Abstract
Maintenance of organ separation is one of the essential phenomena for normal plant development. We have identified and analyzed ONION3 (ONI3), which is required for avoiding organ fusions in rice. Loss-of-function mutations of ONI3, which were identified as mutants with ectopic expression of KNOX genes in leaves and morphologically resembling KNOX overexpressors, showed abnormal organ fusions in developing shoots. The mutant seedlings showed fusions between neighboring organs and also within an organ; they stopped growing soon after germination and subsequently died. ONI3 was shown to encode an enzyme that is most similar to Arabidopsis HOTHEAD and is involved in biosynthesis of long-chain fatty acids. Expression analyses showed that ONI3 was specifically expressed in the outermost cell layer in the shoot apex throughout life cycle, and the oni3 mutants had an aberrant outermost cell layer. Our results together with previous studies suggest that long-chain fatty acids are required for avoiding organ fusions and promoting normal shoot development in rice.
Keywords: Epidermis, Long-chain fatty acid, Organ fusion, Rice, Shoot, Very-long-chain fatty acid
Introduction
The epidermis, which is derived from the outermost cell layer of the shoot apex (L1 or layer 1), is an indispensable barrier to protect organisms from stressed environmental conditions such as drought and pathogen attack. Gas exchange for respiration and photosynthesis is also controlled by a pair of epidermis cells known as guard cells that form stomata. Pollination is also mediated by papillae, a specialized epidermis tissue at the tip of a pistil in a flower. A phytohormone brassinosteroid signal is transduced through epidermis cells (Savaldi-Goldstein et al. 2007), and polar auxin transport is also mainly mediated through epidermis cells in the shoot apex (Gallavotti et al. 2008, Miyashita et al. 2010). Thus, epidermis is indispensable for plant survival, growth, reproduction and phytohormone signaling.
In addition to these vital functions, the epidermis plays an essential role in normal shoot development by preventing inappropriate fusions between neighboring organs and within an organ. For example, Arabidopsis L1-specific genes HOTHEAD (HTH) and FIDDLEHEAD (FDH) showed organ fusions in leaves and floral organs (Yephremov et al. 1999, Pruitt et al. 2000, Krolikowski et al. 2003, Kurdyukov et al. 2006). These plants can grow to maturity, but they were sterile or semi-sterile. Both of these genes are predicted to encode enzymes involved in biosynthesis of fatty acids. HTH was suggested to encode ω-alcohol dehydrogenase catalyzing biosynthesis of long chain α-, ω-dicarboxylic fatty acids, and FDH was suggested to encode fatty acid elongase (β-ketoacyl CoA synthase) catalyzing the elongation reaction of very-long-chain fatty acids (VLCFAs) (Yephremov et al. 1999, Pruitt et al. 2000, Krolikowski et al. 2003, Kurdyukov et al. 2006). In addition, double mutants of LACS1 and LACS2, which encode long-chain acyl-CoA synthase, also showed organ fusions and formed unopened flowers (Schnurr et al. 2004, Weng et al. 2010), and pas2, a mutant of the 3-hydroxy-acyl-CoA dehydratase gene involved in the elongation reaction of VLCFAs, showed impaired embryo and seedling development associated with cell proliferation (Bach et al. 2008, Nobusawa et al. 2013). These mutant analyses clearly demonstrate the importance of the fatty acids in preventing ectopic organ fusions and normal shoot development at vegetative and reproductive stages in Arabidopsis.
Rice organ fusion mutants also showed severe defects in shoot development and seedling lethality. Rice mutants of ONION1 (ONI1), which encodes fatty acid elongase most similar to Arabidopsis FDH and is specifically expressed in an outermost cell layer of the shoot apex throughout the life cycle, had an abnormal composition of VLCFAs in the epicuticular wax, and showed organ fusions during embryogenesis and the vegetative stage (Ito et al. 2011). In addition, oni1 mutants lacked normal epidermis and failed to maintain the shoot apical meristem (SAM), which resulted in seedling lethality (Ito et al. 2011). oni2, another mutant of a fatty acid elongase gene, also had an abnormal composition of VLCFAs in the epicuticular wax and showed organ fusions (Tsuda et al. 2013). These findings indicate that a normal composition of VLCFAs is required for L1 development and maintenance of the SAM in rice. However, little is known about the role of VLCFAs in rice and the genes involved in their biosynthesis. To understand the physiological roles of VLCFAs and the functions of the genes involved in their biosynthesis for plant development, analyses of rice mutants associated with VLCFA biosynthesis will be necessary.
We previously identified three onion mutants (oni1, oni2 and oni3) that resembled the overexpressor of the SAM-specific KNOX gene (Tsuda et al. 2009). In this study, we carried out detailed morphological analyses of oni3 and molecular characterization of ONI3. oni3 showed organ fusions and abnormal embryogenesis. ONI3 was shown to encode a putative ω-alcohol dehydrogenase most similar to Arabidopsis HTH, and was expressed specifically in the outermost cell layer in the shoot apex during embryogenesis, vegetative and reproductive phases. Differing from Arabidopsis hth, oni3 showed abnormal vegetative development, and was seedling lethal. Our results suggest that VLCFAs play an essential role in early-stage shoot development in rice.
Results
We previously reported isolation of oni3 mutants (Tsuda et al. 2009). In this study, we isolated additional oni3 mutant alleles, and carried out detailed analyses of the mutants and molecular characterization of the ONI3 gene. Since oni3 mutants were seedling lethal (Tsuda et al. 2009), we could not examine phenotypes at later developmental stages including reproductive stages, although ONI3 was expressed throughout the life cycle (see ‘Expression pattern of ‘ONI3’).
