however, is that fluorophores often ‘blink’, yielding multiple observations of the same molecule (Annibale et al., 2011a), so caution must be taken to identify unique molecules.
Multicolour imaging
Multicolour superresolution imaging is an extremely powerful tool for investigating protein–protein or protein–DNA interactions in their native cellular environment. Proteins that appear to colocalize with each other using conventional fluorescence light microscopy may not necessarily be in direct contact because the size of a diffraction-limited spot is much larger than that of a single protein (1–10 nm). Superresolution imaging determines the spatial relationship between two proteins with much higher precision that allows more confident assessment of protein interactions. For example, Ptacin et al. investigated chromosome segregation in C. crescentus by using PALM to simultaneously determine the density of the ParA bundle that spans the cell length and the location of parS loci that serve as chromosomal anchors for ParA (Fig. 2B) (Ptacin et al., 2010). By comparing images of cells at different stages of chromosome segregation (where one parS locus moves across the cell), the authors determined that segregation occurs via retraction of a subset of ParA fibres. Using two-colour imaging in E. coli, Wang et al. found a high degree of colocalization between H-NS and two genes that it regulates (hdeA and hchA) but low colocalization between H-NS and lacZ, which it does not regulate, suggesting that H-NS-regulated genes may be organized into close clusters (Fig. 2C) (Wang et al., 2011). As a diffraction-limited spot is similar in size to the E. coli nucleoid, the different H-NS colocalization patterns of these genes would be indistinguishable with conventional microscopy.
Multicolour superresolution imaging also has the potential to provide unprecedented detail into host–pathogen interactions. The first demonstration of this was by Lehmann et al., who performed two-colour superresolution imaging of assembling human immunodeficiency virus (HIV) particles in HeLa cells (Lehmann et al., 2011). They determined molecule distributions of Gag, the HIV-1 structural protein, and tetherin, a human protein that inhibits release of budding virus particles, and found that most budding HIV-1 particles were colocalized with a single cluster of tetherin, rather than recruiting multiple clusters.
SIM
Concept
Structured illumination microscopy achieves superresolution by extracting fine structural details from the interference of a structure with predetermined illumination patterns. When a sinusoidal illumination pattern (Fig. 1B, green shading) is applied to a fluorescent sample, an interference pattern results. The diffraction-limited fringes of this interference pattern, called moiré fringes, contain information about the underlying structural pattern of the sample that cannot by observed with conventional light microscopy. By applying a set of illumination patterns of different spacing and rotation angles to the same sample (Fig. 1B), sub-diffraction-limited structural information of the sample can be extracted from Fourier transforms of the resulting interference patterns (Lukosz and Marchand, 1963; Heintzmann and Cremer, 1999; Frohn et al., 2000; Gustafsson, 2000). Standard implementations of SIM achieve twofold improvement in lateral resolution (Frohn et al., 2000; Gustafsson, 2000), but finer resolution can be achieved by taking advantage of non-linear fluorescence responses (Gustafsson, 2005; Rego et al., 2012). Thus far, applications of SIM in bacterial cells have employed the twofold improvement afforded by commercial systems: Applied Precision DeltaVision OMX (Eswaramoorthy et al., 2011; Wheeler et al., 2011) and Nikon N-SIM (Szwedziak et al., 2012). The main advantage of SIM is that superresolution can be achieved with conventional fluorescent proteins because the resolution enhancement comes solely from the patterned illumination and image processing.
