Abstract
Polycyclic aromatic hydrocarbons and their nitro derivatives are culprits of the detrimental health effects of environmental pollution. These hydrophobic compounds metabolize to reactive species and attach to DNA producing bulky lesions, such as N-[deoxyguanosine-8-yl]-1-aminopyrene (APG), in genomic DNA. The bulky adducts block DNA replication by high-fidelity polymerases and compromise replication fidelities and efficiencies by specialized lesion bypass polymerases. Here we present three crystal structures of the DNA polymerase Dpo4, a model translesion DNA polymerase of the Y family, in complex with APG-lesion-containing DNA in pre-insertion and extension stages. APG is captured in two conformations in the pre-insertion complex; one is highly exposed to the solvent, whereas the other is harbored in a shallow cleft between the finger and unique Y family little finger domain. In contrast, APG is in a single conformation at the extension stage, in which the pyrene ring is sandwiched between the little finger domain and a base from the turning back single-stranded template strand. Strikingly, a nucleotide intercalates the DNA helix to form a quaternary complex with Dpo4, DNA, and an incoming nucleotide, which stabilizes the distorted DNA structure at the extension stage. The unique APG DNA conformations in Dpo4 inhibit DNA translocation through the polymerase active site for APG bypass. We also modeled an insertion complex that illustrates a solvent-exposed pyrene ring contributing to an unstable insertion state. The structural work combined with our lesion replication assays provides a novel structural mechanism on bypass of DNA adducts containing polycyclic aromatic hydrocarbon moieties.
Keywords: DNA damage, translesion DNA replication, polycyclic aromatic hydrocarbons, Y family polymerase, environmental pollution
Introduction
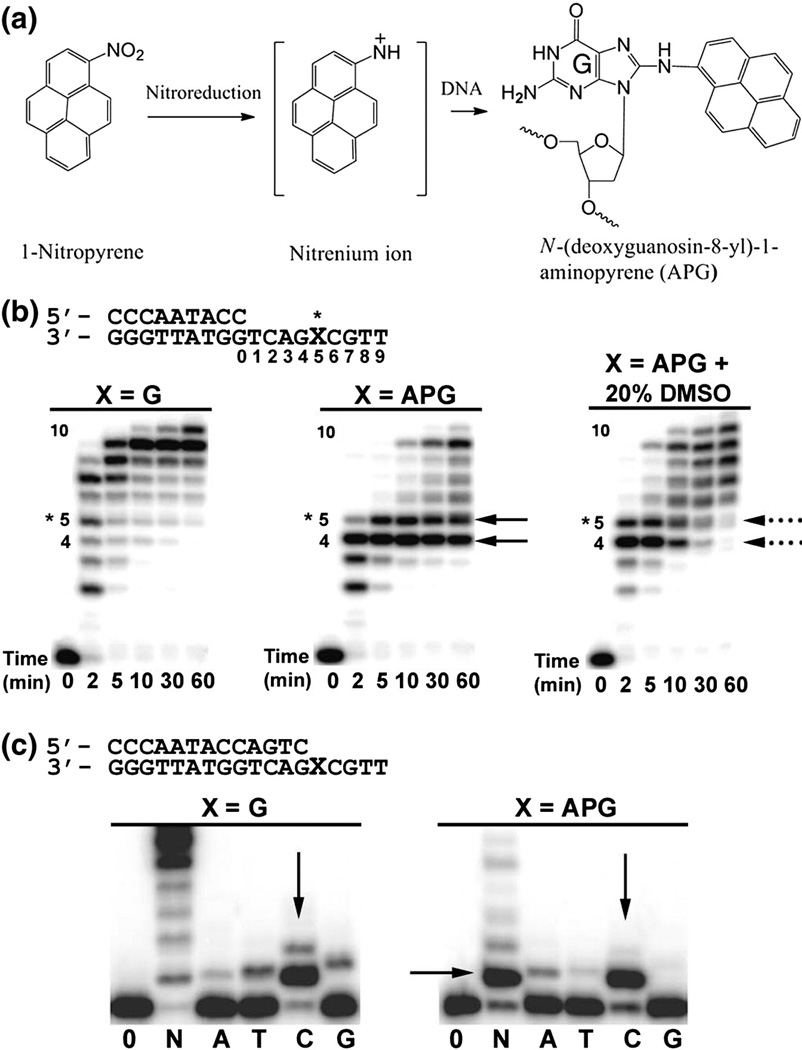
Exposure to atmospheric pollution from industrial emissions and automobile exhaust is a significant contributor to increased morbidity and mortality in human populations [1]. Nitrated polycyclic aromatic hydrocarbons (NPAHs) are toxic and carcinogenic environmental pollutants produced from the combustion of carbon-containing materials including fossil fuel. Human exposure to NPAH compounds is primarily from industrial emissions, automobile exhaust, and cigarette smoke. 1-Nitropyrene (1-NP) is the most abundant and global NPAH in the environment due to its high concentration in diesel fuel exhaust [2]. 1-NP and related NPAH compounds are metabolically reduced in cells to reactive nitrenium ions, which predominately react with the C8 atom of guanine nucleotides producing bulky hydrophobic lesions in the genome [3–5]. These NPAH-generated DNA lesions are shown to significantly block DNA replication and induce mutagenesis [6]. The reduced 1-NP compound forms the bulky N-[deoxyguanosine-8-yl]-1-aminopyrene (APG) adduct (Fig. 1a), which causes cellular toxicity and induces G-to-T transversions in human cell lines. 1-NP-induced tumor formation in animal models are believed to be initiated by the mutagenic effects of this adduct [3,7–10].
Fig. 1.
Formation of the APG adduct and Dpo4 bypass activity. (a) The 1-NP compound is reduced in the cell to generate one or more reactive species that react with the C8 position of guanine nucleotides, producing the APG lesion. (b) Running-start primer extension assays with Dpo4 and undamaged G or the APG lesion. Dpo4 was incubated with DNA substrates and reacted in the presence of all four nucleotides at various time points, indicated under each lane. Replication stalling at APG pre-insertion (position 4) and APG insertion (position 5) are denoted with black arrows. The resolving of stalling bands in the presence of 20% (v/v) DMSO is indicated with black broken lines. (c) Standing-start primer extension assays with Dpo4 and undamaged G or the APG lesion at the first replication position. Dpo4 was incubated with DNA substrates and reacted with all four nucleotides (N) or individual nucleotides (A, T, C, and G) for either 2 min for undamaged G or 30 min for the APG lesion. Vertical arrows indicate nucleotide preferences opposite G and the APG lesion. DNA substrates are shown above the gels.
