Summary
The RecX protein inhibits RecA filament extension leading to net filament disassembly. The RecF protein physically interacts with the RecX protein and protects RecA from the inhibitory effects of RecX. In vitro, efficient RecA filament formation onto SSB-coated circular single-stranded DNA in the presence of RecX occurs only when all of the RecFOR proteins are present. The RecOR proteins contribute only to RecA filament nucleation onto SSB-coated single-stranded DNA and are unable to counter the inhibitory effects of RecX on RecA filaments. RecF protein uniquely supports substantial RecA filament extension in the presence of RecX. In vivo, RecF protein counters a RecX-mediated inhibition of plasmid recombination. Thus, a significant positive contribution of RecF to RecA filament assembly is to antagonize the effects of the negative modulator, RecX, specifically during the extension phase.
Introduction
Recombinational DNA repair pathways play an important role in the restoration of stalled replication forks in Escherichia coli (Cox et al., 2000; Kowalczykowski, 2000; Kuzminov, 1999; Lusetti and Cox, 2002). RecA protein, the central recombinase in E. coli, is a DNA-dependent ATPase that catalyzes homologous DNA pairing and strand exchange reactions in vitro that are thought to mimic its role in recombinational DNA repair (Cox, 2003; Lusetti and Cox, 2002). RecA protein also regulates induction of the SOS response by mediating autocatalytic cleavage of the LexA repressor (Kim and Little, 1993; Nastri and Knight, 1994). The active species in both of these processes is a RecA filament formed on single-stranded DNA (ssDNA). Assembly of a nucleoprotein filament occurs in two distinct stages (Lusetti and Cox, 2002; Roca and Cox, 1997). A slow RecA protomer nucleation event is followed by rapid filament extension in the 5' to 3' direction (Register and Griffith, 1985; Shan et al., 1997). RecA filament disassembly also occurs in the 5' to 3' direction upon ATP hydrolysis (Bork et al., 2001a; Shan et al., 1997), such that monomers are added to one filament end and subtracted from the other. The dynamic nature of RecA nucleoprotein filaments makes assembly and disassembly prime targets for RecA regulation. RecA regulator proteins generally modulate when and where RecA filaments form as opposed to manipulating the catalytic properties of the filament.
One modulator of RecA filament dynamics is the single-stranded DNA binding protein (SSB). SSB strongly inhibits the nucleation phase of RecA filament assembly (Kowalczykowski et al., 1987), but stimulates the extension phase, removing secondary structure in ssDNA that would otherwise impede filament growth (Kowalczykowski and Krupp, 1987; Lavery and Kowalczykowski, 1992).
The RecF, RecO, and RecR proteins promote the establishment of RecA filaments at daughter strand gaps where bound SSB limits RecA nucleation (Clark and Sandler, 1994; Kuzminov, 1999; Madiraju et al., 1988; Sawitzke and Stahl, 1992; Sawitzke and Stahl, 1994; Smith, 1989; Whitby and Lloyd, 1995). The E. coli RecF protein (40.5 kDa) binds both ssDNA and double-stranded DNA (dsDNA) in vitro (Madiraju and Clark, 1992; Webb et al., 1995) and exhibits a weak dsDNA-dependent ATPase activity (Webb et al., 1995; Webb et al., 1999). RecF protein physically interacts with the RecR protein in a manner dependent on both DNA and ATP (Webb et al., 1995). RecR protein (22 kDa) has mainly been studied jointly with the RecF protein or with its other interacting partner, RecO (Bork et al., 2001b; Shan et al., 1997; Umezu and Kolodner, 1994). RecO protein (26 kDa) binds both ssDNA and dsDNA in vitro (Luisi-DeLuca and Kolodner, 1994; Umezu et al., 1993; Umezu and Kolodner, 1994) and promotes renaturation of homologous DNA strands (Kantake et al., 2002; Luisi-DeLuca and Kolodner, 1994).
Decades of research on the recF, recO and recR genes has demonstrated that they constitute an epistasis group intimately involved in the establishment of RecA filaments on DNA (Ennis et al., 1995; Knight et al., 1984; Lavery and Kowalczykowski, 1992; Madiraju et al., 1992; Madiraju et al., 1988; Thomas and Lloyd, 1983; Volkert and Hartke, 1984; Wang et al., 1993; Whitby and Lloyd, 1995). Nevertheless, there is no physical evidence for a complex containing all three proteins. Biochemical data has confirmed that the RecOR complex functions to reduce the long lag in RecA binding to ssDNA prebound by the SSB protein (Bork et al., 2001b; Shan et al., 1997; Umezu and Kolodner, 1994). However, the role of RecF in this process is unclear. RecF protein is completely dispensable in RecOR-mediated SSB displacement by RecA and inhibits RecA activities under many conditions (Bork et al., 2001b; Umezu et al., 1993). RecF does enhance the RecOR-mediated loading of RecA at the end of a DNA gap under at least one set of conditions (Morimatsu and Kowalczykowski, 2003). In vivo, in at least some contexts, RecF functions on its own (Grompone et al., 2004; Kidane et al., 2004; Rangarajan et al., 2002; Sandler et al., 1996). RecF protein may have multiple functions, some of which may be independent of the RecO and RecR proteins.
Work to date on the RecFOR proteins has focused on their effects on RecA filament nucleation. In principle, RecA regulatory proteins could affect either phase of filament assembly, nucleation or extension. The extension phase of RecA filament assembly is blocked by the RecX protein (19.3 kDa) in vitro (Drees et al., 2004a). RecX is thus a negative regulator of RecA in vivo (Stohl et al., 2003) and in vitro (Drees et al., 2004a; Stohl et al., 2003). The RecA and RecX proteins directly interact (Stohl et al., 2003; VanLoock et al., 2003). We set out to further define the role of RecF in mediating RecA filament formation by examining RecF function in the presence of a RecA filament extension inhibitor, RecX.
Results
Experimental Design
Using purified proteins, we investigated the integrated effects of the E. coli RecF, RecO and RecR proteins on RecA filaments assembling on SSB-coated DNA when challenged by the RecX protein. The effects on RecA filaments were examined by monitoring the DNA-dependent ATP hydrolysis activity of the RecA protein, providing a real-time measurement of the amount of RecA bound to DNA (Arenson et al., 1999; Lavery and Kowalczykowski, 1992; Lindsley and Cox, 1990; Shan et al., 1997). The kinetic experimentation was complemented by electron microscopy, and by in vivo studies of the recombination efficiency of pertinent mutants.
Many of the ATPase assays described below include the RecF protein. RecF is a dsDNA-dependent ATPase (kcat <1 min−1; (Webb et al., 1999)). There is no measurable ATP hydrolysis by the RecF protein under our conditions when the RecA protein is omitted (data not shown). We have thus ignored the potential contribution of RecF protein to the observed ATP hydrolysis in our data analysis.
