Abstract
Increasing the production of fatty acids by microbial fermentation remains an important step towards the generation of biodiesel and other portable liquid fuels. In this work, we report an Escherichia coli strain engineered to overexpress a fragment consisting of four dehydratase domains from the polyunsaturated fatty acid (PUFA) synthase enzyme complex from the deep-sea bacterium, Photobacterium profundum. The DH1-DH2-UMA enzyme fragment was excised from its natural context within a multi-enzyme PKS and expressed as a stand-alone protein. Fatty acids were extracted from the cell pellet, esterified with methanol and quantified by GC-MS analysis. Results show that the E. coli strain expressing the DH tetradomain fragment was capable of producing up to a 5-fold increase (80.31 mg total FA/L culture) in total fatty acids over the negative control strain lacking the recombinant enzyme. The enhancement in production was observed across the board for all the fatty acids that are typically made by E. coli. The overexpression of the DH tetradomain did not affect E. coli cell growth, thus showing that the observed enhancement in fatty acid production was not a result of effects associated with cell density. The observed enhancement was more pronounced at lower temperatures (3.8-fold at 16 °C, 3.5-fold at 22 °C and 1.5-fold at 30 °C) and supplementation of the media with 0.4% glycerol did not result in an increase in fatty acid production. All these results taken together suggest that either the dehydration of fatty acid intermediates are a limiting step in the E. coli fatty acid biosynthesis machinery, or that the recombinant dehydratase domains used in this study are also capable of catalyzing thioester hydrolysis of the final products. The enzyme in this report is a new tool which could be incorporated into other existing strategies aimed at improving fatty acid production in bacterial fermentations towards accessible biodiesel precursors.
Keywords: dehydratase domain, fatty acids overproduction, bioengineering, biofuel, biodiesel, Escherichia coli.
Introduction
The development of commercially available transportation and jet fuels from renewable sources will be needed in the coming decades in order to offset the high demand for environmentally deleterious and costly petroleum-derived fuels [1]. Towards this worthy goal, there have been a number of efforts from industry and academia aimed at developing the production of different forms of biofuels which include ethanol from maize or sugarcane, butanes from yeast fermentations and biodiesel derived from the esterification of fatty acids [2–10].
According to the 2012 Report from the U.S. Energy Information Administration (www.eia.gov) from 2010 to 2011, the US consumption of biodiesel increased from 263 to 878 million gallons of fuel, while the consumption of ethanol remained nearly constant between these two years. Currently, biodiesel constitutes about 2.2% of the diesel fuel used in the US and most of it comes from recycled vegetable oils and animal fats (7.3 billion pounds in 2011). With higher demand for biodiesel, there has been an increase in the proportions of soybean oil in biodiesel preparations (4.1 billion pounds in 2011 and 5.2 billion pounds projected for 2012). This diversion of food crops, such as corn and soybeans, towards the production of biofuels has the effect of increasing global prices for these crops. Thus, it is apparent that there will be an increasing pressure to foster the production of oils from non-food crops as the industry grows [11].
An alternative for the production of fatty acids and other biodiesel precursors without directly using food crops, is by microbial fermentation. There are numerous reports demonstrating the application of yeast, fungi and bacteria for the production of free fatty acids as biodiesel precursors [8, 12–16]. One of the most widely used industrial hosts is the gram-negative bacterium Escherichia coli. This organism is approximately 9 % lipid, produces fatty acid metabolites at a commercial productivity (~ 0.2 g l−1 hr−1 per gram of cell mass) and, can achieve product-dependent mass yields of 30 – 35% and is suitable for genetic manipulation [17].
There are a number of reported biochemical strategies for the enhancement of fatty acid production in E. coli (Table 1) [2, 6, 12, 17–22]. Most of them involve either (i) the overexpression of thioesterases to increase fatty acid release during biosynthesis or (ii) the deletion of genes for fatty acid degradation by the beta-oxidation pathway [2, 5–6, 17, 22]. In some studies, both strategies have been combined to achieve up to 100-fold increases in the production of fatty acids in E. coli [17]. Additionally, the heterologous expression of key enzymes involved in alcohol production, such as pyruvate dehydrogenase, alcohol dehydrogenase and acyltransferases, have also been shown to enhance the production of acetate units required for the production of fatty acids [3]. Similarly, the overexpression of regulatory transcription factors such as FadR has been shown to enhance fatty acid production globally by tuning the expression levels of many genes involved in fatty acid pathways to optimal levels (abB, fabF, and accA) [21].
Table 1.
Reports of single genetic modifications of Escherichia coli which result in the enhanced production of fatty acids.
