SUMMARY
HSP90 is a molecular chaperone that associates with numerous substrate proteins called clients. It plays many important roles in human biology and medicine, but determinants of client recognition by HSP90 have remained frustratingly elusive. We systematically and quantitatively surveyed most human kinases, transcription factors, and E3 ligases for interaction with HSP90 and its cochaperone CDC37. Unexpectedly, many more kinases than transcription factors bound HSP90. CDC37 interacted with kinases, but not with transcription factors or E3 ligases. HSP90::kinase interactions varied continuously over a 100-fold range and provided a platform to study client protein recognition. In wild-type clients, HSP90 did not bind particular sequence motifs, but rather associated with intrinsically unstable kinases. Stabilization of the kinase in either its active or inactive conformation with diverse small molecules decreased HSP90 association. Our results establish HSP90 client recognition as a combinatorial process: CDC37 provides recognition of the kinase family, whereas thermodynamic parameters determine client binding within the family.
INTRODUCTION
HSP90 is one of the most abundant chaperone proteins in the cell. It interacts with a large and diverse group of substrate proteins, commonly referred to as “clients,” promoting their folding and function. HSP90 seems to be focused on metastable proteins that are regulatory hubs in biological networks. This makes HSP90 a driving force in the evolution of new forms and functions in organisms as diverse as yeast, fruit flies, and thale cress. Recent work has established the importance of HSP90 in many aspects of human biology, ranging from cancer to infectious diseases to neurodegeneration (Taipale et al., 2010). Identifying the full spectrum of HSP90 client proteins has, therefore, gained increased urgency. So, too, has the need to determine the mechanistic basis of HSP90 client protein interactions.
In contrast to Hsp70 and Hsp60 chaperones, for which the mechanism of client protein recognition is understood in general terms, we still have only a rudimentary understanding of what determines client protein binding by HSP90. Three features of HSP90 client interactions illustrate the complexity of the problem: the perplexing diversity of Hsp90 clients, the metastable character of their folds, and the complexity of its chaperone cycle. First, HSP90 clients encompass a wide range of proteins with unrelated amino acid sequences and functions, including protein kinases and steroid hormone receptors (Taipale et al., 2010). Although the kinase domain and the ligand-binding domain of steroid hormone receptors are structurally unrelated, these domains must have specific sequence or structural features that HSP90 recognizes. Second, in each client protein family some members robustly interact with the chaperone, while others apparently do not associate with it at all. For example, mature epidermal growth factor receptor (EGFR) does not associate with HSP90, whereas its closest homolog, ERBB2, tightly interacts with it (Xu et al., 2005). Thus, despite the overall conservation of the protein fold, there must be specific sequence or structural features that distinguish client proteins from nonclients. Finally, HSP90 associates with a large cohort of auxiliary proteins, known as cochaperones, which regulate its function and seem to provide specificity to client protein interactions. Yet, the role and specificity of cochaperones in client protein recognition is not well understood.
Previous work on steroid hormone receptors and kinases has suggested some features that could determine HSP90 client protein associations (Ricketson et al., 2007; Xu et al., 2005). For some kinases, mutations in the loop between αC helix and b4 strand in the N-lobe of the kinase domain have strong effects on HSP90 binding, leading to the hypothesis that the aC-b4 loop is the HSP90 recognition site (Citri et al., 2006; Xu et al., 2005). Yet, a survey of known HSP90 client kinases and nonclients did not uncover client-specific sequence motifs in the loop (Citri et al., 2006). It is often speculated that oncogenic kinases (and other clients) associate with HSP90 because the chaperone recognizes something related to their conformational instability, but the data are insufficient to reach any unifying conclusions and reconcile diverse observations (Caplan et al., 2007; Taipale et al., 2010).
A quantitative high-throughput assessment of HSP90 client interactions could transform our understanding of these issues but has proved challenging for several reasons. First, HSP90’s interactions with its clients are transient and might not be detected by standard interaction assays that require the formation of stable complexes. Second, binary assays, such as yeast two-hybrid assays, may not recapitulate all HSP90 interactions in human cells due to the absence of correct cochaperones or posttranslational modifications. Third, client proteins tend to be expressed at very low levels. Consequently, methods based on mass spectrometry have primarily revealed chaperone:: cochaperone interactions; only a handful of bona fide client proteins have been found by this method (Falsone et al., 2005, 2007). Finally, most assays do not allow relative quantification of interactions with HSP90.
To circumvent these limitations, we adapted and further developed the LUMIER assay (Barrios-Rodiles et al., 2005) to systematically measure HSP90 interaction with the majority of human protein kinases, transcription factors and ubiquitin ligases in vivo. Surprisingly, HSP90 bound only ~7% of transcription factors, but 30% of ubiquitin ligases and 60% of kinases. The conserved nature of the kinase fold, together with the sheer numbers of clients, then provided a robust platform to address client protein features that determine interaction with HSP90. In addition to providing a comprehensive view of the HSP90::kinase interactome, our results reveal two principles of HSP90 client interactions. A cochaperone provides recognition of particular types of fold, whereas thermal and conformational stability determines the extent of interaction.
RESULTS
Quantitative Measurement of Protein-Protein Interactions In Vivo by LUMIER with BACON
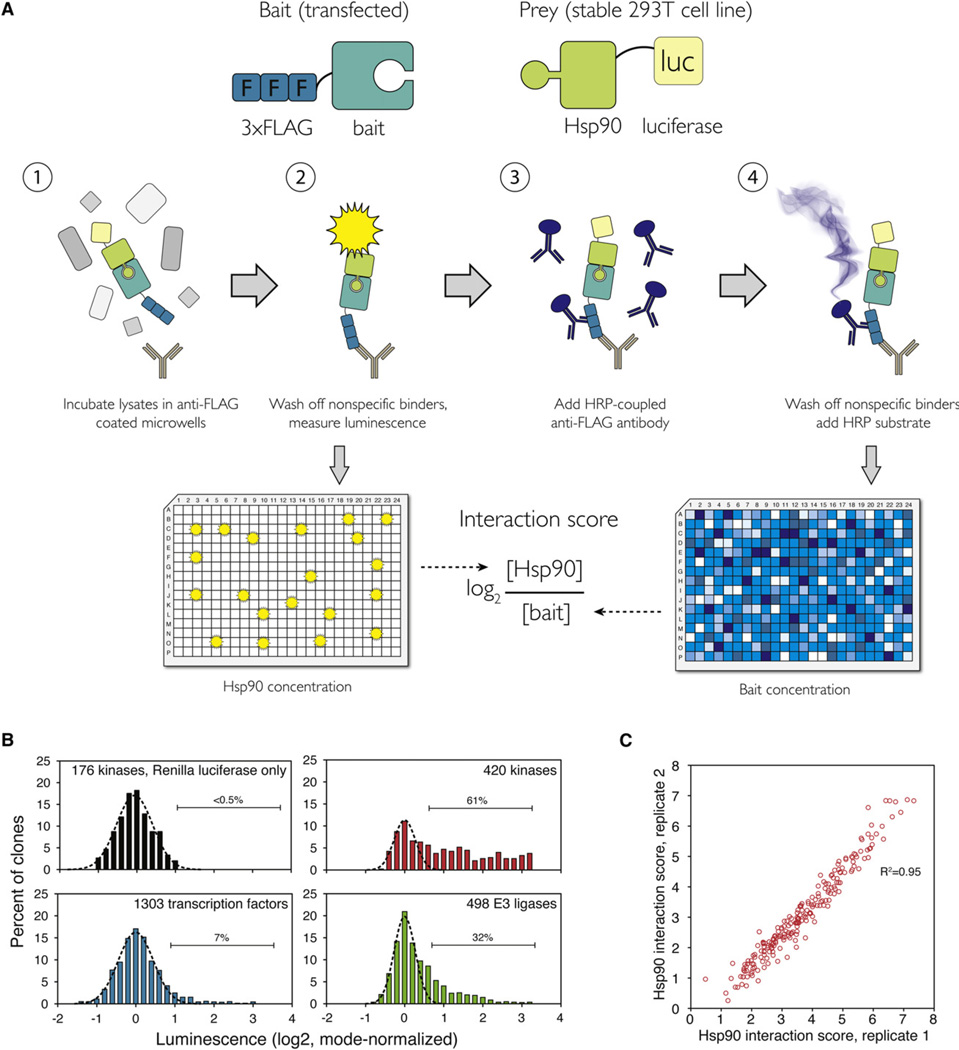
In the LUMIER assay, one test protein (bait) is tagged with a FLAG tag and the other (prey) is tagged with Renilla luciferase (Barrios-Rodiles et al., 2005). Proteins are transiently coexpressed in mammalian cells by transfection, and cell lysates are applied to 96-well plates containing anti-FLAG-coated beads. Interaction of bait and prey proteins is detected by luminescence after immunoprecipitation. We improved the original LUMIER assay to make it more quantitative and to increase its throughput. For example, we used a stable 293T cell line expressing the luciferase-tagged prey protein to minimize well-to-well variation. We also introduced a step that controls for bait abundance: after reading luminescence (i.e., prey abundance), the abundance of the 3 × FLAG -tagged bait is measured with ELISA, using a different, polyclonal antibody against the FLAG tag (Figure 1A). We named the modified assay LUMIER with bait control (LUMIER with BACON).
Figure 1. LUMIER with BACON Assay Reveals the Quantitative Nature of HSP90::Client Interactions.
(A) Principle of the assay. 3 × FLAG-tagged bait constructs (putative HSP90 clients) are transfected into a 293T cell line stably expressing Renilla luciferase-tagged HSP90 (prey). Cell lysates are incubated in 384-well plates coated with anti-FLAG antibody. After washing off nonspecific proteins, luminescence is measured. Interaction of HSP90 with the bait can be detected as luminescence. In the second step, the amount of bait is measured with anti-FLAG ELISA. The log2 ratio between bait and prey concentration is the interaction score.