oni3 mutant morphology
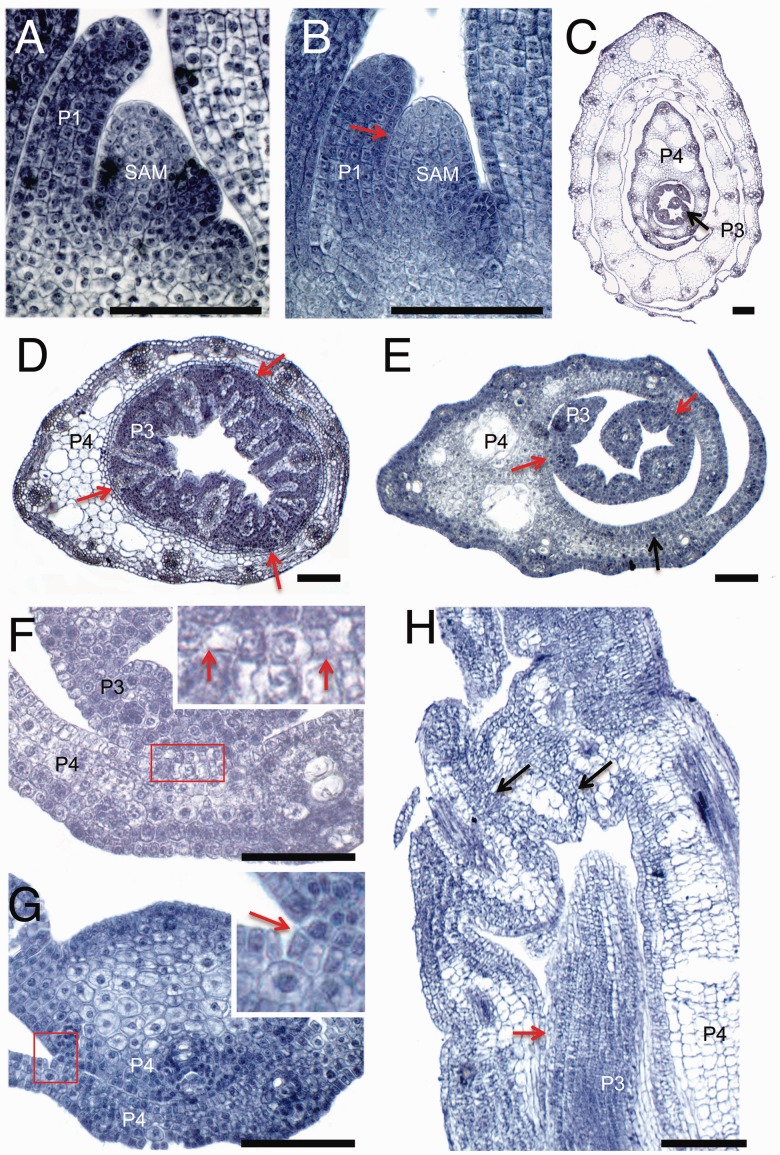
oni3 could germinate and elongate a shoot to various degrees (Fig. 1A, B). In more severe cases, an oni3 shoot resembled an edible onion-like gross morphology (Fig. 1C) (Tsuda et al. 2009). In most cases, shoot growth stopped within a few weeks after germination (Tsuda et al. 2009). The oni3 shoots had a shortened leaf (Fig. 1). Leaf elongation was insufficient and the tip of the leaf blade was missing (Fig. 1D). A lamina joint, at which a leaf blade is connected to a leaf sheath, was unclear, and structures that formed at a lamina joint such as ligules and an auricle were not observed (Fig. 1D). We also observed organ fusions in paraffin sections of shoots (Fig. 2). In the wild type, a P1 leaf was clearly separated from the SAM, but in oni3-6 they were fused to each other (Fig. 2A, B). Organ fusions between two successive leaves were also observed in oni3 (Fig. 2C–H). In some oni3 seedlings, the abaxial surface of a P3 leaf was adhered to the adaxial surface of a P4 leaf, and margins of the P4 leaf were fused to each other (Fig. 2D). In most oni3 seedlings, the abaxial side of the leaf parts, including the leaf margins and the midrib, was fused to the adaxial side of the same leaf or the preceding leaf (Fig. 2E–G). In addition, a distal region of the leaf was folded, and the folded regions were fused to each other (Fig. 2H). The physical separation of the fused leaves in oni3 sometimes allowed emergence of the inner leaves, although the emerging leaves did not grow normally. This indicates that abnormality of the leaf morphology in the inner leaves may be caused by not only ‘organ fusion’ but also ‘physical stress’. However, we cannot verify this hypothesis quantitatively.
Fig. 1.
Gross morphology of oni3. (A) A wild-type seedling at 15 d after germination. (B) An oni3-7 seedling at 15 d after germination. (C) An oni3-1 shoot. (D) A leaf tip of oni3-7. (E) A leaf tip of the wild type. Asterisks indicate a leaf tip that was lost in oni3. Bars = 5 cm in (A), 1 cm in (B) and 1 mm in (C–E).
Fig. 2.
Morphology of leaves. The oni3 mutant leaves were examined using paraffin sections. (A) A longitudinal section of the wild-type shoot apex at 1 week after germination. (B) A longitudinal section of the oni3-6 shoot apex showing a fusion of the P1 leaf primordium and SAM (red arrow) at 1 week after germination. (C) A cross-section of the wild-type shoot apex showing no fusion of leaves at 2 weeks after germination. (D) A cross-section of oni3-6 leaves showing fusions of P3 and P4 leaves (red arrow) at 2 weeks after germination. (E) A cross-section of the oni3-7 shoot apex showing a fusion of P4 leaf margins (black arrow) and fusions between P3 and P4 leaves (red arrows) at 2 weeks after germination. (F) A close-up view of a fusion site of two successive leaves of oni3-7 (red arrow). A close-up view of an inset is shown. (G) A close-up view of a fusion site of two successive leaves of oni3-7. The abaxial side of the midrib is fused with the adaxial surface of the preceding leaf (red arrow). A close-up view of an inset is shown. (H) A longitudinal section of a distal region of the oni3-6 shoot. Black arrows indicate fusions within the same leaf, and a red arrow indicates fusions between successive leaves. SAM, shoot apical meristem; P1, P1 leaf primordium; P3, P3 leaf primordium; P4, P4 leaf primordium. Bars = 100 µm.
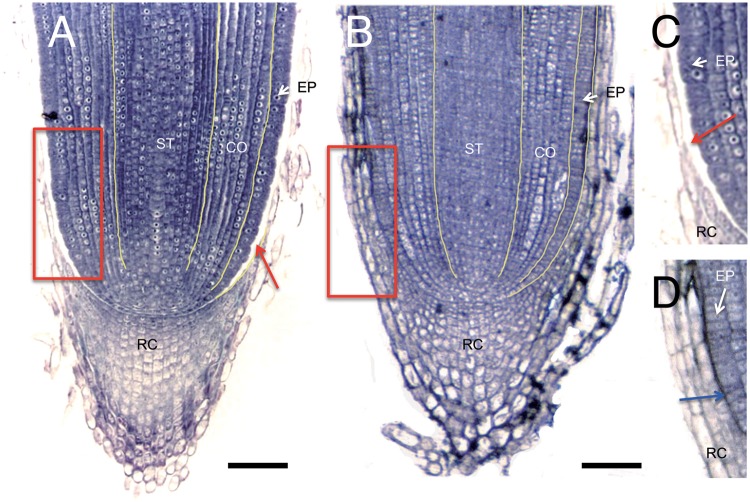
Organ fusions were also observed in roots. In the wild type, a space between the root epidermis and root cap was clearly observed (Fig. 3A, C), whereas the epidermis and root cap were tightly fused to each other and no space was observed in oni3-7 (Fig. 3B, D).