| Plasmid | Description | Fold-increase in FA production | Reference |
|---|---|---|---|
| pACYC-TE | pACYC harboring A. thaliana thioesterase | 1.7 | Cao, Y. et al Appl Microbiol Biotechnol (2010) 87:271–280.5 |
| pET-fabAB and pACYC-TE | pET30a harboring E. coli fabA and fabB, and pACYC harboring A. thaliana thioesterase. | 1.6 | |
| pXL.49 | pET28b harboring plant thioesterase(U31813) from Cinnamomum camphorum | 2.0 | Lu. X. et al Metabolic Engineering (2008) 10:333–339.22 |
| pBAD33-BTE | pBAD33 harboring Umbellularia californica thioesterase (BTE) | 3.6 | Lennen. R. Biotechnology and Bioengineering (2010) 106(2):193–202.6 |
| pBAD33-BTE-ACC | pRL2 harboring accDABC. The accD gene encoding the (β-subunit of acetyt-CoA carboxyltransferase, the accA gene encoding the a-subunit of acetyl-CoA carboxyltransferase, and the accBC operon encoding biotin carboxyl carrier protein and biotin carboxylase. | 2.3 | |
| placUV5:’tesA | E. coli ‘tesA. thioesterase without leader sequence. | 10.7 | Steen. E.J. et al Nature (2010) 463: 559–563.17 |
| pDH1-DH2-UMA (Palmitica Bio Inc IP) | pET200 harboring the DH1-DH2-UMA fragment from the PUFA synthase. | 3.6 | This report. |
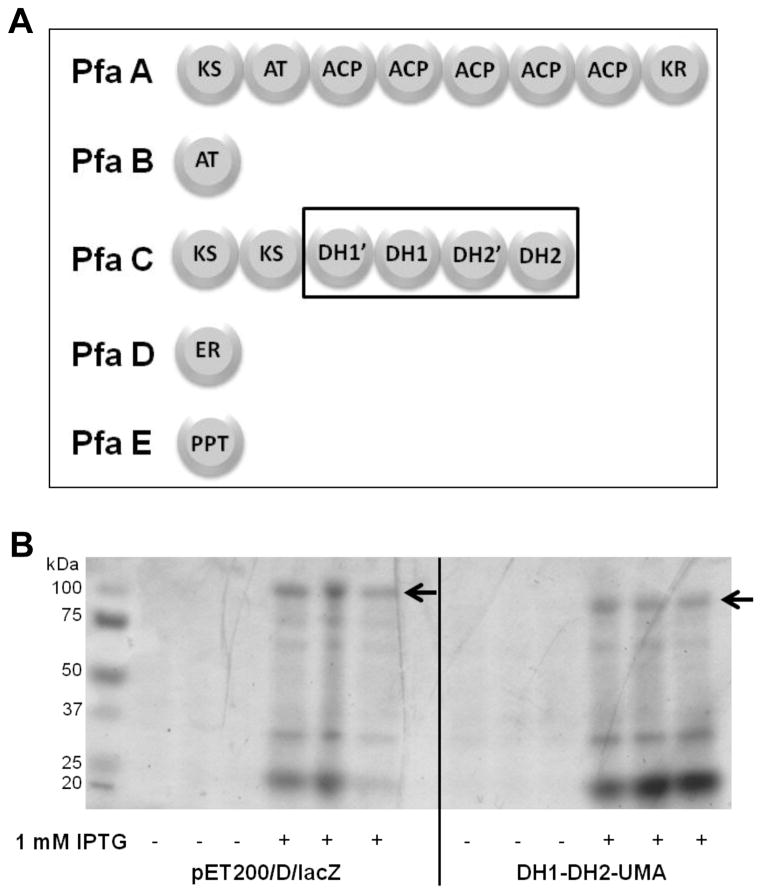
The biosynthesis of polyunsaturated fatty acids (PUFA) in deep-sea bacteria employs a polyketide synthase-like multienzyme system which is widely conserved in marine environments [24–26] (Figure 1A). This conserved PUFA synthase multidomain system contains all the enzyme domains required for the elongation, the reduction and double bond formation in the resulting fatty acid. Our group had previously characterized a tetradomain protein fragment (DH1-DH2-UMA) from deep-sea bacterium Photobacterium profundum which was expressed, purified and shown to have enzymatic activity in vitro [27]. The DH1-DH2-UMA recombinant protein fragment included all four hotdog-fold domains associated with the dehydratase (DH) activity in the PUFA synthase (Figure 1A) [27]. The DH1-DH2-UMA fragment was found to be competent to catalyze the hydration of several surrogate substrates but its applicability in the enhancement of fatty acid biosynthesis has not been assessed [27].
Figure 1. DH1-DH2-UMA overexpression.
(A) The PKS multienzyme for the anaerobic production of eicosapentaenoic acid (EPA) in Photobacterium profundum consists of five different proteins (Pfa A, B, C ,D and E). The four dehydratase (DH) domains are housed within the PfaC multienzyme. The fragment DH1-DH2-UMA contains all four conserved domains. (B) SDS-PAGE analysis of recombinant DH1-DH2-UMA protein before (−) and after (+) induction of expression with IPTG (1mM final concentration) for three replicates. The control protein LacZ shows as a band of >100 kDa while the dehydratase DH1-DH2-UMA shows as a band just below 100 kDa corresponding to DH1-DH2-UMA protein of 96 kDa.
In this work, we report the enhancement of fatty acid production in E. coli which overexpresses this active fragment, DH1-DH2-UMA, which has been excised from its natural context as part of the PUFA synthase complex of Photobacterium profundum [27]. Our results clearly show that the expression of DH1-DH2-UMA in E. coli results in a five-fold increase in fatty acid production for all the typical fatty acids vs. the control. This production enhancement seems to be independent on the presence of carbon supplementation of the media with glycerol but highly dependent on temperature.
Materials and methods
All reagents such as kanamycin, chloramphenicol, IPTG (isopropyl β-D-1-thiogalactopyranoside), yeast extract, NaCl, tryptone, methyl heneicosanoate and glycerol were purchased from Sigma.
General procedures
Mass spectral data was acquired using a GC-MS (Hewlett-Packard 5972A MSD Chemstation; Hewlett-Packard, Palo Alto, CA, USA) at 70 eV equipped with a 30 m x 0.25 mm special performance capillary column (HP-5MS) of polymethylsiloxane cross-linked with 5 % phenyl methylpolysiloxane. For liophilizatation of samples a FreeZone Freeze Dry Systems was used.
Cloning, cell transformation, media and growth
DH fragments were cloned as previously described by Oyola-Robles et al. [27]. The pET200 expression vector containing the cloned genes encoding either the control pET200/D/lacZ (Invitrogen) or the experimental pDH1-DH2-UMA constructs were transformed in E. coli strain BL21-CodonPlus (DE3)-RIL Competent Cells (Stratagene). Transformants were selected and cultured overnight in LB medium and antibiotics (kanamycin 100 mg/L and chloramphenicol 25 mg/L) at 37 °C, 270 rpm. Overnight culture was used to inoculate 1 L of LB medium (supplemented with 0.4 % glycerol when necessary) with antibiotic (kanamycin 100 mg/L and chloramphenicol 25 mg/L) at 37 °C, 250 rpm until the OD600 reach 0.2 and then, cultured at 30 °C, 22 °C or 16 °C, 250 rpm until the OD600 reach 0.5–0.6. Protein expression was induced by adding isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 1.0 mM, incubation continued overnight at 30 °C, 22 °C or 16 °C respectively, 250 rpm. A control experiment was performed with no IPTG induction in a culture at 22 °C. OD600 was monitored for up to 80 hours for cell growth. Protein expression was corroborated by SDS-PAGE using 4–15% Mini-PROTEAN® TGX gels (BioRad®). Cells were collected by centrifugation at 4,400 rpm, 10 min, 4 °C, freeze-dried and pellets stored at −80 °C.