(B) Distribution of luminescence in HSP90 interaction assays with 420 kinase clones (red), 498 E3 ligase clones (green), and 1,093 transcription factor clones (blue). As a control, 176 kinase clones were tested against a cell line expressing Renilla luciferase only (black). Gaussian curve was fitted to each data set to establish a cutoff for true interactions (dashed black line).
(C) Quantitative interaction score was calculated for all 193 kinases that interacted with HSP90. Scatter plot shows the interaction scores from two biological replicates.
HSP90::Client Interaction Assay
We collected 420 ORF clones representing 355 unique protein kinases and their variants (69% of the human kinome), 1,303 transcription factor (TF) clones (1,093 unique TFs/79% of all), and 498 E3 ligase clones (426 unique; 69% of all). These were transferred into an expression vector with a C-terminal 3 × FLAG-V5 tag. We then created a stable cell line expressing codon-optimized Renilla luciferase fused to the HSP90β N terminus, based on earlier observations that N-terminal tagging of HSP90 does not significantly affect its function (Boulon et al., 2010). The tagged HSP90β protein was expressed at less than 10% of the endogenous HSP90β (data not shown) and should therefore not significantly affect client interactions.
Renilla-Hsp90 cells were transfected with bait constructs in 96-well format. Cells were lysed 48 hr later and the lysates were applied to anti-FLAG-coated 384-well plates. After incubation and copious, but very rapid washes, luminescence in each well provided a measure of how much HSP90 had been captured. Immediately thereafter, ELISA was used to quantify bait protein abundance. To establish a robust cutoff for true interactions, we tested 176 kinases for interaction with Renilla luciferase itself, using a stable cell line expressing just the luciferase. We set a cutoff for true interactions such that at least 99.5% of nonspecific interactions were excluded (i.e., all 176 kinase::Renilla luciferase interactions fell below the cutoff; Figure 1B).
Altogether, 314 of the kinases were expressed at levels detectable by ELISA, and of those, 193 (61%) passed the criteria for true interaction (Figure 1B; Table S1 available online). Therefore, HSP90 associates with more than half of the human kinome. We calculated a quantitative interaction score (log2 [HSP90]/[kinase]) for the 193 kinases that interacted with HSP90 (Figure 1C). These scores were highly reproducible (R2 = 0.95) and covered more than a 100-fold range of HSP90/kinase ratios, illustrating that steady-state HSP90 association with client proteins varies over a continuum. Thus, the commonly used binary classification of kinases as HSP90 clients or nonclients does not at all capture the breadth of HSP90::substrate interactions in the cellular context. However, to simplify downstream analysis, we divided kinases into three classes based on the LUMIER interaction score. Kinases with an interaction score above 3 (n = 98) were classified as “strong” clients and those with an interaction score below 3 (n = 95) as “weak” clients. The rest (n = 121) were classified as nonclients.
To assess the quality of our data set, we validated the interaction of ten FLAG-tagged kinases from each class with endogenous HSP90 by conventional coimmunoprecipitation. All strong and weak interactions were reproduced (Figure S1A). Furthermore, confirming the quantitative nature of the LUMIER assay, strong clients indeed interacted more strongly with endogenous HSP90 than weak clients did. Only two of the ten kinases we classified as nonclients interacted with endogenous HSP90, and both interactions were extremely weak (Figure S1A). Thus, the LUMIER data set contains high-confidence kinase clients (true positives), but this does not lead to a large number of true clients classified as nonclients (false negatives).
Finally, because HSP90::client interactions are transient in vivo, it was important to ensure that we did not lose such associations during the washing steps. All tested interactions were abolished when cells were treated for 1 hr with 500 nM ganetespib, a potent HSP90 inhibitor (Ying et al., 2011). However, ganetespib had only a very marginal effect on HSP90::kinase interactions when added after lysis, even at a 20-fold higher concentration (Figure S1B). These results establish that interactions captured by LUMIER are true chaperone::client interactions and that these interactions are stable in the assay conditions.
Few Transcription Factors, but Many E3 Ligases, Interact with HSP90
In stark contrast to kinases, less than 7% (58 of 843 unique TFs detectable by ELISA) of human transcription factors (Figure 1B; Table S1) interacted with HSP90. The low number of TF clients is surprising, since it is commonly thought that TFs represent a substantial fraction of HSP90 clients (Taipale et al., 2010). Based on our results, it appears that steroid hormone receptors, a previously well-established class of HSP90 clients, actually constitute an exceptional TF family with respect to HSP90 binding. Among ubiquitin ligases, 31% (117/372) interacted with HSP90 (Figure 1B; Table S1). Interestingly, most E3 ligases with Kelch or WD40 domains interacted with HSP90. This trend was highly significant (Kelch: 19 interactors out of 32 proteins, p < 0.0004; WD40: 8/11, p < 0.0055, Fisher’s exact test). Thus, beta-propeller domains such as Kelch and WD40 may comprise a novel HSP90 client protein fold. Furthermore, all of the Cullin scaffold proteins we tested (Cul1, Cul2, Cul3, Cul4B, Cul4A) interacted with HSP90 except for Cul4B. Previously, Cul5 was shown to interact with HSP90 and regulate client protein degradation (Ehrlich et al., 2009). Our results suggest that other Cullins may also regulate the proteasomal degradation of client proteins.
Kinase Specificity of HSP90 Is Provided by CDC37
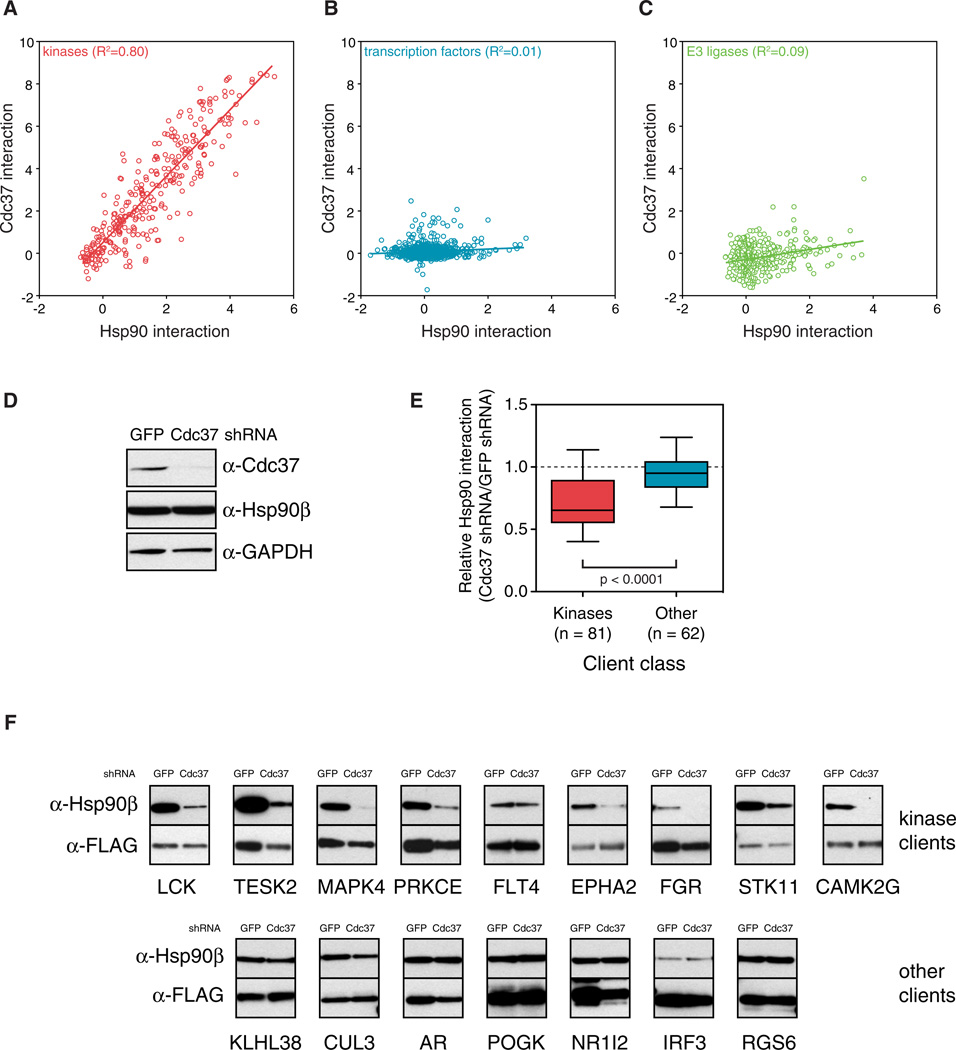
To address what determines HSP90 binding at the protein-family level, we analyzed an HSP90 cochaperone, CDC37, known to associate with many kinases (Grammatikakis et al., 1999; Gray et al., 2008). It is not known, however, whether CDC37 is a universal kinase cochaperone or whether it is strictly kinase specific. We constructed a cell line stably expressing CDC37 that was C-terminally tagged with Renilla luciferase to assess interactions between CDC37 and each of the kinases, TFs, and E3 ligases that we had tested for interaction with HSP90. Similar to HSP90, CDC37 interacted with the majority of kinases. Moreover, HSP90::kinase and CDC37::kinase interaction profiles correlated strongly (R2 = 0.81; Figure 2A). In contrast, CDC37 interacted with only 3.3% of TFs (24/720) and 1.3% (5/375) of E3 ligases, and the correlation between HSP90 and CDC37 interactions within these two protein classes was negligible (Figures 2B and 2C; Table S1).
Figure 2. CDC37 Is a Universal Kinase-Specific Cochaperone.
(A–C) HSP90 interaction profiles were compared with CDC37 interaction profiles for kinases (A), transcription factors (B), and E3 ligases (C). Plots show mode-normalized, log2-transformed luminescence values.
(D) 293T cells were infected with a lentivirus expressing an shRNA construct targeting Cdc37 or GFP.
