Abstract
Arabidopsis thaliana is one of the most popular experimental plants. However, only 40% of its genes have at least one experimental Gene Ontology (GO) annotation assigned. Systematic observation of mutant phenotypes is an important technique for elucidating gene functions. Indeed, several large-scale phenotypic analyses have been performed and have generated phenotypic data sets from many Arabidopsis mutant lines and overexpressing lines, which are freely available online. Since each Arabidopsis mutant line database uses individual phenotype expression, the differences in the structured term sets used by each database make it difficult to compare data sets and make it impossible to search across databases. Therefore, we obtained publicly available information for a total of 66,209 Arabidopsis mutant lines, including loss-of-function (RATM and TARAPPER) and gain-of-function (AtFOX and OsFOX) lines, and integrated the phenotype data by mapping the descriptions onto Plant Ontology (PO) and Phenotypic Quality Ontology (PATO) terms. This approach made it possible to manage the four different phenotype databases as one large data set. Here, we report a publicly accessible web-based database, the RIKEN Arabidopsis Genome Encyclopedia II (RARGE II; http://rarge-v2.psc.riken.jp/), in which all of the data described in this study are included. Using the database, we demonstrated consistency (in terms of protein function) with a previous study and identified the presumed function of an unknown gene. We provide examples of AT1G21600, which is a subunit in the plastid-encoded RNA polymerase complex, and AT5G56980, which is related to the jasmonic acid signaling pathway.
Keywords: Arabidopsis, Database, Gene function, Mutant line, Ontology, Phenotype
Introduction
Arabidopsis thaliana is one of the most commonly used experimental plants, and many techniques, tools and detailed genomic data are available for working with it. Many studies have been performed on this species. The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org/) provides a compilation of such information and has enhanced gene annotations by literature curation (Lamesch et al. 2012). TAIR provides controlled vocabulary annotations for Arabidopsis genes, with Gene Ontology (GO) and Plant Ontology (PO) annotations (Cooper et al. 2013) including both experimental and non-experimental evidence (Li et al. 2012). However, despite the efforts of a large number of researchers, only 40% of genes have been assigned at least one experimental GO annotation (Li et al. 2012). To enhance gene function studies, new methods are required to facilitate our understanding of the plant genome.
One direct method for investigating gene function is to examine and characterize the phenotypic changes associated with loss-of-function gene mutations. Insertional mutagenesis with the Activator (Ac)/Dissociation (Ds) transposon system makes it possible to generate mutants with a high proportion of single-copy transposon insertions. This system requires the production of a large number of mutant lines to obtain genome-wide coverage. Nonetheless, the single insertion site in each line can be easily determined, thereby simplifying the production and subsequent genetic analysis of single-gene knockout series (Fedoroff and Smith 1993, Sundaresan et al. 1995, Martienssen 1998). On the other hand, gain-of-function mutational analyses provide a separate set of tools that can be used to dissect the functions of genes, especially those with functional redundancy, as found in gene families. Thus, gain-of-function mutants may represent a different spectrum of mutants that have not been isolated as conventional loss-of-function mutants (Nakazawa et al. 2003). In contrast to loss-of-function mutants that cause a recessive phenotype, gain-of-function mutants behave in a dominant manner in the T1 generation (Weigel et al. 2000, Bouché and Bouchez 2001). One example of large-scale phenotypic analysis of plant lines generated for gain-of-function mutant screening is the full-length cDNA overexpressor (FOX) gene-hunting system, which is a novel alternative activation tagging technology that uses full-length cDNAs (fl-cDNAs) (Ichikawa et al. 2006). In Arabidopsis, it is possible to conduct gene-based large-scale phenotype analysis from gain-of-function studies using the FOX hunting system (Ichikawa et al. 2006) and to perform loss-of-function studies by saturation mutagenesis (Parinov and Sundaresan 2000).
Several large-scale phenotypic analyses have recently been undertaken, and phenotypic data sets from many Arabidopsis mutant lines and overexpressing lines have been made freely available online (Kuromori et al. 2009). One of these databases, the RIKEN Arabidopsis Phenome Information Database (RAPID; http://rarge.psc.riken.jp/phenome/), houses a total of 4,000 transposon insertion lines, each of which contains a homozygous Ds transposon mutation in a gene or promoter region; these genetic lines have been examined systematically for visible phenotypes at various growth stages of the plant (Kuromori et al. 2006). Another database, the Trapper Database (http://genetrap.cshl.org/) from Cold Spring Harbor Laboratory, contains data from 16,000 lines that each carries a unique insertion of a gene trap (GT) or enhancer trap (ET) transposable Ds element that both disrupts gene function and can be used to monitor gene expression (Sundaresan et al. 1995, Martienssen 1998). The Arabidopsis FOX (http://nazunafox.psc.database.riken.jp/) and RiceFOX (http://ricefox.psc.riken.jp/) at RIKEN contain a total of 14,000 and 18,000 plants, respectively, that overexpress introduced fl-cDNAs from Arabidopsis and rice based on the FOX hunting system (Ichikawa et al. 2006, Kondou et al. 2009, Sakurai et al. 2011).
Although the Arabidopsis mutant line databases that include phenotypic information are useful, the use of uniquely structured term sets to describe phenotypes in each database makes comparisons of phenotypes among the databases difficult and performing searches across the databases impossible. Therefore, the integration of such databases with a controlled structured vocabulary (ontology) is an effective way to enrich the gene annotation. Furthermore, mapping phenotypic descriptions onto ontology terms may enable the comparison of phenotypes across different species as well as among genes of a single species (Walls et al. 2012). In fact, two databases doing just that have been constructed and made publicly available: PO (http://www.plantontology.org/), which includes information on plant anatomy and development stage (Cooper et al. 2013), and phenotypic quality ontology (PATO; http://obofoundry.org/wiki/index.php/PATO:Main_Page), which includes phenotypic annotations (Mungall et al. 2010).