Fatty acids extraction and methylation
The fatty acyl components of the cell culture were obtained as their methyl esters by the reaction of 0.10 g of freeze-dried pellet with 10.0 mL of methanolic HCl, refluxed for 2 hr. The crude of the reaction was taken up with hexane (3 × 15 mL), the organic layer dried over MgSO4 and concentrated in vacuo. The fatty acid methyl esters were analyzed by GC-MS. The temperature program was as follows: 130 °C for two minutes, increase at a rate of 3 °C /min to a 270 °C, where the temperature is maintained for 88 min. Methyl heneicosanoate was used as an internal standard for quantification of fatty acid methyl esters as described previously [28].
Statistical analysis: fatty acid composition determination
Individual fatty acids were identified by their retention time and mass spectral fragmentations in the Chemstation software suite (HP Agilent). Quantitative analysis of fatty acids composition was performed by using the area under the curve of the peaks corresponding to the identified fatty acids, normalized by the area under the curve of the internal standard and, converted to the reported units (mg fatty acid/L culture). All experiments were performed in biological duplicates or triplicates. The data analyzed using the following equations:
| Eq (1) |
In which the total number of millimoles of a fatty acids is given by the known concentration of the internal standard (CIS) multiplied by the ratio of the areas of the fatty acid and the internal standard obtained from the gas chromatogram (AFA/AIS). This is multiplied by a dilution factor of 2 and by the total volume of the sample (Vol total).
| Eq (2) |
The total mmol of fatty acid is divided by the mass of dried cells that were used for extraction (gcell) and then multiplied by the cell density (grams of cell/ L culture).
| Eq (3) |
Finally, the mmol/L culture can be multiplied by the molecular weight for that fatty acid to yield the mg of fatty acids per liter of culture.
Theory
The overproduction of fatty acids is an important goal in the search for renewable fuels. In this work we report an enzyme fragment, DH1-DH2-UMA, which has been taken out of its natural context within a multi-enzyme from Photobacterium profundum. Overexpression of this enzyme fragment in E. coli increases the yield of fatty acid in liquid culture by a factor of five. This level of enhancement is competitive and should be tested in strains of E. coli that have been optimized for fatty acid production.
Results
Effect of DH1-DH2-UMA overexpression on fatty acid production
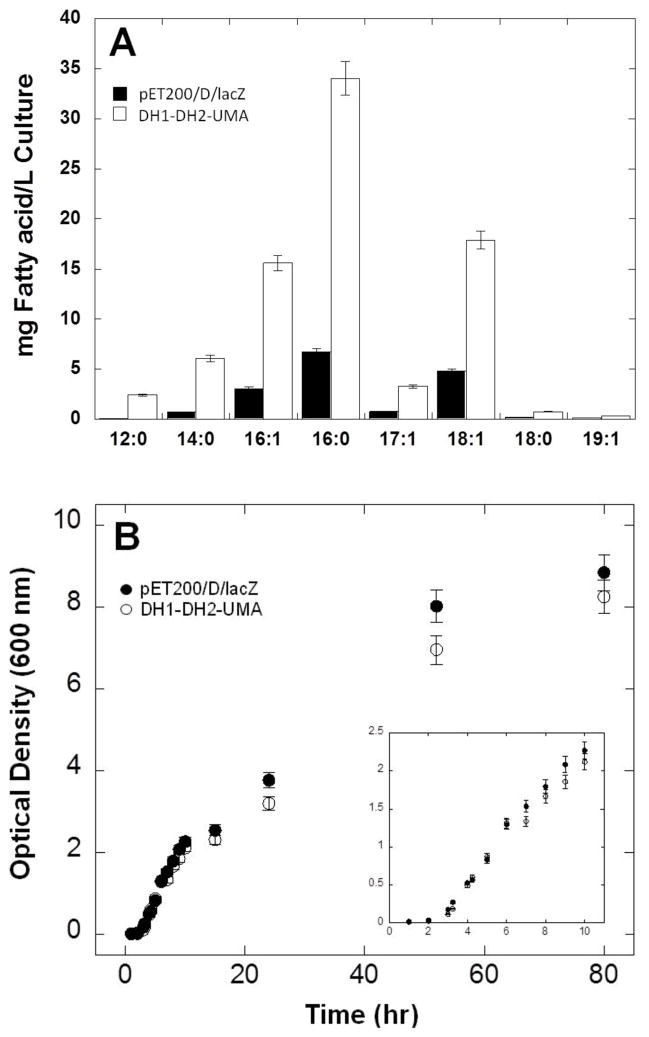
The overexpression of enzymes has been employed as a strategy to enhance fatty acid production in microbial fermentations [5, 17, 22]. In order to investigate whether DH1-DH2-UMA would interact with the endogenous machinery for fatty acid biosynthesis in E. coli, we measured the production of fatty acids in BL21 E coli cells expressing either DH1-DH2-UMA or a negative control protein LacZ (Figure 1B) [27]. No polyunsaturated fatty acids were detected in any of the bacterial extracts. Although the expression of DH1-DH2-UMA did not affect the fatty acid profile of E. coli, we did observe a 4 to 5-fold increase in the total yield of free saturated and monounsaturated fatty acids (Figure 2A). A mixture of saturated and monounsaturated fatty acids from 12 to 19 carbon chain length were isolated from the bacterial culture as shown by the gas chromatograph of their fatty acid methyl esters (FAME) derivatives (Supplemental figure 1). Palmitic acid (16:0) showed to be the major fatty acid produced in both the experiment and in the negative control. Each fatty acid production experiment is accompanied by a protein expression SDS-PAGE gel which shows that the observed fatty acid enhancement correlates with expression of the DH1-DH2-UMA protein (Figure 1B). The fact that the expression of DH1-DH2-UMA affected the production of all fatty acids in equal proportions suggests that the protein is capable of interacting with the E. coli machinery for fatty acid biosynthesis in a way that does not discriminate based on fatty acid chain length.