(E) CDC37 is required for kinase::HSP90 interactions. Client proteins were transfected in duplicate into Renilla-HSP90 expressing cells infected with either Cdc37 or GFP shRNA. Forty-eight hours after transfection, interaction with each client was measured with LUMIER. Boxes indicate 25th and 75th percentiles and whiskers 10th and 90th percentiles. p value was calculated with Wilcoxon rank sum test. (F) 3 × FLAG-tagged client proteins were transfected into 293T cells infected with either Cdc37 or GFP shRNA. Forty-eight hours later, clients were immunoprecipitated with an anti-FLAG antibody. Coimmunoprecipitation with endogenous HSP90 was assayed with a monoclonal HSP90 antibody.
See also Figure S1.
These data strongly suggested that CDC37 serves as an adaptor for the kinase fold. To investigate more rigorously, we reduced CDC37 levels with shRNA-mediated knockdown. This had no effect on HSP90 levels (Figure 2D). However, almost all tested kinase::HSP90 interactions were reduced after CDC37 knockdown, whereas nonkinase clients still interacted with HSP90, as measured by LUMIER (p < 0.0001; Figure 2E). Next, we tested 17 kinase::HSP90 interactions by conventional coimmunoprecipitation. Fifteen of these interactions were significantly reduced after CDC37 knockdown, while none of the seven nonkinase client interactions we tested were affected (Figure 2F; data not shown).
Taken together, these data establish that CDC37 is a highly specialized cochaperone adaptor for kinases. Further, the remarkably strong correlation between HSP90::kinase and CDC37::kinase interactions demonstrates that HSP90 and CDC37 act in concert in chaperoning client kinases. Thus, the adaptor function of CDC37 explains the family-level specificity of HSP90::kinase interactions.
Inhibition of HSP90 Leads to Dissociation of Most HSP90::Kinase Complexes
Next, we took advantage of the large number of HSP90 kinase clients, all having the same kinase fold, to globally assess the consequences of client protein::HSP90 associations and to investigate why some highly related members of a protein family bind HSP90 while others do not. We first tested eight HSP90 inhibitors based on several different chemical scaffolds for their ability to disrupt the HSP90::ARAF complex. All the inhibitors dissociated ARAF from HSP90 with a 1 hr treatment (Figure S2A). The most potent inhibitor was ganetespib with an IC50 of 26 nM (Figure S2B); therefore, we used it in all subsequent assays.
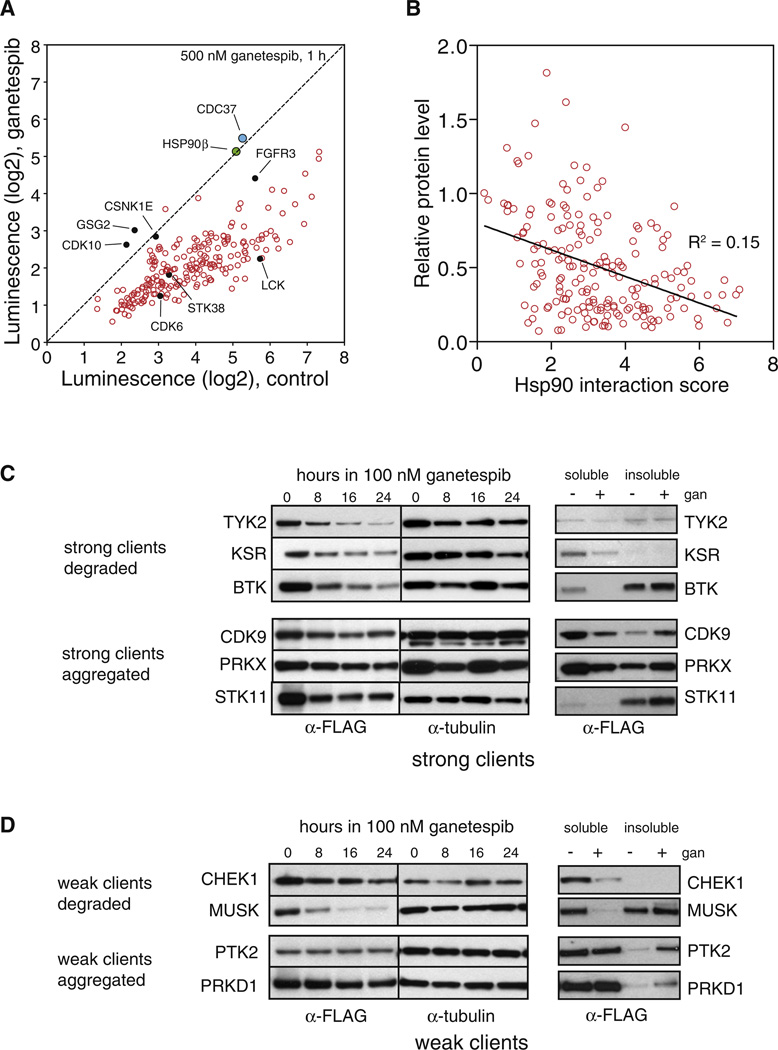
We measured each kinase::HSP90 interaction after treating cells for 1 hr with 500 nM ganetespib or with DMSO. We also tested the effect of ganetespib on HSP90’s interaction with CDC37 and on HSP90β homodimerization, to distinguish client::chaperone interactions from interactions within the chaperone machinery itself. Neither CDC37::HSP90β interactions nor HSP90β::HSP90β dimerization was affected by ganetespib. In contrast, almost all HSP90::kinase interactions were abolished or significantly decreased (Figure 3A).
Figure 3. An HSP90 Inhibitor Induces Dissociation of Most HSP90::Kinase Complexes, but HSP90 Interaction Strength Correlates Poorly with Subsequent Degradation.
(A) HSP90::kinase interactions were measured after treating cells with 500 nM ganetespib or vehicle for 1 hr before cell lysis and LUMIER assay. HSP90β::CDC37 interaction (blue) and HSP90β homodimerization (green) is not affected by ganetespib. Dashed black line indicates no change in HSP90 interaction upon ganetespib treatment. Kinase clones marked with filled black circles were further validated with coimmunoprecipitation with endogenous HSP90 (Figure S2).
(B) Ganetespib-induced degradation of client kinases correlates poorly with HSP90 interaction score. 3 × FLAG-tagged kinases were transfected into 293T cells, which were subsequently treated with 100 nM ganetespib or vehicle for 20 hr. Protein levels were measured with anti-FLAG ELISA. Relative protein level is plotted against Hsp90 interaction score from LUMIER assay.
(C) HSP90 inhibition can lead to either degradation or aggregation of strong client proteins. Left panel: 3 × FLAG-tagged kinases were transfected into 293T cells, which were treated with 100 nM ganetespib for indicated times. Protein levels were measured with an anti-FLAG antibody. Right panel: 3 × FLAG-tagged kinases were transfected into 293T cells, and after 24 hr treatment with 100 nM ganetespib or vehicle, the soluble fraction (0.1% Triton) was separated from the insoluble fraction. Protein levels were measured with an anti-FLAG antibody.
(D) Degradation or aggregation of weak client proteins after HSP90 inhibition. Experiment was performed as in (C) but with weak HSP90 client proteins.
The extent of dissociation from HSP90 did vary, however. For example, CDK10, GSG2, and CSNK1E did not dissociate and FGFR3 dissociated only partially (Figure 3A). We confirmed these results by conventional coimmunoprecipitation (Figure S2C). Next, we used LUMIER to ask if these kinases increased the IC50 of ganetespib for HSP90 or whether they simply have a slower dissociation rate. The IC50 of ganetespib for STK38, CDK6, LCK, and FGFR3 complexes with HSP90 was at the low nanomolar range (15–30 nM; Figures S2D and S2F) similar to that of the ARAF::HSP90 complex. With this more detailed analysis, we also observed partial dissociation of CDK10 and GSG2 from HSP90 with low nanomolar IC50 (Figure S2D).
In contrast to the highly similar IC50 values, there were marked differences in dissociation rates. After treating the cells with 500 nM ganetespib, STK38, CDK6, and LCK (Figure S2C) dissociated from HSP90 with a half-life of ~3 min (Figure S2E). Another 15 kinases we tested had similar dissociation rates (2–6 min; data not shown). In contrast, FGFR3 dissociated with a T1/2 of 29 min. Dissociation of CDK10, GSG2, and CSNK1E showed yet a different pattern. Their dissociation rates were somewhat slower than for most kinases, but more significantly, they only partially dissociated from HSP90 even after prolonged inhibitor treatment (Figure S2E).
Thus, exceptions show that the chaperone cycle for a few kinases, such as FGFR3, is significantly slower than that for most kinases. Furthermore, partial dissociation of other kinases such as CDK10 and GSG2 suggest that these kinase-HSP90 complexes represent two different pools, only one of which is sensitive to HSP90 inhibition. However, the vast majority of HSP90::client kinase interactions had similar sensitivities to disruption of the chaperone cycle, with remarkably similar IC50s for HSP90 inhibitors and very similar HSP90 off-rates.
The Fate of Kinase Clients Varies after Inhibitor-Induced Dissociation
Next, we tested the effect of HSP90 inhibition on the cellular fate of client proteins. We transfected 3 × FLAG-tagged kinases in 96-well format into 293T cells and, 24 hr after transfection, added 100 nM ganetespib or DMSO. After a further 20 hr incubation, cells were lysed and relative protein abundance measured with an anti-FLAG ELISA. As a control, we used c-Src, which is insensitive to HSP90 inhibition, and oncogenic v-Src, which is exquisitely sensitive (Xu et al., 1999).
Protein levels could be confidently assessed for 291 kinases with good reproducibility (Figure S2G). As expected, v-Src was almost completely degraded after ganetespib treatment and c-Src was only modestly affected (Figure S2G). On average, strong clients were degraded to a greater extent after ganetespib treatment (p < 0.01) than weak clients. In turn, weak clients were degraded more than nonclients (p < 0.0001) (Figure S2H). However, the correlation between HSP90 interaction and ganetespib-induced degradation was surprisingly weak (R2 = 0.15, p < 0.0001; Figure 3B). A number of strong client kinases were not efficiently degraded even though they were efficiently dissociated from HSP90 by the inhibitor (Figure S2I). We further validated these results by western blotting (Figures 3C and 3D).