New observations
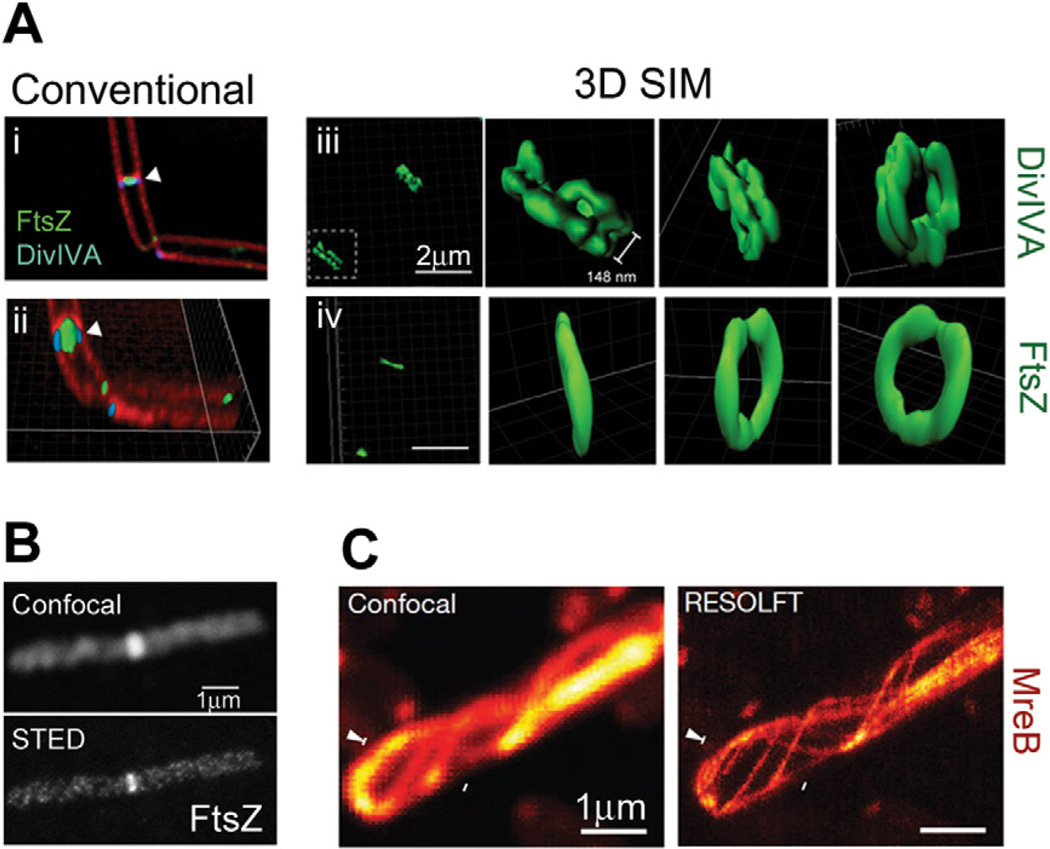
Structured illumination microscopy was recently applied to investigate the three-dimensional superstructures formed by the B. subtilis proteins FtsZ, DivIVA and MinJ in live cells (Eswaramoorthy et al., 2011). While SIM images showed FtsZ in a single-ring conformation, DivIVA and MinJ structures were resolved as double rings that flank division septa (Fig. 3A). These double-ring structures resolve the perplexing question of how MinJ, which recruits the negative FtsZ regulator MinC, could be localized to the midcell but not interfere with cell division. DivIVA and MinJ recruit a local density of MinC to the midcell, but at the same time prevent it from reaching the dividing FtsZ ring until division is complete. Wheeler et al. investigated cell wall biosynthesis in Gram-positive ovococci using fluorescent vancomycin to label peptidoglycan (Wheeler et al., 2011). By observing the localization of newly synthesized peptidoglycan with 50 nm lateral resolution, they were able to follow cell elongation, septum progression and cell constriction with high precision.
Fig. 3.
SIM and STED superresolution images.
A. Ring structures formed by DivIVA and FtsZ in B. subtilis cells. Diffraction-limited images generated by conventional deconvolution (i and ii) show the proximity of DivIVA-GFP and FtsZ-YFP at dividing septa (membrane stained with FM4-64). 3D SIM reconstructions of DivIVA-GFP (iii) and FtsZ-GFP (iv), shown as surface representations, clearly show that DivIVA forms a double-ring structure while FtsZ forms a single-ring structure. Bars, 2 µm.
B. Images of Alexa647N–immunolabelled FtsZ in a B. subtilis cell. Superresolution imaging with STED (bottom) reveals discontinuous helical structures that are unresolvable by confocal microscopy (top). Bar, 1 µm.
C. Images of a live E. coli cell expressing rsEGFP-MreB. RESOLFT imaging (right) can resolve neighbouring MreB filaments that are indistinguishable by confocal microscopy (left). Bars, 1 µm. Images are reproduced from Eswaramoorthy et al. (2011) (A), Jennings et al. (2011) (B) and Grotjohann et al. (2011) (C).
STED
Concept
Stimulated emission-depletion microscopy was the earliest far-field superresolution technique developed (Hell and Wichmann, 1994; Klar et al., 2000). The instrumentation is similar to a confocal microscope with the addition of a depletion laser, which stimulates excited molecules back to the ground state in a donut-shaped region around the central confocal spot (Fig. 1C). In this way, only molecules within 30–80 nm of the centre of the excitation spot are detected. This depletion concept was extended to fluorophore photoswitching by using the donut-shaped depletion beam to switch fluorophores into the off state instead of stimulating emission (Grotjohann et al., 2011). This process, termed Reversible Saturable Optical Fluorescence Transitions (RESOLFT), mitigates the detrimental effect that the high power depletion beam may have on cell viability because the RESOLFT depletion beam is used at much lower power (∼ 1 kW cm−2) than the stimulated depletion laser (100–500 MW cm−2) (Grotjohann et al., 2011).