The RecX protein has no effect on RecA filament assembly onto SSB-coated circular ssDNA in the presence of the RecFOR proteins
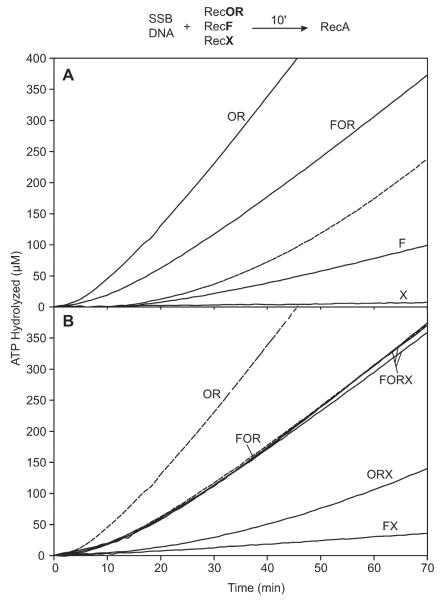
The general effect of various mediator proteins on RecA filament assembly on SSB-coated DNA is shown in Figure 1A. We added RecA protein to ssDNA that was preincubated with SSB or with SSB and RecOR proteins. RecA (1.2 μM) is added in excess over the concentration of available ssDNA binding sites (0.67 μM). The RecO and RecR proteins are added at concentrations (100 nM and 1 μM, respectively) that have been determined to provide maximal enhancement of RecA assembly onto SSB-coated circular ssDNA (M. D. Hobbs and M. M. Cox, unpublished results). The RecOR complex decreases the time required for RecA to nucleate onto SSB coated ssDNA and reach a steady-state rate of ATP hydrolysis from well over 30 min to under 10 min. When RecF protein (200 nM) is included in the RecOR-mediated SSB displacement assay, the time required to reach a steady-state of ATP hydrolysis is increased by ~ 5 minutes; i.e., filament nucleation is inhibited. Furthermore, the final steady-state rate of ATP hydrolysis is consistently lower in the presence of RecF ((Bork et al., 2001b; Madiraju and Clark, 1991); Fig. 1A). The mechanism by which RecF reduces the amount of RecA protein bound to DNA is not understood, but it may be competing with RecA for DNA binding sites.
Figure 1. The effect of RecOR, RecF and RecX on SSB displacement by the RecA protein.
The reaction scheme at the top of the figure details the order of addition for this experiment. SSB protein (0.2 μM) was incubated with 2 μM M13mp8 circular ssDNA under the conditions described in the Experimental Procedures for 10 mins with the mediator protein(s) (RecO, 100 nM; RecR, 1μM; RecF, 200 nM; RecX, 100 nM). The figure labels indicate which mediator protein(s) were preincubated with the ssDNA and the SSB. The reaction was initiated by the addition of RecA to 1.2 μM (t = 0). The dashed line in panel A is a control showing RecA displacement of SSB protein in the absence of any other mediator protein. In panel B, proteins added in addition to RecA and SSB are indicated for each experiment. The three overlapping lines labeled “FORX” include 100, 300 or 500 nM RecF protein.
Relatively low concentrations (100 nM) of RecX protein completely inhibit RecA filament extension even when added to pre-formed RecA filaments (Drees et al., 2004a; Drees et al., 2004b; Stohl et al., 2003). As expected, productive assembly of RecA filaments on SSB-coated ssDNA was blocked by RecX (Fig. 1A). The RecF protein (in the absence of RecOR) also inhibited this process but to a much lesser extent.
We next tested RecA nucleation onto SSB-coated ssDNA using various combinations in which RecX was included with all or a subset of the RecFOR proteins (Fig. 1B). RecX greatly reduced the RecOR-mediated assembly of RecA filaments (compare the OR and ORX data in Fig. 1B). It is possible that RecOR-facilitated loading of RecA protein is futile since filament extension is blocked by the RecX protein. As seen in Figure 1A, RecF protein again inhibits the RecOR–mediated reaction, but to a lesser extent. Remarkably, the addition of RecX to the RecFOR-mediated SSB displacement reaction results in no measurable inhibition relative to the reaction seen with RecFOR alone (Fig. 1B). This protective effect of RecF was observed when as little as 100 nM RecF protein was included (1 RecF monomer for every RecX monomer present) and the effect appears to be saturated since increasing the RecF protein concentration to 500 nM produced the same result as 100 nM RecF (Fig. 1B). Therefore, we have explored further the possibility that RecF acts specifically on RecX, relieving the blockage to RecA filament extension.
The RecF and RecX proteins physically interact
We tested for an interaction between surface-immobilized RecX (ligand) and free RecF (analyte) using biosensor technology (Fig. 2). A direct interaction between the RecF and RecX proteins is demonstrated by the rapid accumulation of RecF to a sensor surface coated by RecX. RecX-dependent accumulation of RecF protein on the sensor surface is observed as an increase in the response reported by the instrument. This response is positively correlated to the density of matter on the sensor surface (Nagata and Handa, 2000).
Figure 2. The RecF and RecX proteins interact.
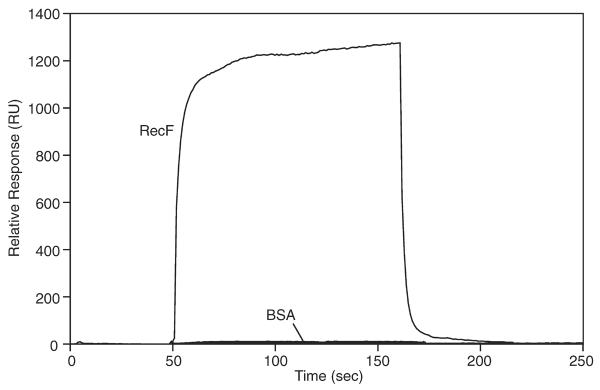
Physical interaction between the RecF and RecX proteins was observed in real-time using a BIAcore2000 as described in the Experimental Procedures. 2701 RU of RecX protein was immobilized and RecF protein (50 μg/mL) was applied for 2 min. RecF binding to RecX is observed by the rapid increase and plateau in the relative response as RecF accumulates. The observed response after injection of BSA is also shown.
Once binding has been saturated, the supply of RecF is turned off. The subsequent dissociation of RecF was rapid, reaching completion within 2 minutes. To determine the specificity of the interaction between RecF and RecX, Bovine Serum Albumin (BSA) was applied to the same sensor surface. In this case, no binding was observed, indicating that the RecF-RecX interaction was specific.
When the reciprocal experiment was attempted, no interaction between RecX and surface immobilized RecF was observed. Interactions between RecF and other known interacting partners such as DNA and RecR protein (Webb et al., 1995; Webb et al., 1999) were also not observed (data not shown). RecF protein appears to be inactivated during the immobilization process, as also seen in previous BIAcore studies carried out on RecF (Umezu and Kolodner, 1994).
RecF protein protects RecA filaments challenged by RecX following RecOR-mediated nucleation
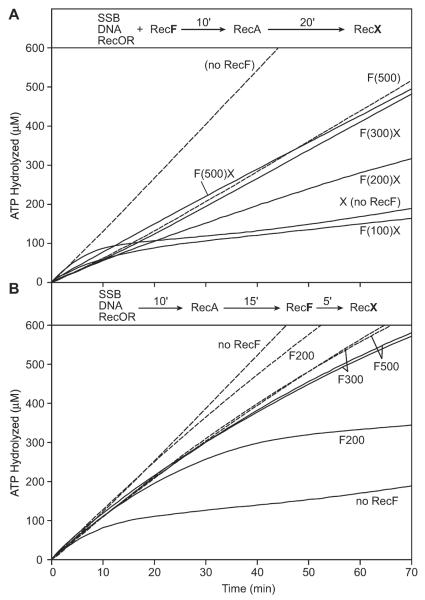
In the experiment described in Figure 1, RecX was present in the reaction prior to the addition of RecA protein. In order to test the effect of RecF when RecX is added to existing RecA filaments, we carried out the RecOR-mediated SSB displacement reaction in the presence and absence of RecF protein and allowed RecA filaments to form for 20 minutes before addition of RecX (Fig. 3A). When RecX was added to pre-formed RecA filaments in the absence of RecF, a decline in the rate of hydrolysis was observed, reflecting RecA filament disassembly. This result is very similar to previous studies where RecX protein is added to preformed RecA filaments in the absence of RecOR (Drees et al., 2004a; Drees et al., 2004b; Lusetti et al., 2004a). The inclusion of RecF protein protected RecA filaments from the negative effects of RecX. However, a higher concentration of RecF (relative to RecX) was required to counter the inhibitory effects of RecX in this experimental situation (Fig. 3A). This suggests that there is a competition between a RecF-RecX interaction and a RecX-RecA interaction.
