Abstract
Plumbagin [5-hydroxy- 2-methyl-1, 4-naphthaquinone] is a well-known plant derived anticancer lead compound. Several efforts have been made to synthesize its analogs and derivatives in order to increase its anticancer potential. In the present study, plumbagin and its five derivatives have been evaluated for their antiproliferative potential in one normal and four human cancer cell lines. Treatment with derivatives resulted in dose- and time-dependent inhibition of growth of various cancer cell lines. Prescreening of compounds led us to focus our further investigations on acetyl plumbagin, which showed remarkably low toxicity towards normal BJ cells and HepG2 cells. The mechanisms of apoptosis induction were determined by APOPercentage staining, caspase-3/7 activation, reactive oxygen species production and cell cycle analysis. The modulation of apoptotic genes (p53, Mdm2, NF-kB, Bad, Bax, Bcl-2 and Casp-7) was also measured using real time PCR. The positive staining using APOPercentage dye, increased caspase-3/7 activity, increased ROS production and enhanced mRNA expression of proapoptotic genes suggested that acetyl plumbagin exhibits anticancer effects on MCF-7 cells through its apoptosis-inducing property. A key highlighting point of the study is low toxicity of acetyl plumbagin towards normal BJ cells and negligible hepatotoxicity (data based on HepG2 cell line). Overall results showed that acetyl plumbagin with reduced toxicity might have the potential to be a new lead molecule for testing against estrogen positive breast cancer.
Keywords: Anticancer, apoptosis, breast cancer, caspase-3/7, plumbagin.
1. INTRODUCTION
Natural products are a significant resource for the development of new drugs. Many of the plants’ natural products/secondary metabolites with interesting biological properties serve as ‘lead’ molecules for drug development processes. Since the 1940s, 48.6% of the all new small molecules discovered in the area of cancer treatment were either natural products or their derivatives [1].
Quinone compounds are widely distributed in nature and comprise the second largest class of anticancer agents [2]. They are known for their role in energy production and the electron transport chain in cells [3]. The plant-derived naphthoquinonoids have been described extensively for their cytotoxic and apoptotic potential against various cancer cell lines [4-10]. Plumbagin (PL) (5-hydroxy-2-methyl-1,4-naphthoquinone), a plant derived naphthoquinone having potent biological activities, has been reported to be extracted from the roots of various species of three major phylogenetic families i.e. Plumbaginaceae, Ebenaceae and Droseraceae. It is well known for its remarkable anticancer activities [6, 11-14]. PL effectively induces apoptosis by a number of mechanisms [11, 15-18] described in the literature. It has been shown that PL induces reactive oxygen species (ROS) production resulting in apoptosis and cell cycle arrest [19-20]. PL has also been found to be an efficient inhibitor of NF-kB activation, which ultimately leads to suppression of downstream NF-kB-regulated gene products and causes apoptosis [16]. It induces apoptosis induction in human BRCA cells through inhibition of NF-kB/Bcl-2 pathway [11, 21] and is also reported to inhibit tumor angiogenesis and tumor growth via VEGFR2 mediated Ras signaling pathway in endothelial cells [22].
Since its first reported apoptotic activities, PL has been envisaged as a “lead” molecule for the development of new bioactive therapeutic agents. Efforts have been undertaken to design novel analogues and derivatives of PL to enhance bioactivity and reduce toxicity, thereby maximizing their therapeutic potential. Several derivatives of PL, including hydroquinonoid, nitro, cyano, methyl ester derivatives and metal complexes, have been tested for their tumor-inhibitory activity [23-25]. Padhye et al., have published a review of the biological activities of PL and its analogs/derivatives [26].
In the present study, we report the cytotoxic activities of five derivatives of PL i.e. acetyl plumbagin (AP), benzoate plumbagin (BP), crotonate plumbagin (CP), isobutyrate plumbagin (IP), and propionate plumbagin (PP), (Fig. 1) synthesized by following the general esterification methods [27]. Prescreening against four human cancer and one normal cell line using growth inhibition assay at various time points led us to choose AP for further investigations in BT20 (estrogen receptor negative) and MCF-7 (estrogen receptor positive), along with normal fibroblasts (BJ) through APOPercentage, caspase-3/7 and ROS assays. The cell cycle analysis and investigation of apoptosis-associated genes such as p53, Mdm2, NF-kB, Bad, Bax, Bcl-2 and Casp-7 using RT-PCR were also performed in MCF-7 cells. We demonstrated that between tested breast cancer cell lines, AP induced apoptotic cell death in estrogen positive MCF-7 cells only. It also displayed negligible hepatotoxicity (HepG2 cell line) and very low toxicity towards normal fibroblasts (BJ).
Fig. (1).
Structures of PL and its derivatives (AP, BP, CP, IP, and PP).
2. MATERIALS AND METHODS
2.1. Materials
Dulbecco's Modified Eagle Medium (DMEM), Fetal Calf Serum (FCS), penicillin-streptomycin, Trizol, Propidium Iodide (PI), 2’, 7’ –dichlorofluorescein diacetate (DCFDA) and TaqMan Universal Master Mix were obtained from Life Technologies, UK. PL and MTT (3-(4,5-Dimethylthiazol-z-yl)-2,5-diphenyltetrazolium bromide) were purchased from Sigma Chemicals, USA. APOPercentage kit was procured from Biocolor (UK) and ApoTox-Glo kit was procured from Promega Corporation, USA.
2.2. Cell Lines and Culture
MCF-7 (Breast Adenocarcinoma), BT20 (Breast carcinoma), DU145 (Prostate carcinoma), HepG2 (Hepatocellular carcinoma) and BJ (normal skin fibroblasts) were procured from the American Type Cell Culture Collection (ATCC, Manassas, VA). All cell lines were cultured in a humidified atmosphere of 5% CO2 in DMEM at 37°C, supplemented with 10% FCS, penicillin (100U/ml) and streptomycin (100 µg/ml).
2.3. Growth Inhibition Assay
Cells were seeded in 384-well culture plates at a density of 2.5 x 103 cells per well and were grown with various (1, 5, 10 and 20 µM) concentrations of PL and its derivatives (AP, BP, CP, IP and PP) for 12, 24, 36 and 48 h (hours). Following incubation with compounds, 5 µl of sterile MTT (5 mg/ml) dissolved in PBS was added and plate was incubated for 4 h followed by the addition of 30 µl of solubilization solution (10% SDS, 10 mM HCl), and further incubation for 16 h at 37˚C. The OD (optical density) of each well was measured at 595 nm using a microtiter plate reader (BMG Labtech PHERAstar FS, Germany).
2.4. Determination of Apoptosis Using APOPercentage Assay
MCF-7, BJ, and BT20 cells at a density of 5 x 103 cells per well were seeded in 96-well plates. After 24 h, 5 µl of desired concentrations of PL and AP (5 and 10 µM) diluted in complete DMEM were added and incubated at 37o C for 12 and 24 h. Cells were incubated with 5mM H2O2 for 30 minutes (min) as a positive control. APOPercentage dye as per manufacturer’s instructions (Biocolor, UK) was used for staining. Each plate was analyzed with HTFC Screening System (IntelliCyt Corporation, Albuquerque, NM) to identify apoptotic cells.
2.5. Measuring Caspase-3/7 Activity
BJ, BT20 and MCF-7 cells were seeded at a density of 2.5 x 103 cells per well in 20 µl of media in 384-well plates. After 24 h, 5 µl of PL or AP (5 and 10 µM) were added. Docetaxel (200nM) was used as a positive control. The ApoTox-Glo kit was used to estimate caspase-3/7 activity at time intervals of 1 h, 2 h, 4 h, 8 h and 16 h following the manufacturer’s instructions and luminescence was measured using a luminescence plate reader (BMG Labtech PHERAstar FS, Germany).
2.6. Reactive Oxygen Species (ROS) Assay
MCF-7 and BT20 cells were seeded at a density of 2.5 x 103 cells per well in 45 µl of media in 96-well plates. After 24 h, 5 µl of 10 or 20 µM of PL or AP was added and incubated for 2 h. ROS activity in cells was determined by staining the cells with 10 µM DCFDA. 10mM H2O2 was used as a positive control. Percentage of cells stained positive for ROS was measured with a high throughput flow cytometer (HTFC) screening system (IntelliCyt Corporation, Albuquerque, NM). Cells were gated for FSC-H, SSC-H and a minimum of 1000 events per well were recorded in FL-1H channel.