Figure 2. Effect of DH1-DH2-UMA overexpression on bacterial fatty acid production.
(A)Fatty acid profile of Escherichia coli expressing either the enzyme in this study, DH1-DH2-UMA (white bars) or the LacZ (black bars) (B) Measurements of the optical density at 600 nm were taken periodically for 80 hours of culture at 22 °C. Errors bars represent standard deviations about the mean of three replicate samples.
In order to verify that the induction of DH1-DH2-UMA overexpression was executed exactly at the logarithmic phase and to rule out the possibility that the observed enhancement in the yield of fatty acids is a reflection of a higher bacterial cell density, we measured the growth by optical density at 600 nm of the BL21 strain expressing DH1-DH2-UMA and compared it to one expressing the control pET200TOPO/D/LacZ. Our results clearly show that induction of the dehydratase domain overexpression was performed at the log phase (4 to 6 hours of cell culture (Figure 2B). Also, the expression of DH1-DH2-UMA did not increase cell density significantly (Figure 2B). Therefore, it is clear that the effects caused by DH1-DH2-UMA are not due to an increase in cell density.
Effect of temperature on the fatty acid production of E. coli
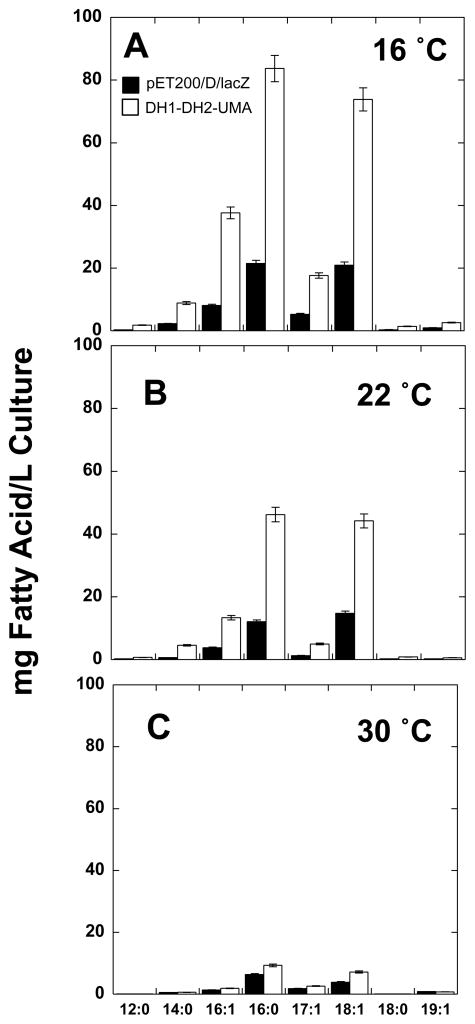
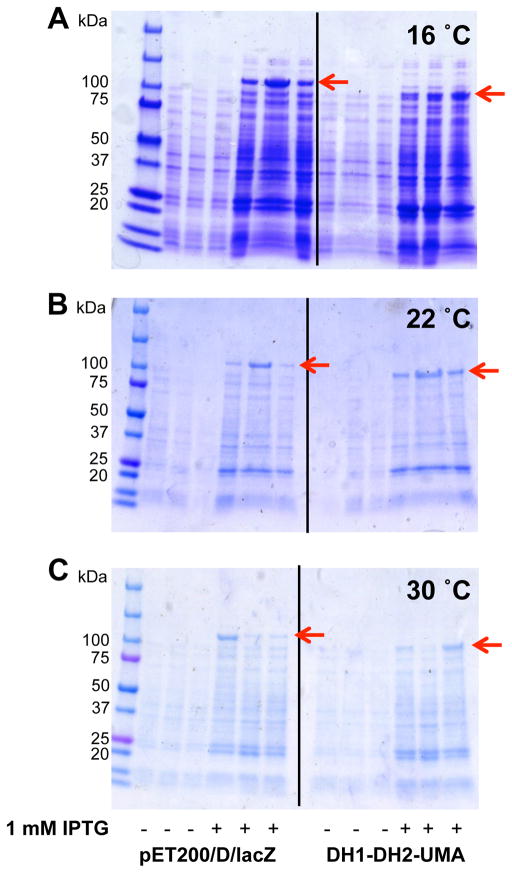
The effect of temperature (16, 22 and 30 °C) on both cell growth and fatty acid production in the E. coli strain overexpressing DH1-DH2-UMA, was measured. As expected, the yield of fatty acids was higher at the lower temperature for both the DH1-DH2-UMA strain and for the control strain [23]. A 10-fold increase in total mg of fatty acids per liter of culture was observed between the cultures grown at 16 °C relative to the cultures grown at 30 °C (Figure 3A and 3C, respectively). The enhancement in fatty acid production by the DH1-DH2-UMA strain was more pronounced at 16 °C than at higher temperatures and the production of fatty acids was correlated to the levels of protein produced as can be seen by SDS-PAGE (Figure 4). The results are summarized in Table 2 and Table S1.
Figure 3. Temperature effects.
Fatty acid profiles were determined for DH1-DH2-UMA (white bars) and pET200/D/lacZ controls (black bars) as described previously. Fatty acid titers were measured at (A) 16 °C (B) 22 °C and (C) 30 °C. Error bars represent standard deviations about the mean of two or three replicate samples.