Figure 3. The effect of RecX on RecAOR filaments pre-formed in the presence or absence of RecF protein.
The reaction schemes at the top of each panel details the order of addition for the experiment. (A) SSB (0.2 μM), RecO (100 nM) and RecR (1 μM) proteins were incubated with 2 μM M13mp8 circular ssDNA for 10 min with the concentration of RecF (in nM) indicated parenthetically. The reaction was initiated by the addition of RecA to 1.2 μM, incubated further for 20 min and the reaction was challenged by the addition (t = 0) of RecX to 100 nM (solid lines) or the equivalent volume of RecX storage buffer (dashed lines). (B) SSB (0.2 μM), RecO (100 nM) and RecR (1 μM) proteins were incubated with 2 μM M13mp8 circular ssDNA for 10 min. The reaction was initiated by the addition of RecA protein to 1.2 μM. After 15 min, RecF was added to the indicated concentration (nM) and the reaction was challenged 5 min later by the addition of (t = 0) of RecX protein to 100 nM (solid lines) or the equivalent volume of RecX storage buffer (dashed lines).
RecF-mediated protection from the negative effects of RecX does not require a contribution of RecF to RecOR-mediated RecA nucleation
The experiments of Figures 1 and 3A show a positive effect when RecF is present during the nucleation phase of the RecOR-mediated SSB displacement reaction. To test whether the RecF protein can protect RecA filaments assembled in the presence of the RecOR complex (as opposed to RecFOR), that are challenged by RecX protein, we added both the RecF and RecX proteins after RecA filaments had formed (Fig. 3B). When RecF is added later in the reaction, the rate of RecA-catalyzed ATP hydrolysis is higher than when RecF is present before the addition of RecA. This is another indication that RecF may be competing with RecA for DNA binding sites when added early in the reaction (during the nucleation step, Figs. 1 and 3A). Once again, the RecX-imposed block to RecA filament extension was offset by the presence of RecF. Therefore, the presence of RecF protein is not required during the RecOR-mediated nucleation step in order for RecF to protect the filaments from RecX. This further argues that RecF protein is specifically acting on RecX protein.
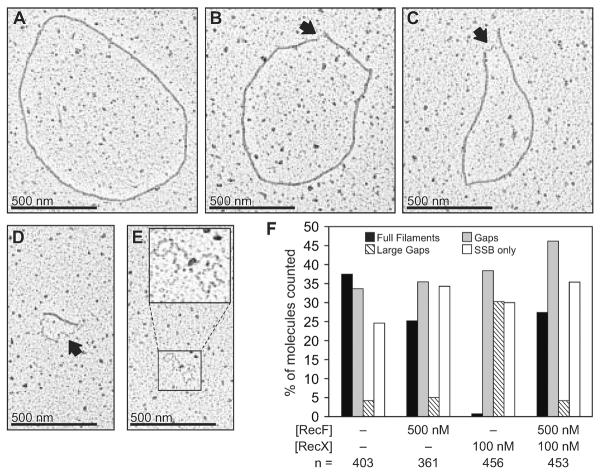
The conclusions derived from the ATPase data in Figure 3B were confirmed under the same conditions by electron microscopy (Fig. 4). Examples of the variety of DNA-bound species observed in all experiments are shown in Figures 4A–E. When RecF and RecX were not present, over 35% of the RecA filaments completely coated the ssDNA as shown in Figure 4A. Most of the remainder of filaments observed had very small gaps in the filaments bound by SSB protein such as the one shown in Figure 4B. In assessing the proportion of RecA filaments that were discontinuous, we distinguished a short filament class in which the RecA filament was unambiguously shorter than 25% of the length of full filaments (large gaps, Fig. 4D). All of the samples contained a significant proportion of DNA molecules bound only by the SSB protein as shown in Figure 4E.
Figure 4. Electron microscopy of RecA filaments formed under the conditions of Figure 3B.
Samples for electron microscopy were prepared using the order of addition detailed in the reaction scheme of Figure 3B. SSB (0.2 μM), RecO (100 nM) and RecR (1 μM) proteins were incubated with 2 μM M13mp8 circular ssDNA for 10 min before the addition of RecA to 1.2 μM. After 15 min, RecF protein (or the equivalent volume of RecF storage buffer) was added to 500 nM and the reaction was challenged 5 min later by the addition of RecX protein (or the equivalent volume of RecX storage buffer) to 100 nM. After 10 min, the filaments were fixed with ATPγS (to 3 mM). The electron micrograph in panel A represents a full RecA filament formed on circular ssDNA. Panels B, C and D show examples of discontinuous RecA filaments, with larger gaps evident as the sequence progresses. Panel E shows a ssDNA molecule bound only with SSB (enlarged in the inset). All molecules are shown at the same magnification. Arrows point to regions of the circular ssDNA bound by SSB protein. In panel F, molecules from 2 different samples for each of the four experiments were counted and scored as fully filamented (black bars) as shown in panel A, circular filaments containing a small to medium sized gaps bound by SSB protein (gray bars) as shown in panels B and C, DNA molecules containing a RecA filament unambiguously encompassing less than 25% of the DNA (striped bars) as shown in panel D, or circular ssDNA bound only by the SSB protein (white bars) as shown in panel E. The counts are expressed as a percentage of the total number of circular molecules counted (n).
Addition of RecF slightly reduced the number of full filaments and increased the number of filaments exhibiting discontinuities. When only RecX protein was added, and the filaments examined 10 min later, complete filaments were rarely observed. A substantial proportion of the gapped filaments fell into the short filament (large gap) class (Fig. 4D). Addition of RecF protein with the RecX had a dramatic effect, retaining a substantial level of full filaments and reducing the proportion of gapped filaments in the short filament class. These observations confirm that RecF counteracts the negative effects of RecX on RecA filament assembly.
RecF protein alone is sufficient to protect RecA from the inhibitory effects of RecX
The data presented above suggests a model in which the RecOR contribution to the RecFOR protection of RecA filaments from RecX is only to mediate the nucleation of RecA. In order to exclude the possibility that RecOR may act with RecF to counteract the effects of RecX, we carried out several experiments with a RecA mutant protein (RecA E38K; also known as RecA730) that readily nucleates onto SSB-coated DNA without the RecOR proteins (Eggler et al., 2003; Lavery and Kowalczykowski, 1992) (Fig. 5). This allowed us to isolate the effects of RecF protein from those of the RecOR complex.
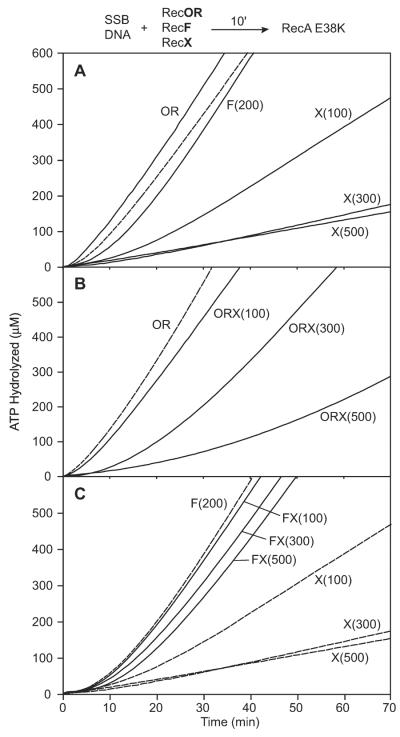
Figure 5. The effect of RecX on RecA E38K filament assembly in the presence of RecF or RecOR.