2.7. FACS for Cell Cycle Analysis
MCF-7 cells were seeded at a density of 1 x 105 cells per well in a twelve well plate and left overnight to settle. Cells were treated with PL or AP (5 and 10 µM) for 12 and 24 h. Cells were then trypsinized and collected into 1.5 ml eppendorf tubes. Samples were centrifuged for 5 min at 1000 rpm and the supernatant was removed and discarded. The resultant pellet was fixed in 800 µl of absolute ethanol and stored at – 80o C overnight. Following fixation, samples were centrifuged at 1000rpm for 5 min, the supernatant was removed and the pellets were resuspended in 500 µl of 1 x PBS. Samples were again centrifuged (1000 rpm for 5 min), the supernatant was removed and the pellets were resuspended in 50 µl RNAseA (50 µg/ml) for 15 min at 37o C or 30 min at room temperature. 200 µl of PI Staining solution (0.1% Triton X-100, 2mM MgCl2, 100mM NaCl, 10mM PIPES ph 6.8 and 10µg/ml PI) was added per sample and incubated for 10–15 min prior to analyzing on HTFC screening system (IntelliCyt Corporation, Albuquerque, NM).
2.8. RNA Extraction
Cells were grown in 6-well plates for 24 h and treated with PL or AP (5 and 10 µM). After desired incubation time, cells were washed twice with 2 ml cold 1 x PBS followed by addition of 0.6 ml of Trizol. Cell lysis was performed by incubating the cells with Trizol for 5 min at room temperature and samples were transferred to 1.5 ml eppendorf tubes and 0.12 ml of chloroform was added. Tubes were then inverted for 15 seconds and incubated on ice for 10 min followed by centrifugation at 8000 rpm for 15 min at 4o C. To the aqueous phase, 0.3 ml of isopropanol was added and RNA was precipitated overnight at -20oC. Samples were centrifuged at 8000 rpm for 30 min at 4o C. The supernatant was removed and the resulting pellet was washed with 0.6 ml of 75% ethanol. Samples were then subjected to centrifugation at 8000 rpm for 20 min at 4oC, the supernatant was discarded and the pellet was air-dried and then resuspended in 30 µl of DEPC water. RNA concentration was determined with a nanodrop (Thermo Scientific, USA).
2.9. cDNA Synthesis
cDNA reaction was set up by taking 2 µg of RNA along with 250 ng of Oligo-dT primer (Promega) and 500 µM of dNTPs. The reaction tube was incubated at 65o C for 5 min followed by a pulse spin and then shifted to ice. Each cDNA reaction was made up to 20 µl containing 1x first strand reaction Buffer (Invitrogen), 5mM DTT, 200 units SuperScript III RT (Invitrogen) and 40 units of RNAse (Invitrogen). The PCR cycles used were: 25o C for 10 min followed by 37o C for 2 h, 85o C for 10 min and finally 4o C hold.
2.10. Quantitative RT-PCR
Each 20 µl PCR reaction in PCR fast reaction tubes (Applied Biosystems), contained 6 µl of DI water, 10 µl of TaqMan Universal Master Mix (Applied Biosystems), 0.6 µl (300 nM) of each forward and reverse primer (Eurofins, Germany), 0.8 µl (200 nM) probe and 2 µl of cDNA. In a StepOnePlus Real-Time PCR machine (Applied Biosystems), the PCR cycles used were: 1 cycle of 95o C for 3 min to activate the enzyme, followed by 40 cycles of 95o C for 1 second (denaturation) and 60o C for 20 seconds (annealing and extension). Fold change in gene expression was calculated using the ∆∆CT method. The sequences of the human genome based primers, probes and details of TaqMan assays used for RT-PCR are tabulated in supplementary file 1. (933.3KB, pdf)
2.11. Statistical Analysis
The data were analyzed using one-way ANOVA and Tukey’s HSD post hoc analysis was used to determine the statistical significance (p<0.05).
3. RESULTS
The cytotoxic potential of five derivatives (AP, BP, CP, IP and EP) was measured against one normal and four cancer cell lines (among the most commonly used cell lines for testing cytotoxicity of PL in published literature), which further led us to choose derivative AP and cell lines for downstream analysis in comparison to PL. Proapoptotic activities of PL and AP were measured using APOPercentage assay in two breast cancer cell lines (MCF-7 and BT20) and one normal cell line (BJ), while caspase-3/7 activity was evaluated for these cell lines using five different time-points. ROS activity of PL and its derivatives was measured at 2 h after treatment with 10 and 20 µM of compounds. To observe the mechanistic effects of AP on MCF-7 cells, cell cycle analysis and mRNA quantification of molecular targets (apoptosis-associated genes) was performed. The detailed results of the investigations are presented below:
3.1. Cytotoxic Potential of PL and its Derivatives
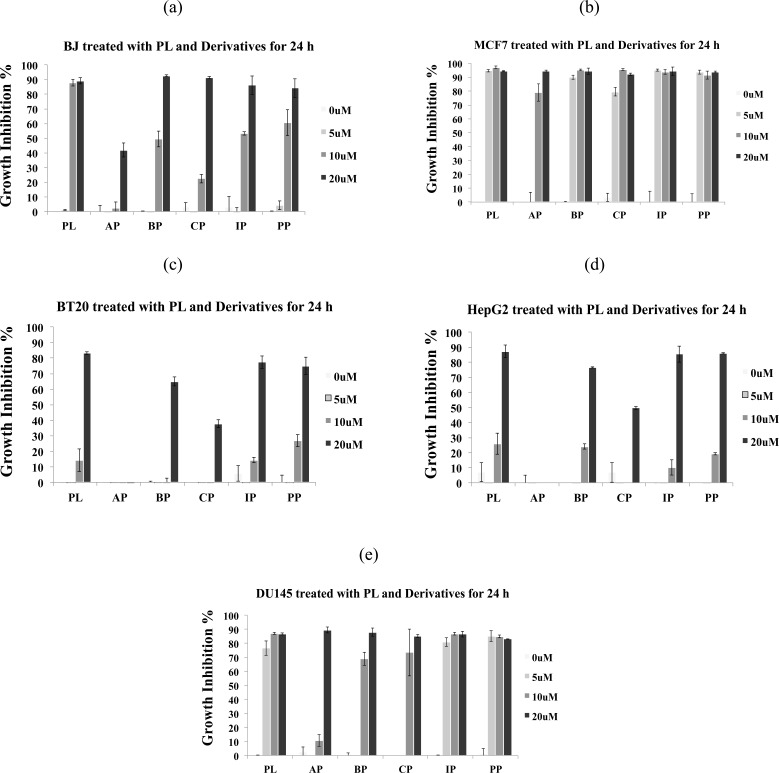
PL and its four derivatives (BP, CP, IP and EP) inhibited the growth of normal BJ cells in a dose- and time-dependent manner; although, the derivatives were found to be less cytotoxic as compared to PL (Fig 2a, supplementary file 2 (933.3KB, pdf) ). AP was found to be least effective in inhibiting the growth of BJ cells at 10 µM (Fig. 2a), which is an indicator of its possible low toxicity to normal cells. Growth of MCF-7 (estrogen positive breast cancer) cells treated with 5, 10 and 20 µM of PL and its derivatives BP, CP, IP and EP was inhibited by 80-90% at 24 h, while AP inhibited the cell growth by 80% at 10 µM (Fig. 2b). BT20 (Estrogen negative breast cancer cells) appeared to be relatively resistant to AP (Fig. 2c) and PL, which demonstrates cell line selective activities of these compounds. In HepG2, PL and its derivatives BP, IP and EP induced growth inhibition of 80-90% cells at 20 µM (Fig. 2d); however, AP was found to be ineffective against HepG2 (hepatocellular carcinoma), which may be another indication of its lower toxicity for liver cells (see discussion for details). For DU145 cell line, all the compounds except AP at 10 µM were showing growth inhibition of >70% at 24 h (Fig. 2e).
Fig. (2).
PL and derivatives (AP, BP, CP, IP, and PP) induced growth inhibition in cancer and normal cells within 24 h of treatment (read 0 µM as untreated). Growth inhibition was assessed using MTT assay. Data are mean ± standard deviation S.D. (n=3). Growth inhibition at 24 h for (a) BJ cells, (b) MCF-7 cells, (c) BT20 cells, (d) HepG2 cells, and (e) DU145 cells.