Here, we describe the integration of the phenotypes of four Arabidopsis mutant lines (including two loss-of-function and two gain-of-function lines) by mapping descriptions into PO and PATO. We have developed an updated version of the RIKEN Arabidopsis Genome Encyclopedia (RARGE), a database of fl-cDNAs and Ds transposon mutant lines (Sakurai et al. 2005), which has been designated RARGE II.
Results and Discussion
Data sets
Mutant line data including flanking sequence information, phenotype description in text form and images of the mutant lines were obtained from four different sources: 17,198 RIKEN Arabidopsis Ds transposon mutant (RATM) lines (Kuromori et al. 2004, Kuromori et al. 2006), 16,337 Cold Spring Harbor Laboratory Arabidopsis Gene trap mutant (TRAPPER) lines (Springer et al. 1995, Martienssen 1998), 14,069 RIKEN Arabidopsis fl-cDNA overexpressed Arabidopsis (AtFOX) lines (Kondou et al. 2010) and 18,605 RIKEN rice full-length cDNA overexpressed Arabidopsis (OsFOX) lines (Kondou et al. 2009, Sakurai et al. 2011) (Table 1).
Table 1.
Sources of data for the mutant lines
| Source | No. of total lines | URL |
|---|---|---|
| RIKEN Arabidopsis Ds transposon mutant lines | 17,198 | http://rarge.psc.riken.jp/ |
| Cold Spring Harbor Laboratory Arabidopsis gene trap mutant lines | 16,337 | http://genetrap.cshl.org/ |
| RIKEN Arabidopsis full-length cDNA overexpressed Arabidopsis lines | 14,069 | http://nazunafox.psc.database.riken.jp/ |
| RIKEN rice full-length cDNA overexpressed Arabidopsis lines | 18,605 | http://ricefox.psc.riken.jp/ |
| Total | 66,209 |
Deduction of disrupted and induced genes
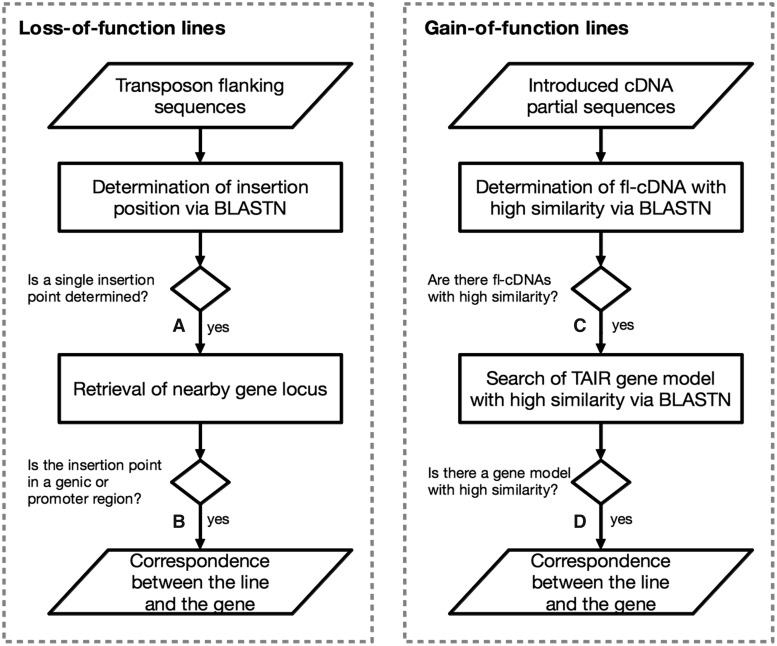
We employed different methods to deduce the genes that were disrupted and induced in the loss-of-function and gain-of-function mutant lines, respectively. Fig. 1 illustrates our workflow to deduce the disrupted and induced genes for each mutant type.
Fig. 1.
Annotation workflow of loss- and gain-of-function lines. To deduce disrupted and induced genes, we employed independent methods for loss- and gain-of-function lines. For loss-of-function lines (A), we determined a single transposon insertion point and then (B) deduced the genes in which the transposon was inserted into the gene or promoter region. For gain-of-function lines (C), we deduced the introduced full-length cDNAs and then (D) searched for highly similar gene models. Detailed values at each step are described in Tables 1 and 2.
For loss-of-function lines, the single genomic insertion point was determined using a similarity search of transposon-flanking sequences against the TAIR10 Arabidopsis whole-genome sequence (Lamesch et al. 2012) using BLASTN (Altschul et al. 1997) (Fig. 1A); then, gene loci in which the determined insertion point was in the gene or promoter region (Fig. 1B) were retrieved to define disrupted genes. We found that 15,690 (92.8%) of the 17,198 RATM lines and 8,540 (52.3%) of the 16,337 TRAPPER lines had a single insertion point in the genome. Among the loss-of-function lines, 72.2% were determined to have a single transposon insertion point, suggesting that the data set was suitable for this study. Finally, gene loci were retrieved from 13,294 (77.3%) of the RATM lines and 7,184 (44.0%) of the TRAPPER lines (Table 2).
Table 2.
Disrupted genes in the loss-of-function lines
| RATM | TRAPPER | |
|---|---|---|
| Total lines | 17,198 (139) | 16,337 (1,409) |
| (A) Lines determined to have a single insertion point | 15,690 (139) | 8,540 (884) |
| (B) Lines with an insertion in a gene or promoter region | 13,294 (139) | 7,184 (773) |
Introduced fl-cDNAs of gain-of-function lines were determined via a similarity search of partial reading sequences against fl-cDNA sequences using BLASTN (Altschul et al. 1997) (Fig. 1C), and the induced genes were deduced via a similarity search of fl-cDNA sequences against the gene models of TAIR10 (Fig. 1D). OsFOX lines are mutant lines into which rice fl-cDNAs were introduced but, for comparison with the other Arabidopsis mutant lines in this study, a TAIR gene with high similarity was defined as the induced gene. We found that 8,365 (59.5%) of a total of 14,069 AtFOX lines and 11,578 (62.2%) of a total of 18,605 OsFOX lines had introduced fl-cDNAs. Induced genes were deduced in 8,357 (59.4%) of the AtFOX and 10,012 (53.8%) of the OsFOX lines (Table 3). The AtFOX and OsFOX lines contained an average of 2.6 and 1.11 fl-cDNA clones, respectively, per line (Ichikawa et al. 2006, Kondou et al. 2009). In the cases in which multiple fl-cDNAs were detected, we assumed that all fl-cDNAs were overexpressed.