The reaction scheme at the top of the figure details the order of addition for this experiment. SSB protein (0.2 μM) was incubated with 2 μM M13mp8 circular ssDNA for 10 min with the mediator protein(s) (RecO, 100 nM; RecR, 1 μM; RecF and RecX concentration in nM indicated). The figure label details which mediator protein(s) were added in a given experiment. The reaction was initiated by the addition of RecA E38K protein to 1.2 μM (t = 0). The dashed line in panel A reflects RecA E38K displacement of SSB protein in the absence of any other mediator protein. RecF was omitted from the assays represented in panel B. In panel C, the RecOR proteins were omitted from the assays and the concentration of RecF and RecX are the same. In panel B and C, the dashed lines are duplicated from panel A and contain no RecX protein. In panel C, the RecX alone data has been duplicated from panel A to provide a convenient point of comparison.
We monitored the RecA E38K SSB displacement reaction in the presence and absence of RecOR, RecF or RecX (Fig. 5A). As expected, RecA E38K filaments assemble efficiently onto SSB-coated ssDNA in the absence of RecOR or RecF. The RecOR proteins stimulated RecA E38K nucleation slightly, since the time required to reach a steady-state rate of hydrolysis is decreased by several minutes when RecOR is present. Inclusion of RecF in the absence of RecOR was less inhibitory to the nucleation of RecA E38K than was observed for wild-type RecA protein (Fig. 1A). The addition of RecF, RecO and RecR proteins in the assay with RecA E38K yielded the same result as RecF alone (data not shown). RecX protein (100 nM and higher) significantly inhibited the assembly of RecA E38K filaments in the absence of RecOR or RecF (Fig. 5A). However, increased concentrations of RecX were required for optimal inhibition of RecA E38K filament assembly (Fig. 5A) as compared to wild-type RecA (Fig. 1A).
The amount of RecA protein bound to the ssDNA in the presence of RecX is a complex function of the RecA nucleation rate (unaffected by RecX protein), the rate of filament capping by RecX protein to block filament extension, and the rate of RecA filament disassembly. The RecA E38K mutant can nucleate relatively quickly to multiple regions of the ssDNA and higher concentrations of RecX are necessary to block the extension of those multiple nucleation events. RecOR-mediated nucleation of RecA E38K is also occurring (Fig 5A), and the two processes together can help offset the effects of RecX (Fig. 5B). Nevertheless, higher concentrations of RecX protein (500 nM) are still sufficient to block the extension of those multiple nucleation events in the RecA E38K-mediated SSB displacement reaction even in the presence of RecOR.
To determine the contribution of RecF protein alone to the RecA filament extension that is challenged by RecX, the RecA E38K SSB displacement reaction was carried out in the absence of RecOR complex (Fig. 5C). RecF and RecX proteins were included at equimolar concentrations. RecX concentrations up to 500 nM had only a modest effect on the assembly of RecA E38K in the presence of RecF. This suggests that RecF protein alone (as opposed to RecFOR) is sufficient to relieve the block to RecA filament extension imposed by the RecX protein.
RecF counters RecX function in vivo
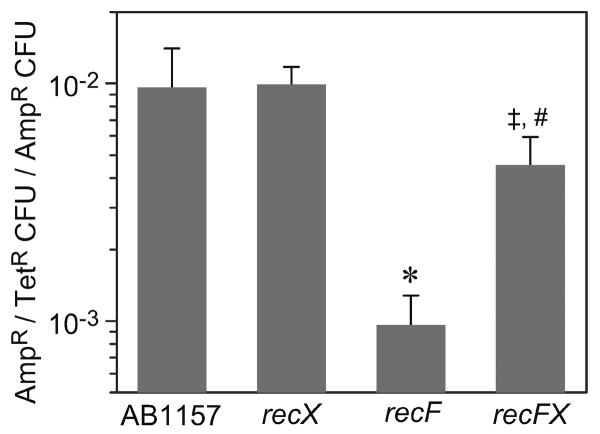
RecF is needed for recombination within plasmids in vivo (Cohen and Laban, 1983; Fishel et al., 1981). To measure the effect of RecX on this process we used a modified method of Fishel et al. (Fishel et al., 1981) to determine the amount of plasmid recombination occurring in strains AB1157, recX, recF, and recFX. Recombination between inactivated tet genes in plasmid pRDK301 was decreased in a recF mutant relative to the parental strain AB1157 (Fig. 6). The recX mutant alone had no effect. However, a strain with null mutants of both recF and recX showed a level of recombination statistically indistinguishable from strain AB1157 and statistically different from the recF strain (Fig. 6). This reversal of the reduced recombination occurring in the recF strain by inactivation of recX indicates that RecX is able to antagonize the action of RecA protein in plasmid recombination, and that the RecF counters the activity of RecX. The interaction between RecX and RecF on RecA activity could occur at two distinct levels, directly on recombination and by differentially activating the SOS response. Measurement of SOS induction of a sulA::lacZ fusion in these strains established that the recF and recFX show similar levels of SOS induction (data not shown), suggesting that the effect of RecF and RecX in vivo is to directly alter RecA function.
Figure 6. Modification of recF-mediated plasmid recombination by recX.
Strains AB1157, recX, recF, and recFX were analyzed for their ability to catalyze recombination of plasmid pRDK301, which confers AmpR/TetR when it recombines and only AmpR when it does not recombine. Plasmid recombination was decreased in a recF mutant relative to the parent strain AB1157 and recX mutant. Strain recFX showed a level of plasmid recombination statistically the same as strain AB1157 and statistically different from strain recF. Error bars represent the standard error of the mean of 6 experiments for all strains except the recX strain (the recX strain was done twice to confirm the published result (Stohl et al., 2003) that elimination of recX function does not affect this recombination reaction). Statistical differences determined by the Students t test are indicated by a star relative to the parental (P<0.01) and a double cross relative to recF (P<0.03). A pound sign indicates a value statistically indistinguishable from the parent strain (P>0.8).
Discussion
The primary conclusion of this paper is that RecF protein has a dramatic and positive effect on RecA filaments that are challenged by the RecX protein. We propose that the RecFOR proteins are required for RecA filament assembly, but that they do not necessarily act together. The primary role of RecOR is to mediate RecA nucleation onto DNA that is bound by the SSB protein. Here we define one role for RecF in the mediation of RecA filament extension by directly interfering with the inhibitory function of RecX protein.
