SUMMARY
Long recognized to be potent suppressors of immune responses, Foxp3+CD4+ regulatory T (Treg) cells are being rediscovered as regulators of nonimmunological processes. We describe a phenotypically and functionally distinct population of Treg cells that rapidly accumulated in the acutely injured skeletal muscle of mice, just as invading myeloidlineage cells switched from a proinflammatory to a proregenerative state. A Treg population of similar phenotype accumulated in muscles of genetically dystrophic mice. Punctual depletion of Treg cells during the repair process prolonged the proinflammatory infiltrate and impaired muscle repair, while treatments that increased or decreased Treg activities diminished or enhanced (respectively) muscle damage in a dystrophy model. Muscle Treg cells expressed the growth factor Amphiregulin, which acted directly on muscle satellite cells in vitro and improved muscle repair in vivo. Thus, Treg cells and their products may provide new therapeutic opportunities for wound repair and muscular dystrophies.
INTRODUCTION
Regulatory T (Treg) cells, particularly those of the Foxp3+CD4+ subset, are critical regulators of immune responses (Josefowicz et al., 2012). They were originally described as controlling the activities of other T cells but were later recognized to regulate B cells and several innate immune system players. There have also been recent reports of Treg cell control over nonimmunological processes. Perhaps the best-characterized example is a unique population of Treg cells residing in the visceral adipose tissue (VAT) and regulating metabolic indices (Feuerer et al., 2009; Cipolletta et al., 2011). The prevalence, transcriptome, and T cell receptor (TCR) repertoire of this population are all distinct from those of their counterparts in lymphoid organs. Surveying a variety of tissues to see whether other nonimmunological processes might be controlled by analogous Treg populations, our attention was drawn to a substantial accumulation of Foxp3+CD4+ T cells in skeletal muscle undergoing repair after acute injury.
Skeletal muscle regeneration follows the same orchestrated plan regardless of the cause of muscle damage. It is driven largely by satellite cells, a pool of quiescent precursors closely associated with muscle fibers (Tabebordbar et al., 2013). In response to injury, these cells become activated, proliferate, differentiate, migrate, and fuse to form new myofibers. This series of events is controlled by the sequential activation and repression of specific transcription factors (Rudnicki et al., 2008). With muscular dystrophies, in which chronic myofiber loss occurs due to genetic defects, the satellite cell pool is called on repeatedly, so it can exhaust or lose function over time, dampening the repair process (Tabebordbar et al., 2013).
Regeneration of skeletal muscle is influenced by inflammatory events that accompany repair (Tidball and Villalta, 2010). Following an early, transient recruitment of neutrophils, myeloid mononuclear cells, mainly derived from a pool of circulating monocytes, infiltrate the injured tissue. Within days, the myeloid infiltrate transitions from a pro- to an anti-inflammatory phenotype, a shift that is critical for proper muscle repair. An initial population of proinflammatory, or M1-type, macrophages is required for clearance of apoptotic or necrotic cells and derivative debris; a subsequent population of anti-inflammatory, or M2-type, macrophages has various proregenerative functions, such as matrix remodeling and promotion of angiogenesis. Ablation or impaired recruitment of macrophages severely compromises muscle repair.
Though far less markedly, lymphocytes also accumulate in skeletal muscle after acute injury, as well as in the dystrophindeficient muscles of mice harboring the mdx mutation or humans with Duchenne muscular dystrophy (DMD) (Tidball and Villalta, 2010). Their function has not been well studied, although both CD4+ and CD8+ T cells seem to promote the mdx pathology. Even less is known about the composition and function of infiltrating T cell populations in models of acute muscle injury. In particular, the contribution of Treg cells is yet to be addressed.
Here, we uncover a unique population of CD4+Foxp3+ Treg cells that accumulates in skeletal muscle shortly after acute injury. We address the impact of this population on muscle repair, its implication in genetically determined muscle aberrancies, and the potential therapeutic effects of modulating this population and one of the proregenerative factors it produces.
RESULTS
Treg Cells Accumulate in Acutely Injured Skeletal Muscle Just as the Myeloid Cell Infiltrate Switches from a Proinflammatory to a Proregenerative Phenotype
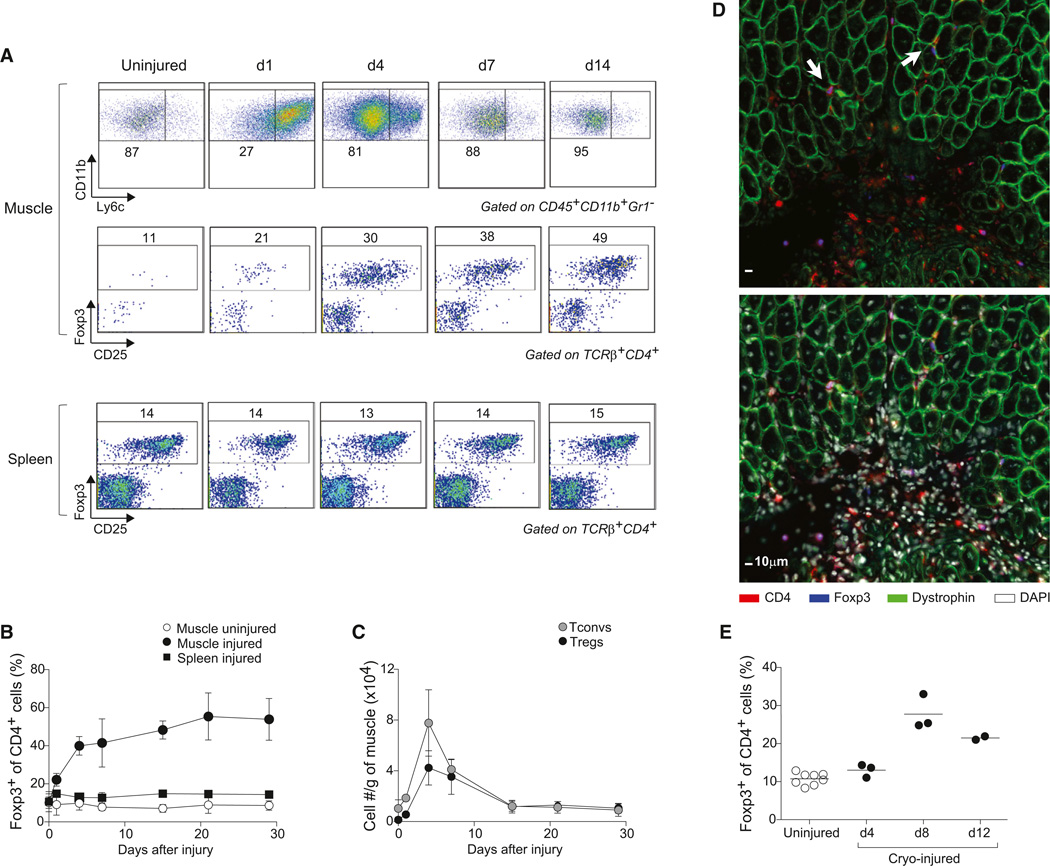
Intramuscular (i.m.) administration of cardiotoxin (Ctx), which rapidly induces myofiber necrosis, provides a convenient and well-studied model of tissue repair after acute injury. We injected the tibialis anterior (TA), gastrocnemius, and quadriceps muscles of C57Bl/6 (B6) mice with Ctx and analyzed the resulting cellular infiltrate various times later; control cells isolated from the uninjured contralateral muscles or the spleen were evaluated in parallel. As anticipated (Arnold et al., 2007), there was a rapid increase in CD11b+Gr1− myeloid mononuclear cells beginning before day 1, initially a proinflammatory Ly6chi population but switching to an anti-inflammatory Ly6clo population by day 4 (Figure 1A, top). Around this time, Treg cells began to accumulate in the injured muscle, their frequency within the CD4+ T cell compartment gradually increasing to ∼50% by 14 days after Ctx injection and remaining at that level until at least day 30 (Figures 1A, middle, and 1B). Numbers of Treg and conventional CD4+ T (Tconv) cells both peaked at day 4 after injury (Figure 1C), mirroring the total number of CD45+ cells and T cells in the infiltrate (Figure S1 available online). However, while Tconv cell numbers had dropped to levels characteristic of uninjured muscle by 28 days after Ctx injection, the number of Treg cells remained elevated by 8-fold (1.05 ± 0.38 × 104 versus 0.13 ± 0.06 × 104 cells/g muscle; p = 0.01; Figure 1C). Staining of frozen sections with a fluorescently tagged anti-Foxp3 monoclonal antibody (mAb) revealed Foxp3+ cells both in heavily infiltrated (probably necrotic) areas and in regions between regenerating fibers (recognizable as centrally nucleated, dystrophin-positive cells) (Figure 1D). An analogous accumulation of Treg cells was observed in a cryoinjury model (Figure 1E).
Figure 1. Treg Accumulation at the Site of Injury.
(A–C)Cytofluorometric analysesofhindlimb muscle infiltratesafter Ctx injury. (A) Top: Ly6c expression by CD11b+ cells. Middle and bottom: Foxp3 expression by CD4+ cells. Numbers depict % of gated cells. Representative of three experiments. (B and C) Summary data on fraction and number of cells. Mean ± SD (n ≥ 4).