New observations
Because of the complex instrumentation required, only two bacterial proteins have been studied using STED and RESOLFT. Using a commercial STED microscope (Leica TCS STED), Jennings et al. observed very discontinuous FtsZ helical structures that spanned the length of B. subtilis cells (Fig. 3B) (Jennings et al., 2011), similar to those observed at the E. coli midcell by Fu et al. using PALM (Fu et al., 2010). Using RESOLFT, Grotjohann et al. were able to distinguish neighbouring MreB filaments in live E. coli cells that were unresolvable by confocal microscopy, and determined that these filaments were 70 nm wide on average (Grotjohann et al., 2011) (Fig. 3C).
Practical considerations
Time resolution
Because each superresolution image is the combined result of multiple frames or scans, the acquisition time for a complete superresolution image relative to the timescale of dynamics associated with a protein of interest is important for live-cell imaging. In this regard, bacterial cells are ideal specimens for superresolution imaging because of their small sizes, which allow very small scanning or illumination areas to increase imaging speed and probe the real-time dynamics of bacterial proteins and cellular structures in live cells.
Structured illumination microscopy has the fastest time resolution of the three categories described in this review. Each 2D SIM image requires only 5–20 frames for reconstruction, so one superresolution image can be generated in a few seconds, and imaging rates of up to 11 Hz (one complete SIM image in 90 ms) have been achieved for fields of view sufficient for bacterial samples (13 µm × 13 µm) (Kner et al., 2009). STED and RESOLFT microscopy have slower time resolutions than SIM because of the requirement for point-scanning. However, the small size of bacterial cells makes this much less of an issue, so STED images can be collected in several seconds, and a 1.8 × 2.5 µm2 area has been imaged at up to 28 Hz (one complete STED image in 36 ms) (Westphal et al., 2008).
Single-molecule localization-based methods have the slowest time resolution because thousands of frames are required to sufficiently sample a structure of interest. The Nyquist sampling criterion states that a desired resolution of X nm will be achieved if the sampling frequency is at least X/2 nm (Nyquist, 1928; Shannon, 1949; Shroff et al., 2008). For example, to achieve a resolution of 30 nm, a structure should be labelled with one molecule every 15 × 15 nm2. Because of this criterion, compact structures are ideal candidates for live-cell superresolution imaging as they require fewer localizations for sufficient sampling (e.g. a 45 × 45 nm2 structure requires nine molecules and a 450 × 450 nm2 structure requires 900 molecules to achieve 30 nm resolution). Consequently, tight clusters and cytoskeletal elements can be sufficiently sampled in less than a minute (Fu et al., 2010; Wang et al., 2011).
Imaging duration
Another component of live-cell imaging is the number of time-lapse superresolution images that can be acquired. For SIM and STED imaging, the number of superresolution images that can be acquired is usually limited by fluorophore photobleaching. This can be very problematic for bacterial structures, which are often made up of only a few hundred molecules. For the SIM studies of DivIVA and MinJ described above (Eswaramoorthy et al., 2011), the authors overcame this dilemma by overexpressing the proteins of interest, but as described below, protein overexpression may limit the biological conclusions that can be drawn. For STED, it was reported that up to 30 superresolution images can be acquired without significant photobleaching in live mammalian cells using the fluorescent protein citrine (Hein et al., 2008). However, this extended imaging time remains to be demonstrated in bacterial cells where protein expression levels are generally lower.
When imaging live cells by single-molecule localization-based techniques, the number of superresolution images that can be acquired is also limited by the protein expression level because each image needs to have sufficient sampling of the visualized structure. An additional constraint for these techniques is the cell viability under continuous laser exposure throughout the imaging sequence. Although the intensity of 405 nm light typically used to induce photoactivation and photoswitching is relatively low (1–5 kW cm−2) (Heilemann et al., 2009), prolonged exposure can result in reactive oxygen species that cause DNA damage. As the transcriptional response to DNA damage begins at approximately 20 min after exposure to UV light in E. coli (Friedman et al., 2005), the duration of live-cell PALM imaging is typically kept to less than 15 min (Fleming et al., 2010; Fu et al., 2010; Ptacin et al., 2010; Wang et al., 2011; Lieberman et al., 2012).
Probe selection
The three methods described here have different fluorophore requirements, and the properties of these probes have been reviewed elsewhere (Fernandez-Suarez and Ting, 2008; Patterson, 2011). Here we will give a brief account on the types of probes suitable for the three imaging categories, then focus on specific probe selection considerations associated with bacterial cells.
Among the three methods described in this review, SIM has the most lax probe requirements, allowing any combination of excitation and emission wavelengths that can be accommodated by a widefield fluorescence microscope. As mentioned above, photostability is crucial for time-lapse and 3D SIM imaging, and consequently most studies utilize bright organic dyes and, more recently, EGFP. STED requires dyes that can be specifically excited and depleted by spectrally separated lasers (e.g. Alexa647N or citrine) and RESOLFT requires fluorophores that can be photoswitched thousands of times (e.g. rsEGFP). For single-molecule localization-based methods, photoactivatable and photoswitchable proteins such as Dendra, Dronpa, mEos2 and PAmCherry are common genetic labels for PALM, while (d)STORM utilizes organic dyes such as the Cy3–Cy5 pair or Alexa647, which have been extensively characterized to determine optimal buffer conditions (Dempsey et al., 2011).