High-fidelity DNA polymerases are unable to replicate through bulky DNA adducts; thus, NPAH lesions in the genome can inhibit replication fork progression [11]. Specialized lesion bypass polymerases belonging to the Y family are required to replicate through bulky DNA adducts such as NPAH lesions in order for DNA duplication to proceed [12–14]. However, Y family polymerases also display significantly reduced bypass efficiency in the presence of NPAH lesions, which is likely a major contributor to cellular toxicity [6,15]. NMR studies have indicated that polycyclic aromatic hydrocarbon adducts usually intercalate into the DNA helix in protein-free solution structures [16–19]. Intercalation of bulky lesions distorts DNA helical structures in Y family polymerases, which inhibits dNTP incorporation [15,20–22]. How NPAH lesions are translated through the active site of Y family DNA polymerases and how the NPAH adducts contribute to replication stalling are currently unknown.
To elucidate the molecular basis of bulky NPAH lesion replication by Y family DNA polymerases, we used biochemical and structural approaches to study the model Y family polymerase Dpo4 from Sulfolobus solfataricus in APG replication. We determined Dpo4 in complex with APG-lesion-containing DNA in two structures that are trapped in different APG lesion bypass states. The first structure reveals how the APG lesion induces replication stalling at the preinsertion state, and the second structure reveals APG lesion stalling at the extension state. In addition, we have modeled the APG into the active site of Dpo4 at the insertion state in order to gain a complete view of how NPAH lesions are translated through a Y family polymerase active site. Our structural and biochemical experiments reveal that the hydrophobic nature of the APG lesion significantly contributes to its unique conformations that cause replication stalling. This work demonstrates the difficulty of NPAH lesion replication by a specialized Y family polymerase and highlights the toxic nature of this group of environmental pollutants.
Results and Discussion
Dpo4 bypasses APG lesion with reduced efficiency and fidelity
To confirm the APG lesion bypass activity of Dpo4, we performed running-start primer extension assays with undamaged G or the APG lesion positioned five nucleotides downstream from the primer-template junction (Fig. 1b). The template strand is identical with what we used for structural analysis in this study. APG lesion present in the template strand dramatically reduces the efficiency of primer elongation by Dpo4. Two major stalling bands are observed during primer extension through the APG lesion (Fig. 1b, black arrows) similar to the previous observation by Sherrer et al. [6]. The first pause site (Fig. 1b, position 4) indicates the difficulty in translocating the lesion into the active site for insertion opposite APG. The second pause site (position 5) indicates the difficulty in extending the primer past the lesion. Both stalling bands remain strong after 60 min of reaction, indicating that these two sites significantly block DNA replication by Dpo4. Interestingly, in the presence of 20% DMSO, the stalling bands resolve after 10 min of reaction (Fig. 1b, black broken lines). Similar increases in primer extension efficiency opposite a different PAH lesion have been observed for Dpo4 in the presence of organic solvents [21]. The bulky DNA lesion exposed to solvent in the major groove causes replication stalling, which is alleviated in the presence of organic solvents [21]. Thus, the observed APG-induced stalling bands likely result from the exposure of the hydrophobic pyrene ring to solvent near the major groove side. In addition, all three assays have a 10th band for one base additional extension beyond the template strand, a typical non-template-dependent extension from a blunt end, described in our previous structure study on the Y family polymerase [23].
To test the fidelity of Dpo4 opposite the APG lesion, we performed standing-start primer extension assays with undamaged G or the APG lesion directly downstream from the primer-template junction (Fig. 1c). For an undamaged template, Dpo4 efficiently extends the primer and selects the correct dC nucleotide for incorporation opposite G (Fig. 1c). In the presence of an APG lesion, significant stalling is observed at the lesion site when all four nucleotides are present, which is consistent with our running-start primer extension assays. Opposite the APG lesion, Dpo4 maintains correct dC incorporation preference; however, the efficiency of incorporation has been reduced as the reaction time was increased 15 times longer for the APG lesion. Pre-steady-state kinetics have revealed a 9-fold decrease in dCTP incorporation efficiency for Dpo4 opposite APG compared to undamaged G [6]. Although Dpo4 prefers the correct dC nucleotide, significant incorporation of mismatched dA is also observed opposite the APG lesion as the second preferred base opposite APG (Fig. 1c). In addition, 5-fold higher misincorporation rate of dA opposite APG has been observed from the pre-steady-state kinetics work compared to undamaged G template [6], which highlights the G-to-T mutagenic and tumor initiating property of this abundant organic chemical pollutant [4].