(D) Immunofluorescence microscopy of muscle sections 7 days after injury. The same section without (top) or with (bottom) DAPI. Arrows show Tregs in close contact with regenerating myofibers. Original magnification ×400.
(E) As in (B), but after cryoinjury.
See also Figure S1.
The Transcriptome of Muscle Treg Cells Is Distinct from that of Other Treg Populations, Especially Those Located in Lymphoid Organs
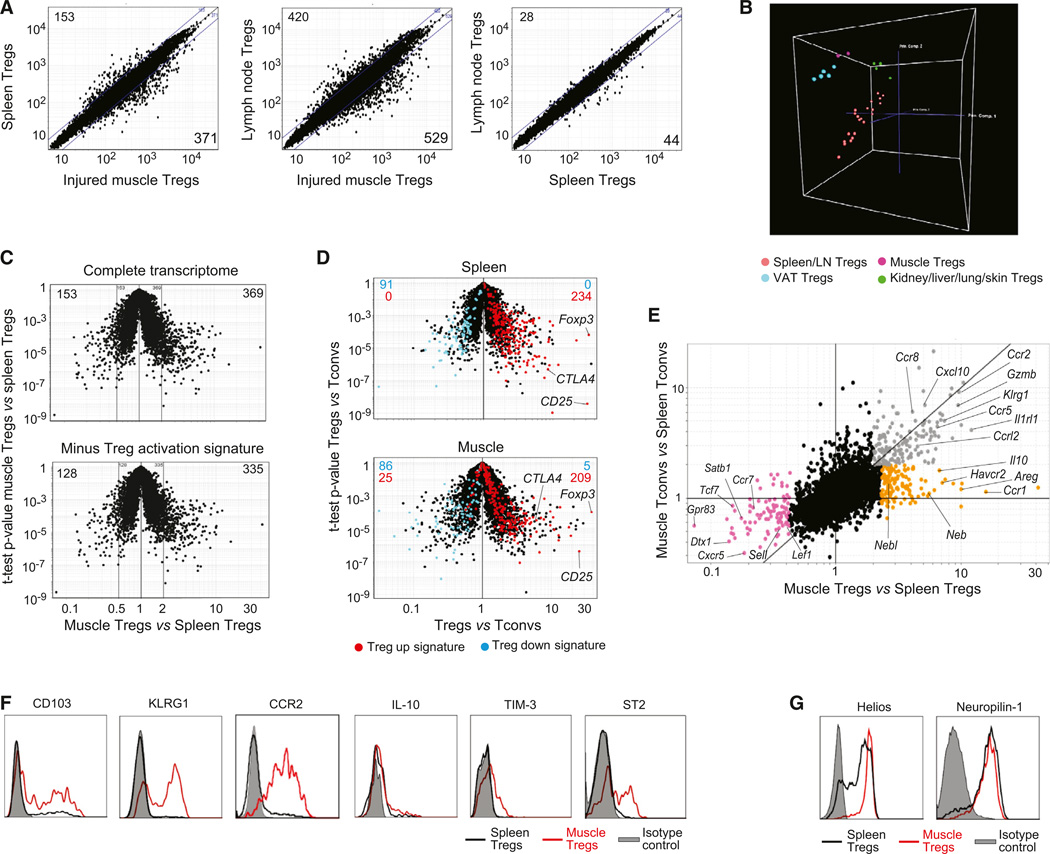
Four or 14 days after i.m. Ctx injection, we isolated Treg and Tconv cells from muscles and lymphoid organs and performed microarray-based gene-expression profiling. (Note that inadequate numbers precluded a comparison with analogous populations from uninjured muscles.) According to both simple comparison plots (Figure 2A) and principal components analysis (PCA) (Figure 2B), the transcriptome of muscle Treg cells differed from that of their spleen or lymph node counterparts much more than the latter two did from each other. Muscle Tregs were most like Treg cells located in adipose tissue (Figure 2B) but were still readily distinguishable; a few hundred transcripts up- or downregulated >2-fold in one vis-à-vis the other. The similarity to another Treg population residing in nonlymphoid tissue, and dissimilarity to lymphoid-organ Treg cells, did not simply reflect a higher activation state in tissues, because few of the distinguishing transcripts were members of a previously determined Treg activation signature (Hill et al., 2007) (Figure 2C). Neither did it reflect a universal “inflammation signature,” since the muscle Treg transcriptome was distinguishable from those of Tregs at several inflamed sites (Figure S2). While exhibiting a distinct gene-expression profile, muscle Treg cells are clearly “Treg,” showing the anticipated pattern of expression of 91% of the canonical Treg signature (Hill et al., 2007); in particular, elevated levels of diagnostic transcripts such as those encoding Foxp3, CD25, and CTLA-4 (Figure 2D).
Figure 2. A Unique Population.
(A–E) Gene-expression analyses on cells 14 days after Ctx injury. (A) Comparison plots of normalized expression values. Numbers indicate the number of genes whose expression differed by more than 2-fold. (B) PCA analysis comparing muscle Tregs 4 or 14 days after Ctx injury with other Treg populations. Each dot represents the meanofthree independentexperiments. (C) Top: volcano plot comparing gene expression ofmuscle versus spleen Tregs. Bottom: same plot after subtraction of the Treg activation signature (Hill et al., 2007). Numbers represent the number of differentially expressed genes (>2-fold). (D) Volcano plots comparing gene expression of Treg versus Tconv cells. Treg signature genes (Hill et al., 2007) are highlighted in red (induced) or blue (repressed). Numbers represent the number of genes from each signature expressed by each population. (E) Fold-change differences in gene expression between Treg and Tconv cells from muscle and spleen. Differentially expressed genes are highlighted in orange, gray, or pink.
(F and G) Cytofluorometric analyses of surface and intracellular markers. Histograms depict expression by Foxp3+ T cells from muscle (red) or spleen (black).
Gray: isotype control.
A fold-change/fold-change (FC/FC) plot afforded a more detailed look at the muscle Treg transcriptome, revealing a set of genes (highlighted in orange) that distinguish muscle Treg from spleen Treg cells and spleen or muscle Tconv cells, and another set (in gray) overexpressed by the two muscle populations vis-à-vis their two spleen counterparts (Figure 2E; Table S1). The first group includes loci encoding an anti-inflammatory cytokine (interleukin [IL]-10), chemokine receptors (e.g., CCR1), and two well-known growth factors (platelet-derived growth factor [PDGF] and Amphiregulin [Areg]] (Pastore et al., 2008). Loci upregulated in both Treg and Tconv cells from injured muscle include those encoding KLRG1, an activation marker; CCR2, important for the recruitment of various leukocyte populations to injured muscle (Warren et al., 2005); and ST2 (encoded by Il1rl1), which is the receptor for the “alarmin” IL-33 (Schmitz et al., 2005). A third group of genes (in pink) was repressed in muscle Treg (and to varying degrees Tconv) cells; notably, those encoding certain chemokine receptors (CXCR5 and CCR7) and several proteins implicated in the Wnt signaling pathway (TCF7, LEF1, SATB1).
We selected several of the genes upregulated in muscle Treg cells and confirmed their induced expression at the protein level by flow cytometry (Figure 2F). Such an analysis also revealed muscle Tregs to be high-level expressors of Helios and Neuropi-lin (Figure 2G) and were therefore likely to be Treg cells exported as such from the thymus.
Treg Cells in Injured Muscle Are Clonally Expanded and Display a Unique Repertoire of TCRs
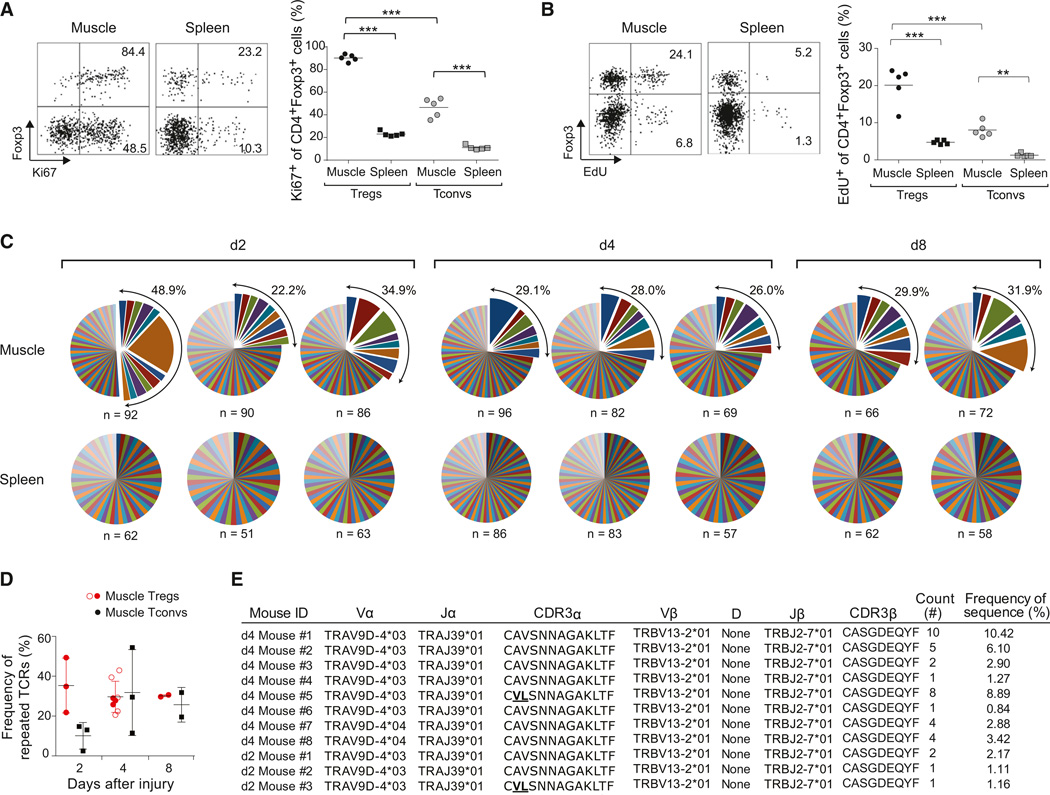
The rapid accumulation of Treg cells at the injury site could reflect their influx, proliferation, or some combination of the two. Four days after Ctx injection, almost all muscle Treg cells stained for Ki67 (Figure 3A), which is expressed in all stages of the cell cycle except for the resting, G0, phase. EdU (5-ethynyl-2′-deoxyuridine), incorporated by cells in S phase, was detected in 20% of muscle Tregs after 24 hr of labeling (Figure 3B). A much lower fraction of splenic Treg cells was proliferating according to both staining criteria. The muscle Tconv population also exhibited an increased frequency of Ki67+ and EdU+ cells compared with the spleen Tconv subset, although the values remained significantly lower than those observed for muscle Treg cells (Figures 3A and 3B).