Fig. 3.
Root morphology. (A) A wild-type root apex in which a space is observed between the root epidermis and root cap (red arrow) at 2 weeks after germination. (B) oni3-7 root apex at 2 weeks after germination. (C and D) Close-up views of insets in (A) and (B), respectively. An air space between the root epidermis and root cap (red arrow) is apparent in (C), whereas the root epidermis and root cap are tightly fused in oni3-7 (blue arrow in D). RC, root cap; ST, stele; CO, cortex; EP, epidermis. Bars = 100 µm.
Epidermis of oni3
Since the gross morphology of oni3 was similar to those of oni1 and oni2, both of which had defects in differentiation and/or functionality in epidermis (Ito et al. 2011, Tsuda et al. 2013), and since ONI3 was specifically expressed in an outermost cell layer in the shoot apex (see ‘Expression pattern of ONI3’), we examined expression of ROC1 in oni3 (Fig. 4). ROC1 is an ortholog of an Arabidopsis L1-specific gene ATML1 required for L1 identification and is specifically expressed in the outermost cell layer in rice (Ito et al. 2002, Abe et al. 2003). In situ hybridization detected ROC1 expression in the epidermis of the wild-type leaf, but in the oni3-6 mutant leaf ROC1 expression was hardly detected (Fig. 4). Perturbed expression of ROC1 suggested that differentiation and/or functionality of the epidermis was compromised in oni3. Thus, oni3 seemed to lack normal epidermis in the leaf.
Fig. 4.
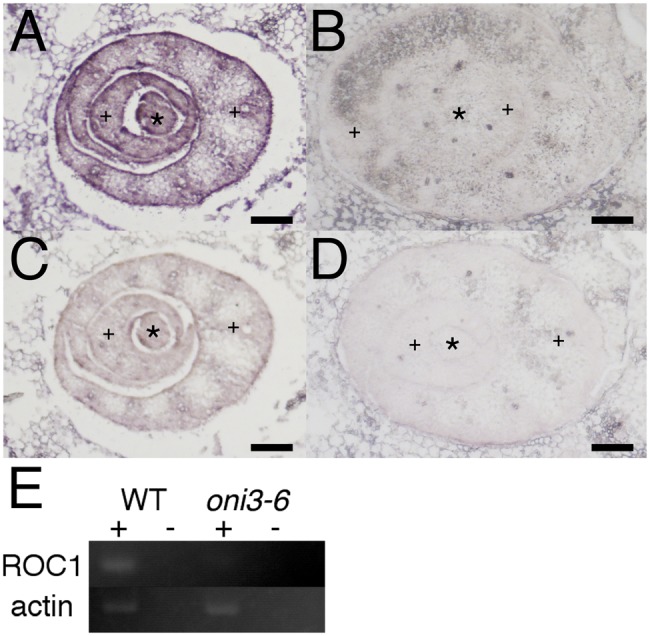
Expression of ROC1 in oni3. Cross-sections of the wild-type (A and C) or oni3-6 (B and D) shoots were hybridized with the antisense probe (A and B) or sense probe (C and D) of ROC1. Bars = 100 µm. *, SAM/stem; +, developing leaves. (E) RT–PCR analysis of ROC1 in the wild type and oni3-6. Actin was used as a control. + and – indicate whether reverse transcriptase was added to or omitted from the reaction mixture, respectively.
Molecular cloning of ONI3
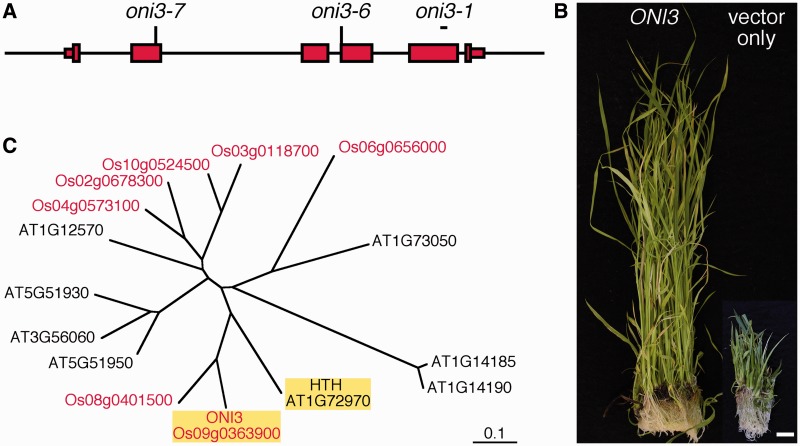
ONI3 was roughly mapped at around 36 cM of chromosome 9 (Tsuda et al. 2009). Near to this position, we found a homolog (Os09g0363900) of Arabidopsis HTH. Because hth showed organ fusions (Krolikowski et al. 2003) like oni3, we examined genomic DNAs from oni3 mutants and identified mutations in the HTH homolog. oni3-1 had a 49 bp deletion in the fifth exon, oni3-6 had a nucleotide substitution at a splicing acceptor site, and oni3-7 had a nucleotide substitution that results in an amino acid substitution from alanine to threonine (Fig. 5A). These results showed that Os09g0363900 was a strong candidate for ONI3.
Fig. 5.
Cloning of ONI3. (A) Genome structure of ONI3. Thin and thick red boxes indicate 5′- and 3′-untranslated regions and coding regions, respectively. The line indicates the 5′ and 3′ upstream regions and introns. Positions of the mutations are shown above the genome structure. oni3-6 and oni3-7 had a nucleotide substitution at the indicated position, and oni3-1 had a deletion in the region shown by a short bar. (B) Complementation of oni3. oni3-6 mutant callus transformed with the wild-type ONI3 genome construct showed regeneration of normal shoots, and oni3-6 mutant callus transformed with an empty vector showed regeneration of mutant shoots. Bar = 1 cm. (C) A phylogenetic tree of ω-alcohol dehydrogenase of rice and Arabidopsis drawn on the basis of entire amino acid sequences. Rice proteins are shown in red, and ONI3 and HTH are highlighted by yellow shading.
