Abstract
Using computational methods, which allow mechanistic insights at a molecular level, we explored the olfactory receptor (OR)–odor interactions for 2 mouse ORs, S79 and S86. Both ORs have been previously experimentally, functionally characterized. The odors used were mostly carboxylic acids, which differed in chain length, substituents on the primary carbon atom-chain and degree of unsaturation. These odors elicited varied activation responses from both ORs. Our studies revealed that both receptors have 2 distinct binding sites. Preferential binding in 1 of the 2 sites is correlated with OR activation. The activating odorants: nonanedioic acid, heptanoic acid, and octanoic acid for OR S79 and nonanoic acid for OR S86 preferentially bind in the region bound by transmembranes (TMs [helical domains]) III, IV, V, and VI. The non excitatory odorants heptanol for S79 and heptanoic acid for S86 showed a greater likelihood of binding in the region bound by TMs I, II, III, and VII. Nanosecond-scale molecular dynamics simulations of the physiologically relevant conditions of docked OR–odorant complexes enabled us to quantitatively assess the roles of individual OR amino acids in odor binding. Amino acid–odorant contact maps and distance determinations over the course of the simulations lend support to our conclusions.
Key words: docking, molecular dynamics, odorant binding, olfactory receptor
Introduction
Olfactory receptor (OR) repertoires constitute the largest gene families in mammalian genomes (Glusman et al. 2000a, 2000b; Rouquier et al. 2000; Crasto et al. 2001; Mombaerts 2001; Zozulya et al. 2001; Young and Trask 2002; Gilad et al. 2003; Olender et al. 2004). Sequence variations among relatively fewer ORs, while retaining an invariant Class-A (rhodopsin-like) GPCR structure, are responsible for discriminating several thousand odorants (Muller 2000; Palczewski et al. 2000; Crasto 2009; Kufareva et al. 2011; Rosenbaum et al. 2011; Rasmussen et al. 2011). The mechanism of odor binding by an OR is not well understood. There exists no experimentally validated predictive method by which specific odorants might be identified a priori as eliciting excitatory responses from specific ORs. The consequences of understanding OR–odorant interactions are also far-reaching from a clinical standpoint: several neurological disorders find their origins, in part, in olfactory dysfunction (Doty et al. 1991; Kovacs 2004; Hawkes 2006; Barresi et al. 2012).
Experimental efforts to deorphanize ORs (Boekhoff et al. 1997; Touhara et al. 1999; Araneda et al. 2000; Kajiya et al. 2001; Touhara 2001, 2002; Bozza et al. 2002; Katada et al. 2003, 2005; Matarazzo et al. 2005; Oka et al. 2004, 2006; Abaffy et al. 2006; Schmiedeberg et al. 2007; Grosmaitre et al. 2009; Kato and Touhara 2009; Repicky and Luetje 2009; Li et al. 2012; Ondachi et al. 2012 ) that is identify odorants that are likely to bind and excite specific ORs are challenging: 1) in heterologous expression systems, the expressed proteins remain embedded in the plasma membrane and 2) the promiscuous nature of OR–odorant binding. The required scale of odor ligand libraries necessary to screen for OR activity therefore becomes daunting.
The structural difference between an excitatory or non excitatory ligand in a single receptor may be as small as the presence or absence of a single heavy atom (Zhao et al. 1998). Gaining mechanistic insights into OR–odor binding would aid in predicting a focused, rational panel of test-odorants for a specific OR. In addition to functional characterization of single ORs, combinatorial studies involving several ORs tested against panels of several odorants have also been carried out. The first combinatorial functional analysis tested 14 mouse ORs and several carboxylic acids and their derivatives (Malnic et al. 1999). A decade later, an extensive study involving more than 60 ORs was completed (Saito et al. 2009).
Computational methods have often been employed to provide a molecular, and, by extension, mechanistic basis for OR interactions with odorants. (Bertsch et al. 1998; Singer 2000; Floriano et al. 2000, 2004; Hall et al. 2004; Trabanino et al. 2004; Lai et al. 2005; Crasto 2009; Lai and Crasto 2012) These studies have largely focused on OR–odorant systems that have been previously experimentally, functionally characterized. The lack of an experimentally derived structure of an OR—a rhodopsin-like GPCR—however, results in uncertainties in making broad assumptions as to the nature of OR–odorant binding.