Figure 4. Effect of temperature on DH1-DH2-UMA expression.
SDS-PAGE gels of the E. coli culture at different temperatures (A) 16 °C (B) 22 °C and (C) 30 °C shows that lower temperatures promote higher expression levels.
Table 2.
Summary of results of biomass and fatty acid yield. The temperature experiments were carried out with 0.4% glycerol and 1 mM IPTG. The glycerol experiment was carried out at 22°C and with 1 mM IPTG. The IPTG experiment was carried out at 22°C and with 0.4% glycerol. Thus, the 22°C data, the 0.4% Glycerol data and the 1 mM IPTG data are exactly the same data.
| Total biomass production (g cells/ L Culture) | Total mg Fatty Acid/ L Culture | |||
|---|---|---|---|---|
| Experimental Condition | pET200/D/lacZ | DH1-DH2-UMA | pET200/D/lacZ | DH1-DH2-UMA |
| 16 °C | 0.43 ± 0.08 | 0.37 ± 0.16 | 59.65 ± 8.88 | 227.58 ± 33.39 |
| 22 °C | 0.57 ± 0.06 | 0.53 ± 0.06 | 33.18 ± 5.87 | 115.33 ± 19.44 |
| 30 °C | 0.66 ± 0.24 | 0.84 ±0.23 | 15.00 ± 2.21 | 22.57 ± 3.50 |
| No Glycerol | 0.29 ± 0.01 | 0.23 ± 0.01 | 16.48 ± 2.54 | 80.31 ± 11.75 |
| 0.4 % Glycerol | 0.57 ±0.06 | 0.53 ± 0.06 | 33.18 ± 5.87 | 115.33 ± 19.44 |
| No IPTG | 1.00 ± 0.08 | 1.03 ± 0.17 | 117.77 ± 7.32 | 182.55 ± 16.23. |
| 1 mM IPTG | 0.57 ± 0.06 | 0.53 ± 0.06 | 33.18 ± 5.87 | 115.33 ± 19.44 |
As expected, cell growth was slower at the lower temperatures in this study. Dried cell measurements confirmed that less biomass is produced at lower temperatures (Table 2). At 16°C, a slight decrease (−0.06 g/L) in biomass was observed in the engineered DH1-DH2-UMA strain compared to the E. coli strain carrying the control vector. Increasing the temperature to 30°C resulted in a ~ 2 fold increase in biomass.
Effect of carbon supplementation on fatty acid production
It has been reported that the distribution of fatty acids can vary according to the composition of the E. coli culture media [23]. We therefore cultured the DH1-DH2-UMA E. coli strain in 1 L of LB media supplemented with 0.4 % (v/v) glycerol as a carbon source. Supplementation with 0.4% glycerol causes a slight elevation in fatty acid production (1.4 – 2 fold) in both the DH1-DH2-UMA and in the control strain (Table 2, Figure 5A and B). The addition of glycerol to the culture media did not cause a significant changes in UFA:SFA ratios or in the general distribution of fatty acids (Table S2). However, a 2-fold increase in biomass production was observed when glycerol is added to the culture media, indicating that the fatty acid production increase resulting from carbon supplementation is due to a general biomass effect (Table 2).
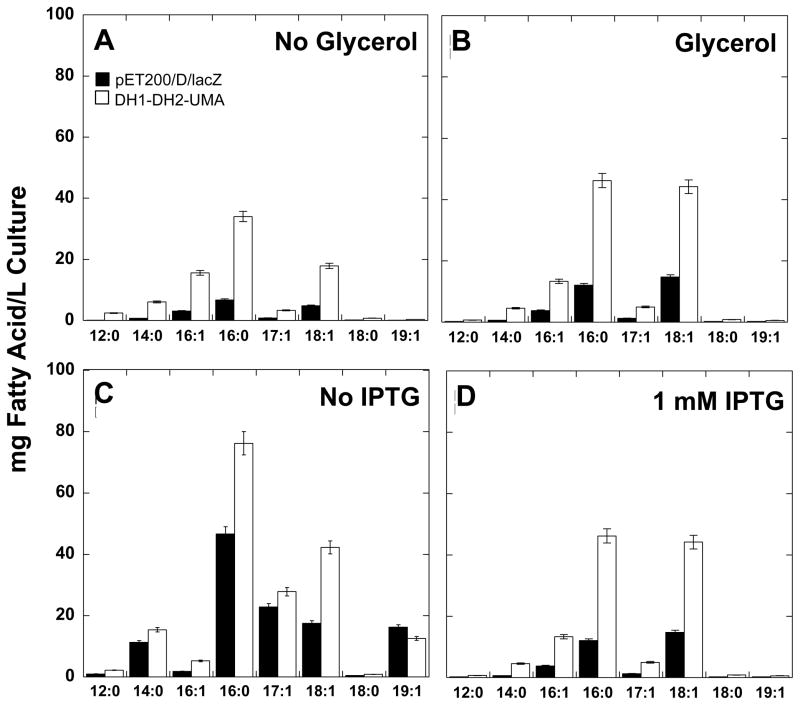
Figure 5. Effect of culture media supplementation.
Fatty acid profiles were determined for DH1-DH2-UMA (white bars) and pET200/D/lacZ controls (black bars) as described previously. Fatty acid titers were measured (A) without supplementation and (B) with supplementation of 0.4 % glycerol as a carbon source. These experiments were carried out at 22°C and with 1 mM IPTG. In separate experiments, fatty acid titers were measured (C) without IPTG and (D) with 1mM IPTG added to the culture media. These experiments were carried out at 22°C and with 0.4% glycerol. Error bars represent standard deviations about the mean of two or three replicate samples.