Next, for kinases that dissociated from HSP90 without being degraded, we asked if dissociation led to aggregation. After the same incubations described above, the soluble fraction (0.1% Triton X-100) was separated from the insoluble fraction. Indeed, kinases that were poorly degraded upon HSP90 inhibition accumulated in the insoluble fraction, presumably because they aggregated (Figures 3C and 3D, right panel). Thus, HSP90 inhibition very broadly affects the fate of its client proteins. The balance between aggregation and degradation varies with each client, however.
Determinants for HSP90 Association Are Complex and Widely Distributed
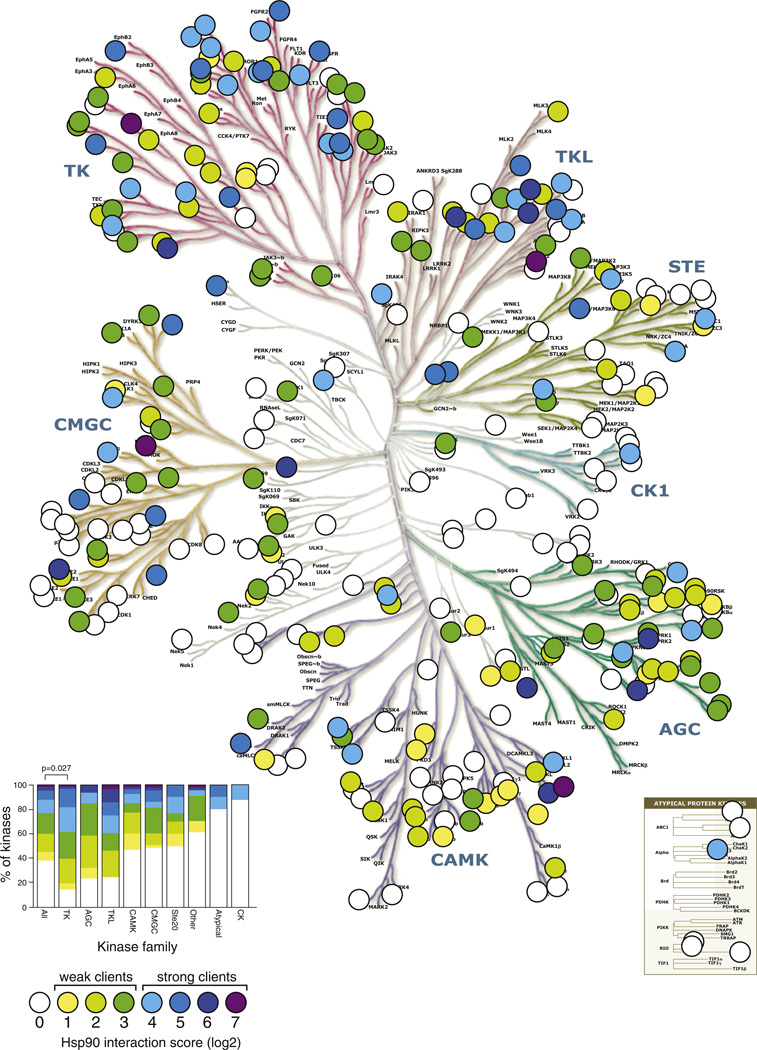
To investigate why some normal cellular kinases interact with HSP90 and others do not, we first plotted interaction scores on the phylogenetic tree of human kinase domains (Manning et al., 2002) (Figure 4). Strikingly, each major kinase branch had both clients and nonclients and, even on different branches, the clients did not cluster. Furthermore, kinases with strong interaction scores often had very close relatives that did not interact.
Figure 4. HSP90 Associates with the Majority of the Human Kinome.
Quantitative HSP90 interaction scores are plotted on the human kinome tree (courtesy of Cell Signaling Technologies and Science Magazine). Interactions were divided into eight bins and color coded. Client proteins were further divided into weak clients (interaction score 1–3) or strong clients (interaction score 4–7). Inset left, distribution of clients within kinase families. Tyrosine kinases are the only family that contains more clients than expected by chance (Bonferroni-corrected p value 0.027, Wilcoxon rank sum test). Kinase tree from Manning et al. (2002). Reprinted with permission from the American Association for the Advancement of Science (AAAS).
See also Figure S3.
The LUMIER assay detected previously reported differences between HSP90’s interaction with EGFR and ERBB2 (nonclient versus weak client; Xu et al., 2005), and between ARAF and BRAF (strong client versus weak client; da Rocha Dias et al., 2005). It also uncovered many other, similarly discordant pairs. For example, transforming growth factor (TGF)-β receptor ACVR1B/ALK4 was a strong client protein, whereas its closest homolog, TGFBR1/ALK5 did not bind HSP90, despite sharing 95% aa similarity over the kinase domain (Figure S3A). The Csk-type protein kinase CTK/MATK was a strong client, whereas CSK, with 77% aa similarity in the kinase domain, did not bind HSP90 at all. All of these differences were validated directly with coimmunoprecipitation (Figure S3A, lower panel). Thus, HSP90 specificity is not determined by the evolutionary history of the kinase.
Several studies have tried to identify regions in the kinase domain that might explain why some kinases associate with HSP90 whereas others do not (Citri et al., 2006; Xu et al., 2005). No consensus motifs have emerged from the limited number of kinases studied, but our comprehensive data set provides unprecedented opportunity to search for them. We used mutual information analysis, OMES covariance, SCA covariance, and McBasc (Fodor and Aldrich, 2004) to identify residues that might covary with HSP90 binding status. We created a multiple alignment of all kinase domains, manually refining the alignment where appropriate. Loops were excluded from the analysis due to poor alignment confidence, and weak clients were excluded to eliminate ambiguity.
Nine residues significantly (p < 0.01) covaried with HSP90 interaction status using all algorithms (Figure S3B). Interestingly, six of the covarying residues were located in the long αE helix of the kinase C-lobe (Figure S3C). Two of these (residues 147 and 151, numbering with reference to protein kinase A) face the hydrophobic core of the C-lobe, and were more hydrophobic in nonclient kinases than in clients, which preferred alanine in these positions (Figure S3B). The neighboring residue 153 is usually a glycine or alanine in kinases. In strong clients, the distribution was significantly biased toward glycine, whereas nonclients had an alanine more frequently in this position. We separately analyzed weak clients and found them to have an intermediate bias in all these residues (Figure S3B). Alanine has the strongest propensity to form α helices, whereas glycine is, after proline, the least favored amino acid in α helices (Pace and Scholtz, 1998). Thus, weaker hydrophobic packing and destabilization of the αE helix by glycine might explain some of the observed differences between nonclients and clients.
However, the differences in covarying residues between clients and nonclients were, at best, modest and could explain only a small fraction of variation in HSP90 client status. This was corroborated by the fact that most of the closely related kinase pairs that had discordant HSP90 interaction scores had identical or nearly identical residues in covarying positions of the αE helix. Thus, the major determinants for HSP90 must reside in other features of these kinases.
Determinants for HSP90 Association Can Be Widely Distributed
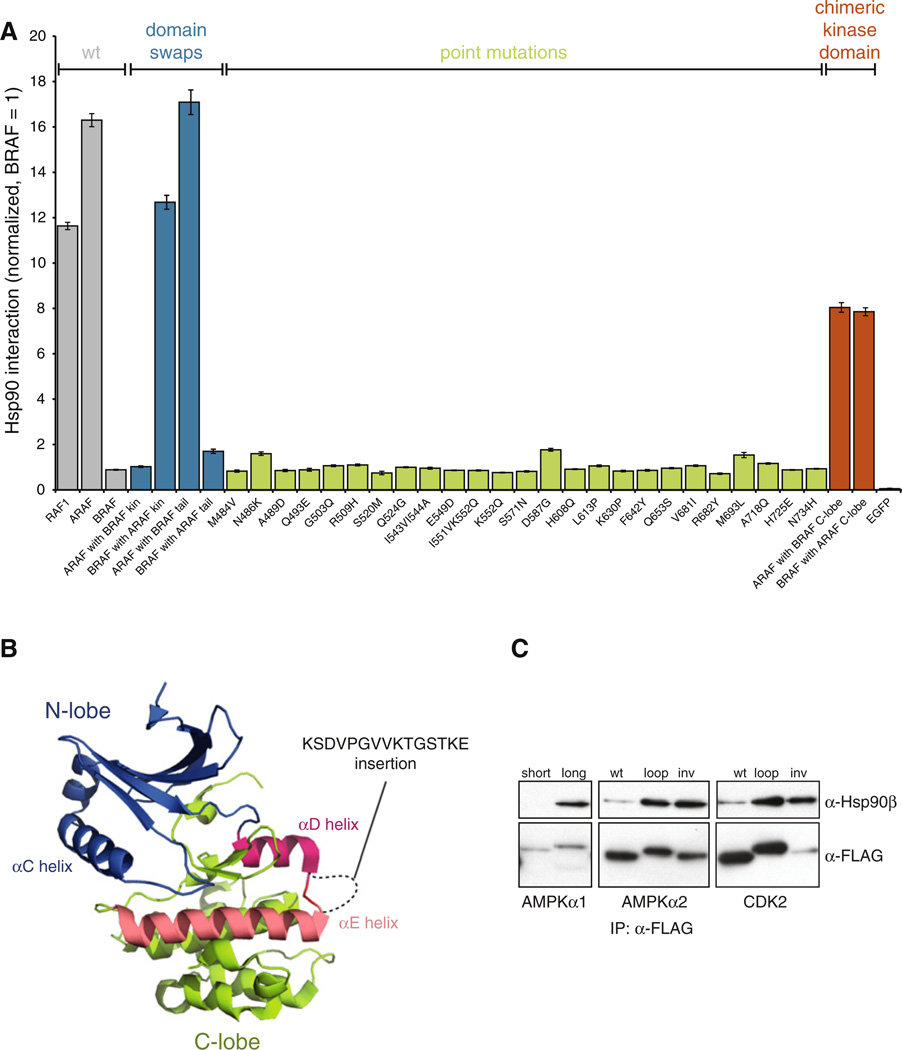
To investigate binding determinants, we focused on the three RAF family kinases, ARAF, CRAF, and BRAF, because they are highly similar over the kinase domain, yet the difference in their association with HSP90 was extreme. Of the 314 kinases we tested, ARAF was the sixth strongest interactor and CRAF was the 11th strongest, whereas the binding of BRAF was barely detectable. First, we created several chimeric constructs to validate that the HSP90 binding determinants are located in the kinase domain (Figure S4A). Chimeric proteins containing the regulatory domain or the C-terminal tail of BRAF but the ARAF kinase domain interacted strongly with HSP90. Reciprocally swapped constructs did not (Figure 5A, blue bars).