Computational methodologies have involved building a model of the OR. The X-ray crystal structure of bovine rhodopsin (Palczewski et al. 2000; Okada et al. 2004) has most often been used as the template on which a (in our protocol, preliminary) structure of the OR is built. Other GPCRs have also recently been crystallized (or co-crystallized with molecules that enhance solubility) and remain viable alternatives as templates for OR structures (Rasmussen et al. 2011; Rosenbaum et al. 2011; Xu et al. 2011; Wu et al. 2012). The next step has been to computationally dock molecular models of odor ligands in the OR’s solvent accessible spaces. Several docking programs are available (Vakser 1997; Ewing et al. 2001; Vakser and Jiang 2002; Tovchigrechko and Vakser 2006; Morris et al. 2009). Docking conformations are scored based on energy of conformation in accessible spaces within the protein’s binding region. The docking programs also make allowances for electrostatic interactions involving the ligand and the protein (Singer et al. 1995, 1998; Singer 2000; Arata-Kawai et al. 2011). Following computational docking, conjectures as to amino acid involvement in binding have been made based on the closeness of the side chains to the docked odor ligand.
Some of our computational studies have gone beyond static computational docking. We have established a dynamic basis for OR–odorant interactions. (Lai et al. 2005; Lai and Crasto 2012) Our protocol (discussed in the next section) is based on the notion that realistic OR–odorant interactions are not static. We have previously shown that the amino acids responsible for facilitating binding and possible OR activation are likely to change as the interactions progress. Molecular dynamics-based simulation studies have been carried out to study odor behavior within the OR binding pocket and the structural response of the OR. The first such study (Lai et al. 2005) showed how within the binding pocket, an odor’s transit through the binding pocket was facilitated (a pathway from inside to outside) by a combination of electrostatic and Van der Waals forces. These studies which attempted to computationally replicate previously published experimental OR activation results (Araneda et al. 2000) showed a strong correlation between ligands that experimentally activated the OR and the existence of a transit event. (Lai et al. 2005)
To further efforts towards elucidating the mechanism by which an odor will activate an OR, we present here the results of a computational study involving 2 ORs and 6 odorants. We advance 2 salient points. One, that the preference of (1 of 2) binding region may be a determinant of whether an odor will excite an OR; two, that it is possible to quantify the role of an amino acid residue in OR binding and subsequent activation. We studied 2 ORs—S79 (MOR42-3 or Olfr544) and S86 (MOR8-2 or Olfr586) for odorants that elicited an excitatory as well as non excitatory response. Although our computational studies use the results of functional analysis from the first OR-function combinatorial studies (Malnic et al. 1999), we will use the nomenclature for the ORs used in that publication—S79 and S86.
Materials and methods
The protocol described here includes: 1) creating computational models of ORs S79 and S86 and the odor ligands; 2) computationally docking ligands that are known to excite these ORs (heptanoic acid, octanoic acid, and nonanedioic acid for S79, and nonanoic acid for S86) and those that inhibit OR excitation or otherwise do not activate the ORs (heptanol for S79 and heptanoic acid for S86)—the choice of ORs and odors are based on experimental functional analyses in the solvent-accessible extracellular region of S79 and S86 (Malnic et al. 1999); 3) embedding the OR-odor complex in a hydrated, lipid bilayer model system; and 4) performing all-atom, molecular dynamics simulations to study the dynamic interactions of the OR and the odor over the duration of the simulation. Here, we describe our protocol in brief. An explicitly detailed protocol has been published previously (Lai and Crasto, 2012).
Building models of S79 and S86
Secondary structure prediction
Experimentally determined structures of rhodopsin from electron diffraction (Schertler et al. 1993; Schertler and Hargrave 1995; Davies et al. 1996) and X-ray crystallography (Palczewski et al. 2000; Okada et al. 2004) at varying resolutions have been used as templates over which an initial OR TM helical scaffold is built. Previous protocols to build TM helices have included: manual orientation of canonical helices with rhodopsin helices, or through homology modeling, often following a sequence alignment of the OR (or GPCR) with rhodopsin, using the high resolution structure for rhodopsin (Okada 2004).
Our structural alignment methodology consists of predicting TM regions using Hidden Markov Models from the OR sequence. The 2 best performing programs for TM predictions are TMHMM (Krogh et al. 2001) (http://www.cbs.dtu.dk/services/TMHMM/) and HMMTOP (http://www.enzim.hu/hmmtop) (Tusnady and Simon 2001). Both tools are available over the World Wide Web. A consensus of assigned regions based on the 2 prediction methods were assigned to be the OR helical regions. The 2 programs also predict loop locations (extracellular or cytoplasmic). This allows us to position the helices in configurations that are functionally relevant to GPCRs (for example, ensuring that the conserved E/DRY motif in GPCRs is located at the intracellular end of TM3).