Figure 3. Clonal Expansion of the Treg Population at the Injury Site.
(A and B) Ki67 expression (A) and EdU uptake (B) 4 days after Ctx-induced injury. Left: representative dot plotsofcells. Numbers refer to % marker+ Treg or Tconv cells. Right: summary plots. n = 5; **p < 0.01; ***p < 0.001.
(C) TCR CDR3 α and CDR3 β sequences for individual cells. Each pie chart represents a single mouse. n, number of analyzed sequences per mouse. Total frequency of expanded clones shown at the top right.
(D) Summary of clonal TCR frequencies over time. Open circles correspond to mice analyzed later in independent experiments for Figure 7. Mean ± SD (n ≥ 3).
(E) Identical paired CDR3-α and β sequences found in multiple individuals in independent experiments. Two CDR3 α sequences from independent mice bear two conserved substitutions.
See also Table S2.
To assess clonality of the Treg population expanding in acutely injured skeletal muscle, we studied TCR repertoires by sequencing the complementarity-determining region (CDR) 3 of the TCR α and TCR β chains of individually sorted cells. Already at day 2 after injury, and continuing out at least until day 8, a substantial proportion (20%–40%) of muscle Treg cells were clonally expanded (Figure 3C and 3D; Table S2). This sequence profile contrasted starkly with that of splenic Treg cells, for which there was not one repeat sequence in the 692 sampled. As expected from the Ki67 and EdU stainings, we also found clonal expansion of muscle Tconv cells (Figure 3D; Table S2). However, there was not a single example of TCR-sequence sharing between muscle Treg and Tconv cells.
In the muscle Treg data sets, there was a striking case of a repeatedly detected TCR: identical paired CDR3 α and β (and corresponding nucleotide) sequences picked up in multiple mice in completely independent experiments (Figure 3E). Interestingly, there were a few Treg cells (from multiple individuals) that had the same TCR β chain paired with an α chain whose CDR3 was modified only conservatively (AV→VL). These repeat sequences were found in mice 2 or 4, but not 8, days postinjury. They never showed up in the bulk spleen Treg or muscle Tconv cell pools.
Ablation of Treg Cells Compromises Muscle Repair According to Multiple Criteria
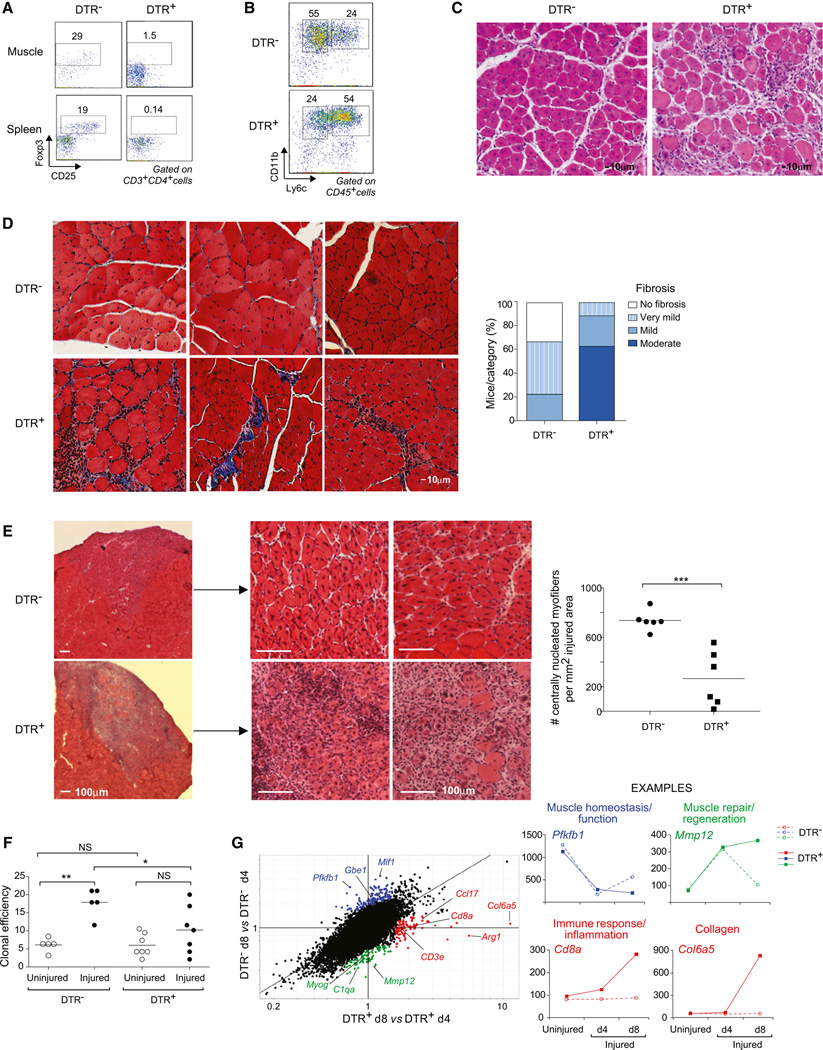
Having uncovered a unique population of Treg cells in the infiltrates of skeletal muscle undergoing repair in response to acute injury, we asked whether they play a role in muscle regeneration. We punctually ablated Treg cells at the time of muscle injury, exploiting a mouse line wherein the diphtheria toxin receptor (DTR) is expressed under the control of Foxp3 regulatory elements (Kim et al., 2007). B6.Foxp3-DTR+ and B6.Foxp3-DTRT− littermates (hereafter referred to as DTR+ and DTR−, respectively) were given DT intraperitoneally (i.p.) starting from the time of im Ctx injection and were analyzed 1 week later, a time chosen to avoid the multiorgan autoimmunity provoked by long-term ablation of Treg cells (Kim et al., 2007). This protocol resulted in effective depletion of Tregs in the injured muscle of the DTR+ mice (Figure 4A, top) as well as in the lymphoid organs (Figure 4A, bottom).
Figure 4. Treg Ablation Impairs Muscle Regeneration after Wounding.
Eight-week-old Foxp3−DTR+ or DTRT− littermates were injured with Ctx and treated with DT to induce Treg depletion.
(A and B) Flow cytometry of Treg and myeloid-lineage population, 7 days postinjury. Numbers in (A) refer to % CD4+ T cells that are Foxp3+, and numbers in (B) refer to % Ly6c−/lo or Ly6chi of CD11b+ cells.
(C) Hematoxylin and eosin (H&E) staining of muscle sections 7 days after Ctx injury. Representative of at least three experiments. Original magnification ×100.
(D) Left: fibrosis detection by Gomori trichrome staining of muscle sections 13 days after injury. Collagen stained in blue. Representative examples of seven DTR+ and seven DTR− mice. Original magnification ×200. Right: summary quantification.
(E) Left: H&E staining of muscle sections 7 days after cryoinjury. Original magnification ×25 (left) and ×100 (middle and right). Representative examples of six samples. Right: quantification of regenerative fibers per mm2 of injured area. ***p < 0.001.
(F) Clonal myogenesis assays of satellite cells from uninjured or day 4 Ctx-injured muscles. Percent of wells seeded that showed a myogenic colony at day 5. Summary of six clonal assays *p < 0.05; **p < 0.01.
(G) Left: FC/FC plot of gene expression values in muscle 4 versus 8 days after Ctx-induced injury in mice without (DTR+) or with (DTR−) Tregs. Highlighted in different colors are sets of genes described in the text. Right, representative examples of genes in each highlighted set.