To confirm that Os09g0363900 is ONI3, a complementation test of oni3-6 with its wild-type genomic fragment was carried out. When a genomic fragment covering Os09g0363900 with 2.2 kb upstream and 1.3 kb downstream regions was introduced into the oni3-6 mutant callus, the transformed callus showed regeneration of normal shoots with a wild-type morphology, whereas when the oni3-6 callus was transformed with an empty vector, the transformed callus showed regeneration of mutant shoots (Fig. 5B). Based on these results, we concluded that Os09g0363900 is ONI3.
Sequence analysis of ONI3
ONI3 encodes a protein similar to Arabidopsis HTH (Fig. 5C). HTH encodes an enzyme similar to long-chain fatty acid (LCFA) ω-alcohol dehydrogenases, and was shown to be involved in biosynthesis of long-chain α-, ω-dicarboxylic fatty acids (Krolikowski et al. 2003, Kurdyukov et al. 2006). Since ONI3 and HTH showed high sequence identity (56% identity in their entire amino acid sequences) (Supplementary Fig. S1) and was categorized into the same clade in a phylogenetic tree (Fig. 5C), ONI3 also seemed to encode LCFA ω-alcohol dehydrogenase and plays a role in biosynthesis of long-chain α-, ω-dicarboxylic fatty acids, but this remains to be characterized biochemically.
Biosynthesis of fatty acids in plants is known to take place in the chloroplast. In agreement with this, a prediction program WoLF PSORT (http://wolfpsort.seq.cbrc.jp/) predicted the localization of ONI3 in the chloroplast.
A database search identified six homologs of ONI3 in the rice genome. Among them, Os08g0401500 was most similar to ONI3, and was categorized into the same clade that includes HTH (Fig. 5C). The amino acid sequences of ONI3 and Os08g0401500 were 72% identical to each other. ONI3 showed 32–50% amino acid identities with the remaining five homologs.
Expression pattern of ONI3
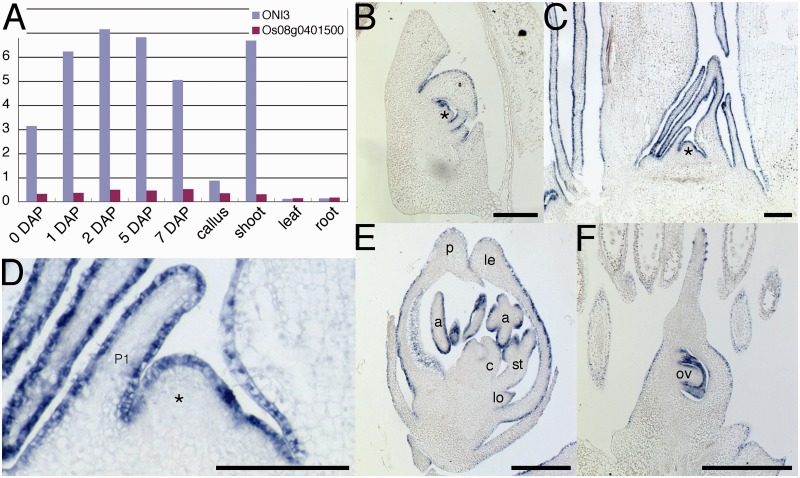
To examine the overall expression pattern of ONI3, we picked up and analyzed ONI3 expression data from a gene expression atlas obtained by microarray experiments (Fig. 6A) (Fujita et al. 2010). We found that ONI3 was expressed in shoots and developing embryos but not in leaves, roots or calli. We also analyzed expression of Os08g0401500, which is the most similar gene to ONI3 in rice. The expression of Os08g0401500 was very low and at a background level in all the organs analyzed (Fig. 6A).
Fig. 6.
Expression pattern of ONI3. (A) Expression of ONI3 and its close homolog Os08g0401500 by microarray analysis. The data were extracted from Oryza_Express (Fujita et al. 2010). (B–F) In situ RNA hybridization of ONI3. (B) 7 DAP embryo. (C) Shoot apex of a seedling 1 month after germination. (D) Close-up view of the shoot apex in (C). ONI3 expression was restricted in L1 in the SAM and protoderm of young leaves. (E) Developing flower. (F) Developing ovary. Bars = 100 µm. *, SAM; P1, P1 leaf primordium; a, anther; c, carpel; le, lemma; lo, lodicule; p, palea; ov, ovule; st, stamen.
To study ONI3 expression in more detail, we carried out in situ RNA hybridization in the organs where ONI3 expression was detected in the atlas. The results showed that ONI3 was expressed in the outermost cell layer of the SAM, young leaf and floral organs. In the embryo 7 days after pollination (DAP), ONI3 expression was detected in shoot organs such as the SAM, leaves and coleoptile (Fig. 6B). In the vegetative growth phase, at 1 month after germination, ONI3 expression was detected specifically in the outermost cell layer of the SAM and young leaves (Fig. 6C, D). In the reproductive growth phase, ONI3 expression was detected again in the outermost cell layer of developing floral organs (Fig. 6E). In the ovary, ONI3 expression was detected in the outermost cell layer of the ovary and ovule (Fig. 6F). Integuments also showed the expression (Fig. 6F). No signal was detected in the inner cells of any organs or at any developmental stages examined. These expression analyses revealed that ONI3 is an outermost cell layer-specific gene of the SAM and young above-ground organs throughout the life cycle.