Improving the stability or photoswitching kinetics of superresolution fluorophores can vastly improve data throughput and quality. For example, because the precision of single-molecule localization is determined by the number of photons collected (Thompson et al., 2002), the frame rate of PALM/STORM measurements is limited by the exposure time required to collect sufficient photons from a single molecule. In addition, the prevalence of photoblinking among most fluorophores, which can cause false measurements of molecule density, must be taken into consideration or accounted for by the superresolution image reconstruction algorithm (Annibale et al., 2011a,b). Hence, the ideal fluorescent protein for PALM would have a high quantum yield during a single fluorescence burst, and photobleach very quickly so that new molecules can be activated at a faster rate.
Most superresolution studies in bacteria have used genetically encoded fluorescent proteins. Although organic dyes often have superior photophysical properties, it is very difficult to achieve high-efficiency labelling of intracellular proteins because of low cell wall permeability. Thus, most dye-labelling schemes require cell wall permeabilization via lysozyme and detergent treatment, and are only compatible with fixed cells. Schoen et al. demonstrated this method using PicoGreen, which becomes 1000-fold brighter upon binding DNA (Schoen et al., 2011).
Functionality of labelled protein
Fusing a fluorescent protein to a protein of interest may affect its function and localization pattern, especially if the fluorescent protein has a tendency to oligomerize (Zhang et al., 2012). To test for functionality, the fusion protein should be expressed and examined for full recovery of physiological activity in a null mutant of the native protein (Greenfield et al., 2009; Lehmann et al., 2011; Lieberman et al., 2012). If full activity recovery cannot be achieved, the fusion protein can be expressed ectopically so that it decorates the endogenous structure as long as the subcellular localization of the fusion protein is identical to that of unlabelled protein (Fu et al., 2010; Eswaramoorthy et al., 2011; Lehmann et al., 2011; Wang et al., 2011). Because each fluorescent protein may have a different interaction with a given protein or its cellular surroundings, probe selection may require testing several different fluorescent proteins (Lehmann et al., 2011) or linkers (English et al., 2011).
Expression level
In general, superresolution techniques are more successful for proteins with naturally high expression levels. For both STED and SIM, higher expression levels provide higher contrast and allow acquisition of more image stacks for 3D or time-lapse imaging. For PALM and STORM, expression levels determine the degree of sampling within a structure and thus directly influence the effective imaging resolution. Overexpression of a protein of interest can enhance image quality, but can also result in aberrant structures or measurements because most biological systems are fine-tuned to operate within a given concentration range. Whenever possible, superresolution studies should be conducted at physiological expression levels (Greenfield et al., 2009; Biteen and Moerner, 2010; Fu et al., 2010), and careful controls should be performed if overexpression is required.
Fixation
Although live cells provide the most physiological conditions for imaging, fixation is often necessary for some samples and measurements. For structures that move on the timescale of imaging, fixation may be the only way to ‘freeze’ the movement and capture true structural dimensions without motion-induced blur. Fixation may also be required to localize fast-moving cytoplasmic proteins, which are usually undetectable in live PALM experiments. Furthermore, the most suitable way to measure molecule density is from fixed cells where molecule movement cannot obstruct counting measurements.
As with high-resolution EM samples, caution must be taken to avoid fixation-induced distortions to cellular structures, especially for membrane-associated proteins. Although formaldehyde and glutaraldehyde are common fixatives for EM, formaldehyde is most common for fluorescence imaging because glutaraldehyde often generates a high autofluorescence background (Fischer et al., 2008). Because fixation is reversible, fixed samples should be stored at 4°C and imaged as soon as possible (Fischer et al., 2008). If the imaged structures do not move on the timescale of imaging, comparison of fixed and live samples can provide assurance that fixation has not introduced noticeable aberrations (Fu et al., 2010; Lee et al., 2011).
Sample immobilization
Preparation of bacterial samples is similar among all superresolution techniques, which have utilized two main methods of cell immobilization: adherence via Poly-L-Lysine (PLL) and compression via agarose gel pad. PLL adherence is very convenient, but has been shown to affect the proton-motive force (Katsu et al., 1984; Strahl and Hamoen, 2010) and protein localization (Colville et al., 2009; Strahl and Hamoen, 2010) in E. coli. As a result, most live-cell studies immobilize cells with 1.5–3% agarose gel pads (Biteen et al., 2008; 2012; Fu et al., 2010; Ptacin et al., 2010; English et al., 2011; Eswaramoorthy et al., 2011; Lee et al., 2011; Lew et al., 2011; Wang et al., 2011; Szwedziak et al., 2012). Detailed instructions regarding sample preparation, imaging and data analysis for PALM studies in bacterial cells have been described for C. crescentus (Biteen and Moerner, 2010) and E. coli (Buss et al., 2012).