According to multiple criteria, the loss of Treg cells had profound effects on the muscle repair process. First, the size of the cellular infiltrate was increased in the absence of Treg cells, assessed either as numbers of total CD45+ cells or as the fraction of T cells (Figure S3A). In addition, the myeloid cell compartment failed to undergo the expected switch from a primarily proinflammatory, Ly6chi to a primarily anti-inflammatory, Ly6clo phenotype (Figure 4B and Figure S3A). Similar results were obtained when DT was administered i.m., which specifically depleted muscle Treg cells without detectably affecting their counterparts in lymphoid organs (data not shown).
DTR-mediated in vivo ablation of a designated cell population is known to be apoptotic and noninflammatory (Bennett and Clausen, 2007). Nonetheless, as detailed in Figure S3B and its legend, we performed an experiment on female heterozygous DTR-positive mice to show that the more inflammatory flavor of the infiltrate in mice lacking Treg cells was not a simple artifact related to their death, but rather a reflection of their functional absence.
Second, Treg cell ablation altered the histological features of skeletal muscle repair (Figure 4C). Although centrally nucleated fibers indicative of regeneration could be detected in muscle from both DTRT− and DTR+ mice, in the latter case, the tissue structure showed a disorganized pattern, with several foci of inflammation. As anticipated, no infiltrate or regenerating fibers were found in the contralateral, uninjured muscles from mice that did or did not have Treg cells (data not shown). One of the later consequences of impaired muscle repair is fibrosis: Gomori’s Trichrome staining showed Treg-less mice to have a substantial accumulation of collagen in the injured muscle compared with their Treg-positive littermates (Figure 4D). To provide a more quantitative view, we returned to the cryo-injury model, wherein the area of injury is clearly delimited. Global examination confirmed the impaired reparative capacity in Treg depleted mice; a quantitative evaluation indicated that the number of centrally nucleated fibers was significantly lower in Treg-depleted than in normal muscles, with some muscles from DTR+ mice showing an almost complete absence of regenerative fibers (Figure 4E).
Third, the absence of Treg cells during muscle repair had an impact on muscle progenitor cells. Satellite cells are the predominant, if not sole, source of regenerated muscle fibers after acute injury (Tabebordbar et al., 2013). Satellite cells were isolated from uninjured or Ctx-injured muscle of DT-treated DTR+ or DTRT mice by double-sorting CD45−Sca-1−Mac-1−CXCR4+ |31-integrin+ myofiber-associated cells (Cerletti et al., 2008), and their functionality was evaluated in clonal myogenesis assays, as described in (Cerletti et al., 2012) (Figure 4F). Injury substantially enhanced the fraction of satellite cells from DTR− mice that formed myogenic colonies; in contrast, there was far less (not significant) injury-induced enhancement of myogenic activity in mice lacking Treg cells.
Fourth, to obtain a broad, unbiased view of the repair pathways impacted by Treg ablation, we performed microarray-based gene-expression profiling of whole, unfractionated muscle tissue. In general, for standard (DTR−) mice, sets of genes were up- or downregulated early after injury (day 4), and expression values began to return to normal as the wound started to repair (day 8). The pattern of expression of many genes was altered in the absence of Treg cells (DTR+) (Figure 4G, Figure S3C, and S3D; Table S3), with three main clusters meriting discussion: One group (highlighted in blue) is composed of genes encoding proteins with important roles in muscle homeostasis and function. These loci were highly expressed in uninjured muscle and were downregulated in both DTR− and DTR+ mice at day 4 after injury owing to the loss of mature muscle fibers. In muscle of DTR− mice, transcript levels began to increase by day 8, as efficient repair ensued; however, in muscle of DTR+ individuals, expression of these loci remained low or in decline at day 8, confirming that the lack of Treg cells compromised the recovery of muscle homeostasis after injury. Another group (green) includes genes encoding proteins necessary for muscle repair, like MyoG (myogenin) and Mmp12 (metallopeptidase-12), but also some factors related to the immune response, in particular a number of chemokines and chemokine receptors, some cytokine receptors, and C1qa, a complement cascade trigger. In DTR− mice, expression of these loci was increased at day 4 but rapidly crashed thereafter, approaching the level in muscles of uninjured mice by day 8. However, in DTR+ mice, this drop did not occur or was greatly attenuated, again suggesting an ineffective and prolonged repair process, especially given the recent report that C1qa and related molecules strongly inhibit muscle regeneration (Naito et al., 2012). Strikingly, the expression pattern of C1qa in injured muscle was mirrored by that of C1qb, C1qc, C1r, C1rb, and C1s as well (Figure S3D). A final group (highlighted in red) is composed of two types of genes: those encoding molecules characteristic of immune cells (e.g., CD8a, CD2) and those specifying matrix proteins (e.g., Col6a5). Expression of these loci was upregulated at day 4 and even more so at day 8 only in mice lacking Treg cells, reflecting their more pronounced muscle infiltrate (Figure S3A) and fibrotic collagen deposition. Microarray expression values for multiple examples of each of these groups are plotted in Figure 4G and Figure S3D, and confirmatory quantitative PCR data for representative group members are presented in Figure S3C. Monitoring expression of these groups of genes should provide a novel and convenient means to quantitatively assess the fidelity of events underlying muscle repair.
As illustrated in Figure S3E and S3F, Rag-deficient mice, lacking all lymphoid cells, showed more pronounced fibrosis than did wild-type individuals, measured histologically or by quantifying collagen transcripts. However, fibrosis was milder and repair progressed more effectively in Rag-1-deficient mice than in Treg-depleted mice.
In summary, evaluations of myeloid cell numbers and phenotype, histology, satellite cell function, and global patterns of gene expression all argue that Treg cells are important for effective repair after acute injury of skeletal muscle. These observations raise the question of whether Treg cells impact muscle aberrancies in other contexts.
An Analogous Treg Population Is Expanded in the Damaged Muscles of Mouse Models of Muscular Dystrophy
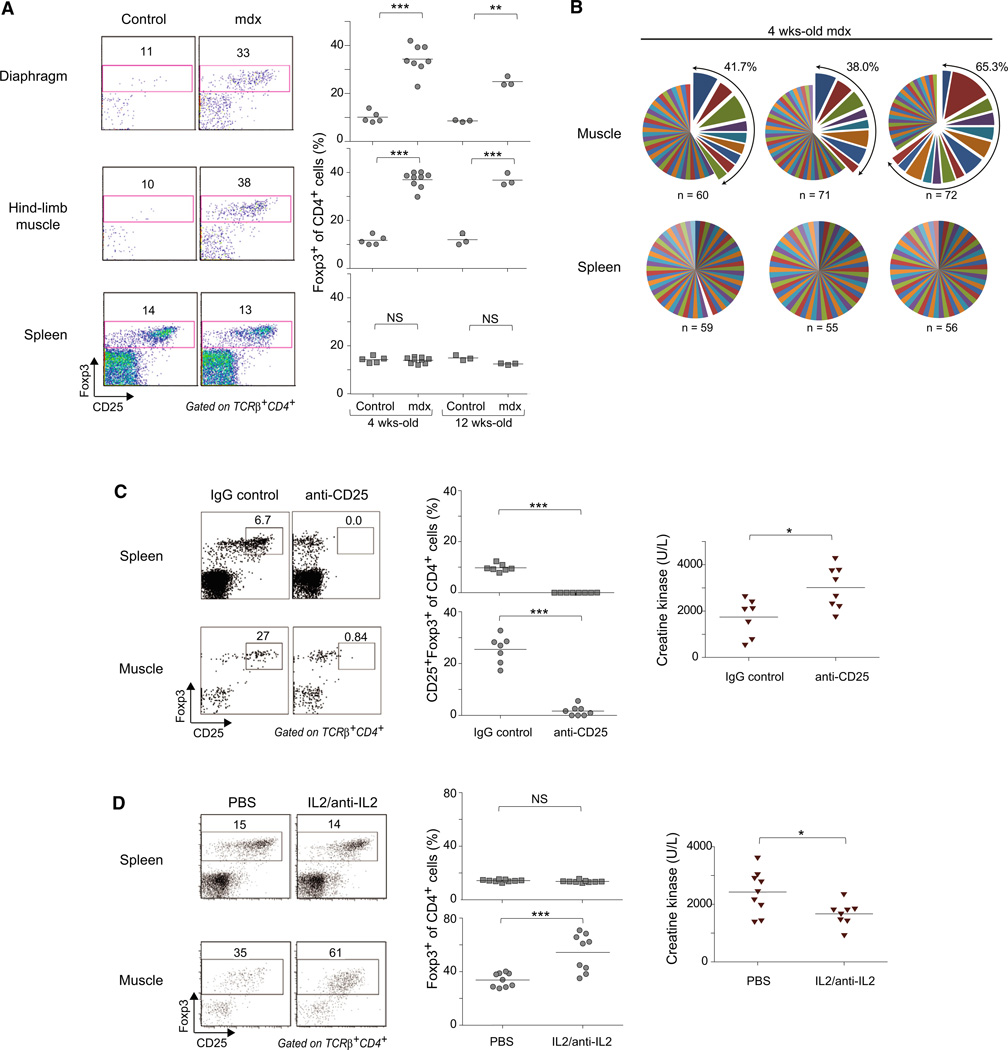
In muscular dystrophies, chronic destruction of muscle fibers is accompanied by an inflammatory response and, in general, a permanent infiltrate. Mdx mice carry a nonsense mutation in the Dmd gene (encoding Dystrophin), which is analogous to those found in DMD patients. In the mouse model, as in the human disease, the most affected sites are the diaphragm and hindlimb muscles. We analyzed the muscle infiltrate in 4-week-old mdx mice, when the disease is in its acute phase, and at 12 weeks, during the chronic phase. In both muscle groups at both ages, the fraction of Treg cells was significantly elevated vis-à-vis the frequency in corresponding muscles of wild-type mice; such an increase was not observed in the spleen (Figure 5A). Likewise, the muscle infiltrates of Dysferlin-deficient mice, which model limb-girdle muscular dystrophy type 2B or Miyoshi myopathy (Tabebordbar et al., 2013), were enriched in Treg cells, while their frequency in the spleen was the same as in wild-type controls (Figure S4A).