Composition VLCFAs in oni3
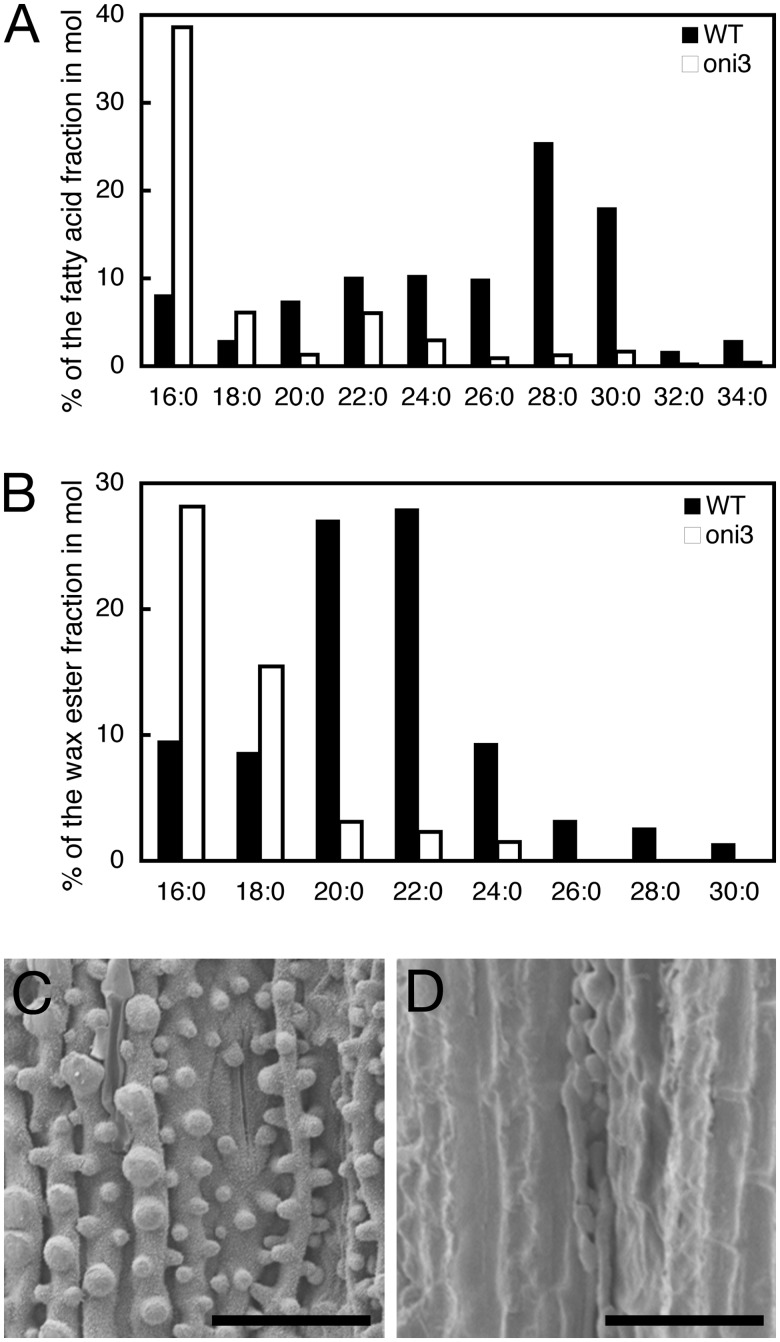
Although ONI3 encodes a putative LCFA ω-alcohol dehydrogenase and seemed not to be directly involved in the biosynthesis of VLCFAs, we examined the composition of VLCFAs in 2-week-old oni3 shoots, because rice organ fusion mutants showed an abnormal composition of VLCFAs (Ito et al. 2011, Tsuda et al. 2013). In the oni3-6 shoots, the amount of free saturated VLCFAs with a carbon number of 20 (C = 20) or more was reduced compared with that of the wild-type shoots (Fig. 7A). In particular, VLCFAs of C = 32 and C = 34 were barely detected (Fig. 7A).
Fig. 7.
Cuticle wax. (A) Amount of free saturated VLCFAs. Percentages of saturated VLCFAs per total fatty acids in a molar ratio are shown. The values are the average of two independent experiments. (B) Amount of saturated VLCFAs in the alkyl ester fraction of epicuticular wax. Percentages of methyl esters of saturated VLCFAs per methyl esters of total fatty acids in a molar ratio are shown. (C and D) Scanning electron microscopy observation of the abaxial surface of the third leaf of the wild type (C) and oni3-6 (D). The surface was rough in the wild type due to cuticle protrusions, but it was rather smooth in oni3-6. Bars = 50 µm.
We further examined the composition of VLCFAs in the alkyl ester fraction of epicuticular wax of 2-week-old oni3-6 shoots. We found that the amount of saturated VLCFAs of C = 20 or more was reduced in the oni3-6 shoots compared with those of the wild-type shoots (Fig. 7B). In particular, VLCFAs of C = 26 and longer were under the level of detection (Fig. 7B). These results indicate that VLCFAs in the alkyl ester fraction of epicuticular wax were reduced in the oni3-6 shoots.
Since VLCFAs are the main components of the cuticle wax, we examined protrusions of cuticle on the abaxial surface of the leaf sheath. Scanning electron microscopic observation showed cuticular protrusions that were well developed on the surface of the leaf epidermis in the wild type, but such protrusions were not observed on the surface of oni3-6 epidermis (Fig. 7C, D). These results indicate that oni3 had a reduced amount of VLCFAs.
Expression of auxin-related genes in oni3
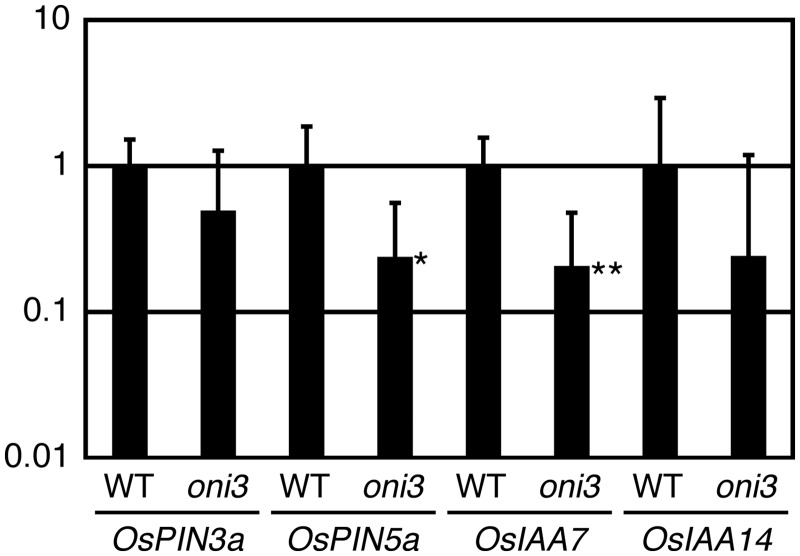
Expression of KNOX genes, which are ectopically expressed in oni3, is known to be negatively regulated by auxin (Hay et al. 2006, Tsuda et al. 2009, Perez-Perez, 2010, Tabata et al. 2010), and oni1 and oni2 showed altered expression of auxin-related genes, suggesting altered distribution of auxin in its shoot apex (Takasugi and Ito 2011, Tsuda et al. 2013). In addition, auxin is suggested to be transported mainly through the outermost cell layer that is affected in oni3 at the shoot apex (Gallavotti et al. 2008, Miyashita et al. 2010). Therefore, we examined expression of OsPIN3a and OsPIN5a, which encode an auxin efflux carrier protein, and OsIAA7 and OsIAA14, which are inducible by auxin, in the wild type and oni3-6 (Jain et al. 2006, Miyashita et al. 2010). Quantitative reverse transcription–PCR (qRT–PCR) analysis showed reduced expression of these genes in oni3-6 (Fig. 8). These results show that the expression of auxin-related genes is perturbed in oni3 shoots.