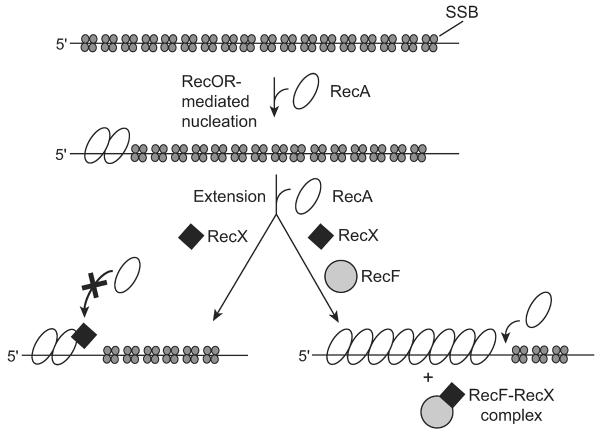
The role of the RecOR proteins in mediating the establishment of a RecA filament onto ssDNA has been well-established (Bork et al., 2001b; Umezu and Kolodner, 1994). SSB protein prebound to ssDNA is a clear inhibitor of RecA filament nucleation. The RecOR complex relieves the SSB inhibition by facilitating the RecA nucleation process. Based on genetic evidence, the RecF protein is also involved in RecA assembly. However, RecF is inhibitory to the RecOR-mediated nucleation of RecA filaments under most conditions, including those used in this study (see Fig. 1A). Given that assembly of RecA filaments requires both nucleation and filament extension, it seemed possible that RecF protein is involved in the extension step of assembly. RecA filament extension is sufficiently fast under standard reaction conditions in vitro that promotion of extension by a modulator is unnecessary. By inhibiting RecA filament extension with the RecX protein, we were able to determine that RecF provides a positive effect to the RecA assembly process by interfering with the negative modulator, RecX. A model for the assembly of RecA filaments in the presence of the SSB and RecX proteins is presented in Figure 7. The RecOR complex mediates the nucleation of RecA filaments. RecX protein inhibits the subsequent filament extension unless RecF sequesters the RecX through a direct interaction.
Figure 7. Model for RecOR and RecF-mediated RecA filament assembly in the presence of SSB and RecX.
We propose that the RecF protein and the RecOR complex enhance the assembly of RecA filaments. The nucleation step is mediated by the RecOR complex. The subsequent extension step of assembly is blocked by RecX. RecF protein provides a positive contribution to RecA assembly by specifically interfering with RecX binding to RecA filaments.
RecX protein is unable to measurably block RecA filament extension when RecF protein is also present at sufficient levels. RecF acts at relatively low levels, and directly interacts with RecX. It seems likely that the RecF-RecX interaction is relevant to the mechanism by which RecF antagonizes RecX function, although this has not been formally demonstrated. The mechanism by which RecF antagonizes RecX function is thus fundamentally different from that observed for the DinI protein. High levels of DinI (stoichiometric with RecA or more) are required for DinI function, and it acts by competing with RecX for binding sites on the RecA filament (Lusetti et al., 2004a; Lusetti et al., 2004b).
The competition between RecX bound to RecA filaments and the RecF-RecX interaction is illustrated by various order of addition experiments. When RecF and RecX are allowed to interact prior to the addition of RecA (Fig. 1B), less RecF is required to eliminate the negative effects of RecX than when RecX is added later to RecA filaments formed in the presence (Fig. 3A) or absence (Fig. 3B) of RecF protein. There is no evidence that RecX interacts with free RecA protein in solution. RecX can, however, bind both to the ends of RecA filaments and along the length of the filament (Drees et al., 2004a; Lusetti et al., 2004a; VanLoock et al., 2003). It appears that once RecA filaments are available for RecX to bind, more RecF protein is required to challenge away the RecA-bound RecX protein.
The protective effects of RecF protein are seen whether RecF is present with RecOR at the initiation of RecA filament formation, or when RecF is added much later than the RecOR. There is thus no evidence in this work for a complex containing RecF, RecO, and RecR proteins together.
The effect of RecF protein alone on RecX-challenged RecA filament extension was investigated using a RecA mutant, RecA E38K (RecA730) that readily competes with the SSB protein without the intervention of the RecOR complex (Lavery and Kowalczykowski, 1992). As Figure 5C clearly demonstrates, RecF protein is both necessary and sufficient for the assembly process in the presence of RecX when the RecOR complex is not required for the nucleation step of filament assembly.
The RecA filament assembly reaction, assayed here in vitro, has particular relevance to the events leading to RecA filament assembly at a daughter strand gap in vivo. Due to its involvement with the replisome, SSB protein would be complexed with the ssDNA gap resulting from replication fork collapse (Kuzminov, 1999). The role of RecX in the events leading to the DNA damage response is not well understood. Low basal levels of RecX protein (< 50 molecules/cell) have been detected and those levels rise significantly during the SOS response (Stohl et al., 2003). Because RecX appears to functionally interact only with RecA filaments (Drees et al., 2004a), and given the low concentrations of RecX relative to RecA that are sufficient for inhibiting RecA in vitro, it is possible that the RecX could act to regulate RecA filaments that assemble inappropriately in the absence of DNA damage. When DNA damage necessitates an SOS response, the RecFOR proteins assemble RecA at the site of the damage. If RecX were allowed to act on RecA filaments in this context, the result would be less RecA protein bound at stalled replication forks. Interestingly, one of the measurable phenotypes associated with recFOR mutants is the delayed induction of the SOS response (Madiraju et al., 1988; Whitby and Lloyd, 1995). RecOR complex mediates the nucleation step of RecA filament formation, countering the effects of SSB. At least in part, RecF protein mediates the extension step, countering the effects of RecX. Based on our in vivo studies, RecF protein is needed to counter the effects of RecX in plasmid recombination (Fig. 6).
The effects of the RecF protein on the activities of RecA are complex. The RecF protein inhibits RecA activities under many conditions (Bork et al., 2001b; Madiraju and Clark, 1991), in the presence or absence of the RecOR complex. RecF is dispensable in the RecOR-mediated nucleation of RecA filaments onto SSB-coated ssDNA (Umezu et al., 1993) in vitro. However, RecF protein enhances the RecOR-mediated loading of RecA protein in the presence of a gap DNA substrate under at least some conditions (Morimatsu and Kowalczykowski, 2003). We have replicated the results of Morimatsu and Kowalczykowski (2003), showing positive effects of RecF on RecA loading onto gapped DNAs, in detail (data not shown). This may indicate that RecF has multiple roles in RecA filament assembly, both assisting with filament nucleation specifically at the ends of DNA gaps (Morimatsu and Kowalczykowski, 2003), and facilitating filament extension via the interference with RecX function as documented here.
The effects of RecF protein in blocking RecX function can be seen in plasmid recombination (Fig. 6), but are not readily apparent in the effects of the relevant mutants on SOS induction in vivo or in measurements of UV sensitivity (data not shown). Several points are relevant. First, UV sensitivity is a complex phenotype and the reported effects of recX null mutants on UV sensitivity have generally been very small (Pages et al., 2003; Stohl et al., 2003). Second, RecF may have multiple roles in recombinational DNA repair, such that elimination of recX function is not sufficient to compensate for the loss of recF function in every context. Third, the known network of proteins involved in regulating RecA function is currently expanding. The full effects of these proteins may not become apparent until their genes are studied in the absence of potentially redundant and/or complementary protein functions.
Experimental Procedures
Bacterial Strains
The recF4115 mutation (Sandler, 1994) was transduced into strains AB1157 and AB1157ΔrecX (Stohl et al., 2003) using P1 transduction (Sambrook et al., 1989). E. coli strains were grown on Luria-Bertani (LB) broth or agar at 37°C. Ampicillin (Amp) and tetracycline (Tet) were used at final concentrations of 100 μg/ml and 12 μg/ml, respectively.