Table 3.
Overexpressed genes in gain-of-function lines
| AtFOX | OsFOX | |
|---|---|---|
| Total lines | 14,069 (3,429) | 18,605 (5,353) |
| (C) Lines determined to have introduced fl-cDNAs | 8,365 (1,670) | 11,578 (2,940) |
| (D) Lines deduced to have induced genes | 8,357 (1,668) | 10,012 (2,555) |
Next, we calculated the number of the genes corresponding to these lines. The RATM and the TRAPPER lines showed 9,424 (28.3%) and 5,628 (16.9%) disrupted genes, respectively, and the AtFOX and the OsFOX lines showed 2,976 (8.9%) and 4,177 (12.5%) induced genes, respectively. Consequently, data sets were obtained in which loss-of-function lines had 12,558 (37.7%) disrupted genes and gain-of-function lines had 6,382 (19.2%) induced genes. There were 16,015 (48.1%) genes that were disrupted or induced by either loss-of-function or gain-of-function mutations, respectively, and 2,925 (8.8%) genes that were disrupted and induced by both loss-of-function and gain-of-function mutations, respectively (Table 4).
Table 4.
Summary of the genes disrupted or induced in loss- and/or gain-of-function lines
| Loss-of-function |
Gain-of-function |
|||
|---|---|---|---|---|
| RATM | TRAPPER | AtFOX | OsFOX | |
| Genes disrupted or induced per resource | 9,424 (159) | 5,628 (849) | 2,976 (1,176) | 4,177 (1,870) |
| Genes disrupted or induced per mutant type | 12,558 (990) | 6,382 (2,906) | ||
| Genes disrupted or induced in loss- or gain-of-function lines | 16,015 (3,974) | |||
| Genes disrupted and induced in loss- and gain-of-function lines | 2,925 (102) | |||
The numbers of genes disrupted in loss-of-function lines and/or overexpressed in gain-of-function lines are shown.
The numbers in parentheses indicate the number of lines showing any of the observable phenotypes.
Mapping of phenotype descriptions to ontology terms
All four resources used in this study include data obtained from the observation of plants. To integrate these phenotypic data, we mapped each phenotype description to a pair of PO and PATO terms (Supplementary Tables S1–S4).
Although PO is structured as a multilevel hierarchy, we expressed information on each tissue or developmental stage using up to two PO level terms. As a result, the phenotypic descriptions in the data set were mapped to PO terms organized within 12 classes [whole plant (PO:0000003), seedling (PO:0008037), root (PO:0009005), vascular leaf (PO:0009025), stem (PO:0009047), flower (PO:0009046), fruit (PO:0009001), seed (PO:0009010), trichome (PO:0000282), 0 seed germination stage (PO:0007057), whole plant flowering stage (PO:0007016) and sporophyte vegetative stage (PO:0007134)] and 10 subclasses [cotyledon (PO:0020030), hypocotyl (PO:0020100), cauline leaf (PO:0000013), rosette leaf (PO:0000014), stipule (PO:0020041), stem internode (PO:0020142), petal (PO:0009032), sepal (PO:0009031), stamen (PO:0009029) and gynoecium (PO:0009062)]. Note that throughout the paper, ontology terms are in italics. PATO terms were organized into 39 classes (Supplementary Table S5). For example, cauline leaf, rosette leaf, and stipule belong to vascular leaf as a higher class.
The numbers of mutant lines observed with phenotypes mapped onto ontology terms are summarized in correspondence with the workflow shown in Fig. 1. Phenotypes were observed for 139 of the RATM lines; all of these were determined as single insertion points of the transposon into the gene or promoter region (Table 2). Phenotypes were observed for 1,409 of the TRAPPER lines, 884 of which were determined to have single insertion points, and the disrupted genes were deduced for 773 of these (Table 2). Phenotypes were observed for 3,429 and 5,353 of the AtFOX and OsFOX lines, respectively, with 1,670 and 2,940 of these lines, respectively, found to contain introduced fl-cDNAs with high similarity. Highly similar Arabidopsis gene models were assigned to 1,668 and 2,555 of the AtFOX and OsFOX lines, respectively (Table 3).
Subsequently, we calculated the number of genes in the same manner as described in the previous section. A total of 159 and 849 of the genes obtained from the RATM and TRAPPER lines, respectively, showed phenotypes by gene disruption; thus, 990 of the genes obtained from the loss-of-function lines had disrupted genes. Similarly, 1,176 and 1,870 genes obtained from the AtFOX and OsFOX lines, respectively, showed phenotypes related to the introduction of fl-cDNAs; thus, 2,906 of the obtained gene gain-of-function lines had these introductions. A total of 3,794 genes showed mutant phenotypes because of either gene disruption or induction; 102 genes showed mutant phenotypes attributable to both gene disruption and induction (Table 4; Supplementary Table S6).