Overall, the derivatives displayed either similar or less toxicity as compared to PL against various cell lines. However, AP exhibits selective cytotoxic activity against estrogen positive MCF-7 cells at 10 µM and remarkably low toxicity towards normal fibroblasts BJ cells and HepG2 cell line, which led us to focus our further investigations into the understanding of mechanism of apoptosis induction in MCF-7 cells by AP and PL using various assays.
3.2. AP Induces Apoptosis and Caspase-3/7 Activity in MCF-7 Cells
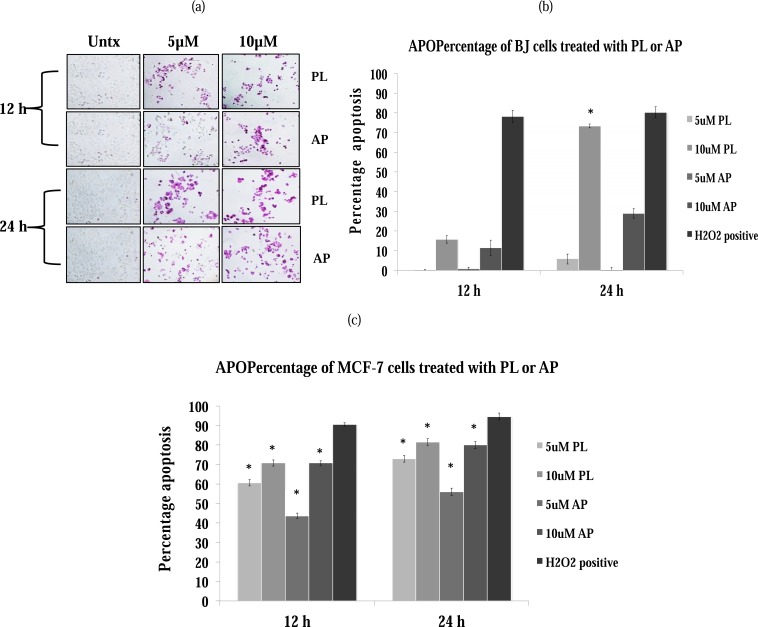
APOPercentage assay was performed by using flow cytometry (Fig. 3) after staining the cells with APOPercentage dye (Fig. 3a). In normal fibroblasts (BJ cells), PL displayed >70% apoptosis at 10 µM while AP at the same concentration showed only 20-30% positive staining at 24 h time point (Fig. 3b). In MCF-7 cells, the apoptotic effect of both compounds is similar at 10 µM, whereas PL was found to be slightly more active at 5 µM (Fig. 3c). In BT20 cell line, both compounds were unable to induce apoptosis (data not shown).
Fig. (3).
PL-and AP- dependent induction of apoptosis in BJ and MCF-7 cells. Cells were incubated with 5 and 10 µM of PL and AP for 12 and 24 h. (a) A representative image of at least two independent experiments each having quadruplicates is shown. H2O2 is used as a positive control to show that assay is working. Percentages of apoptotic cells (based on dye uptake) at 12 and 24 h in: (b) BJ cells, and (c) MCF-7 cells. Data are mean±S.D. (n=4), *p<0.05 significant difference to untreated control.
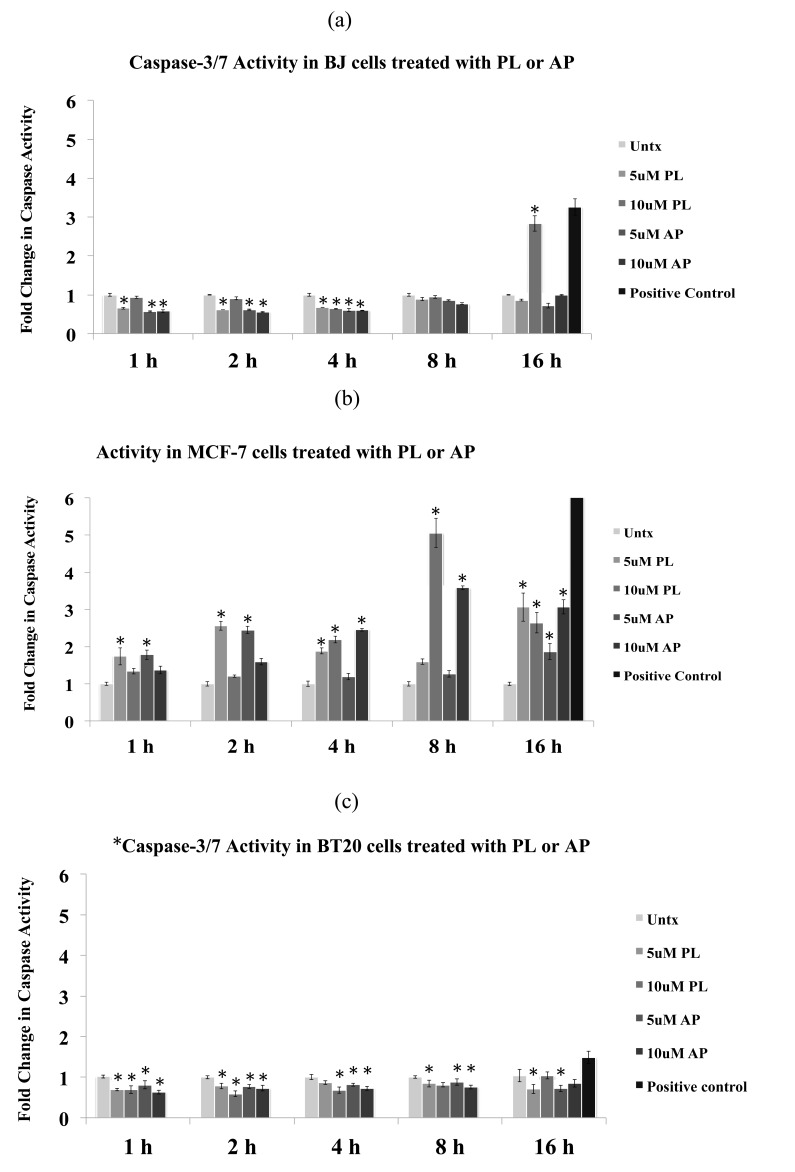
The results of the caspase-3/7 analysis are presented in Fig. 4. In BJ cells, no significant increase in caspase-3/7 activity was observed after 8 h of treatment with PL and AP at 10 µM and the activity was found to be even less than the untreated cells. At 16 h, caspase-3/7 activity after treatment with PL was increased more than 2.5-fold, whereas AP did not show any increase in caspase-3/7 activity (Fig. 4a). In MCF-7 cells, an increase in caspase-3/7 activity was observed within 1 h of treatment with both compounds at 5 µM. The peak caspase-3/7 activities after treatment with PL and AP were observed at 8 h (Fig. 4b). It is worthwhile to mention here that MCF-7 lacks caspases-3, thus the observed activities in MCF-7 cells are contributed by caspase-7. BT20 cells did not display any increase in caspase-3/7 activity at tested concentrations of both compounds at all time-points (Fig. 4c).
Fig. (4).
PL- and AP- dependent induction of caspase-3/7 activity in BJ, MCF-7 and BT20 cells. Cells were incubated with 5 and 10 µM of PL and AP for 1, 2, 4, 8 and 16 h. Fold change in caspase-3/7 activity after treatment with PL and AP in a time-course experiment in (a) BJ cells, (b) MCF-7 cells, and (c) BT20 cells. Docetaxel (200nM) is used as positive control at all time points to show that assay is working and representative activity is shown for 16 h only. Data are mean±S.D. (n=4), *p<0.05 significant difference to untreated (Untx) control.