APG conformations in Dpo4 prevent DNA translocation at the pre-insertion stage
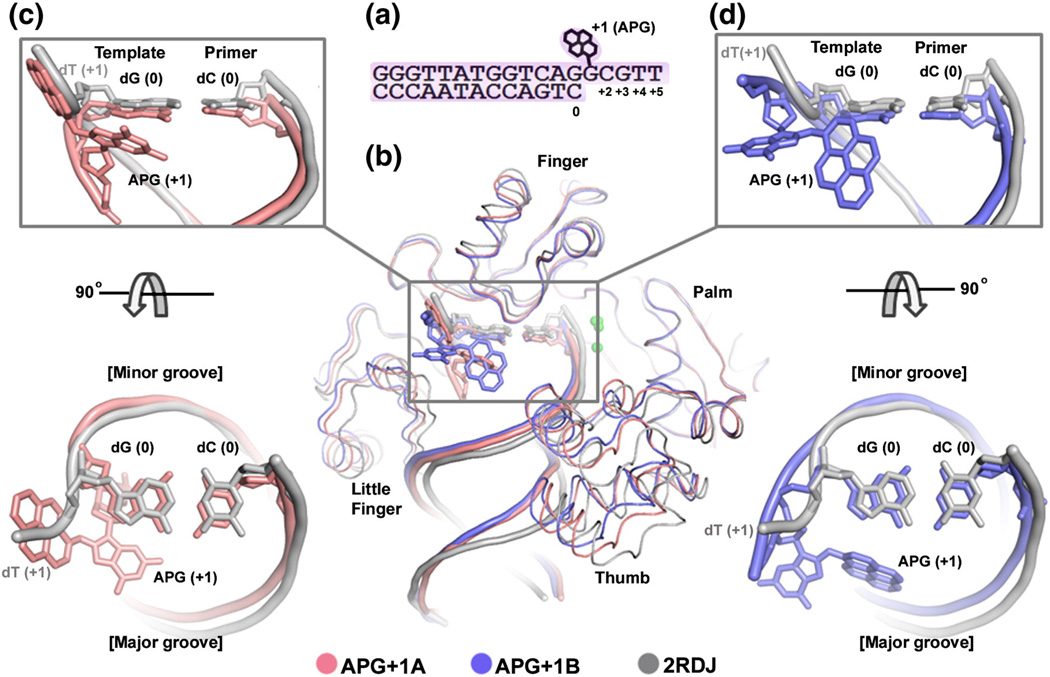
To elucidate the structural mechanism of Dpo4 replication stalling at the APG insertion step, we tried to co-crystallize Dpo4 with DNA containing the APG lesion at the template position next to the template-primer junction (Fig. 2a) and dCTP for a ternary complex at an insertion stage. However, the resulting crystals contain only Dpo4 and the lesion DNA in a binary form, indicating the instability of the insertion ternary complex. The crystals belonged to space group P21 and diffracted to 2.6 Å resolution (Table 1). The binary structure is referred to as APG+1 as the APG lesion is downstream to the template-primer junction at the active site (Fig. 2a). There are two binary complexes in the asymmetric unit, with similar protein structures with a backbone root-mean-square deviation (r.m.s.d.) of 0.6 Å on all Cα atoms (Fig. 2b). Overall, the structures of Dpo4 and the double-stranded part of DNA are similar in two APG lesion complexes and a previously reported Dpo4 binary structure (2RDJ) [24]. Superimposing two structures in APG+1 with 2RDJ shows small domain shifts in the thumb and little finger domains and their associated duplex DNA (Fig. 2b). In contrast, the single-stranded template with the APG lesion takes different conformations in two structures of APG+1 and 2RDJ (Fig. 2). The lesion base in one copy (designated as APG+1A, pink in Fig. 2) has shifted dramatically from that of undamaged dT at the +1 position in 2RDJ. The single-stranded template containing APG base has turned back to the duplex DNA with the pyrene ring system occupying the position of the undamaged template DNA backbone packed against the protein (Fig. 2c). Another conformation (APG+1B, blue in Fig. 2) changes the backbone conformation slightly from undamaged DNA, but its bulky APG ring exposes one side to solvent and packs the other side against the major groove site of the DNA duplex (Fig. 2d). The single-stranded template residues beyond the lesion site are completely disordered in both binary complexes. The back-turned template conformation in APG+1A is analogous to the U-turn template conformation observed in the human polι, which causes replication stalling at T bases [26]. The unusual DNA substrate conformations are either inhibitory to translocation (U-turned) or energetically unfavorable (solvent exposure), which usually causes replication stalling and a reduction in DNA replication efficiency [21,26].
Fig. 2.
APG+1 complex in comparison of with undamaged Dpo4 binary structure. (a) DNA substrate used for crystallization. (b) Superposition of the APG+1 structure (pink/blue) with a previously solved Dpo4 binary complex with undamaged DNA (PDB ID: 2RDJ; gray). Individual domains are labeled and arrows indicate movement relative to Dpo4 in complex with undamaged DNA. DNA (c). (d) Zoom in views of the superimposed active sites showing how the APG base projects into the major groove and the pyrene ring moves to the single-stranded undamaged template DNA position. Top view shows the major and minor groove sides of the DNA helix.
Table 1.
Summary of crystallographic data.
| Data collection | APG+1 | APG-1 |
|---|---|---|
| Space group | P21 | P61 |
| Molecules per asymmetric unit |
2 | 1 |
| Unit cell dimensions | ||
| a, b, c (Å) | 52.8, 181.5, 52.7 | 56.2, 56.2, 290.6 |
| α, β, γ (°) | 90, 109.5, 90 | 90, 90, 120 |
| Resolution (Å)a | 50.0–2.60 | 50.0–2.00 |
| (2.64–2.60) | (2.12–2.00) | |
| Unique reflections | 29,148 | 34,904 |
| Completeness (%)a | 95.8 (92.2) | 99.7 (95.6) |
| Redundancya | 1.9 (1.8) | 4.1 (2.3) |
| I/σIa | 25.56 (5.34) | 33.8 (5.15) |
| Rmergea | 4.2 (15.7) | 4.9 (22.3) |
| Refinement statistics | ||
| Rwork/Rfree | 21.8/26.1 | 19.1/22.6 |
| No. of atoms | ||
| Protein | 5467 | 2755 |
| DNA | 1136 | 609 |
| dNTP | — | 31 |
| Ions | 2 | 3 |
| Water | 248 | 482 |
| Average B-factor | ||
| Protein | 63.6 | 26.4 |
| DNA | 90.8 | 30.2 |
| dNTP | — | 38.1 |
| Ions | 55.9 | 29.3 |
| Waters | 63.6 | 37.7 |
| r.m.s.d. | ||
| Bonds (Å) | 0.007 | 0.008 |
| Angles (°) | 1.22 | 1.32 |
Data in highest-resolution shells are in parentheses.
The pyrene ring that takes the backbone site of a regular template nucleotide in APG+1A is partially protected in the cleft between the finger and little finger domain for template strand binding (Fig. 3a). To be shielded from solvent, the hydrophobic pyrene ring system localizes to mostly hydrophobic residues, such as Ser34, Phe37, Ser40, Gly41, Ala42, and Pro60 from the finger domain and Thr250, Leu293, and Arg331 from the little finger domain (Fig. 3a). Since the single-stranded template DNA needs to be translocated into the active site, the hydrophobic ring system has to move out of the solvent-shielded cleft. Thus, the interaction between the APG lesion and the protein side chains would prevent the lesion translocation into the active site. The hydrophobic ring is stabilized by the protein cleft during the pre-insertion stage, which may account for the increased binding affinity of Dpo4 for DNA substrate with an APG lesion at the +1 position (pre-insertion stage) observed previously by Sherrer et al. [6]. In our primer extension assays, adding 20% DMSO relieves the stalling effect of APG (Fig. 1b), which also supports the notion that the hydrophobic nature of APG lesion in aqueous solution contributes to its inhibition to replication. The protein protection of the hydrophobic ring in the APG+1A structure between the finger and little finger domains keeps the template DNA in a conformation that prevents translocation through the active site.