Figure 5. HSP90 Interaction Is Determined by a Distributed Set of Residues and Is Affected by Alternative Splicing.
(A) Multiple residues regulate ARAF interaction with HSP90. 3 × FLAG-tagged kinase constructs were transfected into Renilla-HSP90 cells and their interaction with HSP90 was measured with LU-MIER. Wild-type BRAF interacts weakly with HSP90, whereas RAF1 (CRAF) and ARAF are among the strongest HSP90 clients (gray). Constructs containing the kinase domain of ARAF but regulatory domains of BRAF interact strongly with HSP90 (blue), but single point mutations in BRAF do not confer robust interaction with HSP90 (green). Clones with chimeric kinase domains display intermediate phenotypes (orange). Error bars indicate SDs (n = 4).
(B) Location and the sequence of the alternatively spliced αD-αE loop in AMPKα1 (PRKAA1) are shown in the structure of the AMPKα2 isoform (PDB 2H6D). Kinase N-lobe is colored blue, C-lobe in green, αD helix in red, and αE helix in salmon pink.
(C) Alternative splicing of AMPKα1 regulates HSP90 association. 3 × FLAG-tagged kinase constructs were transfected into 293T cells and interaction with endogenous HSP90 was assayed by coimmunoprecipitation. AMPKα1 loop was inserted into AMPKα2 or CDK2 that do not have loops between the two helices. As a control, the DNA sequence encoding the loop sequence was also inserted inverted (inv) into the same location.
See also Figure S4.
Next, we mutated each residue in the BRAF kinase domain that was different from the corresponding residues in both ARAF and CRAF (Figure S4B). In total, 25 mutant constructs were tested. Strikingly, no single mutation conferred robust HSP90 association for BRAF (Figure 5A, green bars). In contrast, ARAF and BRAF kinase domain chimeras, in which the N-lobe of one kinase was fused to the C-lobe of the other kinase domain, resulted in intermediate binding for both (Figure 5A). These results establish that HSP90 binding determinants in these wild-type proteins are widely distributed in both lobes of the kinase domain and act to determine HSP90 interaction in a combinatorial way.
HSP90 Association Can Also Be Modulated by Highly Localized Elements
Results with RAF family proteins, as well as amino acid residue covariation analysis, suggest that a distributed set of residues is required for robust HSP90 association. To our surprise, however, we also found that alternative splice isoforms of the very same kinase sometimes had striking differences in HSP90 binding. For example, the AMP-activated kinase AMPKα1 had two alternative splice isoforms in our library. The longer contains an additional exon that adds 15 amino acids to the αD-αE loop in the kinase C-lobe (Figure 5B). This isoform strongly associated with HSP90; the shorter isoform did not. The closely related AMPKα2 isoform, encoded by a distinct gene and sharing 96% amino acid similarity over the kinase domain, does not contain a long loop and is not an HSP90 client.
These data would seem to indicate that the αD-αE loop contains a sequence that determines HSP90 association, but this proved not to be the case. We first confirmed the LUMIER finding by immunoprecipitation with the endogenous HSP90 protein (Figure 5C). Next, we constructed and analyzed new variants of the AMPKα1 paralog AMPKα2. Inserting the 15 aa sequence from AMPKα1 into the same position on AMPKα2 significantly increased its association with HSP90 (Figure 5C). However, insertion of the same DNA sequence but inverted and encoding an unrelated amino acid sequence, similarly increased HSP90 binding. Finally, we inserted these same loops into CDK2, a nonclient from a different kinase family. Again, longer αD-αE loop promoted HSP90 association regardless of the loop sequence. Thus, the mere presence of a long αD-αE loop, regardless of the amino acid sequence, can drastically affect HSP90 association.
Similarly, we found that alternative splicing of the GRK6 kinase regulated its association with HSP90 in a sequence-independent manner (Figures S4C and S4D).
HSP90 Client Kinases Are Intrinsically Less Stable
Our findings presented a paradox. While determinants for HSP90 interaction can be widely distributed, alternative splicing can produce highly localized changes with marked effects. How might these results be reconciled? To address this question, we globally analyzed the general properties of each class of kinases. Client and nonclient kinases had similar subcellular localizations. Strong clients and nonclients had no difference in hydrophobicity. Nonclient proteins were, on average, smaller than weak or strong clients but this difference was slight and not significant when only kinase domains were considered (data not shown). Somewhat more surprisingly, given the importance of HSP90 in protein homeostasis networks, clients and nonclients had similar numbers of interaction partners and, moreover, did not differ in the number or the length of intrinsically disordered regions (Table S1; data not shown). Given the role of Hsp90 in evolutionary processes, however, it is notable that the amino acid sequences of the client kinases have evolved more quickly than those of nonclients (Figure S5A).
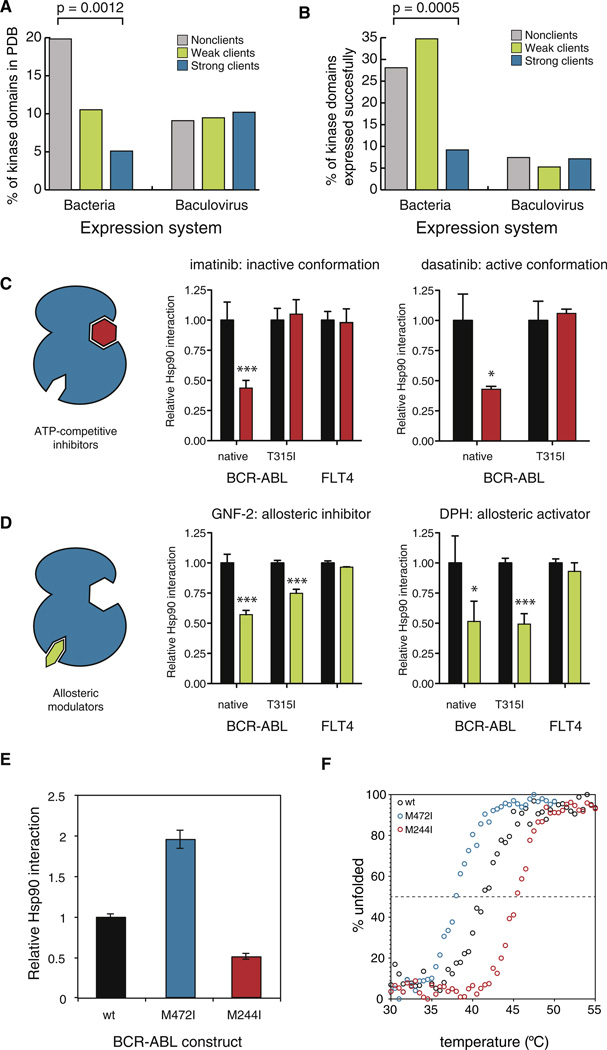
Suggesting HSP90 clients might be less stable than nonclients, fewer clients had a solved crystal structure for the kinase domain deposited in the Protein Database (p = 0.0012, Fisher’s exact test; Figure 6A). (There was no publication bias between the kinase classes (Table S1), excluding the trivial explanation of research priorities.) To corroborate this finding, we analyzed protein expression data from the Structural Genomics Consortium (SGC) (kindly provided by Stefan Knapp). In their efforts to determine the three-dimensional structure of every kinase in the human genome, SGC has expressed a large number of human kinase domains in heterologous expression systems. They have successfully expressed, as determined by protein solubility, 163 human kinase domains. Soluble protein was obtained for 36% of all nonclients but for only 16% of strong clients (p = 0.0005, Fisher’s exact test; Figure 6B). Importantly, neither of these differences was significant for kinases that had been expressed in eukaryotic systems. This is consistent with known differences in the HSP90 machinery of prokaryotic and eukaryotic cells—prokaryotes do not have CDC37 and the prokaryotic HSP90 does not chaperone kinases (Buchner, 2010)—as well as with our observation that CDC37 is universally employed by HSP90 for all kinases.
Figure 6. HSP90 Associates with Thermodynamically Unstable Kinases.
(A) Distribution of kinase domain crystal structures deposited to PDB by client class and expression system extracted from the PDB coordinate file. p value was calculated with Fisher’s exact test.
(B) Strong HSP90 clients are less soluble than nonclients or weak clients. Kinase domains that have been expressed in a soluble manner by Structural Genomics Consortium are divided into client classes. p value was calculated with Fisher’s exact test.
(C) ATP-competitive inhibitors decrease HSP90:: BCR-ABL interactions. 3 × FLAG-tagged kinases were transfected into Renilla-HSP90 cells, and HSP90 interaction was measured by LUMIER after 1 hr treatment with 5 µM inhibitor or DMSO. Average relative interaction with HSP90 (DMSO = 1) from four independent replicates is shown with SD. Statistical significance is shown as asterisks (*p < 0.05; ***p < 0.001).
(D) Stabilization of the kinase fold with allosteric modulators decreases HSP90::BCR-ABL interactions. Experiment was done as in Figure 5C, using 5 µM GNF-2 or DPH.
(E) BCR-ABL point mutations have opposing effects on HSP90 interaction. Interaction of wild-type BCR-ABL (black), M472I mutant (blue) and M244I mutant (red) with HSP90 was measured with LUMIER. Average relative interaction (wild-type = 1) from four independent replicates is shown with standard deviation.