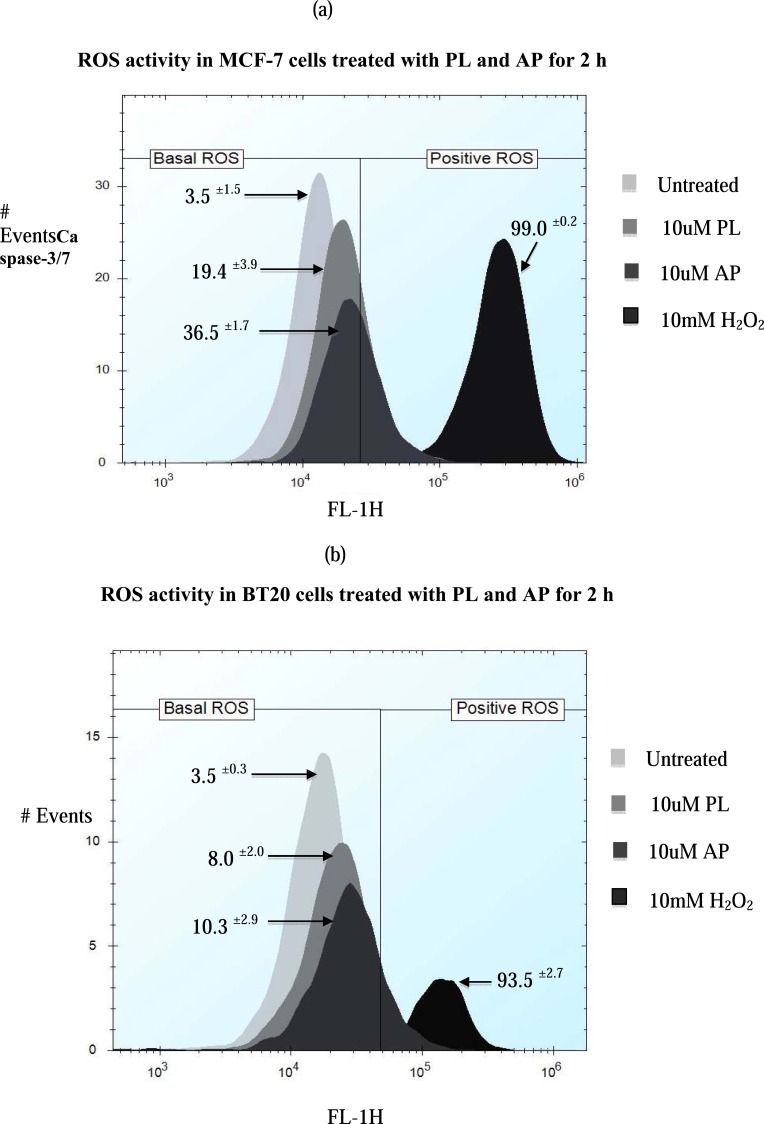
3.3. ROS Activity
Previous reports have linked PL with ROS mediated apoptosis [18, 28]. To test whether derivatives of PL have the ability to activate ROS production in cancer cells, we employed the DCFDA based ROS detection assay using flow cytometry to determine ROS levels in MCF-7 and BT20 cells. ROS activities were observed at 10 and 20 μM of PL and its derivatives (AP, BP, CP, IP and EP) at 2 h time point. It was found that AP induced more ROS production as compared to PL in MCF-7 cells (Fig. 5a); however, minimal ROS production was observed in BT20 cells (Fig. 5b) after treatment with PL and AP when compared with untreated cells. Similar results were observed for derivatives BP, CP, IP and EP (supplementary file 3 (933.3KB, pdf) ).
Fig. (5).
Effect of 10 and 20 µM PL and derivatives on ROS production in MCF-7 and BT20 cells. ROS activity in cells was determined by staining with the DCFDA dye and cells were using flow cytometry. The percentages of stained cells are shown on the histograms. Data are mean ± standard deviation S.D. (n=3). Cells were treated with 10mM H2O2 for 1 h as a positive control to show that assay was working.
3.4. Cell Cycle Analysis and Molecular Markers of Apoptosis
The effects of PL and AP on cell cycle distribution in MCF-7 cells were analyzed by using PI and flow cytometry analysis (Table 1). Neither of the two compounds showed distinct cell cycle arrest at 6 h (results not shown), 12 h (Table 1a) and 24 h time points (Table 1b). A slight increase, i.e. 3-4% of cells in G2-phase was noticed for both PL (10 µM) and AP (5 and 10 µM) at 12 h (Table 1), while a significant decrease in percentage of cells in G2-phase of cell cycle was observed at 24 h for PL and AP at all tested concentrations (Table 1b). Additionally, a significant increase in sub-G1 apoptotic fraction was also detected at 24 h for PL and AP at all tested concentrations (Table 1b).
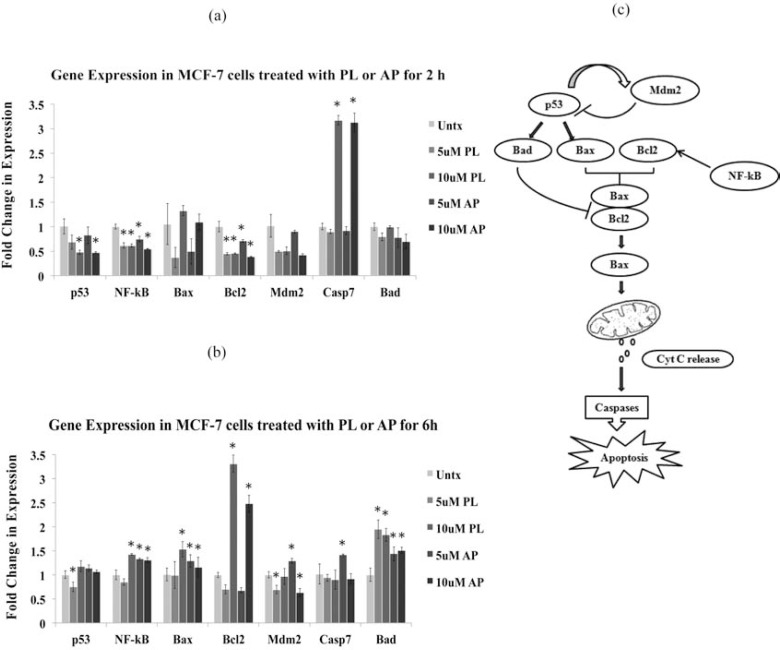
Since it has been reported earlier that PL-mediated cell cycle arrest is partly regulated by p53 [29], which is known to regulate expression of proapoptotic genes such as, Bad [30], Bax and antiapoptotic Bcl-2 [31], our further investigations focused on the changes in expression of apoptosis-associated molecular markers, namely Bad, Bax, Bcl-2, p53, NF-kB, Mdm2 and casp-7 genes in MCF-7 cells in response to treatment with PL and AP. More than 3-fold increase in the expression of casp-7 was observed within 2 h of treatment with both compounds at 10 µM, whereas all other genes except Bax showed decrease in expression in comparison to untreated control (Fig. 6a). At 6 h treatment, a notable increase in the expression of most of the tested genes was observed (Fig. 6b), however, caspase-7 expression returned to its basal levels. This data also confirms that molecular changes start within 1-2 h of treatment and are dose-dependent. The changes in mRNA expression levels were prominent at 10 µM concentration for both compounds. The exception is Bad, which showed slightly higher expression at 5 µM.
Fig. (6).
Molecular profiling by measuring mRNA levels of key apoptosis-associated genes using RT-PCR. The MCF-7 cells were treated with 5 and 10 µM of PL and AP for 2 and 6 h in duplicate experiments. Bar graph representing the expression profiles of p53, NF-Kb, Bax, Bcl2, Mdm2, Casp7, and Bad genes after treating the MCF-7 cells for (a) 2 h, and (b) 6 h. Data are mean±S.D. (n=3), *p<0.05 significant difference to untreated (Untx) control. (c) Pathway diagram showing the interplay of the tested genes during the apoptotic cell death.
4. DISCUSSION
Several efforts in the past have been made to synthesize the analogs and derivatives of PL to further increase its anticancer potential and to decrease its toxic effects on normal cells. It is reported in literature that the presence of active hydrogen in phenolic hydroxyl group of PL makes it more prone to oxidation [32]. Therefore, to block the potential oxidation and to generate more stable bioactive compounds, various derivatives of PL were synthesized by substituting the 5-OH group with acetate, benzoate, crotonate, isobutyrate and propionate groups and were evaluated for their ability to cause cell proliferation inhibition in various cancer cell lines. Overall, the derivatives showed similar or less toxicity against various cancer cell lines as compared to PL, however, AP selectively inhibited growth of MCF-7 cells, while growth of normal cells (BJ) and HepG2 cells (representative model of hepatotoxicity) was unaffected. This selectivity could be attributed to the presence of acetyl group (Fig. 1), which might be responsible for reduced toxicity in normal cells. HepG2 has been proved to be a good in vitro model to estimate the hepatotoxicity in the screening process [33-37] and is most commonly used as a model cell line to assess the hepatotoxicity, however it also has limitations and efforts have been made to upgrade HepG2 [38] and to identify more specific models for hepatotoxicity testing [39]. Based on growth inhibition data, AP was further selected to test its apoptotic effects on both estrogen positive (MCF-7) and estrogen negative (BT20) breast cancer types. To further confirm the apoptotic mode of cell death, APOPercentage assay was performed. Caspase-3/7 activity, ROS activity, cell cycle distribution and gene expression analysis were also included in the investigations to understand the dynamics of molecular changes and their relation to the observed phenotypic changes (apoptosis) in cancer cells.