Enzymes and Biochemicals
The concentrations of all purified proteins described below were determined from the absorbance at 280 nm using the given extinction coefficient (ε). E. coli wild-type RecA and RecA E38K proteins were purified as described (Eggler et al., 2003; Lusetti et al., 2003); ε = 2.23 × 104 M−1 cm−1 (Craig and Roberts, 1981). SSB protein was purified as described (Shan et al., 1996); ε = 2.83 × 104 M−1 cm−1 (Lohman and Overman, 1985). Native RecX protein was purified as described (Drees et al., 2004b) and is stored in 20 mM Tris-HCl (80% cation, pH 7.5), 1 mM DTT, 0.1 mM EDTA, 100 mM potassium glutamate, and 50% (w/v) glycerol; ε = 2.57 × 104 M−1 cm−1 (Drees et al., 2004b). RecF and RecR proteins were purified as described (Webb et al., 1995); ε = 3.87 × 104 M−1 cm−1 (Webb et al., 1999) and 5.60 × 103 M−1 cm−1 (Shan et al., 1997), respectively. RecO protein was purified as described (Shan et al., 1997); ε = 2.3 × 104 M−1 cm−1 (Kantake et al., 2002). RecF, RecR, and RecO proteins are stored in 20 mM Tris-HCl (80% cation, pH 7.5), 1 mM DTT, 0.1 mM EDTA, 100 mM NaCl, and 60% (w/v) glycerol.
Unless otherwise noted, reagents were purchased from Fisher. Antibiotics and all components of ATP regeneration and coupling systems were obtained from Sigma. DTT was from Research Organics.
DNA Substrate
Circular ssDNA from bacteriophage M13mp8 (7229 nucleotides) was prepared as described (Neuendorf and Cox, 1986). The concentration of ssDNA was determined by absorbance at 260 nm, using 36 μg ml−1 A260−1, as the conversion factor. All DNA concentrations are given in μM nucleotides.
ATPase Assay
A coupled spectrophotometric enzyme assay (Lindsley and Cox, 1990; Morrical et al., 1986) was used to measure DNA-dependent ATPase activities of the RecA protein (Lusetti et al., 2003). Reactions were carried out at 37°C in Buffer A (25 mM Tris-OAc (80% cation, pH 7.4), 1 mM DTT, 3 mM potassium glutamate, 10 mM Mg(OAc)2, and 5% (w/v) glycerol), an ATP regeneration system (10 units/ml pyruvate kinase (PK) and 3 mM phosphoenolpyruvate (PEP)), a coupling system (1.5 mM NADH and 10 units/ml lactate dehydrogenase), and 2 μM M13mp8 circular ssDNA. All assays contain 1.2 μM RecA protein, 0.2 μM SSB protein and 3 mM ATP. The concentration of RecO, RecR, RecF and RecX as well as order of addition are indicated in the figure legends. Whenever a particular protein was omitted from a reaction, an equivalent volume of that protein's storage buffer was added in its place. All experiments were carried out at least twice with consistent results.
Surface Plasmon Resonance
Protein–protein interactions were observed using a BIAcore2000 surface plasmon resonance biosensor (BIAcore, Piscataway, NJ). Random amine coupling was used to immobilize RecX protein to the sensor surface of a CM-5 sensor chip. Surface activation with NHS/EDC (Sigma) was followed by injection of RecX protein (50 μg/ml) or immobilization buffer. For the immobilization step, RecX protein was dialyzed into SPR running buffer (20 mM Hepes HCl pH 7.5, 150 mM NaCl, 10% (w/v) glycerol) and diluted 1:5 with 10 mM NaOAc pH 4.5 prior to injection. Unliganded sites on the chip were then blocked using ethanolamine. A 70 μL injection of RecX resulted in immobilization of 2701 RU of RecX. Subsequent interaction experiments were carried out in SPR running buffer at a flow rate of 10 μL min−1. The response measured in a reference surface, treated identically with the RecX surface except that no protein was injected, was subtracted from the response obtained from the RecX surface. Sensogram analysis was carried out using BIAevaluation (Biacore Inc. Piscattaway, NJ). Due to the rapid dissociation of RecF from both the RecX and reference surfaces no regeneration step was required.
Electron Microscopy
A modified Alcian method was used to visualize RecA filaments. Activated grids were prepared as described previously (Lusetti et al., 2003). Samples were prepared by preincubating 100 nM RecO, 1 μM RecR, 0.2 μM SSB and 2 μM M13mp8 circular ssDNA, Buffer A (DTT omitted), 3 mM ATP and an ATP regeneration system (10 units/ml PK and 3 mM PEP) for 10 min. All incubations were at 37 °C. RecA was added to 1.2 μM, followed by a 15-min incubation. RecF and RecX were added as described in the figure legend. Equivalent volumes of RecF or RecX protein storage buffer were added to compensate for the omission of the corresponding proteins. Filaments were stabilized with ATPγS (to 3 mM) for 3 min. The reaction mixture described above was diluted five fold with 200 mM ammonium acetate, 10 mM Hepes (pH 7.5), and 10% glycerol. The sample was prepared for analysis as described (Lusetti et al., 2004b) except that 2 grids per experiment were spread, one of which was spread omitting the ethyl alcohol wash. Imaging and photography were carried out with a Tecnai G2 12 Twin electron microscope (FEI Co.) equipped with a Gatan 890 CCD camera.
Representative molecules are shown in Figure 4A–E. To determine the proportion of the molecules observed that were either fully or partially coated by RecA protein or bound only by the SSB protein, at least 5 separate regions of at least 2 grids (encompassing at least 350 DNA molecules) were counted at the identical magnification for each sample. A molecule was considered discontinuous, or gapped, if it had a detectable region of SSB-coated DNA of any size. Molecules with very short RecA filaments (as in Fig. 4D) were assigned to a special class (large gaps, Fig. 4F) if the filaments were unambiguously less than 25% of the length of a full filament (judgments made visually). This conservative estimate was confirmed by manual measurements of small samples (5–6) of typical short filaments on digital micrographs of the same magnification, which showed that molecules scored in this class were consistently less than 10% of the length of a full filament.
Plasmid Recombination Assay
We used a modified method of Fishel et al. (Fishel et al., 1981) to determine the relative amounts of plasmid recombination occurring in different strains. Strains AB1157, ΔrecX (designated recX), recF, and recFX, were transformed with plasmid pRDK301, plated on LB-Amp, and allowed to grow for 22 h. Colonies on plates containing ~1000 CFU were collected in 6 ml LB and total plasmid DNA was isolated from 1 ml of the homogenous mixture using Qiaquick kits (Qiagen). 1 μl of the plasmid DNA purified from each strain was subsequently transformed into TOP10 cells (Invitrogen), and the resulting transformants were plated on LB/Amp and LB/Amp-Tet. In this assay, plasmids that had not undergone recombination conferred Amp resistance, and plasmids that had recombined conferred Amp-Tet resistance.