(F) Thermal stability of ABL mutants correlates with HSP90 association. Thermal denaturation curves of wild-type ABL and M472I and M244I mutant proteins were determined by circular dichroism. See also Figure S5.
Low solubility in vivo can be caused by multiple mechanisms—electrostatics, aggregation propensity, conformational (kinetic) instability or thermal instability. To further evaluate biophysical differences between nonclient and client kinases, we compared HSP90 interaction profiles with 56 published in vitro thermal stability profiles for kinase domains (Fedorov et al., 2007). Our quantitative interaction scores for Hsp90 clients correlated with thermal instability (R2 = 0.23, p = 0.019; Figure S5B). The correlation between these very different types of assays (in vivo full-length kinases versus in vitro kinase domains) is all the more striking considering that most of the successfully purified proteins were nonclients (and are therefore not plotted on this graph). Only three measurements were available from moderately strong clients (LUMIER interaction scores of 3–4) and none from very strong clients (interaction scores 5–7). Notably, an independent analysis of mutated oncogenic v-Src (which scored as a strong client in our assay) established that it was much less stable in vitro than c-Src (Falsone et al., 2004), a nonclient.
Stabilization of the Kinase Domain by Small Molecules Decreases HSP90 Association
If HSP90 association is determined by the intrinsic stability of the kinase domain, it should be possible to alter HSP90::kinase interactions with stabilizing factors. To test this, we focused on BCR-ABL. A number of potent small-molecule inhibitors, of distinct chemotypes and with different modes of binding, have been developed against BCR-ABL. The binding of such inhibitors increases the stability of kinase domains in vitro (Fedorov et al., 2007).
First, we tested the effects of imatinib (Gleevec) and dasatinib (Sprycel). Both inhibit BCR-ABL very potently and bind the ATP binding pocket in an ATP-competitive manner. When these two inhibitors bind BCR-ABL, however, they stabilize distinct conformations. Imatinib binds the inactive conformation and dasatinib the active one. Both inhibitors significantly decreased HSP90::BCR-ABL interaction (Figure 6C). As a control, interaction between HSP90 and FLT4, a tyrosine kinase not targeted by imatinib, was not affected by the drug. Next we tested the clinically relevant imatinib-resistant BCR-ABL mutation T315I, which blocks the binding of both inhibitors to the ATP-binding site. The mutation abolished the effect of both inhibitors on HSP90 binding.
To address the possibility that HSP90 and the small-molecule inhibitors simply compete for binding to the ATP-binding pocket, we used GNF-2. This molecule is an allosteric Abl inhibitor that binds a unique myristate-binding pocket in the C-lobe far away from the ATP binding site (Adrián et al., 2006). Similar to imatinib and dasatinib, GNF-2 significantly decreased BCR-ABL::HSP90 interaction but had no effect on the FLT4::HSP90 complex (Figure 6D). However, in striking contrast to the ATP-competitive inhibitors, GNF-2 also decreased the interaction between the BCR-ABL T315I mutant and HSP90. This is consistent with the observation that this class of inhibitors binds to both the wild-type and the T315I mutant protein (Iacob et al., 2011).
Finally, we asked if the decrease in HSP90’s interaction with BCR-ABL might simply be the result of reduced kinase activity. DPH, another small-molecule modulator of ABL function, binds the same allosteric pocket as GNF-2 but activates the kinase rather than inhibiting it (Yang et al., 2011). The effect of DPH on the HSP90::BCR-ABL interaction was identical to that of GNF-2: both native BCR-ABL and the T315I mutant were displaced from HSP90 by the drug (Figure 6D). Thus, a decrease in kinase activity is not necessary for HSP90 displacement. Stabilization of the kinase fold, either in its active or inactive conformation, is sufficient.
BCR-ABL Mutations Modulating HSP90 Interaction Affect Protein Stability
Our results with kinase inhibitors suggested that thermal stability is a major determinant of HSP90::kinase interactions. The metastable folds of Hsp90 chaperone clients make them difficult to analyze in vitro. Indeed, only two proteins (oncogenic v-Src and its cellular counterpart c-Src) have been specifically assessed both for differences in HSP90 interaction in vivo and differences in thermodynamic stability in vitro (Falsone et al., 2004). This pair presents an extreme in terms of Hsp90 interactions (very strong versus very weak client) and in terms of conformational equilibrium (active, open conformation versus inactive, closed conformation) (Falsone et al., 2004). To further investigate the relationship between HSP90 association and thermodynamic stability, we focused on two additional BCR-ABL variants, both methionine-to-isoleucine substitutions, that had opposing but relatively subtle effects on the BCR-ABL::HSP90 interaction. The M244I mutation in the N-terminal lobe decreased Hsp90 interaction by about 50%; the M472I mutation in the C-terminal lobe increased it 2-fold (Figure 6E).
We expressed both mutants and the wild-type Abl in bacteria and purified them to homogeneity. The constructs contained Abl residues 65–534, including the SH3, SH2 and kinase domains. Importantly, due to the lack of an inhibitory myristoylation site in the N terminus, recombinant Abl adopts an active conformation similar to that of the kinase domain in the context of BCR-ABL fusion (Iacob et al., 2011). Both mutants had in vitro kinase activities comparable to that of the wild-type kinase (data not shown) and their secondary structure content, as measured by circular dichroism spectroscopy was very similar (Figure S5C). However, the thermal stability of the kinase constructs was significantly different. The Tm of wild-type Abl was 41.5°C. For the M472I mutant it was 38.0°C and for the M244I mutant it was 45.4°C (Figure 6F). Thus, the increase and decrease in HSP90 interactions for these mutants reflected differences in the thermodynamic stability of their kinase folds. Taken together, the breadth of our analyses reveal that thermodynamic stability, not primary amino acid sequence, is a key determinant in the association of kinase clients with HSP90.
DISCUSSION
High-throughput quantitative analysis of genetic interactions in vivo, pioneered in the budding yeast, has greatly increased our understanding of cellular circuitries (Costanzo et al., 2010). LUMIER with BACON provides a method for revealing the physical interaction networks in a quantitative manner on a comparable scale. We utilized the assay to quantitatively characterize the in vivo interactions of HSP90 and CDC37 with the majority of wild-type human kinases, transcription factors, and E3 ligases. HSP90 is known to play a key role in regulating cellular circuitry and, together, the assayed proteins form the core of that circuitry. The analysis uncovered interactions between HSP90 and almost 400 client proteins, more than all previous studies combined, considerably expanding our picture of this essential chaperone’s function in human cells.
More importantly, this quantitative and systematic approach allowed us to place HSP90 interactions into a broader context and uncover global principles that could not be established with studies of individual clients, nor with previous high-throughput approaches. First, the CDC37 cochaperone serves as an adaptor and provides specificity for protein kinases at the family level. Our assay should allow the identification of adaptors for other types of fold, such as leucine-rich repeat proteins (Kadota et al., 2010) and the β-propeller WD40 and Kelch domains (reported here).
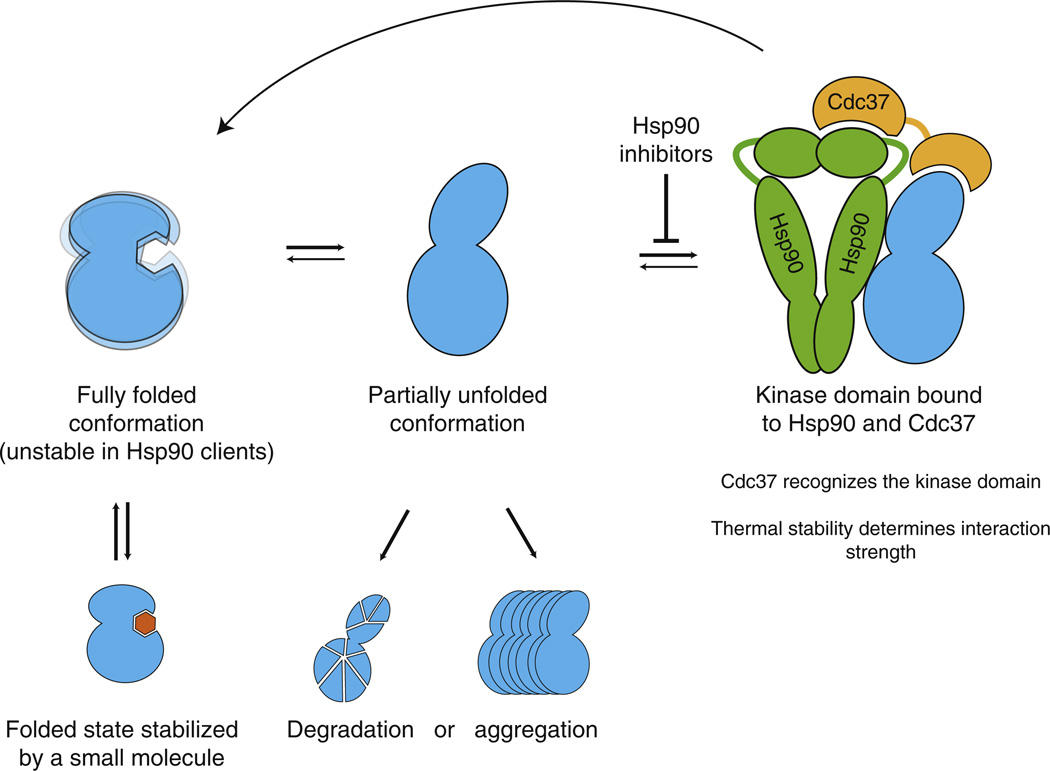
Second, within the kinase family, Hsp90 clients are ultimately distinguished from nonclients on the basis of their intrinsic stability. Agents that stabilize a client kinase in either its active or inactive conformation reduce HSP90 association, demonstrating that HSP90 does not bind the kinase in either of its folded states. We suggest that client kinases are unstable in their fully folded forms, with a higher tendency to adopt a conformation that HSP90 and CDC37 can recognize. Associations with HSP90, then, would be determined by the equilibrium between the fully folded and chaperone-susceptible conformations of each client. This conformational model can explain why kinase::HSP90 interactions fall into a continuum spanning over a 100-fold range, in contrast to the binary model of clients and nonclients. In keeping with this interpretation of our high-throughput analysis, in an electron microscopic analysis of a particular client complex, HSP90/CDC37/CDK4, both lobes of the CDK4 kinase are visible, clearly indicating that this client is not in its tightly packed closed conformation (Vaughan et al., 2006). As HSP90 client kinases undergo repeated rounds of binding and release, inhibition of HSP90’s ATPase activity prevents reloading of clients onto the chaperone (Krukenberg et al., 2011). This redirects the clients to one of two alternative fates: degradation or aggregation (Figure 7).