Fig. 8.
Expression of auxin-related genes in oni3 shoots. RNAs were isolated from shoots of 1-week-old seedlings of oni3-6 and the wild type grown in a growth chamber at 30°C with continuous light, and subjected to qRT–PCR. The relative expression level is shown using actin as a reference. Error bars indicate standard deviations. * and ** indicate the significant P-values (P < 0.05 and P < 0.01, respectively) compared with the wild type by Student t-test.
Discussion
In this study we characterized the ONI3 gene and showed that ONI3 encodes a putative ω-alcohol dehydrogenase that is necessary for shoot development in rice. In spite of the outermost cell layer-specific expression of ONI3 in shoot apex, the effects of the oni3 mutation were not restricted to the outermost cells and oni3 did not show a simply epidermis-affected phenotype, but they were expanded to the entire shoot and oni3 produced a very small shoot. Thus, cell fate and/or function in the inner layers was suggested to be controlled non-cell autonomously by intercellular signaling from the epidermis.
Sequence and phylogenetic analyses showed that ONI3 is most similar to HTH in Arabidopsis. HTH is suggested to encode LCFA ω-alcohol dehydrogenase that catalyzes an oxidation reaction from long-chain ω-hydroxy fatty acids to ω-oxo fatty acids in the ω-oxidation pathway (Kurdyukov et al. 2006). However, analysis of the composition of fatty acids in hth showed that the amount of VLCFAs was reduced in hth compared with the wild type in addition to the change of the composition of LCFAs (Kurdyukov et al. 2006). oni3 also had a reduced amount of VLCFAs. Thus, although details of the biosynthesis pathways and their regulating mechanisms are not well known, it is likely that the composition of LCFAs somehow affects the composition of VLCFAs indirectly, probably due to the reduction of the amount of their precursors.
Previous studies in Arabidopsis showed that VLCFAs play an indispensable role in preventing ectopic organ fusions. Mutants of FIDDLEHEAD, which encodes a fatty acid elongase catalyzing an elongation reaction of VLCFAs with the carbon number of ≥20, showed organ fusions during vegetative and reproductive development. hth mutants showed organ fusions during reproductive development. Since oni3 also showed organ fusions, organ separation associated with VLCFAs may be a common phenomenon shared by various plant species.
A phylogenetic analysis and sequence identity showed that ONI3 is an ortholog of Arabidopsis HTH. Although mutants of both of these two genes showed organ fusions, the organ fusion of hth was minor in vegetative organs and was mainly observed in floral organs, whereas in oni3 organ fusions were observed in vegetative organs. We could not examine organ fusion in the reproductive phase in oni3 due to its seedling lethality. In addition to organ fusions, oni3 showed various abnormalities in shoot development. Leaves were short and lacked most of the leaf blades. A lamina joint did not clearly separate the leaf sheath and leaf blade. Furthermore, a leaf that was covered with a preceding leaf was folded on the adaxial side, and protruding portions at the adaxial side were fused to each other to form a thick leaf. The abaxial side was fused to the adaxial side of the preceding leaf. These phenotypes were not reported in hth, even in floral organs. Thus, ONI3 and HTH seemed to have some functional differences even though they are orthologous to each other, and ONI3 may have additional functions that are not retained by HTH. Otherwise, fatty acids produced by ONI3 or HTH may have more critical physiological activities in rice than in Arabidopsis. Differences in embryogenesis between rice and Arabidopsis may also explain their phenotypic difference. Rice produces three leaves during embryogenesis, and failure of normal differentiation of the epidermis in leaf may affect later development, because it is well known that normal epidermal development is critical for subsequent developmental events.
oni3 showed severe developmental defects and seedling lethality. oni3 leaves were small and lacked a large part of the leaf blade. Considering that ONI3 is specifically expressed in the outermost cells in the shoot apex, these cells may be necessary for entire organ development. Consistent with this notion, several Arabidopsis mutants of L1-specific genes such as ALE2, ACR4, ATML1 and PDF2 showed abnormal shoot development not limited to the epidermis (Abe et al. 2003, Watanabe et al. 2004, Tanaka et al. 2007). oni1 and oni2 mutations in rice also affected the entire shoot development, albeit that both of these genes are specifically expressed in the outermost cell layer (Ito et al. 2011, Tsuda et al. 2013). Thus, L1 and/or epidermis cells might produce or spread a signal that induces normal development of inner cells. Because auxin is suggested to be transported mainly through the outermost cell layer at the shoot apex (Gallavotti et al. 2008, Miyashita et al. 2010) and oni3 showed altered expression of PIN auxin efflux carrier genes and auxin-inducible IAA genes, auxin would be one of the candidates for such a signaling molecule. However, this possibility needs to be elucidated.
A database search identified a close homolog (Os08g0401500) of ONI3 in the rice genome. ONI3 and Os08g0401500 had 72% amino acid identity. This high sequence identity suggests a similar substrate specificity of these two enzymes and functional overlaps between these two genes. However, expression analysis showed that ONI3 was expressed in the developing embryo and shoot apex, whereas the expression of Os08g0401500 was hardly detected in these organs by the microarray analysis. Thus, Os08g0401500 may not function or may play a dispensable role in organs where ONI3 plays an essential role. This may explain why a mutation in ONI3 brought about the abnormal morphologies, even though there is a close homolog in the genome.
Materials and Methods
Plant materials
oni3-1 was identified from Tos17 mutant populations derived from Oryza sativa L. cultivar Nipponbare (Tsuda et al. 2009), and oni3-6 and oni3-7 were identified from N-methyl-N-nitrosourea-mutagenized M2 populations of cultivars Kinmaze and Taichung 65, respectively. Self-pollinated progeny of the heterozygous plants were maintained and used for the mutant analyses. Nipponbare was used as the wild type.