Hardware and software complexity
Although the localization-based methods were the latest to be developed, their incorporation into microbiological study has far outpaced the other two methods as evidenced by the number of studies reviewed here. This is most likely because of the minimal hardware modifications and expertise required to implement a PALM or STORM system. Construction of a STED microscope requires considerable investment and expertise, but the availability of commercial systems for both SIM (Applied Precision DeltaVision OMX and Nikon N-SIM) and STED (Leica TCS STED) makes these methods viable options for shared facilities.
Image processing is conversely much simpler for STED than for localization methods or SIM. Because the superresolution information of STED is encoded in the scanning beams, almost no image processing is required and superresolution images can be observed in real time. SIM images can only be viewed after deconvolving the high-resolution information from each set of fringes, and PALM/STORM experiments require detection and localization of hundreds to thousands of spots per bacterial cell. Improvement of the analysis algorithms for both SIM and PALM/STORM is ongoing. For PALM and STORM, improvements have focused on accurate localization of overlapping emitters (Holden et al., 2011; Zhu et al., 2012) to improve imaging speed and correcting for overcounting artefacts caused by photoblinking (Annibale et al., 2011b; Sengupta et al., 2011; Veatch et al., 2012).
Future directions
Superresolution studies are often accompanied by supporting EM or atomic force microscopy data, which provide high-resolution information about the cellular surroundings of a given structure (Betzig et al., 2006; Wheeler et al., 2011; Szwedziak et al., 2012). Correlative imaging of the same specimen with both superresolution light microscopy and EM, which has been demonstrated for STED (Watanabe et al., 2011) and PALM (Betzig et al., 2006; Watanabe et al., 2011; Kopek et al., 2012), has the potential to provide substantial insight into subcellular architecture, but is still difficult to implement. Explorations into fluorophores and conditions that facilitate the merging of these techniques will therefore greatly advance the fields.
Superresolution imaging of microbiological specimens would also benefit from several areas of fluorescent probe development. First, development of dyes that efficiently penetrate the cell wall would allow implementation of dSTORM investigations in live bacterial cells and the use of smaller genetic tags as binding targets. Furthermore, fluorescent probes that can be efficiently controlled with wavelengths of light longer than the commonly used 405 nm will extend the time-lapse, live-cell imaging capability of localization-based methods. Finally, labelling strategies involving unnatural amino acids (Plass et al., 2011; 2012; Milles et al., 2012) or protease cleavage (Chattopadhaya et al., 2008), which entail single amino acid substitutions, may provide ways to perform superresolution imaging with minimal perturbations to protein function.
In this review we have described superresolution microbiology studies that pave the way for investigations of other complex biological questions. We have also pointed out features that may hinder application of each technique to microbiological specimens, but as improvements to hardware, analysis and fluorescent probes continue to increase both spatial and time resolution, further exciting avenues of superresolution investigation will no doubt vastly increase our understanding of microbiology.
References
- Abbe E. Beiträge zur Theorie des Mikroskops und der mikroskopischen Wahrnehmung. Arch Mikrosk Anat. 1873;9:413–418. [Google Scholar]
- Annibale P, Vanni S, Scarselli M, Rothlisberger U, Radenovic A. Identification of clustering artifacts in photoactivated localization microscopy. Nat Methods. 2011a;8:527–528. doi: 10.1038/nmeth.1627. [DOI] [PubMed] [Google Scholar]
- Annibale P, Vanni S, Scarselli M, Rothlisberger U, Radenovic A. Quantitative photo activated localization microscopy: unraveling the effects of photoblinking. PLoS ONE. 2011b;6:e22678. doi: 10.1371/journal.pone.0022678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakshi S, Siryaporn A, Goulian M, Weisshaar JC. Superresolution imaging of ribosomes and RNA polymerase in live Escherichia coli cells. Mol Microbiol. 2012;85:21–38. doi: 10.1111/j.1365-2958.2012.08081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates M, Huang B, Dempsey GT, Zhuang X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 2007;317:1749–1753. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Biteen JS, Moerner WE. Single-molecule and superresolution imaging in live bacteria cells. Cold Spring Harb Perspect Biol. 2010;2:1–14. doi: 10.1101/cshperspect.a000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteen JS, Thompson MA, Tselentis NK, Bowman GR, Shapiro L, Moerner WE. Super-resolution imaging in live Caulobacter crescentus cells using photoswitchable EYFP. Nat Methods. 2008;5:947–949. doi: 10.1038/NMETH.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biteen JS, Goley ED, Shapiro L, Moerner WE. Three-dimensional super-resolution imaging of the midplane protein FtsZ in live Caulobacter crescentus cells using astigmatism. Chemphyschem. 2012;13:1007–1012. doi: 10.1002/cphc.201100686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette DT, Sengupta P, Dai Y, Lippincott-Schwartz J, Kachar B. Bleaching/blinking assisted localization microscopy for superresolution imaging using standard fluorescent molecules. Proc Natl Acad Sci USA. 2011;108:21081–21086. doi: 10.1073/pnas.1117430109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss J, Coltharp C, Xiao J. Super-resolution imaging of the bacterial division machinery. J Vis Exp. 2012 doi: 10.3791/50048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhaya S, Abu Bakar FB, Srinivasan R, Yao SQ. In vivo imaging of a bacterial cell division protein using a protease-assisted small-molecule labeling approach. Chembiochem. 2008;9:677–680. doi: 10.1002/cbic.200700647. [DOI] [PubMed] [Google Scholar]
- Colville K, Tompkins N, Rutenberg AD, Jericho MH. Effects of poly(L-lysine) substrates on attached Escherichia coli bacteria. Langmuir. 2009;26:2639–2644. doi: 10.1021/la902826n. [DOI] [PubMed] [Google Scholar]
- Dempsey GT, Vaughan JC, Chen KH, Bates M, Zhuang X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat Methods. 2011;8:1027–1036. doi: 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English BP, Hauryliuk V, Sanamrad A, Tankov S, Dekker NH, Elf J. Single-molecule investigations of the stringent response machinery in living bacterial cells. Proc Natl Acad Sci USA. 2011;108:E365–E373. doi: 10.1073/pnas.1102255108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaramoorthy P, Erb ML, Gregory JA, Silverman J, Pogliano K, Pogliano J, Ramamurthi KS. Cellular architecture mediates DivIVA ultrastructure and regulates min activity in Bacillus subtilis. MBio. 2011;2 doi: 10.1128/mBio.00257-11. mBio. 00257-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Suarez M, Ting AY. Fluorescent probes for super-resolution imaging in living cells. Nat Rev Mol Cell Biol. 2008;9:929–943. doi: 10.1038/nrm2531. [DOI] [PubMed] [Google Scholar]
- Fischer AH, Jacobson KA, Rose J, Zeller R. Fixation and permeabilization of cells and tissues. CSH Protoc. 2008;2008 doi: 10.1101/pdb.top36. opdb.top36. [DOI] [PubMed] [Google Scholar]
- Fleming TC, Shin JY, Lee S-H, Becker E, Huang KC, Bustamante C, Pogliano K. Dynamic SpoIIIE assembly mediates septal membrane fission during Bacillus subtilis sporulation. Genes Dev. 2010;24:1160–1172. doi: 10.1101/gad.1925210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folling J, Bossi M, Bock H, Medda R, Wurm CA, Hein B, et al. Fluorescence nanoscopy by ground-state depletion and single-molecule return. Nat Methods. 2008;5:943–945. doi: 10.1038/nmeth.1257. [DOI] [PubMed] [Google Scholar]
- Friedman N, Vardi S, Ronen M, Alon U, Stavans J. Precise temporal modulation in the response of the SOS DNA repair network in individual bacteria. PLoS Biol. 2005;3:e238. doi: 10.1371/journal.pbio.0030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohn JT, Knapp HF, Stemmer A. True optical resolution beyond the Rayleigh limit achieved by standing wave illumination. Proc Natl Acad Sci USA. 2000;97:7232–7236. doi: 10.1073/pnas.130181797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu G, Huang T, Buss J, Coltharp C, Hensel Z, Xiao J. In vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM) PLoS ONE. 2010;5:e12680. doi: 10.1371/journal.pone.0012680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield D, McEvoy AL, Shroff H, Crooks GE, Win-green NS, Betzig E, Liphardt J. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 2009;7:e1000137. doi: 10.1371/journal.pbio.1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotjohann T, Testa I, Leutenegger M, Bock H, Urban NT, Lavoie-Cardinal F, et al. Diffraction-unlimited all-optical imaging and writing with a photochromic GFP. Nature. 2011;478:204–208. doi: 10.1038/nature10497. [DOI] [PubMed] [Google Scholar]