Figure 5. Treg Cells Are Enriched in Muscles of Dystrophic Mice and Impact Muscle Damage.
(A) Frequency of Foxp3+ cells for mdx and control C57Bl/10 mice. Left: representative dot plots. Right: summary data. n = 5; **p < 0.01; ***p < 0.001.
(B) Treg clonal expansion in mdx muscle measured as described in the legend to Figure 3C.
(C) Loss-of-function experiments. Mdx mice treated with anti-CD25 mAb (clone PC61) over 10 days.
(D) Gain-of-function experiments. Mice treated with complexes of recombinant IL-2 and anti-iL-2 mAb (clone JES6-1A12).
(C and D) Left: representative dot plot numbers refer to % of cells in the indicated gates in total CD4+ cells. Middle: summary data. Right: quantification of serum CKe. *p < 0.05; ***p < 0.001.
That the Treg cells accumulating in dystrophic muscle of mdx mice were bona fide muscle Tregs was indicated by their elevated expression of phenotypic markers such as Areg, ST2, KLRG1, and TIM-3 (Figure S4B). (Unfortunately, we could not isolate adequate numbers of Treg cells from the muscles of mdx mice to perform a transcriptome analysis [<500 Tregs/mouse].) In addition, the Treg populations of mdx muscle exhibited clonal expansions, with an even greater fraction of the population derived from expanded clones and an even larger average clone size than found for Ctx-injured muscles of wild-type mice (compare Figure 5B and Figure 3C; also see Table S2).
Next, we performed loss- and gain-of-function studies to evaluate the role(s) of muscle Treg cells in mdx mice. It was not feasible to use the B6.Foxp3-DTR system for the loss-of-function experiments because both the Foxp3 and Dmd genes are located on the X chromosome. Therefore, we manipulated levels of Treg cells by treatment with an anti-CD25 mAb, an approach that has been exploited by a number of investigators to study Treg function (e.g., Suvas et al., 2004). Administration of anti-CD25 to 2.5-week-old mdx mice did not reduce overall Treg cell numbers, whether in skeletal muscle or the spleen, but did largely eliminate the fraction expressing high levels of CD25 (Figure 5C). This change was accompanied by a significant increase in serum levels of creatine kinase (CK), an enzyme released into the blood upon muscle damage and a standard indicator of muscle damage in dystrophic mouse models.
For the gain-of-function experiments, we made use of the published observation that complexes of recombinant IL-2 and a particular anti-IL-2 mAb (clone JES6-1A12) can preferentially expand Treg populations after injection into mice (Boyman et al., 2006). A few days before the peak of acute disease (3 weeks of age), we administered IL-2:JES6-1A12 complexes to mdx mice for 6 consecutive days. As anticipated, this treatment induced a significant increase in the frequency of Treg cells in the muscle, which, in addition, displayed higher levels of CD25 (Figure 5D, bottom). An elevated fraction of Treg cells was also observed in the spleen immediately after the cessation of treatment (14% and 42% of CD4+ cells in control and complex-administered mice, respectively; data not shown), but this increase was not sustained (Figure 5D, top). Accompanying the boost in Treg cells was a significant reduction in serum CK levels (Figure 5D), indicative of less muscle damage.
To gain further insight into the effects of Treg depletion in dystrophic muscle, we performed whole-muscle transcriptional analyses, comparing muscles from control and Treg-depleted mdx mice (Figure S4C). The genes encoding osteopontin (Spp1; 2.1-fold) and connective-tissue growth factor (Ctgf; 1.8-fold), both of which promote skeletal muscle fibrosis and mdx pathology (Morales et al., 2011; Vetrone et al., 2009), were upregulated in the absence of Tregs. More generally, it became clear that, on the one hand, Treg cells were protecting mdx mice from muscle pathology, as their ablation downregulated most of the aforementioned gene set related to muscle homeostasis and function (highly expressed in healthy wild-type muscle), but that, on the other hand, Treg cells were promoting muscle repair because their removal upregulated expression of the set of genes whose inhibition was normally an accompaniment to healthy repair (Figure S4C). (In addition, there was a striking correspondence between the genes induced in muscle from Treg-ablated mdx mice and those repressed in muscle from amphiregulin-treated mice [discussed below].)
Amphiregulin, a Growth Factor Overexpressed by Muscle Treg Cells, Enhances Muscle Regeneration
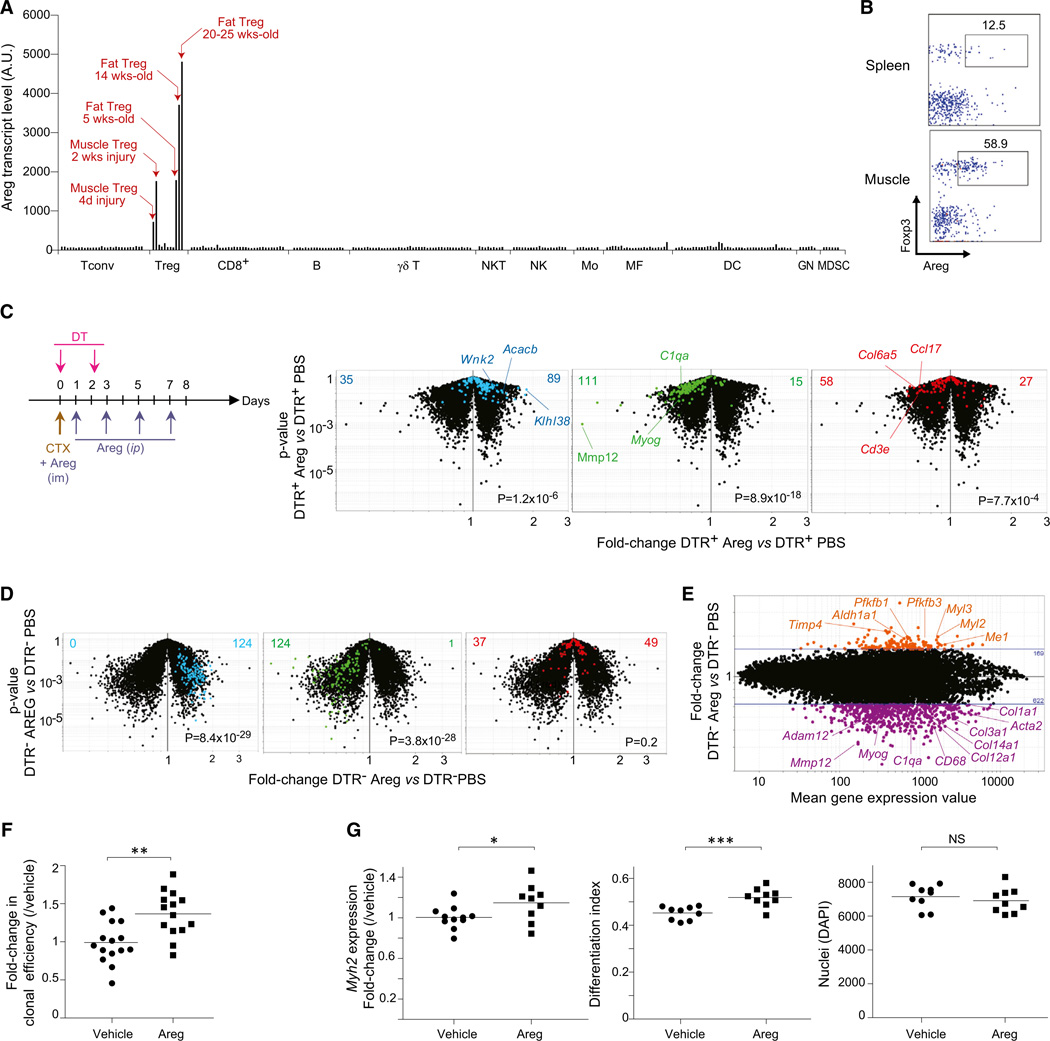
The regulation that muscle Treg cells imposed on infiltrating myeloid cells was probably responsible, at least in part, for the impaired muscle repair observed in the absence of Tregs. However, analogous to the situation with VAT (Feuerer et al., 2009; Cipolletta et al., 2012), it is likely that other mechanisms also play a role, including a direct effect of muscle Tregs on muscle progenitors, nascent myofibers, or other nonhematopoietic cell types within muscle. Among the transcripts preferentially expressed by muscle Tregs vis-à-vis lymphoid-organ Tregs, Areg stood out as encoding a candidate factor capable of directly impacting muscle regeneration. Areg belongs to the epithelial growth factor (EGF) family and signals through the EGF receptor (EGFR) system (Shoyab et al., 1989). EGFR is expressed by a variety of cells, including muscle satellite cells and a myoblast cell line, in which it appears to have antiapoptotic/survival functions (Golding et al., 2007; Horikawa et al., 1999). Examination of the ImmGen database (http://www.immgen.org) indicated that most hematopoietic cell-types express no or only low levels of Areg transcripts (Figure 6A). Nor is Areg expressed at significant levels in the nonhematopoietic cell-types examined by ImmGen (Figure S5A). In addition, the base-line levels of Areg transcriptsininjuredoruninjuredwhole-musclesamples, wherein muscle-lineage cells were in great preponderance, argues that muscle cells do not make significant Areg in this context either (Figure S5A). However, Areg transcripts were readily detectable in a few Treg populations. They were found at high levels in muscle and adipose-tissue Treg cells, although hardly, if at all, in lymphoid-organ Tregs. Substantial expression of Areg protein in muscle Tregs, but only limited expression by their splenic counterparts, was confirmed by flow cytometry (Figure 6B). The fraction of muscle Treg cells expressing Areg protein began to increase at day 4 and peaked at day 7 after Ctx injury (Figure S5B).