Acknowledgements
This work was supported by National Institutes of Health grants GM52725 (to MMC) and AI044239 (to HSS), and by the National Institutes of Health Pre-doctoral Training Grant T32 GM007215 in Molecular Biosciences from the National Institute of General Medical Sciences (to MDH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arenson TA, Tsodikov OV, Cox MM. Quantitative analysis of the kinetics of end-dependent disassembly of RecA filaments from ssDNA. J. Mol. Biol. 1999;288:391–401. doi: 10.1006/jmbi.1999.2705. [DOI] [PubMed] [Google Scholar]
- Bork JM, Cox MM, Inman RB. RecA protein filaments disassemble in the 5' to 3' direction on single-stranded DNA. J. Biol. Chem. 2001a;276:45740–45743. doi: 10.1074/jbc.M109247200. [DOI] [PubMed] [Google Scholar]
- Bork JM, Cox MM, Inman RB. The RecOR proteins modulate RecA protein function at 5 ' ends of single-stranded DNA. EMBO J. 2001b;20:7313–7322. doi: 10.1093/emboj/20.24.7313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AJ, Sandler SJ. Homologous genetic recombination: the pieces begin to fall into place. Crit. Rev. Microbiol. 1994;20:125–142. doi: 10.3109/10408419409113552. [DOI] [PubMed] [Google Scholar]
- Cohen A, Laban A. Plasmidic recombination in Escherichia coli K-12: the role of recF gene function. Mol. Gen. Genet. 1983;189:471–474. doi: 10.1007/BF00325911. [DOI] [PubMed] [Google Scholar]
- Cox MM. The bacterial RecA protein as a motor protein. Ann. Rev. Microbiol. 2003;57:551–577. doi: 10.1146/annurev.micro.57.030502.090953. [DOI] [PubMed] [Google Scholar]
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- Craig NL, Roberts JW. Function of nucleoside triphosphate and polynucleotide in Escherichia coli recA protein-directed cleavage of phage lambda repressor. J. Biol. Chem. 1981;256:8039–8044. [PubMed] [Google Scholar]
- Drees JC, Lusetti SL, Chitteni-Pattu S, Inman RB, Cox MM. A RecA filament capping mechanism for RecX protein. Mol. Cell. 2004a;15:789–798. doi: 10.1016/j.molcel.2004.08.026. [DOI] [PubMed] [Google Scholar]
- Drees JC, Lusetti SL, Cox MM. Inhibition of RecA protein by the Escherichia coli RecX protein - Modulation by the RecA C terminus and filament functional state. J. Biol. Chem. 2004b;279:52991–52997. doi: 10.1074/jbc.M409050200. [DOI] [PubMed] [Google Scholar]
- Eggler AL, Lusetti SL, Cox MM. The C terminus of the Escherichia coli RecA protein modulates the DNA binding competition with single-stranded DNA-binding protein. J. Biol. Chem. 2003;278:16389–16396. doi: 10.1074/jbc.M212920200. [DOI] [PubMed] [Google Scholar]
- Ennis DG, Levine AS, Koch WH, Woodgate R. Analysis of recA mutants with altered SOS functions. Mutat. Res. 1995;336:39–48. doi: 10.1016/0921-8777(94)00045-8. [DOI] [PubMed] [Google Scholar]
- Fishel RA, James AA, Kolodner R. recA-independent general genetic recombination of plasmids. Nature. 1981;294:184–186. doi: 10.1038/294184a0. [DOI] [PubMed] [Google Scholar]
- Grompone G, Sanchez N, Ehrlich SD, Michel B. Requirement for RecFOR-mediated recombination in priA mutant. Mol. Microbiol. 2004;52:551–562. doi: 10.1111/j.1365-2958.2004.03997.x. [DOI] [PubMed] [Google Scholar]
- Kantake N, Madiraju M, Sugiyama T, Kowalczykowski C. Escherichia coli RecO protein anneals ssDNA complexed with its cognate ssDNA-binding protein: A common step in genetic recombination. Proc. Natl. Acad. Sci. U.S.A. 2002;99:15327–15332. doi: 10.1073/pnas.252633399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidane D, Sanchez H, Alonso JC, Graumann PL. Visualization of DNA double-strand break repair in live bacteria reveals dynamic recruitment of Bacillus subtilis RecF, RecO and RecN proteins to distinct sites on the nucleoids. Mol. Microbiol. 2004;52:1627–1639. doi: 10.1111/j.1365-2958.2004.04102.x. [DOI] [PubMed] [Google Scholar]
- Kim B, Little JW. LexA and lambda Cl repressors as enzymes: specific cleavage in an intermolecular reaction. Cell. 1993;73:1165–1173. doi: 10.1016/0092-8674(93)90645-7. [DOI] [PubMed] [Google Scholar]
- Knight KL, Aoki KH, Ujita EL, McEntee K. Identification of the amino acid substitutions in two mutant forms of the RecA protein from Escherichia coli: RecA441 and RecA629. J. Biol. Chem. 1984;259:11279–11283. [PubMed] [Google Scholar]
- Kowalczykowski SC. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC, Clow J, Somani R, Varghese A. Effects of the Escherichia coli SSB protein on the binding of Escherichia coli RecA protein to single-stranded DNA. Demonstration of competitive binding and the lack of a specific protein-protein interaction. J. Mol. Biol. 1987;193:81–95. doi: 10.1016/0022-2836(87)90629-2. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC, Krupp RA. Effects of Escherichia coli SSB protein on the single-stranded DNA-dependent ATPase activity of Escherichia coli RecA protein. Evidence that SSB protein facilitates the binding of RecA protein to regions of secondary structure within single-stranded DNA. J. Mol. Biol. 1987;193:97–113. doi: 10.1016/0022-2836(87)90630-9. [DOI] [PubMed] [Google Scholar]
- Kuzminov A. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 1999;63:751–813. doi: 10.1128/mmbr.63.4.751-813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery PE, Kowalczykowski SC. Biochemical basis of the constitutive repressor cleavage activity of recA730 protein. A comparison to recA441 and recA803 proteins. J. Biol. Chem. 1992;267:20648–20658. [PubMed] [Google Scholar]
- Lindsley JE, Cox MM. Assembly and disassembly of RecA protein filaments occurs at opposite filament ends: relationship to DNA strand exchange. J. Biol. Chem. 1990;265:9043–9054. [PubMed] [Google Scholar]
- Lohman TM, Overman LB. Two binding modes in Escherichia coli single strand binding protein-single stranded DNA complexes. Modulation by NaCl concentration. J. Biol. Chem. 1985;260:3594–3603. [PubMed] [Google Scholar]
- Luisi-DeLuca C, Kolodner R. Purification and characterization of the Escherichia coli RecO protein. Renaturation of complementary single-stranded DNA molecules catalyzed by the RecO protein. J. Mol. Biol. 1994;236:124–138. doi: 10.1006/jmbi.1994.1123. [DOI] [PubMed] [Google Scholar]
- Lusetti SL, Cox MM. The bacterial RecA protein and the recombinational DNA repair of stalled replication forks. Ann. Rev. Biochem. 2002;71:71–100. doi: 10.1146/annurev.biochem.71.083101.133940. [DOI] [PubMed] [Google Scholar]
- Lusetti SL, Drees JC, Stohl EA, Seifert HS, Cox MM. The DinI and RecX proteins are competing modulators of RecA function. J. Biol. Chem. 2004a;279:55073–55079. doi: 10.1074/jbc.M410371200. [DOI] [PubMed] [Google Scholar]
- Lusetti SL, Voloshin ON, Inman RB, Camerini-Otero RD, Cox MM. The DinI protein stabilizes RecA protein filaments. J. Biol. Chem. 2004b;279:30037–30046. doi: 10.1074/jbc.M403064200. [DOI] [PubMed] [Google Scholar]
- Lusetti SL, Wood EA, Fleming CD, Modica MJ, Korth J, Abbott L, Dwyer DW, Roca AI, Inman RB, Cox MM. C-terminal deletions of the Escherichia coli RecA protein - Characterization of in vivo and in vitro effects. J. Biol. Chem. 2003;278:16372–16380. doi: 10.1074/jbc.M212917200. [DOI] [PubMed] [Google Scholar]