Effect of inducing enzyme expression on fatty acid production
Since the increase in the production of fatty acids was found to be accompanied by an increase in DH1-DH2-UMA protein expression, we wanted to know whether inducing the overexpression of the enzyme using isopropyl β-D-1-thiogalactopyranoside (IPTG) would result in further enhancement in fatty acid production. We measured fatty acid yield with and without added IPTG (to induce protein expression levels). GC/MS analysis of the FAME showed the same principal eight monounsaturated and saturated C12 to C19 fatty acids are produced (Figure 5C and D). In the absence of IPTG, the fatty acid yield was 1.6 higher in both control and experimental strains perhaps because lower protein expression means that more of the carbon source can be available for making fatty acids (Table 2). No changes in the UFA:SFA ratio were reported (Table S2). The addition of IPTG suppressed overall fatty acid biosynthesis, but it accentuated the fatty acid enhancement in the DH1-DH2-UMA strain which registered a 3.5 fold increase of FA enhancement under these conditions (Figure 5D, Table 2).
The addition of IPTG causes a 2-fold increase in biomass when compared to the cultures where no IPTG is added (Table 2). However, there were no differences in cell density between the control and experimental strains (Table 2).
Discussion
In recent years, there has been a substantial interest in the identification of new enzymes that increase the yield of fatty acids produced in microbial cultures [2, 5–6, 17, 22]. There are numerous reports of strategies to increase the production of fatty acids in E. coli with enhancements fluctuating between 3 and 5-fold for individual modifications (Table 1) [2, 5–6, 17]. In this report we have measured the ability of an active dehydratase tetradomain protein fragment to increase the production of fatty acids in E .coli by as much as 5-fold. This level of enhancement is within the range observed for a single modification in a strain of E. coli which has not been optimized for fatty acid production. We can confidently project that the yields of fatty acids can be pushed upwards by overexpressing DH1-DH2-UMA in a strain with an impaired beta-oxidation pathway (ΔfadD, ΔfadE) or by combining with other orthogonal strategies for enhancement, such as FadR co-expression [20].
The observed enhancement in fatty acid production by DH1-DH2-UMA was more pronounced at lower temperatures (16°C). This was not unexpected for a variety of reasons. Firstly, it is well-established that E. coli makes or accumulates a higher proportion of free fatty acids at lower temperatures, perhaps as an adaptive mechanism to the stress induced at cold temperatures [20, 23, 30]. Also, the exogenous enzyme being introduced in our study comes from P. profundum, a piezophilic deep-sea bacterium adapted to low temperatures [25]. Thus, it is possible that the enzyme itself is more active or that its structure is more stabilized at the lower temperatures. Thirdly, our results show that the expression of DH1-DH2-UMA was higher at the lower temperature. Therefore it is possible that the fatty acid enhancement could be reflecting the increase in enzyme production. The most likely explanation is that a combination of these three effects (enzyme expression, enzyme activity and enzyme stability) could be contributing towards the optimization of fatty acid enhancement at 16 °C.
Carbon supplementation of the media typically results in an improvement of fatty acid production in bacterial cultures [6]. In this study, we assessed the effect of adding 0.4 % v/v glycerol to the culture media on the production of fatty acids. The addition of glycerol allowed the cells to grow up to 80 hours without entering the death phase thus achieving higher cell density (Figure 2). This increase in biomass resulted in a modest increase in fatty acid production per unit of volume. The fact that carbon supplementation did not greatly affect fatty acid production, suggests that the LB media without supplementation contained enough carbon-containing nutrients for the bacteria to grow and make fatty acids.
The effect of enzyme expression on fatty acid production was assessed by the addition of the inducer IPTG. Our results showed that the overexpression of the tetradomain fragment resulted in a lower overall fatty acid production, but a higher degree of enhancement of fatty acid production over the control strain (3.5-fold increase when IPTG is added versus 1.5-fold increase without IPTG) (Table 2). These results are consistent with those from Zheng et al., who report that E. coli expressing a thioesterase from a medium copy number vector, which would be expected to make less of the enzyme, actually makes more fatty acids than the same gene cloned in a high copy number vector [31]. Also, Hoover et al. reported an optimal IPTG concentration of 50 μM for the induction of fatty acid production, suggesting that higher concentrations of the inducer results in the expression of too much enzyme, which is not always ideal for the production of fatty acids [32].
Our finding that a dehydratase, DH1-DH2-UMA, was capable of enhancing fatty acid production was somewhat surprising, especially since the dehydration reaction has not been identified as a bottleneck in bacterial fatty acid biosynthesis. In fact, previous work by others showed that overexpression of the native dehydratase from E. coli, FabA, does not increase the production of fatty acids [5]. One possible explanation is that this dehydratase tetradomain domain also catalyzes other types of reactions, such as thioester hydrolysis. The tetradomain DH1-DH2-UMA is composed of four contiguous hotdog fold domains [27] and this family of structural domains has been implicated in both dehydratase and thioesterase activity [33]. Therefore, it is possible that DH1-DH2-UMA also contains an additional hydrolase activity within its four different hotdog domains. Further work will need to be carried out to ascertain the mechanism by which DH1-DH2-UMA enhances fatty acid biosynthesis in E. coli.
Conclusions
In this study the production of fatty acids in E. coli was increased by the expression of an enzyme fragment with dehydratase activity from P. profundum. Others have reported higher yields in the production of fatty acids in E. coli, but these high yields are the result of combining several genetic manipulations within the same strain. We report an enhancement in fatty acid production which is comparable to that reported by others, for a single genetic manipulation of E. coli. Thus, this work lays the groundwork for further exploration of the applicability of enzymes from marine organisms towards increasing the yields, purity or quality of fatty acids in microbial fermentations.
Supplementary Material
Table S1. Fatty acid composition of Escherichia coli strains carrying control and recombinant plasmids at different temperatures of cellular growth.
Table S2. Effect of carbon and IPTG supplementation to the culture media on the fatty acid composition of Escherichia coli.
Figure S1. Representative GC chromatogram and mass spectra (A) Representative gas chromatogram for a FAME mixture isolated from the bacterial culture showed the principal fatty acid methyl esters identified to be monounsaturated or saturated fatty acids ranging from 12 to 19 carbon chain lengths. (B) to (I) Mass spectra data for each of the peaks identified in the GC chromatogram showing the molecular weight of the methyl ester
Highlights.