We considered various parameters of apoptosis, such as; caspase-3/7 activation, phosphatidylserine exposure (APOPercentage staining) and ROS production to explain differential cell death inducing capabilities of AP in MCF-7 and BT-20 cells. Caspase activation (a hallmark of apoptosis) was observed as early as 1 h of treatment in MCF-7 cells, whereas BT20 showed no increase in caspase-3/7 activity even at 16 h. The results of our study point out that AP and PL induced apoptosis through caspases in MCF-7 cells, but failed to induce apoptosis in BT20 cells. It is interesting to note, while no apoptosis was observed after treatment with AP and PL in BT20 cells, the growth inhibition induced by PL was quite high. The inability to induce apoptosis in BT20 cells could possibly be due to either specific mechanisms (caspases inhibition or absence of estrogen receptors (ER)) that prevent the cells to undergo apoptosis or cells might be undergoing a non-apoptotic death. PL has been described to bind to the active site of ER-alpha in BRCA1-blocked BG1 ovarian cancer cells [40]. One of the key mechanisms of apoptosis induction by PL has been linked to ROS production in cells [18-19, 28, 41]. It has been shown that the ROS scavengers or inhibitors e.g. NAC, catalase, glutathione, superoxide dismutase completely abrogated PL induced apoptosis and DNA fragmentation [20, 42, 43]. Similar to parent compound PL, all the derivatives (AP, BP, CP, IP and EP) induced ROS production in MCF-7 cells, which might be contributing to observed caspase-3/7 activation and apoptosis. On another note, the resistance of BT20 cells to apoptosis and caspase-3/7 activation when treated with PL and its derivatives could be due to lack of ROS production (Fig. 5b).
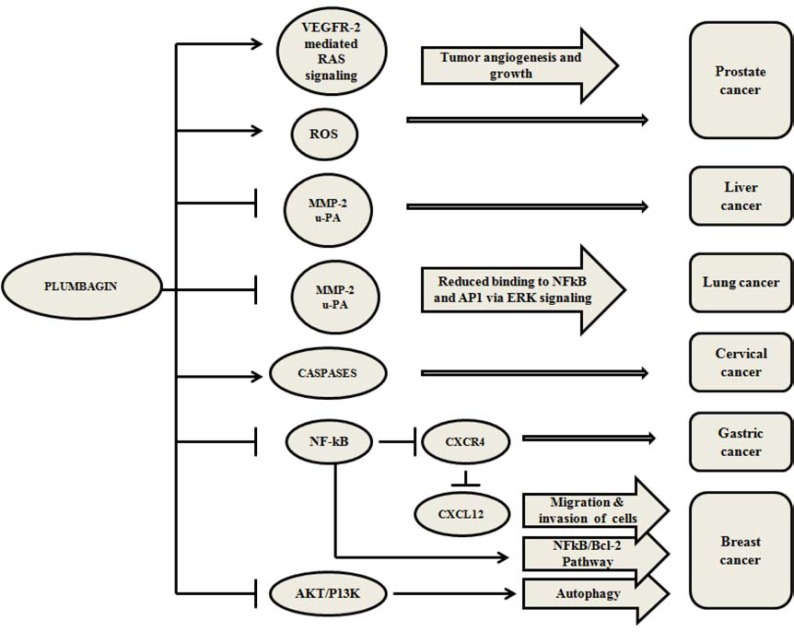
Furthermore, we also observed that AP at 5 µM concentration activated the caspase-3/7 in MCF-7 within 1 h similar to PL, but the percentage apoptosis induced was at least 20% less than that of PL. This may be either due to high toxicity of PL or the cell death mediated by other mechanisms independent of caspases. Kuo et al., reported that PL activates autophagy in breast cancer cells by AKT/PI3K inhibition [11]. It has been reported that caspase inhibition leads to selective degradation of catalase thus promoting generation of ROS and leads to autophagic cell death [44]. There are several known anticancer agents such as tamoxifen, arsenic trioxide, rapamycin, histone deacetylase inhibitors, temozolomide, ionizing radiation [45], vitamin D analogues [46] and etoposide [47] that induced cell death by autophagy. Previously published reports on caspase-independent cell death in L1210 cells [48] suggested that staurosporine induced cell death by two parallel pathways, where the early cell death was caspase-dependent (activated within <3 h of staurosporine treatment) and the late cell death was caspase-independent (>12 h of drug treatment). However, both mechanisms led to apoptotic cell death [49]. Several mechanisms have been proposed for the mode of action of PL in different types of cancers [6, 13-14, 16-17, 22, 26, 50-53]. An overview of these mechanisms is presented in Fig. (7).
Fig. (7).
An overview of the cell death mechanisms induced by PL in various cancer cells. Black arrows represent the activation, black bars represent the inactivation, blue ovals represent the genes/mechanisms targeted by PL and pink arrows represent the biological processes or pathways affected by PL in various types of cancers [6, 13-14, 16-17, 22, 26, 50-53].
PL is known to mediate G2-phase cell cycle arrest in A549 cells through p53 accumulation [29] and in MCF-7 cells by inhibiting the PI3K/AKT/mTOR pathway [11]. However, our results did not show G2-arrest in MCF-7 cells after treatment with PL or AP. Unexpectedly, more number of cells was observed in sub-G1 phase at 5 µM as compared to 10 µM concentrations for both PL and AP at 24 h (Fig. 5b). Similar observations have been reported earlier [54], where higher concentration (5 µM as compared to 1 µM) of doxorubicin showed less number of cells in sub-G and G1-phases with continuous treatment of 24 h as compared to a bolus treatment of 3 h [45]. Authors correlated this behavior to cellular senescence where cells show senescence-like phenotype and re-enter cell cycle after recovery [55, 56]. This study [54] also suggested that in vitro behavior of drugs may differ considerably due to the concentration and duration of treatment.
P53 is known to play a vital role during G1- and G2-arrest in cell cycle [57] and is controlled by Mdm2 through an autoregulatory feedback loop [58]. PL upregulates p53 in non-small cell lung cancer (H460) cells [59] and osteosarcoma (UTOS) cells [60] and hence affects cell cycle arrest. In our investigations, a slight increase in p53 expression has been observed in MCF-7 cells after treatment with PL and AP at 10 µM and could be the possible explanation for observing a higher percentage of cells in sub-G1 phase. Transcription factor p53 is also a regulator of Bax and Bax/Bcl2 heterodimers.The relative levels of these proteins determine the cell survival/death, with Bax being proapoptotic and Bcl2 being an antiapoptotic gene. PL is known to alter Bax/Bcl2 ratio [60], thus causing cytochrome c release and activation of caspases, which eventually lead to apoptosis (Fig. 6c). Another key component in the process of apoptosis is NF-kB, which is activated by p53 and is an essential component of p53-mediated apoptosis in cells retaining wild type p53 [61]. On the contrary, the role of NF-kB in cell proliferation or inhibition of apoptosis has also been described [62]. The conditions in which NF-kB acts pro- or antiapoptotic are not fully understood. The downregulated NF-kB further downregulates Bcl2 gene and favors apoptosis in cancer cells. This mechanism has been confirmed in H460 cells [59], MCF-7 cells [21] and UVR-induced squamous cell carcinoma [63] in response to PL treatment. In the light of interplay of molecular markers of apoptosis described above, our study revealed that Bcl2 is downregulated at 2 h, which indicates the initiation of apoptosis in MCF-7 cells. However, the increase in expression of Bcl2 at 6 h could represent an effort of molecular machinery of the cell to survive and escape apoptotic cell death. At 6 h, proapoptotic genes, Bad and Bax were also observed to be upregulated along with their regulator p53. The predisposition to cell death is determined by the relative levels and interactions of Bcl-2, Bax, and Bad proteins [64]. Antiapoptotic gene Bcl2 is known to inhibit Bax and thus preventing the cell from undergoing apoptosis [65]. However, the presence of Bad (transcriptionally activated by p53 [30]) determines whether the Bcl2 will be effective in repressing apoptosis. It has been reported that Bad inhibits Bcl2 by directly binding to it and rescues Bax [64]. Bax induces permeabilization of the outer mitochondrial membrane causing cytochrome c release, which further leads to the caspase activation and finally to apoptosis. The changes in the expression of various pro- and antiapoptotic genes observed in the present study can be explained in the light of above mentioned studies. The molecular dynamics of these apoptosis-associated genes are depicted in Fig. 6c.