Fig. 3.
APG positioning in stalled Dpo4 complexes. Dpo4 is shown with finger, thumb, and palm colored cyan and labeled “core”. The little finger domain is colored light blue, the DNA helix is in yellow, the APG lesion is in black, and the nucleotides are in pink. Metal ions are shown as green spheres. (a) Structure of APG+1A; (b) APG-1. Zoom in view of the APG+1A active site showing the pyrene contacts with residues from the finger (Ser34, Phe37, Ser40, Gly41, Ala42, and Pro60) and little finger (Thr250, Leu293, Arg331) domains. The pyrene ring in APG-1 is sandwiched between R332, V289, L293, I295, and the +1 G base. The Fo – Fc difference electron density for the APG lesions (generated from simulated annealing omit maps) is colored green and contoured at 2σ.
In the APG+1B structure, the hydrophobic bulky ring has one face packing against major grooves and another face fully exposed to solvent (Fig. 2d). This conformation has been observed in benzo[a]pyrene adducted DNA in complex with Dpo4 [21]. It seems a common characteristic to position major groove attached bulky lesion rings into the open major groove side to avoid intercalated conformations. Intercalated bulky lesions are usually conformationally stable by shielding themselves from solvent in the DNA helix. However, intercalated bulky lesions often deform DNA helical structures, which results in non-productive adducted DNA/polymerase complexes [21,22]. In contrast, the solvent-exposed bulky pyrene ring in APG+1B is very flexible with high B-factor and weak electron density, similar to the benzo[a]pyrene dA adduct structure [21]. The instability of the solvent-exposed pyrene ring in APG+1B could contribute to the APG stalling band at the pre-insertion state and consequently induce low replication efficiency at this stage (Fig. 1b). Diminishment of the stalling band in the presence of DMSO in our running-start primer extension assays (Fig. 1b) supports the DNA conformation with the exposed bulky ring observed in the structure. Protein protection in APG+1A and less polar environments provided by organic solvent helps to stabilize the hydrophobic bulky ring in the APG lesion. The hydrophobicity of the pyrene ring and its conformation in Dpo4 play important roles in inhibition of nucleotide insertion against this bulky lesion.
Quaternary structure of Dpo4 at the APG post-insertion state
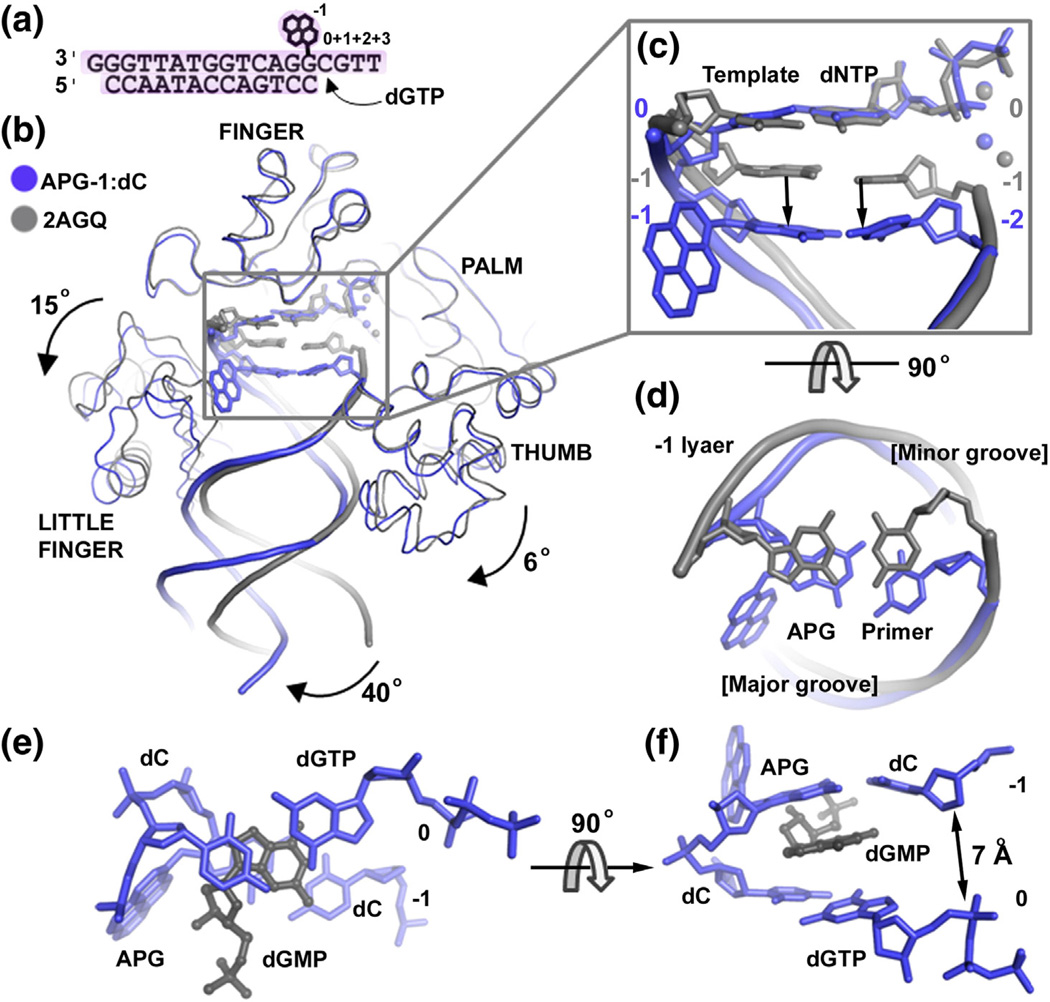
Dpo4 was co-crystallized with DNA containing the APG lesion upstream to the template base and incoming dGTP (Fig. 4a), forming a replicating complex at the extension stage (Figs. 3b and 4). The incoming dGTP nucleotide is paired with a template dC base (the replicating base pair referred to as position 0). The post-insertion structure is referred to as APG-1 according to the position of the APG lesion at the −1 position (Fig. 4a). Crystals belonged to space group P61 and diffracted to 2.0 Å resolution (Table 1). Superimposing APG-1 with a Dpo4 ternary structure with undamaged DNA (2AGQ) [27] reveals a base pair gap between replicating base pair and the lesion base pair beneath (Fig. 4b). The APG-1 structure contains an extra nucleotide dGMP filling the gap between the replicating base pair dC:dGTP and the lesion APG base, forming a quaternary complex (Fig. 3b). The dGMP molecule stacks with both the replicating base pair and the APG:C lesion base pair that shifted into the −2 position (Fig. 4b and c). No dGMP was added in crystallization; thus, this compound likely arises from degradation of the 3′ end G base of the template DNA by Dpo4 (Fig. 4a). The 3′–5′ exonuclease activity has been previously observed in Dpo4 [24], which generates deoxyribonucleotide monophosphates (dNMP).