An engineered E. coli strain is capable of overproducing fatty acids up to 5-fold.
The observed enhancement is more pronounced at lower temperatures (16 °C).
The observed enhancement was not due to changes in E. coli cell density.
The DH domains tested in this study interact with the endogenous E. coli pathways.
The overexpression of DH domains represents an unexplored strategy for biofuels.
Acknowledgments
The authors thank Nashbly Montano and Elsie A. Orellano at the Department of Chemistry, UPR-Río Piedras for help with GC/MS samples preparation and data analysis. In the interest of full disclosure, the authors have applied for patent protection the proprietary inventions described in this manuscript. DO-R and AB-O have financial interest in the commercial venture Palmitica-Bio, licensee of the patent-pending technology. This publication was made possible by NSF grant CHE0953254 to AB-O and NIGMS grant R25GM061838 to DO-R. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. Shared instrumentation was purchased with NIH Grant G12RR03051 (RCMI Program).
List of abbreviations
- FA
fatty acid
- PUFA
polyunsaturated fatty acids
- FAME
fatty acid methyl ester
- DH
dehydratase
- AT
acyl tranferases
- KS
keto-acyl synthase, ACP, acyl carrier protein
- KR
keto-acyl reductase, ER, enoyl reductase
- GC
gas chromatography
- MS
mass spectrometry, UFA, unsaturated fatty acid
- SFA
saturated fatty acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Delise Oyola-Robles, Email: delise.oyola@upr.edu.
Carlos Rullán-Lind, Email: carlos.rullan@upr.edu.
Néstor M. Carballeira, Email: nestor.carballeira1@upr.edu.
Abel Baerga-Ortiz, Email: abel.baerga@upr.edu.
References
- 1.Abelson PH. Renewable Liquid Fuels. Science. 1995;268(5213):955. doi: 10.1126/science.268.5213.955. [DOI] [PubMed] [Google Scholar]
- 2.Liu T, Khosla C. Genetic engineering of Escherichia coli for biofuel production. Annu Rev Genet. 2010;44:53–69. doi: 10.1146/annurev-genet-102209-163440. [DOI] [PubMed] [Google Scholar]
- 3.Kalscheuer R, Stölting T, Steinbüchel A. Microdiesel: Escherichia coli engineered for fuel production. Microbiology. 2006;152:2529–36. doi: 10.1099/mic.0.29028-0. [DOI] [PubMed] [Google Scholar]
- 4.Canilha L, Chandel AK, dos Santos Milessi TZ, Fernandes Antunes FA, da Costa Freitas WL, das Graςias Almeida Felipe M, da Silva SS. Bioconversion of Sugarcane Biomass into Ethanol: An Overview about Composition, PretreatmentMethods, Detoxification of Hydrolysates, Enzymatic Saccharification, and Ethanol Fermentation. J Biomed Biotech. 2012:1–15. doi: 10.1155/2012/989572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cao Y, Yang J, Xian M, Xu X, Liu W. Increasing unsaturated fatty acid contents in Escherichia coli by coexpression of three different genes. Appl Microbiol Biotechnol. 2010;87:271–80. doi: 10.1007/s00253-009-2377-x. [DOI] [PubMed] [Google Scholar]
- 6.Lennen RM, Braden DJ, West RM, Dumesic JA, Pfleger BF. A process for microbial hydrocarbon synthesis: overproduction of fatty acids in Escherichia coli and catalytic conversion to alkanes. Biotech Bioeng. 2010;106:193–202. doi: 10.1002/bit.22660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lennen RM, Pfleger BF. Microbial production of fatty acid-derived fuels and chemicals. Curr Opin Biotechnol. 2013 doi: 10.1016/j.copbio.2013.02.028. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Zhao Z, Bai F. High-density cultivation of oleaginous yeast Rhodosporidium toruloides Y4 in fed-batch culture. Enzyme Microb Tecnhol. 2007;41:312–17. [Google Scholar]
- 9.Balat M. Potential alternatives to edible oils for biodiesel production – A review of current work. Energy Conservation and Management. 2011;52:1479–92. [Google Scholar]
- 10.Voelker TA, Worrell AC, Anderson C, Bleibaum J, Fan C, Hawkins DJ, Radke SE, Davies HM. Fatty acid biosynthesis redirected to medium chains in transgenic oilseed plants. Science. 1992;257:72–4. doi: 10.1126/science.1621095. [DOI] [PubMed] [Google Scholar]
- 11.US Department of Energy (Washington DC) Independent Statistics and Analysis. US Energy Information Administration; 2012. Biofuels uses and trends. [Google Scholar]
- 12.Angerbauer C, Siebenhofer M, Mittelbach M, Guebitz GM. Conversion of sewage sludge into lipids by Lipomyces starkeyi for biodiesel production. Bioresour Technol. 2008;99:3051–6. doi: 10.1016/j.biortech.2007.06.045. [DOI] [PubMed] [Google Scholar]
- 13.Ban K, Kaieda M, Matsumoto T, Kondo A, Fukuda H. Whole-cell biocatalyst for biodiesel fuel production utilizing Rhizopus oryzae cells immobilized within biomass support particles. Biochem Eng J. 2001;8:39–43. doi: 10.1016/s1369-703x(00)00133-9. [DOI] [PubMed] [Google Scholar]
- 14.Tamalampudi S, Talikder MM, Hama S, Tanino T, Susuki Y, Kondo A, Fukuda H. Development of recombinant Aspergillus oryzae whole-cell biocatalyst expressing lipase-encoding gene from Candida Antarctica. Appl Microbio Biotechnol. 2007;75:387–95. doi: 10.1007/s00253-006-0814-7. [DOI] [PubMed] [Google Scholar]