Figure 6. Areg Improves Muscle Repair after Injury.
(A) Expression of Areg in ImmGen microarray data sets from hematopoietic-lineage cells. Mo, monocytes; MF, macrophages; DC, dendritic cells; GN, granulocytes; MDSC, myeloid-derived suppressor cells; AU, arbitrary units.
(B) Expression of Areg 2 weeks after Ctx-induced injury. Numbers represent % Foxp3+ cells that are Areg+.
(C) Volcano plots of transcriptomes of whole muscle from injured DTR+ (Treg-less) mice treated or not treated with Areg. Left: experimental protocol. Right: superimposition of transcript sets highlighted in Figure 4G and identically color-coded. Values refer to the number of genes upregulated (right) ordownregulated (left) in Areg-treated versus PBS-treated muscles. p values from a χ2 test.
(D) As in (C), except DTRT− mice were used.
(E) The same data set shown in (D) plotted as the fold-change for Areg- versus PBS-treated muscles versus the mean expression value for each transcript. Highlighted in orange and purple are genes up- ordownregulated by Areg, respectively.
(F) Myogenic clonal assay in the presence of Areg. Results were plotted as the fold-change over the mean of vehicle-control treated samples per experiment. Data represent mean ± SD and five independent experiments. **p < 0.01.
(G) Induction of satellite cells differentiation by Areg. Satellite cells were bulk-cultured in the presence of Areg or vehicle control for 12 days. Left: MyHC mRNA titered by quantitative PCR, represented as fold-change expression of Myh2 relative to vehicle control. Center: MyHC protein quantified by immunofluorescence microscopy; differentiation index calculated as the mean ratio of MyHC-positive nuclei to the total nuclei per field of view. Right: sum of DAPI-positive nuclei per well. Data represent mean ± SD and four experiments. *p < 0.05; ***p < 0.0001. See also Figure S5.
If Areg is important for the proregenerative function of muscle Treg cells, then treatment of Treg-ablated mice with this growth factor should ameliorate their deficit in muscle repair subsequent to acute injury. To test this notion, we administered Areg to Ctx-injected, DT-treated DTR+ mice according to the protocol illustrated in Figure 6C, and evaluated gene-expression profiles of repairing muscles at days 4 and 8 after injury. (Note that the initial Areg injection was done i.m., promoting the muscle specificity of its effects.) Transcriptome changes were evaluated in terms of the three gene clusters defined in Figure 4G and detailed above. Expression of all three gene groups was largely normalized in Areg-treated Treg-less mice: vis-à-vis buffer-only controls: “muscle homeostasis/function” genes were upregulated; muscle “repair/regeneration” loci were repressed as required for effective repair; and inflammatory transcripts and collagens were also downregulated (Figure 6C). These results were confirmed in representative RT-PCR assays (Figure S5C).
We also tested whether administration of Areg was effective in a context where Tregs were present as normal (DTRT− or B6 mice). Under these conditions, Areg had a strong and rapid impact on the “muscle homeostasis/function” and “repair/regeneration” gene groups but no significant effect on the “immune response/inflammation” groups, at least according to volcano-plot visualization and representative RT-PCR assays (Figure 6D and Figure S5D). More detailed analysis showed that Areg dampened expression of several loci encoding proteins related to fibrosis (e.g., a battery of collagens, Acta2, Adam12), and enhanced expression of loci encoding molecules highly represented in healthy muscle, like Pfkfb1 and 3 and Myl2 (Figure 6E).
Because our previous results indicated that Treg cells can influence the efficacy of myogenic colony formation by muscle precursor cells (Figure 4F), and given the high-level expression of Egfr by the satellite cells employed in our assays (and, parenthetically, minimal expression by muscle Treg cells; Figure S5E), we investigated whether Areg might have a direct effect on satellite cells. Addition of Areg to myogenic assays significantly augmented the colony-forming efficiency of satellite cells from unmanipulated B6 mice (Figure 6F). In addition, Areg enhanced myogenic differentiation in bulk cultures of satellite cells, as indicated both by increased levels of transcripts encoding myosin heavy chain (MyHC) (Figure 6G, left) and by increased numbers of cells that stained for MyHC protein (Figure 6G, center). In contrast, there was no apparent difference in proliferation in satellite cell cultures with or without Areg (Figure 6G, right).
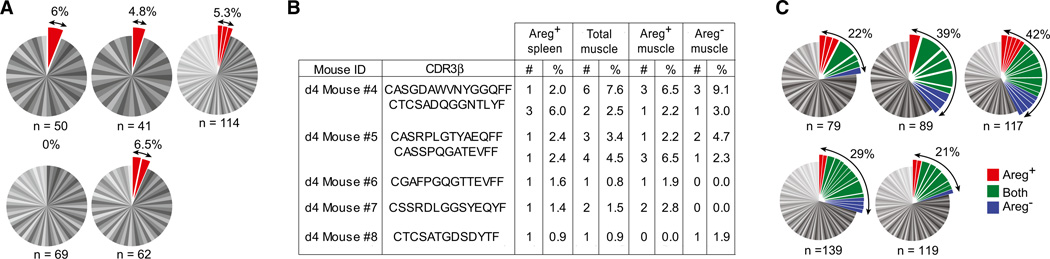
Since Areg was expressed by a small fraction of splenic Treg cells in addition to a considerable proportion of muscle Tregs (Figure 6B), we wondered whether the two Areg+ populations might somehow be related. As opposed to the lack of clonality characteristic of bulk splenic Treg cells (Figure 3C), ~5% of the splenic Areg+ subset was clonally expanded (Figure 7A). There was, indeed, some degree of overlap between the CDR3 α and CDR3 β sequences of splenic Areg+ Treg cells and those of muscle Tregs (independent of their expression of Areg) (Figure 7B). The muscle Areg+ and Areg− subsets showed no difference in their extent of clonal expansion (Figure 7C) or in their representation of the oft-repeated TCR sequence illustrated in Figure 3E (Table S2).
Figure 7. Sharing of TCR Sequences between Areg+ Splenic and Muscle-Infiltrating Treg Cells.
(A) TCRCDR3 α and β sequences for individual Areg+ splenic Treg cells. Each pie chart represents a single mouse. n, number of analyzed sequences per mouse. Total frequency of expanded clones shown at the top right.
(B) Identical paired CDR3 α (data not shown) and β sequences found in splenic Areg+ and muscle Areg+ and Areg− Treg cells from the same individual in independent experiments. #: number of times the sequence was found. %: frequency of Tregs expressing the same sequence.
(C) As in (A), for muscle Treg cells. Clones represented in Areg+, Areg−, or both subsets of muscle Tregs were labeled in red, blue, and green, respectively. See also Table S2.
DISCUSSION
A Unique Population of Treg Cells that Accumulates in Skeletal Muscle
There is growing appreciation that the Treg compartment is heterogeneous, has multiple functions, and exerts influences exceeding the boundaries of the immune system (Josefowicz et al., 2012). Here, we have described a previously unrecognized population of Treg cells that emerges in skeletal muscle subjected to acute or chronic damage and have demonstrated that these cells are important players in the muscle-repair process.
Treg cells began to accumulate in injured muscle within days after an insult. That this enrichment represents a conversion of the Tconv to the Treg phenotype is unlikely given that essentially all of the muscle Treg cells expressed Helios and Neuropilin, considered to be markers of Treg cells exported as such from the thymus, and that none of their TCRs were shared with muscle (or lymphoid-organ) Tconv cells. Rather, the elevated levels of muscle Tregs appear to reflect population expansion: they were proliferating in the muscle, substantially more so than in the spleen, and constituted a number of clonally expanded micropopulations. There might also be preferential recruitment of muscle-type Tregs to injured muscle, perhaps driven by one or more members of the constellation of chemokine receptors up- or downregulated vis-à-vis lymphoid-organ Tregs, certain of which have been implicated in migration of diverse leukocyte subsets to skeletal muscle (Warren et al., 2005). Interestingly, a small population of Areg+ splenic Treg cells appeared to include a muscle Treg constituent, evidenced by shared TCR sequences. But it is not clear at present whether the splenic population seeds or arises from the Treg population accumulating in injured muscle.