According to Kuromori et al. (2009), many mutants have been assigned as having epinastic leaves in phenotypic data sets of activation-tagged lines, whereas only hyponastic leaf mutants have been registered for transposon-tagged lines (Ichikawa et al. 2003, Kuromori et al. 2006). We found similar results in that many gain-of-function lines were detected using the PO and PATO terms leaf::epinastic (PATO:0000945), cauline leaf::epinastic and rosette leaf::epinastic (note: throughout the paper, pairs of PO and PATO terms are joined with a double colon), whereas very few loss-of-function lines were detected by the terms rosette leaf::epinastic (Supplementary Table S5). As another example, several mutants had overgrowth phenotypes (taller plants or high fertility) in the activation-tagged lines, whereas very few such mutants were found in transposon-tagged lines (Ichikawa et al. 2003, Kuromori et al. 2006); several gain-of-function lines were detected by the term whole plant::increased height (PATO:0000570), whereas no loss-of-function lines were detected with this term (Supplementary Table S5). Several gain-of-function lines, but no loss-of-function lines, were detected by the term root::present in greater numbers in organism (PATO:0000470), whereas several loss-of-function lines, but no gain-of-function lines, were detected by the term root::present in fewer numbers in organism (Supplementary Table S5). These examples suggest that some opposite propensities found to date may have depended on the mutation type, such as loss of function and gain of function, rather than the gene category.
Web interface
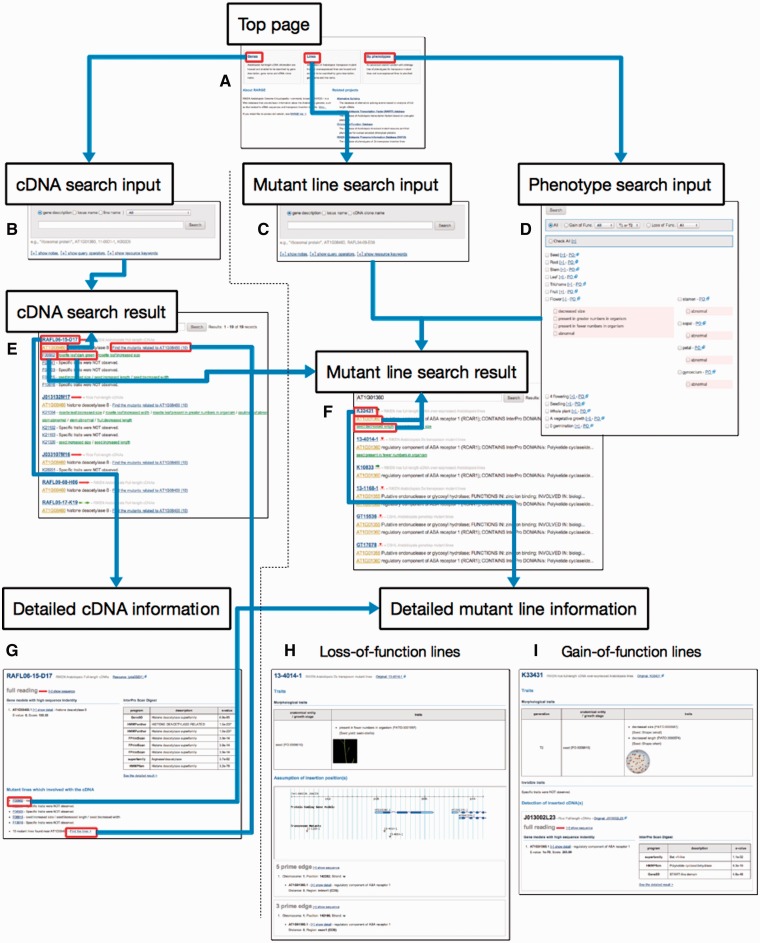
We developed a publicly accessible web-based database, RARGE II (http://rarge-v2.psc.riken.jp/). All data from this study are housed in the database, which provides multifaceted search functions and enables browsing of all mutant lines and phenotype data without restrictions via the Internet using a modern web browser. It was designed to allow seamless, user-friendly viewing of mutant lines and fl-cDNAs (Fig. 2). Users can choose the intended resource, fl-cDNAs or mutant lines, and/or can find mutant lines by phenotype using the ontology tree (Fig. 2A).
Fig. 2.
Operation workflow of the web-based database. (A) On the top page, users can select the desired resource, fl-cDNAs or mutant lines, or can find mutant lines by phenotype using the ontology tree. Then they can (B) search fl-cDNAs by keywords with filtering from the fl-cDNA search input page, (C) search for mutant lines by keywords with filtering from the mutant line search input page and/or (D) search all mutant lines by selecting phenotype items in the alternative mutant line search input page by phenotype tree. (E) The fl-cDNA search result page shows records that include the fl-cDNA clone name, gene locus name, gene description, the number of mutant lines with disruption or induction of the gene and the mutant lines into which the fl-cDNA was introduced. (F) The mutant line search result page shows records that include the line name, resource name, deduced disrupted or induced gene locus name, gene description and phenotypes observed. (G) The detailed fl-cDNA information page shows the fl-cDNA sequence, its InterPro Scan result, gene models with high degrees of similarity and the names of mutant lines into which the cDNA was introduced. (H) The detailed mutant line information page for the loss-of-function lines shows the visible phenotypes, including the original phenotype description, mapped PO information and PATO information, line photographs and deduced transposon insertion point. (I) The detailed mutant line information page for the gain-of-function lines shows the visible and invisible phenotypes including the original phenotype description, mapped PO information and PATO information, line photographs, the fl-cDNA sequence, its InterPro Scan result and gene models with high degrees of similarity.
Users can search for specific mutant lines by gene description, gene locus name or line name, and can also filter results by selecting the resource name (RATM, TRAPPER, AtFOX and OsFOX) or mutant type (loss-of-function lines and gain-of-function lines) on the mutant search input page (Fig. 2C). The search results page shows the records, including line name, resource name, deduced disrupted or induced gene locus name, gene description and phenotypes observed, making it possible to locate the desired lines easily (Fig. 2F). In addition, gene locus name and phenotypes are linked to the mutant search results queried by the term as the search keyword (Fig. 2F). Each mutant line name in the results is linked to detailed mutant information. A detailed information page shows phenotypes of loss-of-function (Fig. 2H) and gain-of-function lines (Fig. 2I). Phenotype information shown in a table includes the original phenotype description, mapped PO and PATO information and photographs of the mutant line (Fig. 2H, I). With the loss-of-function lines, the flanking sequences, determined insertion point and genome map around the point are shown, allowing the user to deduce the disrupted genes (Fig. 2H). With the gain-of-function lines, the introduced fl-cDNA details are listed on the information page (Fig. 2I).