Fig. 4.
APG-1 in comparison to the undamaged DNA ternary structure. (a) DNA substrate used for post-insertion crystallization. (b) Superposition of the APG-1 structure (blue) with a previously solved Dpo4 ternary complex with undamaged DNA (PDB ID: 2AGQ; gray). The dGMP in APG-1 has been removed for clarity. Individual domains are labeled and arrows indicate movement relative to Dpo4 in complex with undamaged DNA. The APG lesion and incoming nucleotides are shown in stick model with coordinated metal ions as spheres. (c) Zoom in view of the superimposed active sites showing how the APG:dC base pair moves down into a non-standard position. Top view shows the major and minor groove sides of the DNA helix. (d) The −1 base pairs (upstream to the replicating base pair) in the APG-1 and undamaged DNA structures. (e and f) The 0 and −1 base pairs with dGMP in the APG-1 structure.
The little finger and thumb domains that bind double-stranded helical DNA also rotate 15° and 6°, respectively (Fig. 4b). This inter-domain flexibility of the little finger and thumb domains is a common structural characteristic of the Y family DNA polymerases as the DNA helix distal from active site can swing around as well [24,25,28,29]. A 40° swing of the helical DNA distance from the active site is likely caused by different crystal packing between 2AGQ (P21212) and APG-1 (P61) as this part of the DNA is out of Dpo4 binding range. However, the DNA substrate near the template-primer junction at the active site adapts a unique conformation in APG-1. The replicating base pair is in a standard position, but the underlying APG lesion base pair (position − 1) has moved down along the helical axis by one base pair distance (∼3.4 Å) to the position −2 of a normal DNA structure (Fig. 4c). The gap in the helix is filled by dGMP (Fig. 4c and d), indicating the instability of the ternary complex due to helical distortion caused by the APG lesion.
The shifted APG lesion in APG-1 has its pyrene ring in one side of the major groove and covered by the G base from the template strand at the +1 position and the little finger domain (Fig. 3b). Instead of flipping away from the active site, the single-stranded template G base at the +1 position turns back toward the major groove to pack with the pyrene ring (Fig. 3b). The pyrene ring system is sandwiched between the +1 G base and Arg332 of the little finger (Fig. 4b). In addition, Val289, Leu293, and Ile295 of the little finger form van der Waals interactions with the pyrene ring and further shield the APG lesion from the solvent (Fig. 3b). The protein–APG interactions shift the APG base off the standard template-primer junction position and cause the 3′ end of the primer strand to move away (∼7 Å) from the α phosphate of the incoming nucleotide (Figs. 3b and 4). Thus, the APG-1 complex is found in a non-productive conformation that may account for the replication stalling observed during the extension stage (band 5 in Fig. 1b). A hydrophobic pocket present in the LF domain, in addition to the +1 template base and dNMP, stabilizes the extra helical conformation and contributes to the inhibition of DNA replication at the post-insertion state.
Insights into insertion opposite to APG by Dpo4
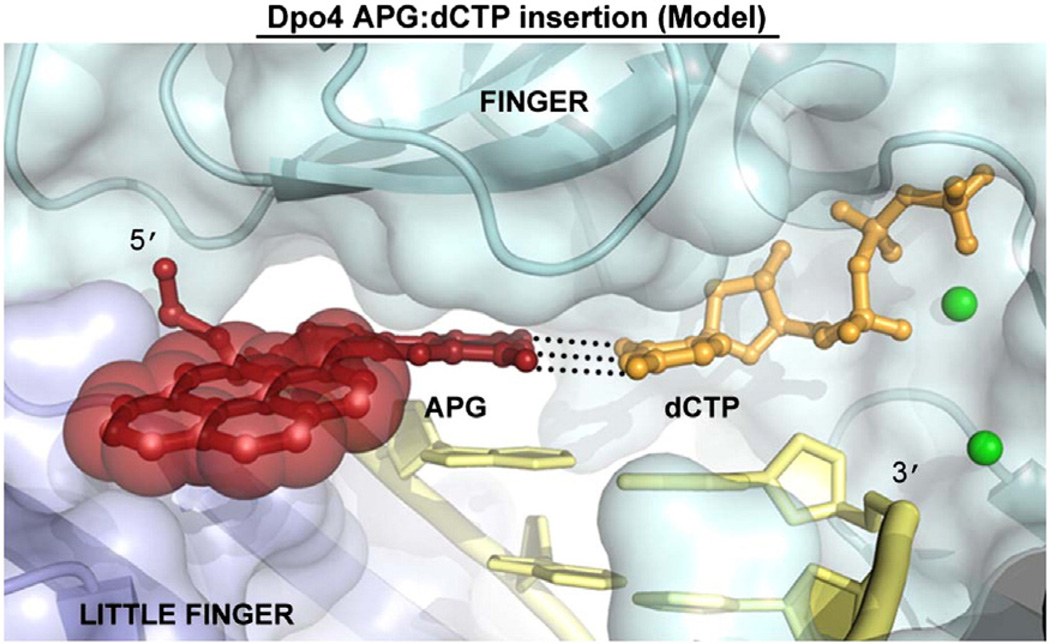
To illustrate how Dpo4 incorporates nucleotides directly opposite the APG lesion, we modeled the APG adduct in the Dpo4 active site in the insertion state. Most PAH lesion DNA complexes with translesion Y family DNA polymerases and, notably, the bulky benzo[a]pyrene adducts are either in binary forms [30] or in ternary forms of non-insertion states [15,21,22,30]. We recently determined two ternary complexes of human polι with APG in the insertion stage with both correct dCTP and incorrect dATP for APG bypass and mutagenic replication [31]. Considering the strong preference for correct dCTP insertion opposite APG by Dpo4, we put an incoming dCTP forming a Watson–Crick base pair with the APG lesion base (Fig. 5). We modeled the anti-APG lesion conformation with the bulky pyrene ring exposed to the major groove, based on the structure of the human Y family polymerase ι with an APG:dCTP Watson– Crick base pair in its active site [31]. The bulky lesion resides well in the active site of Dpo4 with the hydrophobic pyrene ring fully solvent exposed in the modeled structure (Fig. 5).The modeled complex is in a productive conformation without steric hindrances (Fig. 5), supporting the preferred incorporation of dCTP across APG by Dpo4 (Fig. 1c). However, the solvent exposure of the hydrophobic ring would be unstable, which may account for the stalling band in Fig. 1b (position 5). The difficulty to trap the bulky lesion ternary complexes in the insertion state in this study and most of Y family polymerase structures [15,21,22] also supports the unstable complex in this model. As the major groove around the active site of most Y family polymerases is open to solvent [12,13,32–36], using the solvent-exposed major groove to accommodate bulky lesions is likely a common mechanism for Y family polymerases to bypass bulky lesions [21,31].