- 15.Johnson MB, Wen Z. Development of an attatched microalgal growth system for biofuel production. Appl Microbio Biotechnol. 2010;85:525–34. doi: 10.1007/s00253-009-2133-2. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Horsman M, Wang B, Wu N, Lan CQ. Effects of nitrogen spurces on cell growth and lipid accumulation of green alga Neochloris oleoabundands. Appl Microbiol Biotechnol. 2008;81:629–36. doi: 10.1007/s00253-008-1681-1. [DOI] [PubMed] [Google Scholar]
- 17.Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, McClure A, del Cardaye SB, Keasling JD. Microbial Production of Fatty-Acid-Derived Fuels and Chemicals from Plant Biomass. Nature. 2010;463:559–62. doi: 10.1038/nature08721. [DOI] [PubMed] [Google Scholar]
- 18.Stephanopoulos G. Challenges in engineering microbes for biofuels production. Science. 2007;315:801–4. doi: 10.1126/science.1139612. [DOI] [PubMed] [Google Scholar]
- 19.Antoni D, Zverlov VV, Schwarz WH. Biofuels from microbes. App Microbiol Biotechnol. 2007;77:23–35. doi: 10.1007/s00253-007-1163-x. [DOI] [PubMed] [Google Scholar]
- 20.Huffer S, Roche CM, Blanch HW, Clark DS. Escherichia coli for biofuel production: bridging the gap from promise to practice. Trends Biotechnol. 2012;30:538–545. doi: 10.1016/j.tibtech.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 21.Zhang F, Rodriguez S, Keasling JD. Metabolic engineering of microbial pathways for advanced biofuels production. Curr Opin Biotechnol. 2011;22:775–783. doi: 10.1016/j.copbio.2011.04.024. [DOI] [PubMed] [Google Scholar]
- 22.Lu X, Vora H, Khosla C. Overproduction of free fatty acids in E. coli: Implications for biodiesel production. Metab Eng. 2008;10:333–9. doi: 10.1016/j.ymben.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 23.Marr AG, Ingraham JL. Effect of temperature on the composition of fatty acids in Escherichia coli. J Bacteriol. 1962;84:1260–7. doi: 10.1128/jb.84.6.1260-1267.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Metz JG, Roessler P, Facciotti D, Levering C, Dittrich F, Lassner M, Valentine R, Lardizabal K, Domergue F, Yamada A, Yazawa K, Knauf V, Browse J. Production of polyunsaturated fatty acids by polyketide synthases in both prokaryotes and eukaryotes. Science. 2001;293:290–3. doi: 10.1126/science.1059593. [DOI] [PubMed] [Google Scholar]
- 25.Allen EE, Barlett DH. Structure and regulation of the omega-3polyunsaturated fatty a. cid synthase genesfrom the deep-sea bacterium Photobacterium profundum strain SS9. Microbiology. 2002;148:1903–13. doi: 10.1099/00221287-148-6-1903. [DOI] [PubMed] [Google Scholar]
- 26.Shulse CN, Allen EE. Diversity and distribution of microbial long-chain fatty acid biosynthetic genes in the marine environment. Environ Microbiol. 2011;13:684–95. doi: 10.1111/j.1462-2920.2010.02373.x. [DOI] [PubMed] [Google Scholar]
- 27.Oyola-Robles D, Gay DC, Trujillo U, Sánchez-Parés JM, Bermúdez M-L, Rivera-Díaz M, Carballeira NM, Baerga-Ortiz A. Identification of novel protein domains required for the expression of an active dehydratase fragment from a polyunsaturated fatty acid synthase. Protein Science. 2013 doi: 10.1002/pro.2278. E-pub ahead of printing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishida T, Orikasa Y, Ito Y, Yu R, Yamada A, Watanabe K, Okuyama H. Escherichia coli engineered to produce eicosapentaenoic acid becomes resistant against oxidative damages. FEBS Lett. 2006;580(11):2731–5. doi: 10.1016/j.febslet.2006.04.032. [DOI] [PubMed] [Google Scholar]
- 29.Clark D, Cronan J. EcoSal - Escherichia coli and Salmonella: Cellular and Molecular Biology. 3. Washington DC: ASM Press; 2005. [Google Scholar]
- 30.Orikasa Y, Yamada A, Yu R, Ito Y, Nishida T, Yumoto I, Watanabe K, Okuyama H. Characterization of the eicosapentaenoic acid biosynthesis gene cluster from Shewanella sp. strain SCRC-2738. Cell. Mol Biol. 2004;50:625–30. [PubMed] [Google Scholar]
- 31.Zheng Y, Li L, Liu Q, Qin W, Yang J, Cao Y, Jiang X, Zhao G, Xian M. Boosting the free fatty acid synthesis of Escherichia coli by expression of a cytosolic Acinetobacter baylyi thioesterase. Biotechnol Biofuels. 2012;5:76. doi: 10.1186/1754-6834-5-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoover SW, Marner WD, 2nd, Brownson AK, Lennen RM, Wittkopp TM, Yoshitani J, Zulkifly S, Graham LE, Chaston SD, McMahon KD, Pfleger BF. Bacterial production of free fatty acids from freshwater macroalgal cellulose. Appl Microbiol Biotechnol. 2011;91:435–46. doi: 10.1007/s00253-011-3344-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dillon SC, Bateman A. The Hotdog fold: wrapping up a superfamily of thioesterases and dehydratases. BMC Bioinfotmatics. 2004;5:109. doi: 10.1186/1471-2105-5-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Fatty acid composition of Escherichia coli strains carrying control and recombinant plasmids at different temperatures of cellular growth.
Table S2. Effect of carbon and IPTG supplementation to the culture media on the fatty acid composition of Escherichia coli.
Figure S1. Representative GC chromatogram and mass spectra (A) Representative gas chromatogram for a FAME mixture isolated from the bacterial culture showed the principal fatty acid methyl esters identified to be monounsaturated or saturated fatty acids ranging from 12 to 19 carbon chain lengths. (B) to (I) Mass spectra data for each of the peaks identified in the GC chromatogram showing the molecular weight of the methyl ester