- Gustafsson MGL. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc. 2000;198:82–87. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- Gustafsson MGL. Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. Proc Natl Acad Sci USA. 2005;102:13081–13086. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilemann M. Fluorescence microscopy beyond the diffraction limit. J Biotechnol. 2010;149:243–251. doi: 10.1016/j.jbiotec.2010.03.012. [DOI] [PubMed] [Google Scholar]
- Heilemann M, van de Linde S, Schüttpelz M, Kasper R, Seefeldt B, Mukherjee A, et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew Chem Int Ed Engl. 2008;47:6172–6176. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- Heilemann M, van de Linde S, Mukherjee A, Sauer M. Super-resolution imaging with small organic fluorophores. Angew Chem Int Ed Engl. 2009;48:6903–6908. doi: 10.1002/anie.200902073. [DOI] [PubMed] [Google Scholar]
- Hein B, Willig KI, Hell SW. Stimulated emission depletion (STED) nanoscopy of a fluorescent protein-labeled organelle inside a living cell. Proc Natl Acad Sci USA. 2008;105:14271–14276. doi: 10.1073/pnas.0807705105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzmann R, Cremer CG. Laterally modulated excitation microscopy: improvement of resolution by using a diffraction grating. Proc SPIE 3568 (Optical Biopsies and Microscopic Techniques) 1999;3:185–196. [Google Scholar]
- Hell SW. Far-field optical nanoscopy. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- Hell SW. Microscopy and its focal switch. Nat Methods. 2009;6:24–32. doi: 10.1038/nmeth.1291. [DOI] [PubMed] [Google Scholar]
- Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19:780–782. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- Holden SJ, Uphoff S, Kapanidis AN. DAOS-TORM: an algorithm for high-density super-resolution microscopy. Nat Methods. 2011;8:279–280. doi: 10.1038/nmeth0411-279. [DOI] [PubMed] [Google Scholar]
- Huang B, Bates M, Zhuang X. Super-resolution fluorescence microscopy. Annu Rev Biochem. 2009;78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings PC, Cox GC, Monahan LG, Harry EJ. Super-resolution imaging of the bacterial cytokinetic protein FtsZ. Micron. 2011;42:336–341. doi: 10.1016/j.micron.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Katsu T, Tsuchiya T, Fujita Y. Dissipation of membrane potential of Escherichia coli cells induced by macromolecular polylysine. Biochem Biophys Res Commun. 1984;122:401–406. doi: 10.1016/0006-291x(84)90489-3. [DOI] [PubMed] [Google Scholar]
- Klar TA, Jakobs S, Dyba M, Egner A, Hell SW. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc Natl Acad Sci USA. 2000;97:8206–8210. doi: 10.1073/pnas.97.15.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kner P, Chhun BB, Griffis ER, Winoto L, Gustafsson MGL. Super-resolution video microscopy of live cells by structured illumination. Nat Methods. 2009;6:339–342. doi: 10.1038/nmeth.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopek BG, Shtengel G, Xu CS, Clayton DA, Hess HF. Correlative 3D superresolution fluorescence and electron microscopy reveal the relationship of mitochondrial nucleoids to membranes. Proc Natl Acad Sci USA. 2012;109:6136–6141. doi: 10.1073/pnas.1121558109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SF, Thompson MA, Schwartz MA, Shapiro L, Moerner WE. Super-resolution imaging of the nucleoid-associated protein HU in Caulobacter crescentus . Biophys J. 2011;100:L31–L33. doi: 10.1016/j.bpj.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M, Rocha S, Mangeat B, Blanchet F, Uji-I H, Hofkens J, Piguet V. Quantitative multicolor super-resolution microscopy reveals tetherin HIV-1 interaction. PLoS Pathog. 2011;7:e1002456. doi: 10.1371/journal.ppat.1002456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew MD, Lee SF, Ptacin JL, Lee MK, Twieg RJ, Shapiro L, Moerner WE. Three-dimensional superresolution colocalization of intracellular protein superstructures and the cell surface in live Caulobacter crescentus . Proc Natl Acad Sci USA. 2011;108:E1102–E1110. doi: 10.1073/pnas.1114444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman JA, Frost NA, Hoppert M, Fernandes PJ, Vogt SL, Raivio TL, et al. Outer membrane targeting, ultrastructure, and single molecule localization of the enteropathogenic Escherichia coli type IV pilus secretin BfpB. J Bacteriol. 2012;194:1646–1658. doi: 10.1128/JB.06330-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Linde S, Heilemann M, Sauer M. Live-cell super-resolution imaging with synthetic fluorophores. Annu Rev Phys Chem. 2012;63:519–540. doi: 10.1146/annurev-physchem-032811-112012. [DOI] [PubMed] [Google Scholar]
- Lukosz W, Marchand M. Optische Abbildung unter Überschreitung der beugungsbedingten Auflösungs-grenze. Opt Acta. 1963;10:241–255. [Google Scholar]
- Milles S, Tyagi S, Banterle N, Koehler C, VanDelinder V, Plass T, et al. Click strategies for single-molecule protein fluorescence. J Am Chem Soc. 2012;134:5187–5195. doi: 10.1021/ja210587q. [DOI] [PubMed] [Google Scholar]