Several other elements of the muscle Treg transcriptome distinguish it from that of lymphoid-organ Treg cells. Included in the list of repressed genes is Satb1, which encodes a chromatin organizer that inhibits Treg suppressive activity (Beyer et al., 2011). The reduced numbers of Satb1 transcripts, coupled with enhanced expression of transcripts encoding other molecules linked to Treg suppressive function, like IL-10, Gzmb, CTLA-4, and TIM-3 (Josefowicz et al., 2012), support the idea that muscle Treg cells might be endowed with especially strong suppressor activity. Moreover, muscle Treg cells overexpressed certain genes whose products could arm them to perform particularly effectively in the context of regenerating muscle. Areg is a prime example. Interestingly, it was recently reported that Areg might also impact Treg cells directly and enhance their ability to suppress immune responses (Zaiss et al., 2013). However, expression of Egfr was undetectable in Treg cells from lymphoid (spleen, lymph node) or nonlymphoid (muscle, VAT) tissues in our microarray studies.
In addition, the TCR repertoires of injured-muscle and lymphoid-organ Treg cells were clearly distinct. There were striking clonal expansions in the muscle Treg population and one TCR α and TCR β chain pair was found over and over again in different individuals despite Draconian measures to avoid contamination. Interestingly, the small subset of Areg+ splenic Tregs did share TCR sequences with muscle Treg cells, whether Areg positive or negative. These findings raise the possibility that a muscle antigen might be involved in recruiting Treg cells to the site of injury and/or retaining them therein.
How Might Treg Cells Influence Muscle Repair?
Muscle repair subsequent to acute injury was impaired in the absence of Treg cells, an influence no doubt exerted at several levels. First, Tregs regulated the myeloid populations that infiltrated the damaged tissue, appearing to contain their numbers and to promote their switch from a pro- to an anti-inflammatory phenotype. Both “flavors” of myeloid cell are important for proper muscle repair, and any alteration in the tightly orchestrated scenario wherein each accumulates and functions at the site of injury can be detrimental (Tidball and Villalta, 2010). A variety of intrinsic and extrinsic factors are responsible for their phenotypic polarization; we propose that Treg cells represent an additional layer of control.
Second, Treg cells also regulated coinfiltrating conventional T cell populations. Upon Treg depletion, the total number and frequency of T cells in the injured muscle was significantly increased, and many T cell-specific genes were overrepresented in whole-muscle microarrays of injured muscle from standard versus Treg-less mice. Moreover, depletion of Treg cells in Rag-deficient mice had a less severe influence on muscle regeneration after acute injury than did ablation of Tregs alone, suggesting that infiltrating Tconv cells (and/or CD8+ or B cells) normally have a negative impact on muscle repair that is kept in check by Treg cells. Yet, the effect of removing Treg cells was not abrogated by eliminating the other lymphocyte classes, indicating that Tregs can impact muscle regeneration independently of either T or B cells.
Third, Treg cells impacted the activities of muscle-lineage cells, specifically satellite cells. According to clonal myogenic assays, satellite cells from Treg-deficient mice had decreased colony-forming capacity, consistent with the reduced in vivo regenerative potential of Treg-depleted muscle. Thus, Treg cells can regulate the cells that are directly responsible for the repair of injured muscle. In vitro experiments have shown that macro-phages can directly influence muscle progenitors (Bosurgi et al., 2011); it is possible, then, that Treg cells act on satellite cells indirectly, by modulation of the infiltrating myeloid populations. However, two of our results suggest a more direct effect: (1) histological evidence that Treg cells were located in close proximity to regenerating fibers (in addition to being present in heavily infiltrated areas); and (2) the demonstration that muscle Tregs expressed Areg and that this molecule enhanced satellite cell differentiation in vitro and in vivo. Areg is an EGF family member known to promote healing and regeneration of multiple tissues, for example intestinal mucosa and lung epithelium (Shao and Sheng, 2010; Monticelli et al., 2011). Most relevant in the current context, previous studies have suggested that Areg might act directly on muscle cells (Golding et al., 2007; Andrechek et al., 2002). Together with these previous studies, our results suggest that Areg may directly modulate muscle repair.
Treg cells and their products are new players to consider in the orchestrated series of events underlying muscle repair in both acute and chronic contexts. Poor wound healing is a growing problem, especially given its association with diabetes and aging, both of which affect a large and increasing fraction of the population. Moreover, adequate treatments for muscular dystrophies remain elusive. Harnessing the power of Treg cells, whether in cell- or molecule-based strategies, opens novel therapeutic avenues.
EXPERIMENTAL PROCEDURES
Mice and Their Manipulation
C57BL/6, C57BL/10ScSn, RAG1-deficient, and C57BL/10ScSn-Dmdmdx mice were purchased from Jackson Laboratory. Foxp3-DTR, Foxp3-IRES-gfp, and Dysferlin-deficient mice were obtained from A. Rudensky, V. Kuchroo, and R. Bittner, respectively. All mice were bred in our specific-pathogen-free facilities at Harvard Medical School. Protocols were approved by Harvard Medical School’s Institutional Animal Care and Use Committee.
We injected anesthetized mice i.m. with 0.03 ml/muscle of Naja mossambica mossambica cardiotoxin (0.03 mg/ml) (Sigma) in one or more hindlimb muscles. Alternatively, TA muscles were directly exposed to dry ice for 5 s.
A total of 1 mg of EdU was injected intravenously, and 24 hr later cells were processed for detection by the Click-iT EdU kit (Molecular Probes).
Foxp3-DTR+ mice and Foxp3-DTR littermates were injected i.p. with DT (Sigma), at 6 ng/g body weight, every other day for 6 days, starting at the day of injury. Depletion of CD25+ cells was accomplished by i.p. injection of 100 µg of anti-CD25 mAb (clone PC61) or rat IgG (Jackson ImmunoResearch) at days 17 and 20 of age. Muscle infiltrate and serum CK were analyzed 7 days after the last injection, the latter via the Creatine Kinase-SL kit (Genzyme). For expansion of Tregs, we incubated 2.5 µg of anti-mouse IL-2 mAb (JES6-1A12) and 0.25 µg of mouse IL-2 per mouse for 20 min on ice followed by i.p. injection. Then 17-day-old mdx mice were given daily injections for 6 days and were analyzed 10 days after the last injection. Control mice were administered phosphate-buffered saline (PBS).
Recombinant mouse Areg (R&D Systems) was administered i.m. (1 ng/muscle) together with Ctx at day 0. Areg was then injected i.p. (7 µg/mouse) every other day until the time of analysis.
Muscle Leukocytes and Manipulations of Them
Mice were perfused with PBS. Muscles were excised, cut up, and collagenase/DNase digested, and leukocytes were isolated by standard procedures. For RT-PCR analyses and single-cell TCR sequencing, published protocols (Wong et al., 2007; Baker et al., 2002) were followed with some modifications to the primer sets used. Raw sequencing files were filtered for sequence quality, processed in automated fashion, and parsed using IMGT/V-QUEST (Brochet et al., 2008). See detailed protocols in the Extended Experimental Procedures.
Microarray
Cells were double-sorted into TRIzol (Invitrogen). Whole-muscle tissue was flash frozen in liquid nitrogen and homogenized in TRIzol. Data sets used for comparison in the PCA were previously generated in our lab. All samples were generated in duplicate or (usually) triplicate. Sample processing and data analysis were performed as previously described (Cipolletta et al., 2012).
Clonal Myogenesis and Myogenic Differentiation Assays
Fortheclonal assays, myofiber-associated cells were prepared from hind-limb muscles as described (Cerletti et al., 2008). Satellite cells (CD45−Sca-1−Mac-1−CXCR4+β1-integrin+) were double-sorted individually into 96-well plates and cultured for 5–7 days. Wells containing myogenic colonies were scored as described in Extended Experimental Procedures.
For differentiation assays, 3,000 satellite cells were double-sorted onto 24- well plates, cells were cultured ±1 ng/ml recombinant mouse Areg, and after 12 days the cultures were processed for RT-PCR or immunofluorescence microscopy of myosin expression as described in the Extended Experimental Procedures, which also details our method of scoring and calculation of the differentiation index.
Statistical Analyses
Data were routinely presented as means ± SD. Significance was assessed by the Student’s t test or ANOVA. A p value of < 0.05 was deemed statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Florence, K. Rothamel, N. Asinovski, A. Ortiz-Lopez, D. Jepson, K. Hattori, J. Ericson, S. Davis, H. Paik, R. Cruse, J. LaVecchio, and G. Buru-zula for experimental support. Cell sorting was performed at the HSCI/DRC Flow Core (NIH P30DK036836). This work benefited from public data generated by the Immunological Genome Project (http://www.immgen.org) and was funded by NIH grants R37AI051530 and RO1DK092541 (to C.B. and D.M.) and R01AG033053 and UO1HL100402 (to A.J.W.). A.J.W. is an Early Career Scientist at the Howard Hughes Institute. D.B. was supported by a Kaneb Fellowship, D.K. by an NSF fellowship, E.S. by a Boehringer Ingelheim Fonds Fellowship, and T.G.T. by an A*STAR Graduate Scholarship (Singapore).