5. CONCLUSIONS
The present study has revealed that PL and its derivatives (AP, BP, CP, IP and EP) have significant dose- and time-dependent cytotoxicity in vitro against one or more human cancer cell lines. The study also highlights selective inhibitory activity of AP in estrogen positive breast cancer cell line MCF-7 and its remarkably low toxicity in normal BJ and HepG2 cells. . We demonstrated that addition of acetyl group at 5-C position was able to reduce the toxicity of the parent compound (PL) towards normal cells. AP could possibly act as a lead molecule for further drug development targeting estrogen positive breast cancer sub-type. Further in vivo investigations are needed to confirm its antitumor activity and toxicity profiles.
Table 1.
Effects of 5 and 10 M PL and AP on cell cycle distribution in MCF-7 cells is shown.
(a)
| Untreated | 5 µM PL | 10 µM PL | 5 µM AP | 10 µM AP | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | S.D. | Mean | S.D. | Mean | S.D. | Mean | S.D. | Mean | S.D. | |
| G2 | 23.22 | 0.86 | 22.69 | 0.86 | 26.74 | 1.71 | 27.89 | 0.40 | 26.98 | 0.70 |
| S | 28.19 | 0.86 | 28.64 | 1.06 | 27.11 | 2.43 | 26.36 | 2.09 | 27.18 | 1.67 |
| G1 | 46.88 | 0.66 | 47.41 | 0.58 | 42.61 | 2.92 | 43.77 | 0.48 | 43.64 | 2.25 |
| Sub-G1 | 2.95 | 0.07 | 2.83 | 0.46 | 4.88 | 2.88 | 3.42 | 1.02 | 3.62 | 1.92 |
(b)
| Untreated | 5 µM PL | 10 µM PL | 5 µM AP | 10 µM AP | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | S.D. | Mean | S.D. | Mean | S.D. | Mean | S.D. | Mean | S.D. | |
| G2 | 24.37 | 0.42 | 9.62* | 1.91 | 14.57* | 2.09 | 15.57* | 2.47 | 14.52* | 1.84 |
| S | 29.59 | 0.34 | 30.54 | 1.02 | 31.87 | 1.60 | 27.37 | 4.92 | 31.35 | 1.57 |
| G1 | 43.85 | 0.25 | 24.39* | 5.14 | 34.54 | 4.25 | 31.33* | 4.22 | 35.65 | 2.03 |
| Sub-G1 | 2.86 | 0.61 | 35.78* | 6.20 | 21.25* | 5.77 | 26.72* | 1.62 | 20.06* | 3.58 |
Cell cycle was analyzed using PI staining and flow cytometry. In one experiment, at least 10,000 cells were counted based on DNA content after staining with PI. Representative plots of one set of duplicate experiment are shown after (a) 12 h, and (b) 24 h of treatment with PL and AP. Data are mean and S.D. (n=3), *p<0.05 significant difference to untreated control.
ACKNOWLEDGEMENTS
Declared none.
FUNDING SOURCE
SS is supported by the SEDCO grant to VBB. SS and MK are supported by KAUST CBRC Base Funds of VBB. LE is supported by KAUST Research Grant to John AC Archer. VBB is supported by KAUST Base Research Funds.
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflict of interest.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s web site along with the published article.
REFERENCES
- 1.Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanchez-Cruz P, Alegria AE. Quinone-enhanced reduction of nitric oxide by xanthine/xanthine oxidase. Chem. Res. Toxicol. 2009;22:818–823. doi: 10.1021/tx800392j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Powis G. Free radical formation by antitumor quinones. Free Radic. Biol. Med. 1989;6:63–101. doi: 10.1016/0891-5849(89)90162-7. [DOI] [PubMed] [Google Scholar]
- 4.Chen CH, Chern CL, Lin CC, Lu FJ, Shih MK, Hsieh PY, Liu TZ. Involvement of reactive oxygen species, but not mitochondrial permeability transition in the apoptotic induction of human SK-Hep-1 hepatoma cells by shikonin. Planta Med. 2003;69:1119–1124. doi: 10.1055/s-2003-45193. [DOI] [PubMed] [Google Scholar]
- 5.Li CJ, Wang C, Pardee AB. Induction of apoptosis by beta-lapachone in human prostate cancer cells. Cancer Res. 1995;55:3712–3715. [PubMed] [Google Scholar]
- 6.Srinivas P, Gopinath G, Banerji A, Dinakar A, Srinivas G. Plumbagin induces reactive oxygen species, which mediate apoptosis in human cervical cancer cells. Mol. Carcinog. 2004;40:201–211. doi: 10.1002/mc.20031. [DOI] [PubMed] [Google Scholar]
- 7.Sagar S, Green IR. Pro-apoptotic activities of novel synthetic quinones in human cancer cell lines. Cancer Lett. 2009;285:23–27. doi: 10.1016/j.canlet.2009.04.036. [DOI] [PubMed] [Google Scholar]
- 8.Huang L, Pardee AB. Beta-lapachone induces cell cycle arrest and apoptosis in human colon cancer cells. Mol. Med. 1999;5:711–720. [PMC free article] [PubMed] [Google Scholar]
- 9.Yoon Y, Kim YO, Lim NY, Jeon WK, Sung HJ. Shikonin an ingredient of Lithospermum erythrorhizon induced apoptosis in HL60 human premyelocytic leukemia cell line. Planta Med. 1999;65:532–535. doi: 10.1055/s-1999-14010. [DOI] [PubMed] [Google Scholar]
- 10.Sagar S, Kaur M, Minneman KP, Bajic VB. Anti-cancer activities of diospyrin, its derivatives and analogues. Eur. J. Med. Chem. 2010;45:3519–3530. doi: 10.1016/j.ejmech.2010.06.021. [DOI] [PubMed] [Google Scholar]
- 11.Kuo PL, Hsu YL, Cho CY. Plumbagin induces G2-M arrest and autophagy by inhibiting the AKT/mammalian target of rapamycin pathway in breast cancer cells. Mol. Cancer Ther. 2006;5:3209–3221. doi: 10.1158/1535-7163.MCT-06-0478. [DOI] [PubMed] [Google Scholar]
- 12.Aziz MH, Dreckschmidt NE, Verma AK. Plumbagin a medicinal plant-derived naphthoquinone is a novel inhibitor of the growth and invasion of hormone-refractory prostate cancer. Cancer Res. 2008;68:9024–9032. doi: 10.1158/0008-5472.CAN-08-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shih YW, Lee YC, Wu PF, Lee YB, Chiang TA. Plumbagin inhibits invasion and migration of liver cancer HepG2 cells by decreasing productions of matrix metalloproteinase-2 and urokinase- plasminogen activator. Hepatol. Res. 2009;39:998–1009. doi: 10.1111/j.1872-034X.2009.00540.x. [DOI] [PubMed] [Google Scholar]
- 14.Powolny AA, Singh SV. Plumbagin-induced apoptosis in human prostate cancer cells is associated with modulation of cellular redox status and generation of reactive oxygen species. Pharm Res. 2008;25:2171–2180. doi: 10.1007/s11095-008-9533-3. [DOI] [PubMed] [Google Scholar]
- 15.Kawiak A, Zawacka-Pankau J, Lojkowska E. Plumbagin induces apoptosis in her2-overexpressing breast cancer cells through the mitochondrial-mediated pathway. J. Nat. Prod. 2012;75:747–751. doi: 10.1021/np3000409. [DOI] [PubMed] [Google Scholar]
- 16.Sandur SK, Ichikawa H, Sethi G, Ahn KS, Aggarwal BB. Plumbagin (5-hydroxy-2-methyl-1,4-naphthoquinone) suppresses NF-kappaB activation and NF-kappaB-regulated gene products through modulation of p65 and IkappaBalpha kinase activation leading to potentiation of apoptosis induced by cytokine and chemotherapeutic agents. J. Biol. Chem. 2006;281:17023–17033. doi: 10.1074/jbc.M601595200. [DOI] [PubMed] [Google Scholar]