Users can search all mutant lines by selecting phenotype terms that describe PO and PATO in the tree (Fig. 2D). The terms of only 12 classes of the paired PO are listed and all trees are collapsed. Clicking ‘+’ next to the PO description causes the tree to expand, showing the PO subclasses and PATO (Fig. 2D). Selecting terms by using the checkboxes and performing a search will show search results similar to those of a mutant line search, as described above. In addition, the results can be filtered by selecting a mutant type, a resource name and/or the generation of overexpressed plant (Fig. 2D).
Users can also search fl-cDNAs by gene description, gene locus name and/or fl-cDNA clone name (Fig. 2B). The search results include the records, fl-cDNA clone name, gene locus name, gene description and number of mutant lines with disruption or induction of the gene, especially mutant lines into which an fl-cDNA has been introduced, which are listed with the phenotypes. In addition, the gene locus name and number of mutant lines in which the gene is disrupted or induced are linked to the fl-cDNA search results and mutant search results, respectively, and can be queried by terms such as search keywords (Fig. 2E). Each fl-cDNA clone name in the search results links to detailed fl-cDNA information including the fl-cDNA sequence, its InterPro Scan result, gene models with high similarity, names of mutant lines with the introduced cDNA (that link to the detailed mutant information page) and the number of mutant lines with disrupted or induced gene models that link to the mutant search result page.
The information in our database is widely available to a large number of researchers and will provide the basis for a variety of research projects that rely on large-scale Arabidopsis information. The RARGE II database is easy to use and provides additional important information to the plant biology community, enabling searches for visible phenotypes of genes of interest.
Comparison of phenotypes among mutant lines with disruption and/or induction of the same gene
Among the loss-of-function lines, we first examined multiple lines in which the mutations were disrupting the same gene. A total of 71 genes had multiple mutant alleles with loss-of-function phenotypes, 45 of which (63%) had at least two alleles showing the same phenotypes (Supplementary Table S7). The most common phenotype was seed::present in fewer numbers in organism (93%), followed by whole plant::decreased height. On the other hand, when we analyzed multiple mutant lines in which the same gene was induced in gain-in-function lines, 864 genes were obtained, 421 of which (48.7%) showed the same phenotype in more than two independent lines (Supplementary Table S8). Based on past experience, we know that not all individuals in which the same gene was introduced show the same phenotype; in the laboratory, when we investigated the effects of gene transfer, we created many individual overexpressed plants. This study indicated that approximately 50% of genes show gain-of-function phenotypes confirmed by multiple lines; however, the other half seemed to be pseudo-phenotypes.
We wondered what proportion of the mutant lines with related phenotypes among the loss- and gain-of-function lines were affected by the same gene. We compared phenotypes related to 102 of the genes obtained from both loss- and gain-of-function lines that exhibited phenotypes (Supplementary Table S6). In previous phenotype analyses, the fl-cDNAs of desired genes were introduced individually into wild-type plants in both sense and antisense directions. In many instances, the phenotypes of the sense and antisense transgenic lines were discovered to be opposite to those of the gain-of-function and loss-of-function mutants, respectively. In this study, we did not find any exactly opposite phenotypes among loss- and gain-of-function lines except for leaf color. Quantitative traits such as size and length of leaves and roots are caused by multiple factors including the gene expression level. Therefore, these mutant lines had the potential to show other phenotypes even if a disrupted gene was related to a quantitative trait. In other words, it is difficult to define ‘opposite’ and ‘same’ phenotypes. For example, the opposite term for ‘variegated’ cannot be defined as a mutant phenotype because it is likely to be the wild type. The opposite term of ‘large’ is simply ‘small’. However, other phenotypes such as ‘notched’, ‘wrinkled’ and ‘curled’ may appear as opposite phenotypes. The integration of phenotype information in this study does provide a unified phenotype description and a comfortable information environment for deduction of the functions of more unknown proteins. On the other hand, the difficulty in interpreting phenotype descriptions remains.
We sought to determine whether the data set provides new insights for elucidating the functions of genes annotated as ‘unknown protein’. Here, we first present an example that adds supporting information to a previous study on known protein function and then an example of deduction of the function of a gene annotated as an unknown protein.
Example consistent with a previous study on known protein function. The AT1G21600 gene is disrupted and induced in the RATM line 15-0303-1 and OsFOX line K06951, which were obtained as a loss-of-function line and a gain-of-function line, respectively (Supplementary Table S6). The protein product, AT1G21600, is a subunit of the plastid-encoded RNA polymerase complex (PTAC6/PAP8), and homozygous seedlings of this line develop white cotyledons, fail to accumulate Chl even under conditions of low light intensity and do not produce primary leaves (Pfalz et al. 2006). The line with a transposon inserted into AT1G21600, 15-0303-1, showed an albino value phenotype in our data set.
K06951 is an overexpressed line in which the rice fl-cDNA AK102323 was introduced, which is similar to the Arabidopsis AT1G21600 gene. To the best of our knowledge, there have been no studies regarding overexpression of the AT1G21600 gene. Our data for K06951 include several phenotypic descriptions of the cauline leaves, rosette leaves and stem. We found a change in the rosette leaf color to dark green, strongly suggesting a relationship between loss-of-function and gain-of-function phenotypes. Analysis of K06951 would help to elucidate the function of AT1G21600 further.