Fig. 5.
Model of Dpo4 incorporating dCTP opposite the APG lesion. The positioning of the APG lesion and the dCTP nucleotide were based on a previous undamaged Dpo4 ternary complex (2AGQ) and a DNA polymerase iota AGP:dCTP insertion structure. The APG lesion is colored red with transparent spheres on the pyrene ring system. The DNA is in yellow, the dCTP is in orange, the finger domain is in cyan, the little finger domain is in light blue, and the metal ions are shown as green spheres. Black broken lines between APG and dCTP represent Watson– Crick hydrogen bonding.
A key issue for PAH lesion DNA bypass is the stability of hydrophobic bulky attachments at the template-primer junction. Multiple conformations have been observed for PAH DNA lesions in complex with Y family DNA polymerases [15,21,22,30,31]. The conformation preference of lesion DNA is highly dependent on the structural environment. Intercalation of a bulky pyrene ring into base stacking system of DNA helices is one way to stabilize the bulky lesion by shielding the PAH ring from water. However, the intercalation at the template-primer junction usually generates non-productive conformations in different PAH lesion DNA complex structures [21,22]. The reduced incorporation efficiency opposite bulky adducts is likely the result of an equilibrium between productive and non-productive conformations. In the insertion complexes of polι, the unique narrow active site either holds the exposed bulky pyrene ring with the closed little finger domain or intercalates it in a unique productive conformation [31]. With different Y family polymerase binding, APG likely exists in multiple conformations, which may account for the different replication efficiencies.
Conclusions
Exposure to NPAH compounds from automobile exhaust and industrial emissions produces bulky hydrophobic lesions in the genome. Such lesions block replication, and even when translesion polymerases are recruited, replication fork progression by Y family DNA polymerases is severely impeded and the translesion synthesis is highly error prone. We have shown that the hydrophobic NPAH lesion APG causes replication stalling. By interacting with polymerase hydrophobic side chains, the bulky hydrophobic DNA adduct maintains unusual conformations within the Y family polymerase Dpo4. Such unique conformations and unstable solvent-exposed conformations play critical roles in the low replication efficiency and high error rates in the translesion DNA replication.
Materials and Methods
APG-containing DNA synthesis
The monomer 2,8-diisobutyryl-8-(1-aminopyrenyl)-5′-O-(4,4′-dimethoxytrityl)-3′-O-[N,N′-diisopropylamino(2-cyanoethoxy)phosphonyl]-2′-deoxyguanosine was prepared as previously described [37]. It was chemically incorporated by standard DNA synthesis into the oligonucleotide, 5′-TCAG*GGGTCCTAGGACCC-3′, containing APG as the G* position. The APG modification of the 18-mer was confirmed by electrospray ionization mass spectrometry analysis.
Protein and DNA preparation
Dpo4 protein used for crystallization and functional assays was purified as previously described [12]. The APG DNA substrate used for crystallization and activity assays was purified using ion-exchange chromatography on a Source-15Q column [38]. For Dpo4 crystallization, an 18-nucleotide template (5′-TTGCG*GACTGGTATTGGG-3′) containing an APG adduct (G*) was annealed to either a pre-insertion 13-nucleotide primer (5′-CCCAATAC-CAGTC-3′) or a post-insertion 13-nucleotide primer (5′-CCAATACCAGTCCdd-3′) containing a dideoxy 3′ end (Cdd). For running-start primer extension assays, the 18-nucleotide APG template or an 18-nucleotide undamaged G template was annealed to a 9-nucleotide primer (5′-CCCAATACC-3′). For standing-start primer extension assays, the 18-nucleotide APG or undamaged G templates were annealed to the 13-nucleotide pre-insertion primer. The primers used for extension assays were 5′ end labeled using [γ-32P]ATP and T4 polynucleotide kinase. All primers were mixed with template DNA at a 1.5:1 molar ratio and heated to 60 °C, followed by slow cooling to form the annealed DNA substrates.
Replication assays
DNA substrates (10 nM) were incubated with Dpo4 (10 nM) and 100 µM either all four dNTPs or individual dNTPs at 37 °C in reaction buffer containing 40 mM Tris (pH 8.0), 5 mM MgCl2, 250 µg/ml bovine serum albumin, 10 mM DTT, and 2.5% (v/v) glycerol. For the running-start primer extension assays, reactions were carried out in the presence of all four nucleotides at various time points indicated below the gel images. For standing-start fidelity assays, reactions were carried out for 2 min with undamaged DNA and 30 min for the APG lesion DNA. Reactions were terminated with loading buffer [95% (v/v) formamide, 20 mM ethylenediaminetetraacetic acid, 0.025% (w/v) xylene, and 0.025% (w/v) bromophenol blue] and resolved on a 20% (v/v) polyacrylamide gel containing 7 M urea. Gels were visualized using a PhosphorImager.