Figure 7. Model for HSP90::Kinase Interactions.
Kinase domains are in equilibrium between the fully folded and HSP90-binding competent conformations. CDC37 cochaperone recognizes the kinase fold and recruits kinases to HSP90. HSP90 binds the alternative kinase conformation and assists the kinase in adopting the fully folded conformation. Client kinases thus undergo repeated rounds of chaperoning, whereas nonclient kinases are stable when fully folded. Binding of an inhibitor to its target kinase increases the stability of the kinase fold and thus decreases HSP90 interaction. HSP90 inhibitors block the loading of the client to the chaperone, which leads to aggregation of the partially unfolded kinase or degradation by the ubiquitin-proteasome system.
The client protein dissociation rates we observed in vivo (T1/2 = 2–6 min) upon HSP90 inhibition were surprisingly close to the ATP hydrolysis rate of HSP90 in vitro (Richter et al., 2008). Moreover, although the strength of Hsp90’s associations with strong versus weak clients varied over a 100-fold range, most clients dissociated from HSP90 with very similar kinetics. These observations imply that the dominant determinants of HSP90 association are client-binding on-rates.
How can our model be reconciled with the existing αC-β4 recognition loop model (Citri et al., 2006; Xu et al., 2005)? The αC-β4 loop is located immediately adjacent to the kinase hinge region that in many kinases is involved in maintaining the inactive conformation (Chen et al., 2007). In fact, the backbone atoms of the αC-β4 loop in several kinases form hydrogen bonds with critical residues in the hinge. Loss of these hydrogen bonds leads to ligand-independent kinase activation (Chen et al., 2007). This network of stabilizing hydrogen bonds is conserved in several tyrosine kinases, and mutations in the αC-β4 loop in ERBB2 lead to activation of the kinase (Fan et al., 2008). Thus, it is likely that mutations introduced in the loop increase the flexibility of the hinge and lead to increased HSP90 interaction due to structural destabilization, not through the introduction of a new binding motif.
Our model accounts for a number of previous findings. First, some kinases, such as c-Src and EGFR, associate transiently with HSP90 during maturation, but the fully folded proteins do not bind HSP90 (Xu et al., 1999, 2005). Second, some mutations can decrease a kinase’s steady state association with HSP90 without having an effect on the transient association during maturation. It has been suggested that the motif HSP90 recognizes during posttranslational folding of kinases is distinct from that recognized in mature proteins (Xu et al., 2005). A more parsimonious explanation is that HSP90 binds on-pathway conformations. Kinases such as c-Src and EGFR access this conformation during initial folding, but they are stable after reaching their mature states. But strong client kinases repeatedly access the Hsp90-susceptible conformation after their initial folding.
Parallels between Kinase and Steroid Hormone Chaperoning
For 30 years, steroid hormone receptors have been the most thoroughly studied class of HSP90 clients, and our current mechanistic understanding of the chaperone is largely based on detailed in vitro studies with steroid hormone receptors (SHRs) (Picard, 2006). Although we find that transcription factors actually represent a minor fraction of HSP90 clients, our data present striking parallels between the modes of kinase and SHR chaperoning. Consider the steps of progesterone receptor chaperoning by HSP90: without progesterone, the receptor associates with HSP90 through its ligand-binding domain. HSP90 aids the ligand-binding domain in maintaining a conformation that is competent to bind the ligand. Upon ligand binding, the receptor dissociates from HSP90, translocates to the nucleus and regulates target gene expression (Picard, 2006). The binding of a kinase inhibitor stabilizes the kinase fold and shifts the equilibrium away from HSP90 as, presumably, would the binding of ATP and the kinases’ physiological substrates. Might a common HSP90 function be the modulation of ligand-binding clefts in signal transducers, as previously suggested (Pratt et al., 2008)? Notably, other HSP90 clients are also regulated by ligands. Argonaute proteins, which are involved in small RNA biogenesis, require HSP90 for loading of the ligand, a small RNA duplex (Iwasaki et al., 2010). Similarly, inducible nitric oxide synthase (iNOS), another HSP90 client, binds heme as a cofactor, and depletion of heme increases HSP90::iNOS interaction (Ghosh et al., 2011).
HSP90 and the Evolution of Eukaryotic Kinomes
We have found that all kinase families have both client and nonclient kinases, indicating that HSP90 client proteins did not evolve from a common client ancestor, or even a small number of ancestors. Clients are not “chaperone-addicted” in an evolutionary sense, but rapidly evolve into and out of dependence on HSP90. If kinases can rapidly evolve away from chaperone-assisted folding, why haven’t they generally done so? One explanation is that we are simply catching them in the act of making such transitions. But a more interesting possibility is that some kinases specifically exploit association with the HSP90 machinery as a means of regulation. Compared to other regulatory proteins, kinase activity is controlled by a particularly rich spectrum of mechanisms and these protein undergo extensive structural rearrangements upon activation (Taylor and Kornev, 2010). The inherent instability of some kinases, which tunes their interaction with Hsp90, may well be employed as a mechanism to ensure the fidelity of regulatory cascades. Such instability could affect the regulation of signal transduction pathways with more immediacy, and be more easily tuned to changing environmental conditions, than mechanisms acting at the level of transcription, translation or degradation. Indeed, because HSP90’s chaperone activity is ATP-dependent and its affinity for ATP is relatively low, HSP90 interactions could provide a means to sense energy states and coordinate an entire network of kinases with the ATP balance in the cell.
On a longer timescale, considering the role of HSP90 in the evolution of complex traits (Jarosz and Lindquist, 2010; Queitsch et al., 2002; Rutherford and Lindquist, 1998; Specchia et al., 2010), the chaperone has likely facilitated the remarkable diversification of the kinase family. Eukaryotic genomes encode hundreds of different kinases, but the exact composition of the kinome varies strikingly between phyla (Manning et al., 2011). In yeast, CDC37 is required for the stability of most kinases(Mandal et al., 2007). Together with our observation that CDC37 is a universal kinase cochaperone in human cells, this suggests that HSP90 and CDC37 have together sculpted the evolution of kinases for billions of years. As the evolution of novel functions occurs usually through destabilizing mutations, HSP90 may have acted as an evolutionary “backbone,” allowing kinases to evolve through a relaxed mutational landscape. The facilitation of kinome evolution by HSP90 could partly explain why kinases have emerged as some of the most diverse drivers of biological complexity.
EXPERIMENTAL PROCEDURES
LUMIER with BACON
3 × FLAG-tagged bait proteins were transfected into stable 293T cell line in 96-well format with polyethylemine (PEI). Two days after transfection, cells were washed in 1 × PBS and lysed in ice-cold HENG buffer (50 mM HEPES-KOH [pH 7.9], 150 mM NaCl, 20 mM Na2MoO4, 2mM EDTA, 5% glycerol, 0.5% Triton X-100). The combination of low temperature, the absence of ATP and the presence of molybdate freezes the chaperone cycle and stabilizes HSP90::client interactions. The lysate was transferred to 384-well plates coated with anti-FLAG M2 antibody (Sigma-Aldrich). Plates were incubated in cold room for 3 hr, after which plates were washed with the lysis buffer using an automated plate washer. Luminescence in each well was measured with an Envision plate reader using Gaussia FLEX luciferase kit (New England Biolabs). After luminescence measurement, HRP-conjugated anti-FLAG antibody in ELISA buffer (1 × PBS, 2% goat serum, 5% Tween 20) was added to wells. Plates were washed in 1 × PBS/0.05% Tween 20 with a plate washer and ELISA signal was detected with 3,3’,5,5’-tetramethylbenzidine (TMB) substrate.
Computational Analysis of HSP90 Clients
To identify significantly covarying residues, we used mutual information analysis, OMES covariance, SCA covariance, and McBasc as implemented in Java (Fodor and Aldrich, 2004). All programs were used with their default settings. Only strong clients and nonclients were considered in the analysis and loops were excluded due to poor alignment confidence. To evaluate the significance of the scores from each covariance method, we performed a randomization analysis with 10,000 permutations. We sampled without replacement the class labels for all sequences and ran all algorithms on the newly generated randomized data sets to generate 10,000 scores for each algorithm. We generated an empirical p value for every residue from each algorithm. We used a p value cutoff of 0.01 for marking a residue as significant and selected for further analysis only the residues identified as significant by all algorithms. Nonparametric statistical analyses were applied unless specified otherwise.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank D.F. Jarosz, L. Whitesell, and M. Mendillo for valuable comments, K. Salehi-Ashtiani and M. Vidal (Dana-Farber Cancer Institute) for the human ORFeome collection,D. Proia (Synta Pharmaceuticals) for ganetespib, and N.S. Gray (Dana-Farber Cancer Institute) for DPH. M.T. was supported by the Academy of Finland, Sigrid Juselius Foundation and the Human Frontier Science Program. G.I.K. was supported by an EMBO Long-Term Fellowship and the Human Frontier Science Program. S.L. is an investigator of the Howard Hughes Medical Institute. Support for this study was also provided by the NIH Genomics Based Drug Discovery-Driving Medical Projects grant UL1-DE019585, administratively linked to NIH grants RL1-GM084437, RL1-CA133834, and RL1-HG004671. M.T. and S.L. planned the project and designed the experiments. M.T. developed and validated LUMIER with BACON assay and performed all the experiments with the help of I.K. G.I.K. assayed HSP90 interactions against E3 ligases. K.D.W. and M.T. expressed and purified recombinant ABL, and C.K. and M.T. analyzed ABL with circular dichroism. M.T. and M.K. performed computational analyses of HSP90 interactions. M.T. and S.L. wrote the paper, with comments from all the coauthors.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures five figures, and one table, and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2012.06.047.