Example deducing the function of a gene annotated as an unknown protein. First, we defined an unknown protein as a protein encoded by a gene that does not have a high degree of similarity to well-known genes and does not have known protein functions, including interaction, localization and enzyme activity. The AT5G56980 gene was found to encode an unknown protein that is disrupted and induced in the lines GT4800 and F21325, which were obtained as a loss-of-function line and a gain-of-function line, respectively (Supplementary Table S6). GT4800 is a line with a transposon insertion into AT5G56980 and shows various phenotypes, such as fewer numbers of seeds, decreased plant height and seedling lethality. F21325 is the overexpressed line with introduction of Arabidopsis fl-cDNA AF385692, corresponding to AT5G56980; it showed an abnormal morphological feature, i.e. rosette leaves.
The AT5G56980 gene was identified as a pathogen-associated molecular pattern-induced gene (A70), which is rapidly induced in systemic tissue after challenge with Pseudomonas syringae pv. tomato DC3000(avrRpm1) but not with the compatible DC3000 strain (Truman et al. 2007). Although AT5G56980 expression was found to be responsive to jasmonic acid (JA) and wounding, but unaffected by heat, cold or salicylic acid treatment, the function of the protein product of AT5G56980 is unclear (Truman et al. 2007). An example of the JA-related gene, coi1, is a principal component of a JA receptor in Arabidopsis and other plants (Xie et al. 1998, Katsir et al. 2008, Yan et al. 2009), and OsCOI1-RNAi (RNA interference) plants show increased plant height and cellular elongation (Yang et al. 2012). If JA is normally controlled via negative feedback by AT5G56980, the GT4800 phenotype whole plant::decreased height would be consistently described inversely with OsCOI1-RNAi plants. If the above assumptions are correct, further analyses of GT4800 and F21325 would be worthwhile.
Perspective
We integrated phenotype information using ontologies from four large-scale mutant databases. As the results of this strategy helped to deduce the function of a gene annotated as an unknown protein, the integration of additional data regarding mutant phenotypes may facilitate deduction of the functions of more unknown proteins. The majority of Arabidopsis insertion mutant lines are transfer DNA-tagged (T-DNA-tagged) lines. However, a systematic phenotype analysis of such lines has not been available to date. Systematic phenotype analysis targeting T-DNA-tagged lines would markedly improve the accuracy of deduction of unknown protein functions. Other types of phenotypic information are also important. For example, the Chloroplast Function Database focuses on nuclear-encoded chloroplast proteins (Myouga et al. 2010, Myouga et al. 2013), and the RiceFOX database houses invisible phenotype information such as photosynthesis activity, plant hormone accumulation and stress sensitivity (Sakurai et al. 2011). The Plant Organelles Database (Mano et al. 2014) contains an electron micrograph database and an organelles database for various plant species and mutant lines. Comprehensive analyses of metabolites and plant hormones (metabolomics and hormonomics) have become a rapidly developing research field in recent years. For example, comprehensive measurement data for metabolites and plant hormones are publicly available in PRIMe (http://prime.psc.riken.jp/) (Akiyama et al. 2008, Sakurai et al. 2013) and UniVIO (http://univio.psc.riken.jp/) (Kudo et al. 2013), respectively. A database of the relationships between species and metabolites has been developed in KNApSAcK (http://kanaya.naist.jp/KNApSAcK_Family/) (Afendi et al. 2012, Nakamura et al. 2013). We expect that invisible phenotypes will be important resources for mutant analysis in the future. It is significant that the information resources stated above are appended to the RARGE II database. On the other hand, an approach to extract phenotypic descriptions manually from the literature has been developed by TAIR (Li et al. 2012). In addition, the achievements of our exhaustive research and many individual studies reported in the literature should be integrated. Furthermore, although we used only visible phenotypes in this study, we will integrate invisible phenotypes such as chemical and biological phenotypes in future studies as more data on these phenotypes become available every year.
Materials and Methods
Data sets
The data sets of mutant line information were obtained from the public databases detailed in Table 1. RATM, AtFOX and OsFOX data were obtained directly from each database. TRAPPER data were obtained using Web Crawler. The PO data file (version #19) was obtained from http://palea.cgrb.oregonstate.edu/viewsvn/Poc/tags/live/plant_ontology.obo. The PATO data file [21:05:2013/14:51 (date/time last created)] was obtained from http://pato.googlecode.com/svn/trunk/quality.obo.
Deduction of disrupted genes in loss-of-function lines
Transposon flanking sequences were subjected to a BLASTN search against the Arabidopsis genome sequence from the TAIR10 genome release (Lamesch et al. 2012). The position showing the highest similarity score with an E-value <1e-15 was defined as an insertion point. When all insertion points of multiple flanking sequences were mapped within 200 bp, it was defined as a single insertion point. When a transposon was inserted into a gene or promoter region, we picked the gene locus name. The 1,000 bp of upstream sequence from each transcriptional start site was defined as the promoter region. When a transposon was inserted into the gene or promoter regions of multiple genes, both genes were selected.
Deduction of induced genes in gain-of-function lines
The partial sequences of the introduced fl-cDNAs were subjected to a BLASTN search against Arabidopsis fl-cDNA sequences (Seki et al. 2002) or rice fl-cDNA sequences (Kikuchi et al. 2003). The fl-cDNA showing the highest similarity score was defined as an introduced fl-cDNA. The sequences of introduced fl-cDNAs were subsequently subjected to a BLAST search against the Arabidopsis gene model sequences from the TAIR10 genome release (Lamesch et al. 2012). We picked the gene model name showing the highest degree of similarity and an E-value <1e-5.
Mapping of phenotype description to ontology
We manually mapped the original phenotype descriptions to ontology terms paired with PO (Cooper et al. 2013) and PATO (Gkoutos et al. 2005) based on recorded observations about each line as well as photographs and the literature. For example, the phenotype descriptions ‘Seed yield::low’, ‘Seed yield::semi sterile’ and ‘Seed yield::sterile’ in AtFOX; ‘Seed::Number::few’ in OsFOX; ‘Seed yield::low yield’, ‘Seed yield::semi-sterile’ and ‘Seed yield::sterile’ in RATM; and ‘conditional male sterile’, ‘embryo lethal’, ‘semi-sterile’, ‘sterile’ and ‘very few seeds’ in TRAPPER were mapped to the PO term seed (PO:0009010) and the PATO term present in fewer numbers in organism (PATO:0001997).