- 17.Manu KA, Shanmugam MK, Rajendran P, Li F, Ramachandran L, Hay HS, Kannaiyan R, Swamy SN, Vali S, Kapoor S, Ramesh B, Bist P, Koay ES, Lim LH, Ahn KS, Kumar AP, Sethi G. Plumbagin inhibits invasion and migration of breast and gastric cancer cells by downregulating the expression of chemokine receptor CXCR4. Mol. Cancer. 2011;10:107. doi: 10.1186/1476-4598-10-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun J, McKallip RJ. Plumbagin treatment leads to apoptosis in human K562 leukemia cells through increased ROS and elevated TRAIL receptor expression. Leuk. Res. 2011;35:1402–1408. doi: 10.1016/j.leukres.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Srinivas G, Annab LA, Gopinath G, Banerji A, Srinivas P. Antisense blocking of BRCA1 enhances sensitivity to plumbagin but not tamoxifen in BG-1 ovarian cancer cells. Mol. Carcinog. 2004;39:15–25. doi: 10.1002/mc.10164. [DOI] [PubMed] [Google Scholar]
- 20.Wang CC, Chiang YM, Sung SC, Hsu YL, Chang JK, Kuo PL. Plumbagin induces cell cycle arrest and apoptosis through reactive oxygen species/c-Jun N-terminal kinase pathways in human melanoma A375. 2 cells. Cancer Lett. 2008;259:82–98. doi: 10.1016/j.canlet.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Ahmad A, Banerjee S, Wang Z, Kong D, Sarkar FH. Plumbagin-induced apoptosis of human breast cancer cells is mediated by inactivation of NF-kappaB and Bcl-2. J. Cell Biochem. 2008;105:1461–1471. doi: 10.1002/jcb.21966. [DOI] [PubMed] [Google Scholar]
- 22.Lai L, Liu J, Zhai D, Lin Q, He L, Dong Y, Zhang J, Lu B, Chen Y, Yi Z, Liu M. Plumbagin inhibits tumour angiogenesis and tumour growth through the Ras signalling pathway following activation of the VEGF receptor-2. Br. J. Pharmacol. 2012;165:1084–1096. doi: 10.1111/j.1476-5381.2011.01532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen ZF, Tan MX, Liu LM, Liu YC, Wang HS, Yang B, Peng Y, Liu HG, Liang H, Orvig C. Cytotoxicity of the traditional chinese medicine (TCM) plumbagin in its copper chemistry. Dalton Trans. 2009:10824–10833. doi: 10.1039/b910133k. [DOI] [PubMed] [Google Scholar]
- 24.Chen ZF, Tan MX, Liu YC, Peng Y, Wang HH, Liu HG, Liang H. Synthesis characterization and preliminary cytotoxicity evaluation of five lanthanide(III)-plumbagin complexes. J. Inorg. Biochem. 2011;105:426–434. doi: 10.1016/j.jinorgbio.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Hazra B, Sarkar R, Bhattacharyya S, Ghosh PK, Chel G, Dinda B. Synthesis of plumbagin derivatives and their inhibitory activities against Ehrlich ascites carcinoma in vivo and Leishmania donovani Promastigotes in vitro. Phytother. Res. 2002;16:133–137. doi: 10.1002/ptr.867. [DOI] [PubMed] [Google Scholar]
- 26.Padhye S, Dandawate P, Yusufi M, Ahmad A, Sarkar FH. Perspectives on medicinal properties of plumbagin and its analogs. Med. Res. Rev. 2012;32 (6):1131–1158. doi: 10.1002/med.20235. [DOI] [PubMed] [Google Scholar]
- 27.Mathew R, Kruthiventi AK, Prasad JV, Kumar SP, Srinu G, Chatterji D. Inhibition of mycobacterial growth by plumbagin derivatives. Chem. Biol. Drug Des. 2010;76:34–42. doi: 10.1111/j.1747-0285.2010.00987.x. [DOI] [PubMed] [Google Scholar]
- 28.Xu KH, Lu DP. Plumbagin induces ROS-mediated apoptosis in human promyelocytic leukemia cells in vivo. Leuk. Res. 2010;34:658–665. doi: 10.1016/j.leukres.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 29.Hsu YL, Cho CY, Kuo PL, Huang YT, Lin CC. Plumbagin (5-hydroxy-2-methyl-1,4-naphthoquinone) induces apoptosis and cell cycle arrest in A549 cells through p53 accumulation via c-Jun NH2-terminal kinase-mediated phosphorylation at serine 15 in vitro and in vivo. J. Pharmacol. Exp. Ther. 2006;318:484–494. doi: 10.1124/jpet.105.098863. [DOI] [PubMed] [Google Scholar]
- 30.Jiang P, Du W, Wu M. P53 and Bad: Remote strangers become close friends. Cell Res. 2007;17:283–285. doi: 10.1038/cr.2007.19. [DOI] [PubMed] [Google Scholar]
- 31.Miyashita T, Krajewski S, Krajewska M, Wang HG, Lin HK, Liebermann DA, Hoffman B, Reed JC. Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene. 1994;9:1799–1805. [PubMed] [Google Scholar]
- 32.Sakao K, Fujii M, Hou DX. Acetyl derivate of quercetin increases the sensitivity of human leukemia cells toward apoptosis. Biofactors. 2009;35:399–405. doi: 10.1002/biof.53. [DOI] [PubMed] [Google Scholar]
- 33.Jennen DG, Magkoufopoulou C, Ketelslegers HB, van Herwijnen MH, Kleinjans JC, van Delft JH. Comparison of HepG2 and HepaRG by whole-genome gene expression analysis for the purpose of chemical hazard identification. Toxicol. Sci. 2010;115:66–79. doi: 10.1093/toxsci/kfq026. [DOI] [PubMed] [Google Scholar]
- 34.O'Brien PJ, Irwin W, Diaz D, Howard-Cofield E, Krejsa CM, Slaughter MR, Gao B, Kaludercic N, Angeline A, Bernardi P, Brain P, Hougham C. High concordance of drug-induced human hepatotoxicity with in vitro cytotoxicity measured in a novel cell-based model using high content screening. Arch Toxicol. 2006;80:580–604. doi: 10.1007/s00204-006-0091-3. [DOI] [PubMed] [Google Scholar]
- 35.Schoonen WG, de Roos JA, Westerink WM, Debiton E. Cytotoxic effects of 110 reference compounds on HepG2 cells and for 60 compounds on HeLa, ECC-1 and CHO cells.II mechanistic assays on NA (P)H ATP and DNA contents. Toxicol. In Vitro . 2005;19:491–503. doi: 10.1016/j.tiv.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 36.Schoonen WG, Westerink WM, de Roos JA, Debiton E. Cytotoxic effects of 100 reference compounds on Hep G2 and HeLa cells and of 60 compounds on ECC-1 and CHO cells.I mechanistic assays o ROS glutathione depletion and calcein uptake. . Toxicol. In Vitro. 2005; 19:505–516. doi: 10.1016/j.tiv.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 37.Van Summeren A, Renes J, Bouwman FG, Noben JP, van Delft JH, Kleinjans JC, Mariman EC. Proteomics investigations of drug-induced hepatotoxicity in HepG2 cells. Toxicol. Sci. 2011;120:109–122. doi: 10.1093/toxsci/kfq380. [DOI] [PubMed] [Google Scholar]
- 38.Tolosa L, Donato MT, Perez-Cataldo G, Castell JV, Gomez-Lechon MJ. Upgrading cytochrome P450 activity in HepG2 cells co-transfected with adenoviral vectors for drug hepatotoxicity assessment. Toxicol. In Vitro. 2012;26:1272–1277. doi: 10.1016/j.tiv.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 39.Gerets HH, Tilmant K, Gerin B, Chanteux H, Depelchin BO, Dhalluin S, Atienzar FA. Characterization of primary human hepatocytes, HepG2 cells, and HepaRG cells at the mRNA level and CYP activity in response to inducers and their predictivity for the detection of human hepatotoxins. Cell Biol. Toxicol. 2012;28:69–87. doi: 10.1007/s10565-011-9208-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thasni KA, Rakesh S, Rojini G, Ratheeshkumar T, Srinivas G, Priya S. Estrogen-dependent cell signaling and apoptosis in BRCA1-blocked BG1 ovarian cancer cells in response to plumbagin and other chemotherapeutic agents. Ann. Oncol. 2008;19:696–705. doi: 10.1093/annonc/mdm557. [DOI] [PubMed] [Google Scholar]