Histological analysis
Shoot apices were fixed in FAA [formaldehyde : glacial acetic acid : ethanol (1 : 1 : 18)] for 24 h at 4°C and then dehydrated in a graded ethanol series. Dehydrated samples in absolute ethanol were replaced with xylene and embedded in Paraplast plus (McCormick Scientific). Microtome sections (8 µm thick) were stained with Delafield’s hematoxylin and observed with a light microscope.
Scanning electron microscopy analysis
The leaf sheaths of 10-day-old seedlings grown in an incubator at 30°C under 24 h light conditions were used for scanning electron microscope analysis. The samples were freeze-dried without pre-treatment, coated with platinum–palladium and subjected to observation using SU8000 (Hitachi).
Cloning of ONI3
ONI3 was mapped to around 36 cM of chromosome 9 (Tsuda et al. 2009). We noticed that an Arabidopsis HTH homolog (Os09g0363900) is located near this position in RAP-DB build 3 (http://rapdb.dna.affrc.go.jp/). Coding regions of the HTH homolog were amplified by PCR from genomic DNAs isolated from the oni3 mutants with the following combinations of primers: oni3-1 and oni3-2 for exon 1, oni3-3 and oni3-4 for exon 2, oni3-5 and oni3-6 for exon 3, oni3-7 and oni3-8 for exon 4, and oni3-9 and oni3-10 for exon 5, and the PCR products were directly sequenced. Sequences of the primers used for PCR and sequencing are shown in Supplementary Table S1.
Complementation test
An 8.5 kb genomic fragment containing a 2.2 kb upstream region from the 5′ end of full-length cDNA of ONI3, a transcribed region and a 1.3 kb downstream region from the 3′ end of the cDNA was amplified by PCR using primers ONI3-F1 and ONI3-R1 (Supplementary Table S1), and PrimeStar GXL (TAKARA BIO INC.), and the PCR product was cloned into pDONR221 by the BP clonase reaction (Invitrogen) according to the manufacturer’s protocol. The nucleotide sequence of the amplified region was verified by sequencing. Then, the genomic fragment was cloned into a binary vector pGWB1 (Nakagawa et al. 2007) by LR clonase (Invitrogen), and the resultant plasmid pBGONI3 was used for transformation of oni3-6 callus. We used pANDAΔ as an empty vector, which was generated from pANDA (Miki and Shimamoto 2004) by restriction digestion and self-ligation to remove a GATEWAY cassette. pANDAΔ is identical to the backbone of pBGONI3. These vectors have a hygromycin resistance gene as a selectable marker.
Calli were prepared from seeds obtained by self-pollination of oni3-6 heterozygous plants. For selection of oni3-6 callus, DNAs were isolated from a small piece of each callus, and a 83 bp region containing the mutation site was amplified by PCR with the primer combination ONI3-F5 and ONI3-R2 (Supplementary Table S1) followed by digestion with PstI, whose cutting site exists in the wild-type allele but not in the oni3-6 allele in the amplified region. oni3-6 homozygous mutant calli were used for transformation. Agrobacterium-mediated transformation was carried out as described by Hiei et al. (1994). The calli were transformed with Agrobacterium haboring pBGONI3 or pANDAΔ, and the transformed cells were selected on a medium containing hygromycin followed by shoot regeneration.
Expression analysis
Microarray data were obtained from Oryza_Express (Fujita et al. 2010). For in situ RNA hybridization, paraffin sections were prepared as described above or using a microwave apparatus as described previously (Miyashita et al. 2010), and 8 µm or 10 µm thick microtome sections were applied to glass slides coated with aminopropylsilane (Matsunami Glass). Digoxigenin-labeled antisense probes were prepared from the full-length cDNAs of ONI3 (AK072899) or ROC1 (AK120496). In situ hybridization and immunological detection of the hybridization signals were performed as described by Kouchi and Hata (1993).
Conventional RT–PCR and qRT–PCR were carried out using RNAs isolated from 1-week-old shoot apex of oni3-6 and the wild type grown in an incubator at 30°C under 24 h light conditions. Gene-specific primers used for the PCR are shown in Supplementary Table S1. Actin was used as a reference.
VLCFA analysis
Analysis of fatty acids was carried out as described previously using shoots of 2-week-old seedlings grown in an incubator at 30°C under constant light (Ito et al. 2011, Tsuda et al. 2013). For analysis of fatty acids in epicuticular wax, free VLCFAs and alkyl VLCFAs of chloroform-extracted wax were separated by thin-layer chromatography (hexane : ethyl ether : acetic acid = 9 : 1 : 0.1), and derivatized to the methyl ester by an HCl/methanol method (Lepage and Roy 1986). The fatty acid methyl ester composition was analyzed using a gas chromatograph equipped with a flame ionization detector (GC-380, GL Sciences) and a column (ZB-5 ms, 30 m × 0.25 mm internal diameter, 0.2 µm film thickness, Phenomenex). The column temperature program was as follows; 170°C for 1 min, to 200°C at 2°C min−1, to 320°C at 10°C min−1 and hold at 320°C for 10 min.
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the Japan Society for the Promotion of Science [Grants-in-Aid for Scientific Research (B) (grant No. 24380003 to Y.I. and F.K. and grant No. 24658003 to J.I.)]; the Japan Society for the Promotion of Science for Young Scientists [a Research Fellowship (22-1871) to K.T.].
Supplementary Material
Acknowledgments
We thank Dr. Nakagawa (Shimane University) and Dr. Shimamoto (NAIST) for providing pGWB1 and pANDA, respectively.
Glossary
Abbreviations
- DAP
days after pollination
- LCFA
long-chain fatty acid
- RT–PCR
reverse transcription–PCR
- SAM
shoot apical meristem
- VLCFA
very-long-chain fatty acid
Disclosures
The authors have no conflicts of interest to declare.