REFERENCES
- Adrián FJ, Ding Q, Sim T, Velentza A, Sloan C, Liu Y, Zhang G, Hur W, Ding S, Manley P, et al. Allosteric inhibitors of Bcrabl-dependent cell proliferation. Nat. Chem. Biol. 2006;2:95–102. doi: 10.1038/nchembio760. [DOI] [PubMed] [Google Scholar]
- Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS, Shinjo F, Liu Y, Dembowy J, Taylor IW, et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307:1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- Boulon S, Pradet-Balade B, Verheggen C, Molle D, Boireau S, Georgieva M, Azzag K, Robert MC, Ahmad Y, Neel H, et al. HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol. Cell. 2010;39:912–924. doi: 10.1016/j.molcel.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner J. Bacterial Hsp90—desperately seeking clients. Mol. Microbiol. 2010;76:540–544. doi: 10.1111/j.1365-2958.2010.07140.x. [DOI] [PubMed] [Google Scholar]
- Caplan AJ, Mandal AK, Theodoraki MA. Molecular chaperones and protein kinase quality control. Trends Cell Biol. 2007;17:87–92. doi: 10.1016/j.tcb.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Chen H, Ma J, Li W, Eliseenkova AV, Xu C, Neubert TA, Miller WT, Mohammadi M. A molecular brake in the kinase hinge region regulates the activity of receptor tyrosine kinases. Mol. Cell. 2007;27:717–730. doi: 10.1016/j.molcel.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri A, Harari D, Shohat G, Ramakrishnan P, Gan J, Lavi S, Eisenstein M, Kimchi A, Wallach D, Pietrokovski S, Yarden Y. Hsp90 recognizes a common surface on client kinases. J. Biol. Chem. 2006;281:14361–14369. doi: 10.1074/jbc.M512613200. [DOI] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Rocha Dias S, Friedlos F, Light Y, Springer C, Workman P, Marais R. Activated B-RAF is an Hsp90 client protein that is targeted by the anticancer drug 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 2005;65:10686–10691. doi: 10.1158/0008-5472.CAN-05-2632. [DOI] [PubMed] [Google Scholar]
- Ehrlich ES, Wang T, Luo K, Xiao Z, Niewiadomska AM, Martinez T, Xu W, Neckers L, Yu XF. Regulation of Hsp90 client proteins by a Cullin5-RING E3 ubiquitin ligase. Proc. Natl. Acad. Sci USA. 2009;106:20330–20335. doi: 10.1073/pnas.0810571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsone SF, Leptihn S, Osterauer A, Haslbeck M, Buchner J. Oncogenic mutations reduce the stability of SRC kinase. J. Mol. Biol. 2004;344:281–291. doi: 10.1016/j.jmb.2004.08.091. [DOI] [PubMed] [Google Scholar]
- Falsone SF, Gesslbauer B, Tirk F, Piccinini AM, Kungl AJ. A proteomic snapshot of the human heat shock protein 90 interactome. FEBS Lett. 2005;579:6350–6354. doi: 10.1016/j.febslet.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Falsone SF, Gesslbauer B, Rek A, Kungl AJ. A proteomic approach towards the Hsp90-dependent ubiquitinylated proteome. Proteomics. 2007;7:2375–2383. doi: 10.1002/pmic.200600996. [DOI] [PubMed] [Google Scholar]
- Fan YX, Wong L, Ding J, Spiridonov NA, Johnson RC, Johnson GR. Mutational activation of ErbB2 reveals a new protein kinase autoinhibition mechanism. J. Biol. Chem. 2008;283:1588–1596. doi: 10.1074/jbc.M708116200. [DOI] [PubMed] [Google Scholar]
- Fedorov O, Marsden B, Pogacic V, Rellos P, Müller S, Bullock AN, Schwaller J, Sundström M, Knapp S. A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc. Natl. Acad. Sci. USA. 2007;104:20523–20528. doi: 10.1073/pnas.0708800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor AA, Aldrich RW. Influence of conservation on calculations of amino acid covariance in multiple sequence alignments. Proteins. 2004;56:211–221. doi: 10.1002/prot.20098. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Chawla-Sarkar M, Stuehr DJ. Hsp90 interacts with inducible NO synthase client protein in its heme-free state and then drives heme insertion by an ATP-dependent process. FASEB J. 2011;25:2049–2060. doi: 10.1096/fj.10-180554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grammatikakis N, Lin JH, Grammatikakis A, Tsichlis PN, Cochran BH. p50(cdc37) acting in concert with Hsp90 is required for Raf-1 function. Mol. Cell. Biol. 1999;19:1661–1672. doi: 10.1128/mcb.19.3.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray PJ, Jr, Prince T, Cheng J, Stevenson MA, Calderwood SK. Targeting the oncogene and kinome chaperone CDC37. Nat. Rev. Cancer. 2008;8:491–495. doi: 10.1038/nrc2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacob RE, Zhang J, Gray NS, Engen JR. Allosteric interactions between the myristate- and ATP-site of the Abl kinase. PLoS ONE. 2011;6:e15929. doi: 10.1371/journal.pone.0015929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Kobayashi M, Yoda M, Sakaguchi Y, Katsuma S, Suzuki T, Tomari Y. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol. Cell. 2010;39:292–299. doi: 10.1016/j.molcel.2010.05.015. [DOI] [PubMed] [Google Scholar]
- Jarosz DF, Lindquist S. Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science. 2010;330:1820–1824. doi: 10.1126/science.1195487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadota Y, Shirasu K, Guerois R. NLR sensors meet at the SGT1-HSP90 crossroad. Trends. Biochem. Sci. 2010;35:199–207. doi: 10.1016/j.tibs.2009.12.005. [DOI] [PubMed] [Google Scholar]
- Krukenberg KA, Street TO, Lavery LA, Agard DA. Conformational dynamics of the molecular chaperone Hsp90. Q. Rev. Biophys. 2011;44:229–255. doi: 10.1017/S0033583510000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal AK, Lee P, Chen JA, Nillegoda N, Heller A, DiStasio S, Oen H, Victor J, Nair DM, Brodsky JL, Caplan AJ. Cdc37 has distinct roles in protein kinase quality control that protect nascent chains from degradation and promote posttranslational maturation. J. Cell Biol. 2007;176:319–328. doi: 10.1083/jcb.200604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Manning G, Reiner DS, Lauwaet T, Dacre M, Smith A, Zhai Y, Svard S, Gillin FD. The minimal kinome of Giardia lamblia illuminates early kinase evolution and unique parasite biology. Genome Biol. 2011;12:R66. doi: 10.1186/gb-2011-12-7-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace CN, Scholtz JM. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 1998;75:422–427. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard D. Chaperoning steroid hormone action. Trends Endocrinol. Metab. 2006;17:229–235. doi: 10.1016/j.tem.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Pratt WB, Morishima Y, Osawa Y. The Hsp90 chaperone machinery regulates signaling by modulating ligand binding clefts. J. Biol. Chem. 2008;283:22885–22889. doi: 10.1074/jbc.R800023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Sangster TA, Lindquist SL. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417:618–624. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- Richter K, Soroka J, Skalniak L, Leskovar A, Hessling M, Reinstein J, Buchner J. Conserved conformational changes in the ATPase cycle of human Hsp90. J. Biol. Chem. 2008;283:17757–17765. doi: 10.1074/jbc.M800540200. [DOI] [PubMed] [Google Scholar]
- Ricketson D, Hostick U, Fang L, Yamamoto KR, Darimont BD. A conformational switch in the ligand-binding domain regulates the dependence of the glucocorticoid receptor on Hsp90. J. Mol. Biol. 2007;368:729–741. doi: 10.1016/j.jmb.2007.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford SL, Lindquist SL. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- Specchia V, Piacentini L, Tritto P, Fanti L, D’Alessandro R, Palumbo G, Pimpinelli S, Bozzetti MP. Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature. 2010;463:662–665. doi: 10.1038/nature08739. [DOI] [PubMed] [Google Scholar]
- Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Kornev AP. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem. Sci. 2010;36:65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan CK, Gohlke U, Sobott F, Good VM, Ali MM, Prodromou C, Robinson CV, Saibil HR, Pearl LH. Structure of an Hsp90-Cdc37-Cdk4 complex. Mol. Cell. 2006;23:697–707. doi: 10.1016/j.molcel.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yuan X, Xiang Z, Mimnaugh E, Marcu M, Neckers L. Surface charge and hydrophobicity determine ErbB2 binding to the Hsp90 chaperone complex. Nat. Struct. Mol. Biol. 2005;12:120–126. doi: 10.1038/nsmb885. [DOI] [PubMed] [Google Scholar]
- Xu Y, Singer MA, Lindquist SL. Maturation of the tyrosine kinase c-src as a kinase and as a substrate depends on the molecular chaperone Hsp90. Proc. Natl. Acad. Sci. USA. 1999;96:109–114. doi: 10.1073/pnas.96.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Campobasso N, Biju MP, Fisher K, Pan XQ, Cottom J, Galbraith S, Ho T, Zhang H, Hong X, et al. Discovery and characterization of a cell-permeable, small-molecule c-Abl kinase activator that binds to the myristoyl binding site. Chem. Biol. 2011;18:177–186. doi: 10.1016/j.chembiol.2010.12.013. [DOI] [PubMed] [Google Scholar]
- Ying W, Du Z, Sun L, Foley KP, Proia DA, Blackman RK, Zhou D, Inoue T, Tatsuta N, Sang J, et al. Ganetespib, a unique triazolone-containing Hsp90 inhibitor, exhibits potent antitumor activity and a superior safety profile for cancer therapy. Mol. Cancer. Ther. 2011;11:475–484. doi: 10.1158/1535-7163.MCT-11-0755. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.