- 41.Tian L, Yin D, Ren Y, Gong C, Chen A, Guo FJ. Plumbagin induces apoptosis via the p53 pathway and generation of reactive oxygen species in human osteosarcoma cells. Mol. Med. Rep. 2012;5:126–132. doi: 10.3892/mmr.2011.624. [DOI] [PubMed] [Google Scholar]
- 42.Nazeem S, Azmi AS, Hanif S, Ahmad A, Mohammad RM, Hadi SM, Kumar KS. Plumbagin induces cell death through a copper-redox cycle mechanism in human cancer cells. Mutagenesis. 2009;24:413–418. doi: 10.1093/mutage/gep023. [DOI] [PubMed] [Google Scholar]
- 43.Qian Y. A comparative study of the two-dimensional echocardio- graphy ECG and selective coronary arteriography in detecting coronary artery disease. Zhonghua Xin Xue Guan Bing Za Zhi. 1991;19:345–347. [PubMed] [Google Scholar]
- 44.Yu L, Wan F, Dutta S, Welsh S, Liu Z, Freundt E, Baehrecke EH, Lenardo M. Autophagic programmed cell death by selective catalase degradation. Proc. Natl. Acad. Sci. USA. 2006;103:4952–4957. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat. Rev. Cancer. 2005;5:726–734. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 46.Hoyer-Hansen M, Bastholm L, Mathiasen IS, Elling F, Jaattela M. Vitamin D analog EB1089 triggers dramatic lysosomal changes and Beclin 1-mediated autophagic cell death. Cell Death Differ. 2005;12:1297–1309. doi: 10.1038/sj.cdd.4401651. [DOI] [PubMed] [Google Scholar]
- 47.Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa-Kobayashi S, Thompson CB, Tsujimoto Y. Role of Bcl-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat. Cell Biol. 2004;6:1221–1228. doi: 10.1038/ncb1192. [DOI] [PubMed] [Google Scholar]
- 48.Belmokhtar CA, Hillion J, Segal-Bendirdjian E. Staurosporine induces apoptosis through both caspase-dependent and caspase-independent mechanisms. Oncogene. 2001;20:3354–3362. doi: 10.1038/sj.onc.1204436. [DOI] [PubMed] [Google Scholar]
- 49.Belmokhtar CA, Hillion J, Dudognon C, Fiorentino S, Flexor M, Lanotte M, Segal-Bendirdjian E. Apoptosome-independent pathway for apoptosis.Biochemical analysis of APAF-1 defects and biological outcomes. J. Biol. Chem. 2003;278:29571–29580. doi: 10.1074/jbc.M302924200. [DOI] [PubMed] [Google Scholar]
- 50.Buchholz TA, Garg AK, Chakravarti N, Aggarwal BB, Esteva FJ, Kuerer HM, Singletary SE, Hortobagyi GN, Pusztai L, Cristofanilli M, Sahin AA. The nuclear transcription factor kappaB/bcl-2 pathway correlates with pathologic complete response to doxorubicin-based neoadjuvant chemotherapy in human breast cancer. Clin. Cancer Res. 2005;11:8398–8402. doi: 10.1158/1078-0432.CCR-05-0885. [DOI] [PubMed] [Google Scholar]
- 51.Li J, Shen Q, Peng R, Chen R, Jiang P, Li Y, Zhang L, Lu J. Plumbagin enhances TRAIL-mediated apoptosis through up-regulation of death receptor in human melanoma A375 cells. J. Huazhong Univ. Sci. Technolog. Med. Sci. 2010;30:458–463. doi: 10.1007/s11596-010-0449-x. [DOI] [PubMed] [Google Scholar]
- 52.Montoya J, Varela-Ramirez A, Estrada A, Martinez LE, Garza K, Aguilera RJ. A fluorescence-based rapid screening assay for cytotoxic compounds. Biochem. Biophys. Res. Commun. 2004;325:1517–1523. doi: 10.1016/j.bbrc.2004.10.196. [DOI] [PubMed] [Google Scholar]
- 53.Shieh JM, Chiang TA, Chang WT, Chao CH, Lee YC, Huang GY, Shih YX, Shih YW. Plumbagin inhibits TPA-induced MMP-2 and u-PA expressions by reducing binding activities of NF-kappaB and AP-1 via ERK signaling pathway in A549 human lung cancer cells. Mol. Cell Biochem. 2010;335:181–193. doi: 10.1007/s11010-009-0254-7. [DOI] [PubMed] [Google Scholar]
- 54.Lupertz R, Watjen W, Kahl R, Chovolou Y. Dose- and time-dependent effects of doxorubicin on cytotoxicity, cell cycle and apoptotic cell death in human colon cancer cells. Toxicology. 2010;271:115–121. doi: 10.1016/j.tox.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 55.Di Leonardo A, Linke SP, Clarkin K, Wahl GM. DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev. 1994;8:2540–2551. doi: 10.1101/gad.8.21.2540. [DOI] [PubMed] [Google Scholar]
- 56.Sliwinska MA, Mosieniak G, Wolanin K, Babik A, Piwocka K, Magalska A, Szczepanowska J, Fronk J, Sikora E. Induction of senescence with doxorubicin leads to increased genomic instability of HCT116 cells. Mech. Ageing Dev. 2009;130:24–32. doi: 10.1016/j.mad.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 57.Basu A, Haldar S. The relationship between BcI2 Bax and p53: consequences for cell cycle progression and cell death. Mol. Hum. Reprod. 1998;4:1099–1109. doi: 10.1093/molehr/4.12.1099. [DOI] [PubMed] [Google Scholar]
- 58.Moll UM, Petrenko O. The MDM2-p53 interaction. Mol. Cancer Res. 2003;1:1001–1008. [PubMed] [Google Scholar]
- 59.Gomathinayagam R, Sowmyalakshmi S, Mardhatillah F, Kumar R, Akbarsha MA, Damodaran C. Anticancer mechanism of plumbagin, a natural compound, on non-small cell lung cancer cells. Anticancer Res. 2008;28:785–792. [PubMed] [Google Scholar]
- 60.Tian L, Yin D, Ren Y, Gong C, Chen A, Guo FJ. Plumbagin induces apoptosis via the p53 pathway and generation of reactive oxygen species in human osteosarcoma cells. Mol. Med. Report. 2012;5:126–132. doi: 10.3892/mmr.2011.624. [DOI] [PubMed] [Google Scholar]
- 61.Ryan KM, Ernst MK, Rice NR, Vousden KH. Role of NF-kappaB in p53-mediated programmed cell death. Nature. 2000;404:892–897. doi: 10.1038/35009130. [DOI] [PubMed] [Google Scholar]
- 62.Dixit V, Mak TW. NF-kappaB signaling.Many roads lead to madrid. Cell. 2002;111:615–619. doi: 10.1016/s0092-8674(02)01166-2. [DOI] [PubMed] [Google Scholar]
- 63.Sand JM, Bin Hafeez B, Jamal MS, Witkowsky O, Siebers EM, Fischer J, Verma AK. Plumbagin (5-hydroxy-2-methyl-1 4-naphthoquinone) isolated from Plumbago zeylanica inhibits ultraviolet radiation-induced development of squamous cell carcinomas. Carcinogenesis. 2012;33:184–190. doi: 10.1093/carcin/bgr249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang E, Zha J, Jockel J, Boise LH, Thompson CB, Korsmeyer SJ. Bad a heterodimeric partner for Bcl-XL and Bcl-2, displaces Bax and promotes cell death. Cell. 1995;80:285–291. doi: 10.1016/0092-8674(95)90411-5. [DOI] [PubMed] [Google Scholar]
- 65.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s web site along with the published article.