- Madiraju MV, Clark AJ. Evidence for ATP binding and double-stranded DNA binding by Escherichia coli RecF protein. J. Bacteriol. 1992;174:7705–7710. doi: 10.1128/jb.174.23.7705-7710.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madiraju MV, Lavery PE, Kowalczykowski SC, Clark AJ. Enzymatic properties of the RecA803 protein, a partial suppressor of recF mutations. Biochemistry. 1992;31:10529–10535. doi: 10.1021/bi00158a016. [DOI] [PubMed] [Google Scholar]
- Madiraju MV, Templin A, Clark AJ. Properties of a mutant recA-encoded protein reveal a possible role for Escherichia coli recF-encoded protein in genetic recombination. Proc. Natl. Acad. Sci. USA. 1988;85:6592–6596. doi: 10.1073/pnas.85.18.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madiraju MVVS, Clark AJ. Effect of RecF protein on reactions catalyzed by RecA protein. Nuc. Acids Res. 1991;19:6295–6300. doi: 10.1093/nar/19.22.6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimatsu K, Kowalczykowski SC. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: A universal step of recombinational repair. Mol. Cell. 2003;11:1337–1347. doi: 10.1016/s1097-2765(03)00188-6. [DOI] [PubMed] [Google Scholar]
- Morrical SW, Lee J, Cox MM. Continuous association of Escherichia coli single-stranded DNA binding protein with stable complexes of RecA protein and single-stranded DNA. Biochemistry. 1986;25:1482–1494. doi: 10.1021/bi00355a003. [DOI] [PubMed] [Google Scholar]
- Nagata K, Handa H. Real-time analysis of biomolecular interactions : applications of BIACORE. Springer; New York: 2000. [Google Scholar]
- Nastri HG, Knight KL. Identification of residues in the L1 region of the RecA protein which are important to recombination or coprotease activities. J. Biol. Chem. 1994;269:26311–26322. [PubMed] [Google Scholar]
- Neuendorf SK, Cox MM. Exchange of RecA protein between adjacent RecA protein-single-stranded DNA complexes. J. Biol. Chem. 1986;261:8276–8282. [PubMed] [Google Scholar]
- Pages V, Koffel-Schwartz N, Fuchs RP. recX, a new SOS gene that is co-transcribed with the recA gene in Escherichia coli. DNA Repair. 2003;2:273–284. doi: 10.1016/s1568-7864(02)00217-3. [DOI] [PubMed] [Google Scholar]
- Rangarajan S, Woodgate R, Goodman MF. Replication restart in UV-irradiated Escherichia coli involving pols II, III, V, PriA, RecA and RecFOR proteins. Mol. Microbiol. 2002;43:617–628. doi: 10.1046/j.1365-2958.2002.02747.x. [DOI] [PubMed] [Google Scholar]
- Register JC, III, Griffith J. The direction of RecA protein assembly onto single strand DNA is the same as the direction of strand assimilation during strand exchange. J. Biol. Chem. 1985;260:12308–12312. [PubMed] [Google Scholar]
- Roca AI, Cox MM. RecA protein: structure, function, and role in recombinational DNA repair. Prog. Nuc. Acid Res. Mol. Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Sandler SJ. Studies on the mechanism of reduction of UV-inducible sulAp expression by recF overexpression in Escherichia coli K-12. Mol. Gen. Genet. 1994;245:741–749. doi: 10.1007/BF00297281. [DOI] [PubMed] [Google Scholar]
- Sandler SJ, Samra HS, Clark AJ. Differential suppression of priA2::kan phenotypes in Escherichia coli K-12 by mutations in priA, lexA, and dnaC. Genetics. 1996;143:5–13. doi: 10.1093/genetics/143.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawitzke JA, Stahl FW. Phage lambda has an analog of Escherichia coli recO, recR and recF genes. Genetics. 1992;130:7–16. doi: 10.1093/genetics/130.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawitzke JA, Stahl FW. The phage lambda orf gene encodes a trans-acting factor that suppresses Escherichia coli recO, recR, and recF mutations for recombination of lambda but not of E. coli. J. Bacteriol. 1994;176:6730–6737. doi: 10.1128/jb.176.21.6730-6737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Q, Bork JM, Webb BL, Inman RB, Cox MM. RecA protein filaments: end-dependent dissociation from ssDNA and stabilization by RecO and RecR proteins. J. Mol. Biol. 1997;265:519–540. doi: 10.1006/jmbi.1996.0748. [DOI] [PubMed] [Google Scholar]
- Shan Q, Cox MM, Inman RB. DNA strand exchange promoted by RecA K72R. Two reaction phases with different Mg2+ requirements. J. Biol. Chem. 1996;271:5712–5724. doi: 10.1074/jbc.271.10.5712. [DOI] [PubMed] [Google Scholar]
- Smith GR. Homologous recombination in prokaryotes: enzymes and controlling sites. Genome. 1989;31:520–527. doi: 10.1139/g89-100. [DOI] [PubMed] [Google Scholar]
- Stohl EA, Brockman JP, Burkle KL, Morimatsu K, Kowalczykowski SC, Seifert HS. Escherichia coli RecX inhibits RecA recombinase and coprotease activities in vitro and in vivo. J. Biol. Chem. 2003;278:2278–2285. doi: 10.1074/jbc.M210496200. [DOI] [PubMed] [Google Scholar]
- Thomas A, Lloyd RG. Control of recA dependent activities in Escherichia coli: a possible role for the recF product. J. Gen. Microbiol. 1983;129:681–686. doi: 10.1099/00221287-129-3-681. [DOI] [PubMed] [Google Scholar]
- Umezu K, Chi NW, Kolodner RD. Biochemical interaction of the Escherichia coli RecF, RecO, and RecR proteins with RecA protein and single-stranded DNA binding protein. Proc. Natl. Acad. Sci. USA. 1993;90:3875–3879. doi: 10.1073/pnas.90.9.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezu K, Kolodner RD. Protein interactions in genetic recombination in Escherichia coli. Interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein. J. Biol. Chem. 1994;269:30005–30013. [PubMed] [Google Scholar]
- VanLoock MS, Yu X, Yang S, Galkin VE, Huang H, Rajan SS, Anderson WF, Stohl EA, Seifert HS, Egelman EH. Complexes of RecA with LexA and RecX differentiate between active and inactive RecA nucleoprotein filaments. J. Mol. Biol. 2003;333:345–354. doi: 10.1016/j.jmb.2003.08.053. [DOI] [PubMed] [Google Scholar]
- Volkert MR, Hartke MA. Suppression of Escherichia coli recF mutations by recA-linked srfA mutations. J. Bacteriol. 1984;157:498–506. doi: 10.1128/jb.157.2.498-506.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TC, Chang HY, Hung JL. Cosuppression of recF, recR and recO mutations by mutant recA alleles in Escherichia coli cells. Mutat. Res. 1993;294:157–166. doi: 10.1016/0921-8777(93)90024-b. [DOI] [PubMed] [Google Scholar]
- Webb BL, Cox MM, Inman RB. An interaction between the Escherichia coli RecF and RecR proteins dependent on ATP and double-stranded DNA. J. Biol. Chem. 1995;270:31397–31404. doi: 10.1074/jbc.270.52.31397. [DOI] [PubMed] [Google Scholar]
- Webb BL, Cox MM, Inman RB. ATP hydrolysis and DNA binding by the Escherichia coli RecF protein. J. Biol. Chem. 1999;274:15367–15374. doi: 10.1074/jbc.274.22.15367. [DOI] [PubMed] [Google Scholar]
- Whitby MC, Lloyd RG. Altered SOS induction associated with mutations in recF, recO and recR. Mol. Gen. Genet. 1995;246:174–179. doi: 10.1007/BF00294680. [DOI] [PubMed] [Google Scholar]