Crystallization and structure determination
The complex crystals were formed by incubating Dpo4 protein (0.2 mM) with DNA in a 1:1.2 ratio, dCTP or dGTP (2 mM) and MgCl2 (5 mM). Both pre-insertion (APG+1) and extension (post-insertion, APG-1) crystals were obtained in 20% (w/v) polyethylene glycol 3350, 0.2 M CaAC2, 2.5% (v/v) glycerol, and 0.1 M Hepes (pH 7.0) and were cryoprotected with the addition of 20% (v/v) ethylene glycol to the mother liquor. X-ray diffraction data were collected on beamline 24-ID-C at the Advanced Photon Source in Argonne National Laboratory. All data were processed and scaled using HKL2000 [39].
Both complex structures were solved by molecular replacement using PHASER [40] with a previously solved ternary complex (PDB ID: 1JX4) as initial search models. The pre-insertion crystals displayed pseudo-merohedral twinning with a twinning fraction of 0.54. The data were detwined using DETWIN [41]. Structural refinement was performed using Phenix [42], starting with rigid-body refinement followed by positional and B-factor refinement, and finished with TLS refinement [43]. All structures have good stereochemistry with over 95% of the residues in the most favored region of the Ramachandran plot. Model inspection and re-building were performed using Coot [44], and figures were created with PyMOL [45].
Accession number
The atomic coordinates and structure factors have been deposited in the Protein Data Bank† with accession codes 4FBU and 4FBT for APG+1 and APG-1, respectively.
Acknowledgements
We would like to thank beamline support at 24-ID of Advanced Photon Source in Argonne National Laboratory. This work was supported by the Canadian Institutes of Health Research grant MOP93590 (to H.L.) and the National Institute of Environmental Health Sciences grant ES09127 (to A.K.B.).
Abbreviations used
- NPAH
nitrated polycyclic aromatic hydrocarbon
- 1-NP
1-nitropyrene
Footnotes
References
- 1.Samet JM, Dominici F, Curriero FC, Coursac I, Zeger SL. Fine particulate air pollution and mortality in 20 US Cities, 1987–1994. N Engl J Med. 2000;343:1742–1749. doi: 10.1056/NEJM200012143432401. [DOI] [PubMed] [Google Scholar]
- 2.Scheepers PTJ, Velders DD, Martens MHJ, Noordhoek J, Bos RP. Gas-chromatographic mass-spectrometric determination of nitro polycyclic aromatic-hydrocarbons in airborne particulate matter from workplace atmospheres contaminated with diesel exhaust. J Chromatogr A. 1994;677:107–121. [Google Scholar]
- 3.Andersson H, Piras E, Demma J, Hellman B, Brittebo E. Low levels of the air pollutant 1-nitropyrene induce DNA damage, increased levels of reactive oxygen species and endoplasmic reticulum stress in human endothelial cells. Toxicology. 2009;262:57–64. doi: 10.1016/j.tox.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Djuric Z, Fifer E, Yamazoe Y, Beland F. DNA-binding by 1-nitropyrene and 1,6-dinitropyrene in vitro and in vivo: effects of nitroreductase induction. Carcinogenesis. 1988;9:357–364. doi: 10.1093/carcin/9.3.357. [DOI] [PubMed] [Google Scholar]
- 5.Howard PC, Heflich RH, Evans FE, Beland FA. Formation of DNA adducts in vitro and in Salmonella typhimurium upon metabolic reduction of the environmental mutagen 1-nitropyrene. Cancer Res. 1983;43:2052–2058. [PubMed] [Google Scholar]
- 6.Sherrer S, Brown J, Pack L, Jasti V, Fowler J, Basu A, et al. Mechanistic studies of the bypass of a bulky single-base lesion catalyzed by a Y-family DNA polymerase. J Biol Chem. 2009;284:6379–6388. doi: 10.1074/jbc.M808161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watt DL, Utzat CD, Hilario P, Basu AK. Mutagenicity of the 1-nitropyrene-DNA adduct n-(deoxyguanosin-8-yl)-1-aminopyrene in mammalian cells. Chem Res Toxicol. 2007;20:1658–1664. doi: 10.1021/tx700131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asare N, Landvik N, Lagadic-Gossmann D, Rissel M, Tekpli X, Ask K, et al. 1-Nitropyrene (1-NP) induces apoptosis and apparently a non-apoptotic programmed cell death (paraptosis) in Hepa1c1c7 cells. Toxicol Appl Pharmacol. 2008;230:175–186. doi: 10.1016/j.taap.2008.02.015. [DOI] [PubMed] [Google Scholar]
- 9.Elbayoumy K, Rivenson A, Johnson B, Dibello J, Little P, Hecht SS. A study of chemical carcinogenesis 114 Comparative tumorigenicity of 1-nitropyrene, 1-nitrosopyrene, and 1-aminopyrene administered by gavage to Sprague-Dawley rats. Cancer Res. 1988;48:4256–4260. [PubMed] [Google Scholar]
- 10.Hirose M, Lee MS, Wang CY, King CM. Induction of rat mammary-gland tumors by 1-nitropyrene, a recently recognized environmental mutagen. Cancer Res. 1984;44:1158–1162. [PubMed] [Google Scholar]
- 11.Hsu GW, Huang X, Luneva NP, Geacintov NE, Beese LS. Structure of a high fidelity DNA polymerase bound to a benzo [a]pyrene adduct that blocks replication. J Biol Chem. 2005;280:3764–3770. doi: 10.1074/jbc.M411276200. [DOI] [PubMed] [Google Scholar]
- 12.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 13.Biertumpfel C, Zhao Y, Kondo Y, Ramon-Maiques S, Gregory M, Lee JY, et al. Structure and mechanism of human DNA polymerase eta. Nature. 2010;465:1044–1048. doi: 10.1038/nature09196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase-iota occurs by hoogsteen base-pairing. Nature. 2004;430:377–380. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- 15.Schorr S, Schneider S, Lammens K, Hopfner KP, Carell T. Mechanism of replication blocking and bypass of Y-family polymerase eta by bulky acetylaminofluorene DNA adducts. Proc Natl Acad Sci USA. 2010;107:20720–20725. doi: 10.1073/pnas.1008894107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cosman M, de los Santos C, Fiala R, Hingerty BE, Singh SB, Ibanez V, et al. Solution conformation of the major adduct between the carcinogen (+)-anti-benzo[a]pyrene diol epoxide and DNA. Proc Natl Acad Sci USA. 1992;89:1914–1918. doi: 10.1073/pnas.89.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng B, Gorin A, Kolbanovskiy A, Hingerty BE, Geacintov NE, Broyde S, et al. Solution conformation of the (−)-trans-anti-[BP]dG adduct opposite a deletion site in a DNA duplex: intercalation of the covalently attached benzo[a]pyrene into the helix with base displacement of the modified deoxyguanosine into the minor groove. Biochemistry. 1997;36:13780–13790. doi: 10.1021/bi970070r. [DOI] [PubMed] [Google Scholar]