Footnotes
ACCESSION NUMBERS
Microarray data are available from the National Center for Biotechnology Information/Gene Expression Omnibus repository under accession numbers GSE50096, GSE50094, GSE50095, and GSE50097.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, five figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2013.10.054.
REFERENCES
- Andrechek ER, Hardy WR, Girgis-Gabardo AA, Perry RL, Butler R, Graham FL, Kahn RC, Rudnicki MA, Muller WJ. ErbB2 is required for muscle spindle and myoblast cell survival. Mol. Cell. Biol. 2002;22:4714–4722. doi: 10.1128/MCB.22.13.4714-4722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold L, Henry A, Poron F, Baba-Amer Y, van Rooijen N, Plonquet A, Gherardi RK, Chazaud B. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J. Exp. Med. 2007;204:1057–1069. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker FJ, Lee M, Chien YH, Davis MM. Restricted islet-cell reactive T cell repertoire of early pancreatic islet infiltrates in NOD mice. Proc. Natl. Acad. Sci. USA. 2002;99:9374–9379. doi: 10.1073/pnas.142284899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CL, Clausen BE. DC ablation in mice. promises, pitfalls, and challenges. Trends Immunol. 2007;28:525–531. doi: 10.1016/j.it.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Beyer M, Thabet Y, Müller RU, Sadlon T, Classen S, Lahl K, Basu S, Zhou X, Bailey-Bucktrout SL, Krebs W, et al. Repression of the genome organizer SATB1in regulatory T cells is required for suppressive function and inhibition of effector differentiation. Nat. Immunol. 2011;12:898–907. doi: 10.1038/ni.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosurgi L, Manfredi AA, Rovere-Querini P. Macrophages in injured skeletal muscle: a perpetuum mobile causing and limiting fibrosis, prompting or restricting resolution and regeneration. Front. Immunol. 2011;2:62. doi: 10.3389/fimmu.2011.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyman O, Kovar M, Rubinstein MP, Surh CD, Sprent J. Selective stimulation of T cell subsets with antibody-cytokine immune complexes. Science. 2006;311:1924–1927. doi: 10.1126/science.1122927. [DOI] [PubMed] [Google Scholar]
- Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36(Web Server issue):W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerletti M, Jurga S, Witczak CA, Hirshman MF, Shadrach JL, Goodyear LJ, Wagers AJ. Highly efficient, functional engraftment of skeletal muscle stem cells in dystrophic muscles. Cell. 2008;134:37–47. doi: 10.1016/j.cell.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerletti M, Jang YC, Finley LW, Haigis MC, Wagers AJ. Short-term calorie restriction enhances skeletal muscle stem cell function. Cell Stem Cell. 2012;10:515–519. doi: 10.1016/j.stem.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolletta D, Kolodin D, Benoist C, Mathis D. Tissular T(regs): a unique population of adipose-tissue-resident Foxp3+CD4+ T cells that impacts organismal metabolism. Semin. Immunol. 2011;23:431–437. doi: 10.1016/j.smim.2011.06.002. [DOI] [PubMed] [Google Scholar]
- Cipolletta D, Feuerer M, Li A, Kamei N, Lee J, Shoelson SE, Benoist C, Mathis D. PPAR-γ is a major driver of the accumulation and phenotype of adipose tissue Treg cells. Nature. 2012;486:549–553. doi: 10.1038/nature11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerer M, Herrero L, Cipolletta D, Naaz A, Wong J, Nayer A, Lee J, Goldfine AB, Benoist C, Shoelson S, Mathis D. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat. Med. 2009;15:930–939. doi: 10.1038/nm.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding JP, Calderbank E, Partridge TA, Beauchamp JR. Skeletal muscle stem cells express anti-apoptotic ErbB receptors during activation from quiescence. Exp. Cell Res. 2007;313:341–356. doi: 10.1016/j.yexcr.2006.10.019. [DOI] [PubMed] [Google Scholar]
- Hill JA, Feuerer M, Tash K, Haxhinasto S, Perez J, Melamed R, Mathis D, Benoist C. Foxp3 transcription-factor-dependent and -independent regulation of the regulatory T cell transcriptional signature. Immunity. 2007;27:786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Horikawa M, Higashiyama S, Nomura S, Kitamura Y, Ishikawa M, Taniguchi N. Upregulation of endogenous heparin-binding EGF-like growth factor and its role as a survival factor in skeletal myotubes. FEBS Lett. 1999;459:100–104. doi: 10.1016/s0014-5793(99)01213-2. [DOI] [PubMed] [Google Scholar]
- Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: mechanisms of differentiation and function. Annu. Rev. Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 2007;8:191–197. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- Monticelli LA, Sonnenberg GF, Abt MC, Alenghat T, Ziegler CG, Doer-ing TA, Angelosanto JM, Laidlaw BJ, Yang CY, Sathaliyawala T, et al. Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat. Immunol. 2011;12:1045–1054. doi: 10.1031/ni.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales MG, Cabello-Verrugio C, Santander C, Cabrera D, Gold-schmeding R, Brandan E. CTGF/CCN-2 over-expression can directly induce features of skeletal muscle dystrophy. J. Pathol. 2011;225:490–501. doi: 10.1002/path.2952. [DOI] [PubMed] [Google Scholar]
- Naito AT, Sumida T, Nomura S, Liu ML, Higo T, Nakagawa A, Okada K, Sakai T, Hashimoto A, Hara Y, et al. Complement C1q activates canonical Wnt signaling and promotes aging-related phenotypes. Cell. 2012;149:1298–1313. doi: 10.1016/j.cell.2012.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastore S, Mascia F, Mariani V, Girolomoni G. The epidermal growth factor receptor system in skin repair and inflammation. J. Invest. Dermatol. 2008;128:1365–1374. doi: 10.1038/sj.jid.5701184. [DOI] [PubMed] [Google Scholar]
- Rudnicki MA, Le Grand F, McKinnell I, Kuang S. The molecular regulation of muscle stem cell function. Cold Spring Harb. Symp. Quant. Biol. 2008;73:323–331. doi: 10.1101/sqb.2008.73.064. [DOI] [PubMed] [Google Scholar]
- Schmitz J, Owyang A, Oldham E, Song Y, Murphy E, McClanahan TK, Zurawski G, Moshrefi M, Qin J, Li X, et al. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 2005;23:479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Shao J, Sheng H. Amphiregulin promotes intestinal epithelial regeneration: roles of intestinal subepithelial myofibroblasts. Endocrinology. 2010;151:3728–3737. doi: 10.1210/en.2010-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoyab M, Plowman GD, McDonald VL, Bradley JG, Todaro GJ. Structure and function of human amphiregulin: a member of the epidermal growth factor family. Science. 1989;243:1074–1076. doi: 10.1126/science.2466334. [DOI] [PubMed] [Google Scholar]
- Suvas S, Azkur AK, Kim BS, Kumaraguru U, Rouse BT. CD4+CD25+ regulatory T cells control the severity of viral immunoinflamma-tory lesions. J. Immunol. 2004;172:4123–4132. doi: 10.4049/jimmunol.172.7.4123. [DOI] [PubMed] [Google Scholar]
- Tabebordbar M, Wang ET, Wagers AJ. Skeletal muscle degenerative diseases and strategies for therapeutic muscle repair. Annu. Rev. Pathol. 2013;8:441–475. doi: 10.1146/annurev-pathol-011811-132450. [DOI] [PubMed] [Google Scholar]
- Tidball JG, Villalta SA. Regulatory interactions between muscle and the immune system during muscle regeneration. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010;298:R1173–R1187. doi: 10.1152/ajpregu.00735.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetrone SA, Montecino-Rodriguez E, Kudryashova E, Kramerova I, Hoffman EP, Liu SD, Miceli MC, Spencer MJ. Osteopontin promotes fibrosis in dystrophic mouse muscle by modulating immune cell subsets and intramuscular TGF-beta. J. Clin. Invest. 2009;119:1583–1594. doi: 10.1172/JCI37662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren GL, Hulderman T, Mishra D, Gao X, Millecchia L, O’Farrell L, Kuziel WA, Simeonova PP. Chemokine receptor CCR2 involvement in skeletal muscle regeneration. FASEB J. 2005;19:413–415. doi: 10.1096/fj.04-2421fje. [DOI] [PubMed] [Google Scholar]
- Wong J, Obst R, Correia-Neves M, Losyev G, Mathis D, Benoist C. Adaptation of TCR repertoires to self-peptides in regulatory and nonre-gulatory CD4+ T cells. J. Immunol. 2007;178:7032–7041. doi: 10.4049/jimmunol.178.11.7032. [DOI] [PubMed] [Google Scholar]
- Zaiss DM, van Loosdregt J, Gorlani A, Bekker CP, Gröne A, Sibilia M, van Bergen en Henegouwen PM, Roovers RC, Coffer PJ, Sijts AJ. Amphiregulin enhances regulatory T cell-suppressive function via the epidermal growth factor receptor. Immunity. 2013;38:275–284. doi: 10.1016/j.immuni.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.