Database construction
The integrated phenotypic data were stored in the database and implemented in the web application. All programs for manipulating data, such as those used to query the database and generate web pages, were written in Perl (http://www.perl.org/) with the web application framework Catalyst (http://www.catalystframework.org/) and the web server interface PSGI/Plack (http://plackperl.org/). The relational database management system was MySQL (http://www.mysql.com/). This web application system is capable of running on a UNIX-like operating system such as Linux (http://www.linux.org/). The web pages were written in HTML, cascading style sheet and Javascript with the YUI library (http://yuilibrary.com/).
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the Japan Society for the Promotion of Science [a Grant-in-Aid for Young Scientists (B) (18700106 to T.S.)].
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Donghui Li (TAIR) for invaluable advice on some aspects of the curation process of Arabidopsis genes, and Dr. Minami Matsui (RIKEN) for provision of all AtFOX and OsFOX data.
Glossary
Abbreviations
- Ac/Ds
Activator/Dissociation
- AtFOX
RIKEN Arabidopsis full-length cDNA overexpressed Arabidopsis lines
- fl-cDNA
full-length cDNA
- FOX
full-length cDNA overexpressor
- GO
Gene Ontology
- JA
jasmonic acid
- OsFOX
RIKEN rice full-length cDNA overexpressed Arabidopsis lines
- PATO
Phenotype Quality Ontology
- PO
Plant Ontology
- RARGE
RIKEN Arabidopsis genome encyclopedia
- RATM
RIKEN Arabidopsis Ds transposon mutant lines
- RNAi
RNA interference
- TAIR
the Arabidopsis information resource
- TRAPPER
Cold Spring Harbor Laboratory Arabidopsis gene trap mutant lines
Disclosures
The authors have no conflicts of interest to declare.
References
- Afendi FM, Okada T, Yamazaki M, Hirai-Morita A, Nakamura Y, Nakamura K, et al. KNApSAcK family databases: integrated metabolite–plant species databases for multifaceted plant research. Plant Cell Physiol. 2012;53:e1. doi: 10.1093/pcp/pcr165. [DOI] [PubMed] [Google Scholar]
- Akiyama K, Chikayama E, Yuasa H, Shimada Y, Tohge T, Shinozaki K, et al. PRIMe: a web site that assembles tools for metabolomics and transcriptomics. In Silico Biol. 2008;8:339–345. [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché N, Bouchez D. Arabidopsis gene knockout: phenotypes wanted. Curr. Opin. Plant Biol. 2001;4:111–117. doi: 10.1016/s1369-5266(00)00145-x. [DOI] [PubMed] [Google Scholar]
- Cooper L, Walls RL, Elser J, Gandolfo MA, Stevenson DW, Smith B, et al. The plant ontology as a tool for comparative plant anatomy and genomic analyses. Plant Cell Physiol. 2013;54:e1. doi: 10.1093/pcp/pcs163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff NV, Smith DL. A versatile system for detecting transposition in Arabidopsis. Plant J. 1993;3:273–289. doi: 10.1111/j.1365-313x.1993.tb00178.x. [DOI] [PubMed] [Google Scholar]
- Gkoutos GV, Green ECJ, Mallon A-M, Hancock JM, Davidson D. Using ontologies to describe mouse phenotypes. Genome Biol. 2005;6:R8. doi: 10.1186/gb-2004-6-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichikawa T, Nakazawa M, Kawashima M, Iizumi H, Kuroda H, Kondou Y, et al. The FOX hunting system: an alternative gain-of-function gene hunting technique. Plant J. 2006;48:974–985. doi: 10.1111/j.1365-313X.2006.02924.x. [DOI] [PubMed] [Google Scholar]
- Ichikawa T, Nakazawa M, Kawashima M, Muto S, Gohda K, Suzuki K, et al. Sequence database of 1172 T-DNA insertion sites in Arabidopsis activation-tagging lines that showed phenotypes in T1 generation. Plant J. 2003;36:421–429. doi: 10.1046/j.1365-313x.2003.01876.x. [DOI] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl Acad. Sci. USA. 2008;105:7100–7105. doi: 10.1073/pnas.0802332105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, et al. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science. 2003;301:376–379. doi: 10.1126/science.1081288. [DOI] [PubMed] [Google Scholar]
- Kondou Y, Higuchi M, Matsui M. High-throughput characterization of plant gene functions by using gain-of-function technology. Annu. Rev. Plant Biol. 2010;61:373–393. doi: 10.1146/annurev-arplant-042809-112143. [DOI] [PubMed] [Google Scholar]
- Kondou Y, Higuchi M, Takahashi S, Sakurai T, Ichikawa T, Kuroda H, et al. Systematic approaches to using the FOX hunting system to identify useful rice genes. Plant J. 2009;57:883–894. doi: 10.1111/j.1365-313X.2008.03733.x. [DOI] [PubMed] [Google Scholar]
- Kudo T, Akiyama K, Kojima M, Makita N, Sakurai T, Sakakibara H. UniVIO: a multiple omics database with hormonome and transcriptome data from rice. Plant Cell Physiol. 2013;54:e9. doi: 10.1093/pcp/pct003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T, Hirayama T, Kiyosue Y, Takabe H, Mizukado S, Sakurai T, et al. A collection of 11 800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J. 2004;37:897–905. doi: 10.1111/j.1365.313x.2004.02009.x. [DOI] [PubMed] [Google Scholar]