- 18.Zhang N, Lin C, Huang X, Kolbanovskiy A, Hingerty BE, Amin S, et al. Methylation of cytosine at C5 in a CpG sequence context causes a conformational switch of a benzo[a]pyrene diol epoxide-n2-guanine adduct in DNA from a minor groove alignment to intercalation with base displacement. J Mol Biol. 2005;346:951–965. doi: 10.1016/j.jmb.2004.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mao B, Vyas RR, Hingerty BE, Broyde S, Basu AK, Patel DJ. Solution conformation of the N-(deoxyguanosin-8-yl)-1-aminopyrene ([AP]dG) adduct opposite dC in a DNA duplex. Biochemistry. 1996;35:12659–12670. doi: 10.1021/bi961078o. [DOI] [PubMed] [Google Scholar]
- 20.Banerjee S, Christov PP, Kozekova A, Rizzo CJ, Egli M, Stone MP. Replication bypass of the trans-4-hydroxynonenal-derived (6S,8R,11S)-1,N2-deoxyguanosine DNA adduct by the Sulfolobus solfataricus DNA polymerase IV. Chem Res Toxicol. 2012;25:422–435. doi: 10.1021/tx200460j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ling H, Sayer JM, Plosky BS, Yagi H, Boudsocq F, Woodgate R, et al. Crystal structure of a benzo[a]pyrene diol epoxide adduct in a ternary complex with a DNA polymerase. Proc Natl Acad Sci USA. 2004;101:2265–2269. doi: 10.1073/pnas.0308332100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bauer J, Xing G, Yagi H, Sayer JM, Jerina DM, Ling H. A structural gap in Dpo4 supports mutagenic bypass of a major benzo[a]pyrene dG adduct in DNA through template misalignment. Proc Natl Acad Sci USA. 2007;104:14905–14910. doi: 10.1073/pnas.0700717104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fiala KA, Brown JA, Ling H, Kshetry AK, Zhang J, Taylor JS, et al. Mechanism of template-independent nucleotide incorporation catalyzed by a template-dependent DNA polymerase. J Mol Biol. 2007;365:590–602. doi: 10.1016/j.jmb.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong JH, Fiala KA, Suo Z, Ling H. Snapshots of a Y-family DNA polymerase in replication: substrate-induced conformational transitions and implications for fidelity of Dpo4. J Mol Biol. 2008;379:317–330. doi: 10.1016/j.jmb.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 25.Ling H, Boudsocq F, Woodgate R, Yang W. Snapshots of replication through an abasic lesion; structural basis for base substitutions and frameshifts. Mol Cell. 2004;13:751–762. doi: 10.1016/s1097-2765(04)00101-7. [DOI] [PubMed] [Google Scholar]
- 26.Kirouac KN, Ling H. Structural basis of error-prone replication and stalling at a thymine base by human DNA polymerase-iota. EMBO J. 2009;28:1644–1654. doi: 10.1038/emboj.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaisman A, Ling H, Woodgate R, Yang W. Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J. 2005;24:2957–2967. doi: 10.1038/sj.emboj.7600786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xing G, Kirouac K, Shin YJ, Bell SD, Ling H. Structural insight into recruitment of translesion DNA polymerase Dpo4 to sliding clamp PCNA. Mol Microbiol. 2009;71:678–691. doi: 10.1111/j.1365-2958.2008.06553.x. [DOI] [PubMed] [Google Scholar]
- 29.Uljon SN, Johnson RE, Edwards TA, Prakash S, Prakash L, Aggarwal AK. Crystal structure of the catalytic core of human DNA polymerase kappa. Structure. 2004;12:1395–1404. doi: 10.1016/j.str.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 30.Rechkoblit O, Kolbanovskiy A, Malinina L, Geacintov NE, Broyde S, Patel DJ. Mechanism of error-free and semitar-geted mutagenic bypass of an aromatic amine lesion by Y-family polymerase Dpo4. Nat Struct Mol Biol. 2010;17:379–388. doi: 10.1038/nsmb.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirouac KN, Basu AK, Ling H. Replication of a carcinogenic nitropyrene DNA lesion by human Y-family DNA polymerase. Nucleic Acids Res. 2013;41:2060–2071. doi: 10.1093/nar/gks1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silverstein TD, Johnson RE, Jain R, Prakash L, Prakash S, Aggarwal AK. Structural basis for the suppression of skin cancers by DNA polymerase eta. Nature. 2010;465:1039–1043. doi: 10.1038/nature09104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- 34.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Human DNA polymerase iota incorporates dCTP opposite template G via a G·C + Hoogsteen base pair. Structure. 2005;13:1569–1577. doi: 10.1016/j.str.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 35.Lone S, Townson SA, Uljon SN, Johnson RE, Brahma A, Nair DT, et al. Human DNA polymerase kappa encircles DNA: implications for mismatch extension and lesion bypass. Mol Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- 36.Yang W, Woodgate R. What a difference a decade makes: insights into translesion DNA synthesis. Proc Natl Acad Sci USA. 2007;104:15591–15598. doi: 10.1073/pnas.0704219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colis LC, Chakraborti D, Hilario P, McCarty C, Basu AK. Synthesis of oligonucleotides containing 2′-deoxyguanosine adducts of nitropyrenes. Nucleosides Nucleotides Nucleic Acids. 2009;28:67–77. doi: 10.1080/15257770902736426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shanagar J. Purification of a synthetic oligonucleotide by anion exchange chromatography: method optimisation and scale-up. J Biochem Biophys Methods. 2005;64:216–225. doi: 10.1016/j.jbbm.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 39.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 40.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr Sect D Biol Crystallogr. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 41.Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr Sect D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 42.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, et al. Phenix: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Painter J, Merritt E. TLSMD Web server for the generation of multi-group TLS models. J Chromatogr. 2006;39:109–111. [Google Scholar]
- 44.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.DeLano WL. The PyMOL molecular graphics system. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]