References
- Abe M, Katsumata H, Komeda Y, Takahashi T. Regulation of shoot epidermal cell differentiation by a pair of homeodomain proteins in Arabidopsis. Development. 2003;130:635–643. doi: 10.1242/dev.00292. [DOI] [PubMed] [Google Scholar]
- Bach L, Michaelson LV, Haslam R, Bellec Y, Gissot L, Marion J, et al. The very-long-chain hydroxy fatty acyl-CoA dehydratase PASTICCINO2 is essential and limiting for plant development. Proc. Natl Acad. Sci. USA. 2008;105:14727–14731. doi: 10.1073/pnas.0805089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Horiuchi Y, Ueda Y, Mizuta Y, Kubo T, Yano K, et al. Rice expression atlas in reproductive development. Plant Cell Physiol. 2010;51:2060–2081. doi: 10.1093/pcp/pcq165. [DOI] [PubMed] [Google Scholar]
- Gallavotti A, Yang Y, Schmidt RJ, Jackson D. The relationship between auxin transport and maize branching. Plant Physiol. 2008;147:1913–1923. doi: 10.1104/pp.108.121541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A, Barkoulas M, Tsiantis M. ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Development. 2006;133:3955–3961. doi: 10.1242/dev.02545. [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Ito M, Sentoku N, Nishimura A, Hong S-K, Sato Y, Matsuoka M. Position dependent expression of GL2-type homeobox gene, Roc1: significance for protoderm differentiation and radial pattern formation in early rice embryogenesis. Plant J. 2002;29:497–507. doi: 10.1046/j.1365-313x.2002.01234.x. [DOI] [PubMed] [Google Scholar]
- Ito Y, Kimura F, Hirakata K, Tsuda K, Takasugi T, Eiguchi M, et al. Fatty acid elongase is required for shoot development in rice. Plant J. 2011;66:680–688. doi: 10.1111/j.1365-313X.2011.04530.x. [DOI] [PubMed] [Google Scholar]
- Jain M, Kaur N, Garg R, Thakur JK, Tyagi AK, Khurana JP. Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa) Funct. Integr. Genomics. 2006;6:47–59. doi: 10.1007/s10142-005-0005-0. [DOI] [PubMed] [Google Scholar]
- Kouchi H, Hata S. Isolation and characterization of novel nodulin cDNAs representing genes expressed at early stages of soybean nodule development. Mol. Gen. Genet. 1993;238:106–119. doi: 10.1007/BF00279537. [DOI] [PubMed] [Google Scholar]
- Krolikowski KA, Victor JL, Wagler TN, Lolle SJ, Pruitt RE. Isolation and characterization of the Arabidopsis organ fusion gene HOTHEAD. Plant J. 2003;35:501–511. doi: 10.1046/j.1365-313x.2003.01824.x. [DOI] [PubMed] [Google Scholar]
- Kurdyukov S, Faust A, Trenkamp S, Bar S, Franke R, Efremova N, et al. Genetic and biochemical evidence for involvement of HOTHEAD in the biosynthesis of long-chain α-, ω-dicarboxylic fatty acids and formation of extracellular matrix. Planta. 2006;224:315–329. doi: 10.1007/s00425-005-0215-7. [DOI] [PubMed] [Google Scholar]
- Lepage G, Roy CC. Direct transesterification of all classes of lipids in a one-step reaction. J. Lipid Res. 1986;27:114–120. [PubMed] [Google Scholar]
- Miki D, Shimamoto K. Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 2004;45:490–495. doi: 10.1093/pcp/pch048. [DOI] [PubMed] [Google Scholar]
- Miyashita Y, Takasugi T, Ito Y. Identification and expression analysis of PIN genes in rice. Plant Sci. 2010;178:424–428. [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, et al. Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 2007;104:34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- Nobusawa T, Okushima Y, Nagata N, Kojima M, Sakakibara H, Umeda M. Synthesis of very-long-chain fatty acids in the epidermis controls plant organ growth by restricting cell proliferation. PLoS Biol. 2013;11:e1001531. doi: 10.1371/journal.pbio.1001531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Perez JM, Candela H, Robles P, Lopez-Torrejon G, del Pozo JC, Micol JL. A role for AUXIN RESISTANT3 in the coordination of leaf growth. Plant Cell Physiol. 2010;51:1661–1673. doi: 10.1093/pcp/pcq123. [DOI] [PubMed] [Google Scholar]
- Pruitt RE, Vielle-Calzada J-P, Ploense SE, Grossniklaus U, Lolle SJ. FIDDLEHEAD, a gene required to suppress epidermal cell interactions in Arabidopsis, encodes a putative lipid biosynthetic enzyme. Proc. Natl Acad. Sci. USA. 2000;97:1311–1316. doi: 10.1073/pnas.97.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi-Goldstein S, Peto C, Chory J. The epidermis both drives and restricts plant shoot growth. Nature. 2007;446:199–202. doi: 10.1038/nature05618. [DOI] [PubMed] [Google Scholar]
- Schnurr J, Shockey J, Browse J. The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell. 2004;16:629–642. doi: 10.1105/tpc.017608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata R, Ikezaki M, Fujibe T, Aida M, Tian CE, Ueno Y, et al. Arabidopsis auxin response factor6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol. 2010;51:164–175. doi: 10.1093/pcp/pcp176. [DOI] [PubMed] [Google Scholar]
- Takasugi T, Ito Y. Altered expression of auxin-related genes in the fatty acid elongase mutant oni1 of rice. Plant Signal. Behav. 2011;6:887–888. doi: 10.4161/psb.6.6.15306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H, Watanabe M, Sasabe M, Hiroe T, Tanaka T, Tsukaya H, et al. Novel receptor-like kinase ALE2 controls shoot development by specifying epidermis in Arabidopsis. Development. 2007;134:1643–1652. doi: 10.1242/dev.003533. [DOI] [PubMed] [Google Scholar]
- Tsuda K, Akiba T, Kimura F, Ishibashi M, Moriya C, Nakagawa K, et al. ONION2 fatty acid elongase is required for shoot development in rice. Plant Cell Physiol. 2013;54:209–217. doi: 10.1093/pcp/pcs169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Ito Y, Yamaki S, Miyao A, Hirochika H, Kurata N. Isolation and mapping of three rice mutants that showed ectopic expression of KNOX genes in leaves. Plant Sci. 2009;177:131–135. [Google Scholar]
- Watanabe M, Tanaka H, Watanabe D, Machida C, Machida Y. The ACR4 receptor-like kinase is required for surface formation of epidermis-related tissues in Arabidopsis thaliana. Plant J. 2004;39:298–308. doi: 10.1111/j.1365-313X.2004.02132.x. [DOI] [PubMed] [Google Scholar]
- Weng H, Molina I, Shockey J, Browse J. Organ fusion and defective cuticle function in a lacs1 lacs2 double mutant of Arabidopsis. Planta. 2010;231:1089–1100. doi: 10.1007/s00425-010-1110-4. [DOI] [PubMed] [Google Scholar]
- Yephremov A, Wisman E, Huijser P, Huijser C, Wellesen K, Saedler H. Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell. 1999;11:2187–2201. doi: 10.1105/tpc.11.11.2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.