- Kuromori T, Takahashi S, Kondou Y, Shinozaki K, Matsui M. Phenome analysis in plant species using loss-of-function and gain-of-function mutants. Plant Cell Physiol. 2009;50:1215–1231. doi: 10.1093/pcp/pcp078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T, Wada T, Kamiya A, Yuguchi M, Yokouchi T, Imura Y, et al. A trial of phenome analysis using 4000 Ds-insertional mutants in gene-coding regions of Arabidopsis. Plant J. 2006;47:640–651. doi: 10.1111/j.1365-313X.2006.02808.x. [DOI] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, et al. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Berardini TZ, Muller RJ, Huala E. Building an efficient curation workflow for the Arabidopsis literature corpus. Database. 2012;2012 doi: 10.1093/database/bas047. bas047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano S, Nakamura T, Kondo M, Miwa T, Nishikawa SI, Mimura T, et al. The Plant Organelles Database 3 (PODB3) update 2014: integrating electron micrographs and new options for plant organelle research. Plant Cell Physiol. 2014;55 doi: 10.1093/pcp/pct140. (in press) [DOI] [PubMed] [Google Scholar]
- Martienssen RA. Functional genomics: probing plant gene function and expression with transposons. Proc. Natl Acad. Sci. USA. 1998;95:2021–2026. doi: 10.1073/pnas.95.5.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungall CJ, Gkoutos GV, Smith CL, Haendel MA, Lewis SE, Ashburne M. Integrating phenotype ontologies across multiple species. Genome Biol. 2010;11:R2. doi: 10.1186/gb-2010-11-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myouga F, Akiyama K, Motohashi R, Kuromori T, Ito T, Iizumi H, et al. The Chloroplast Function Database: a large-scale collection of Arabidopsis Ds/Spm- or T-DNA-tagged homozygous lines for nuclear-encoded chloroplast proteins, and their systematic phenotype analysis. Plant J. 2010;61:529–542. doi: 10.1111/j.1365-313X.2009.04074.x. [DOI] [PubMed] [Google Scholar]
- Myouga F, Akiyama K, Tomonaga Y, Kato A, Sato Y, Kobayashi M, et al. The Chloroplast Function Database II: a comprehensive collection of homozygous mutants and their phenotypic/genotypic traits for nuclear-encoded chloroplast proteins. Plant Cell Physiol. 2013;54:e2. doi: 10.1093/pcp/pcs171. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Shimura N, Otabe Y, Hirai-Morita A, Nakamura Y, Ono N, et al. KNApSAcK-3D: a three-dimensional structure database of plant metabolites. Plant Cell Physiol. 2013;54:e4. doi: 10.1093/pcp/pcs186. [DOI] [PubMed] [Google Scholar]
- Nakazawa M, Ichikawa T, Ishikawa A, Kobayashi H, Tsuhara Y, Kawashima M, et al. Activation tagging, a novel tool to dissect the functions of a gene family. Plant J. 2003;34:741–750. doi: 10.1046/j.1365-313x.2003.01758.x. [DOI] [PubMed] [Google Scholar]
- Parinov S, Sundaresan V. Functional genomics in Arabidopsis: large-scale insertional mutagenesis complements the genome sequencing project. Curr. Opin. Biotechnol. 2000;11:157–161. doi: 10.1016/s0958-1669(00)00075-6. [DOI] [PubMed] [Google Scholar]
- Pfalz J, Liere K, Kandlbinder A, Dietz K-J, Oelmüller R. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell. 2006;18:176–197. doi: 10.1105/tpc.105.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Kondou Y, Akiyama K, Kurotani A, Higuchi M, Ichikawa T, et al. RiceFOX: a database of Arabidopsis mutant lines overexpressing rice full-length cDNA that contains a wide range of trait information to facilitate analysis of gene function. Plant Cell Physiol. 2011;52:265–273. doi: 10.1093/pcp/pcq190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Satou M, Akiyama K, Iida K, Seki M, Kuromori T, et al. RARGE: a large-scale database of RIKEN Arabidopsis resources ranging from transcriptome to phenome. Nucleic Acids Res. 2005;33:D647–D650. doi: 10.1093/nar/gki014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Yamada Y, Sawada Y, Matsuda F, Akiyama K, Shinozaki K, et al. PRIMe update: innovative content for plant metabolomics and integration of gene expression and metabolite accumulation. Plant Cell Physiol. 2013;54:e5. doi: 10.1093/pcp/pcs184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, et al. Functional annotation of a full-length Arabidopsis cDNA collection. Science. 2002;296:141–145. doi: 10.1126/science.1071006. [DOI] [PubMed] [Google Scholar]
- Springer PS, McCombie WR, Sundaresan V, Martienssen RA. Gene trap tagging of PROLIFERA, an essential MCM2-3-5-like gene in Arabidopsis. Science. 1995;268:877–880. doi: 10.1126/science.7754372. [DOI] [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JD, Dean C, et al. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 1995;9:1797–1810. doi: 10.1101/gad.9.14.1797. [DOI] [PubMed] [Google Scholar]
- Truman W, Bennett MH, Kubigsteltig I, Turnbull C, Grant M. Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proc. Natl Acad. Sci. USA. 2007;104:1075–1080. doi: 10.1073/pnas.0605423104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls RL, Athreya B, Cooper L, Elser J, Gandolfo MA, Jaiswal P, et al. Ontologies as integrative tools for plant science. Amer. J. Bot. 2012;99:1263–1275. doi: 10.3732/ajb.1200222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blázquez MA, Borevitz JO, Christensen SK, Fankhauser C, et al. Activation tagging in Arabidopsis. Plant Physiol. 2000;122:1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998;280:1091–1094. doi: 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- Yan J, Zhang C, Gu M, Bai Z, Zhang W, Qi T, et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell. 2009;21:2220–2236. doi: 10.1105/tpc.109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D-L, Yao J, Mei C-S, Tong X-H, Zeng L-J, Li Q, et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc. Natl Acad. Sci. USA. 2012;109:E1192–E1200. doi: 10.1073/pnas.1201616109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.