- Nyquist H. Certain topics in telegraph transmission theory. Trans AIEE. 1928;47:617–644. [Google Scholar]
- Patterson GH. Highlights of the optical highlighter fluorescent proteins. J Microsc. 2011;243:1–7. doi: 10.1111/j.1365-2818.2011.03505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plass T, Milles S, Koehler C, Schultz C, Lemke EA. Genetically encoded copper-free click chemistry. Angew Chem Int Ed Engl. 2011;50:3878–3881. doi: 10.1002/anie.201008178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plass T, Milles S, Koehler C, Szymanński J, Mueller R, Wieβler M, et al. Amino acids for Diels–Alder reactions in living cells. Angew Chem Int Ed Engl. 2012;51:4166–4170. doi: 10.1002/anie.201108231. [DOI] [PubMed] [Google Scholar]
- Ptacin JL, Lee SF, Garner EC, Toro E, Eckart M, Comolli LR, et al. A spindle-like apparatus guides bacterial chromosome segregation. Nat Cell Biol. 2010;12:791–798. doi: 10.1038/ncb2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayleigh L. On the theory of optical images, with special reference to the microscope. Phil Mag. 1896;42:167–195. [Google Scholar]
- Rego EH, Shao L, Macklin JJ, Winoto L, Johansson GA, Kamps-Hughes N, et al. Nonlinear structured-illumination microscopy with a photoswitchable protein reveals cellular structures at 50-nm resolution. Proc Natl Acad Sci USA. 2012;109:E135–E143. doi: 10.1073/pnas.1107547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh L, Heintzmann R, Leonhardt H. A guide to super-resolution fluorescence microscopy. J Cell Biol. 2010;190:165–175. doi: 10.1083/jcb.201002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoen I, Ries J, Klotzsch E, Ewers H, Vogel V. Binding-activated localization microscopy of DNA structures. Nano Lett. 2011;11:4008–4011. doi: 10.1021/nl2025954. [DOI] [PubMed] [Google Scholar]
- Sengupta P, Jovanovic-Talisman T, Skoko D, Renz M, Veatch SL, Lippincott-Schwartz J. Probing protein heterogeneity in the plasma membrane using PALM and pair correlation analysis. Nat Methods. 2011;8:969–975. doi: 10.1038/nmeth.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon CE. Communication in the presence of noise. Proc Inst Radio Eng. 1949;37:10–21. [Google Scholar]
- Sharonov A, Hochstrasser RM. Wide-field sub-diffraction imaging by accumulated binding of diffusing probes. Proc Natl Acad Sci USA. 2006;103:18911–18916. doi: 10.1073/pnas.0609643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff H, Galbraith CG, Galbraith JA, Betzig E. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat Methods. 2008;5:417–423. doi: 10.1038/nmeth.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl H, Hamoen LW. Membrane potential is important for bacterial cell division. Proc Natl Acad Sci USA. 2010;107:12281–12286. doi: 10.1073/pnas.1005485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwedziak P, Wang Q, Freund SMV, Lowe J. FtsA forms actin-like protofilaments. EMBO J. 2012;31:2249–2260. doi: 10.1038/emboj.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veatch SL, Machta BB, Shelby SA, Chiang EN, Holowka DA, Baird BA. Correlation functions quantify super-resolution images and estimate apparent clustering due to over-counting. PLoS ONE. 2012;7:e31457. doi: 10.1371/journal.pone.0031457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Li G-W, Chen C, Xie XS, Zhuang X. Chromosome organization by a nucleoid-associated protein in live bacteria. Science. 2011;333:1445–1449. doi: 10.1126/science.1204697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Punge A, Hollopeter G, Willig KI, Hobson RJ, Davis MW, et al. Protein localization in electron micrographs using fluorescence nanoscopy. Nat Methods. 2011;8:80–84. doi: 10.1038/nmeth.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal V, Rizzoli SO, Lauterbach MA, Kamin D, Jahn R, Hell SW. Video-rate far-field optical nanoscopy dissects synaptic vesicle movement. Science. 2008;320:246–249. doi: 10.1126/science.1154228. [DOI] [PubMed] [Google Scholar]
- Wheeler R, Mesnage S, Boneca IG, Hobbs JK, Foster SJ. Super-resolution microscopy reveals cell wall dynamics and peptidoglycan architecture in ovococcal bacteria. Mol Microbiol. 2011;82:1096–1109. doi: 10.1111/j.1365-2958.2011.07871.x. [DOI] [PubMed] [Google Scholar]
- Zhang M, Chang H, Zhang Y, Yu J, Wu L, Ji W, et al. Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nat Methods. 2012;9:727–729. doi: 10.1038/nmeth.2021. [DOI] [PubMed] [Google Scholar]
- Zhu L, Zhang W, Elnatan D, Huang B. Faster STORM using compressed sensing. Nat Methods. 2012;9:721–723. doi: 10.1038/nmeth.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]