A more recent crystal structure of the β-2-adrenergic receptor in the active as well as inactive state has been determined. (Rasmussen et al. 2011; Rosenbaum et al. 2011) To justify our choice of rhodopsin as template for the OR structure, over other structurally characterized GPCRs, for example, the β-2-adrenergic receptor, we assessed the sequence similarities of S79 and S86 with rhodopsin and the β-2-adrenergic receptor, using the PRALINE multiple sequence alignment Web-based software (http://www.ibi.vu.nl/programs/pralinewww/). Very poor sequence identities resulted. For S79, the sequence identities with rhodopsin and β-2-adrenergic receptor were 19% and 24%, respectively. For S86, the sequence identities with rhodopsin and the β-2-adrenergic receptor were 24% and 25%, respectively. The PRALINE software ranks amino acid matched-similarities from 1 to 10, one being unconserved and 10 being identical. Manually counting the similarities with a rank of 6 and above, showed between 29% and 33% similarities, for all 4 alignments. Using PRALINE, we also assessed the sequence identities between rhodopsin and the β-2-adrenergic receptor. The sequence identity was 24% with a similarity of 30%. Sequence similarity, therefore, is not a primary contributor of information with which to predict TM locations in ORs. We pursue the notion, in our protocol, that the one conserved aspect of GPCRs is the structure, despite the obvious sequence variability in the GPCRs assessed here, and presumably others.
Performing computational studies by strictly using experimentally characterized structures of GPCRs as templates, on which the OR model would be constructed, would contribute structural artifacts (from rotameric aspects of mostly unconserved residues, especially in the TMs) of the template that are not in the OR, and would hinder our efforts in assessing odor binding. The MEMBSTRUK computational GPCR modeling protocol was successfully able to validate experimental OR and β-2-adrenergic receptor ligand binding results starting from a low-resolution electron diffraction structure of rhodopsin (Vaidehi et al. 2002)
In order to further justify our choice of using the rhodopsin crystallographically obtained structure as the template on which to build the OR model (if only to establish the initial GPCR helical scaffold), we carried out a structural comparison of the X-ray crystallographically determined structures of active and inactive rhodopsin and β-2-adrenergic receptor. The results are illustrated in Figure 1. To create a basis to compare the structures, alpha-carbon positions from conserved residues in the middle of TM3 of all structures were fitted to PDB structure 1U19 (inactive rhodopsin) using the superpose function in Modeller (Eswar 2008). Other than a shift in TM1 between rhodopsin and the β-2-adrenergic receptor, and a slight shift in TM5, no discernible shifts are seen. More importantly, from the TM scaffold position standpoint, there is no discernible difference between the structure of the active and inactive proteins. Our results will show that TM1 is not involved in interactions with the odors. We will also show that the all-atom, constraint free simulation system that we have set up ensures that all the conformations that the OR protein will undergo will necessarily involve the active and inactive structures.
Figure 1.
Structural comparison between the active and inactive structures of rhodopsin and the β-2-adrenergic receptor. TM3 structure is used as the basis for aligning the structures. The figure shows that there are no discernible structural differences between the active and inactive forms of both receptors. There appears to be no translational shifts between receptors. The only structural differences are between rhodopsin and B2AR are in TM1 (not typically involved in ligand interactions) and TM5. The shifts in TM5 are at the helical termini.
The above reasons, we believe, justify our use of the rhodopsin structure, which we use merely to create a starting transmembrane helical scaffold for the OR protein. Discussed later in this section are steps in our protocol, which we use to remove structural artifacts from rhodopsin’s structure. Given the significant differences in the sequences similarities, the specific starting structure of the OR will not have a significant bearing on our conclusions.
Structural alignment
We aligned the helical regions of the rhodopsin template with the sequence regions of the OR that were predicted to be TMs. Using the homology modeling program Modeller (http://salilab.org/modeller/) (Sanchez and Sali 2000; Fiser and Sali 2003; Eswar et al. 2006; Eswar et al. 2007; Eswar et al. 2008) preliminary models of the ORs were created using this secondary structure alignment. Our OR structures derived from homology modeling are preliminary because the homology modeling software causes the resulting structure to inherit certain structure-specific artifacts from rhodopsin, as mentioned earlier. For example, there is a helical region in the C-terminus of rhodopsin that is orthogonal to the TM bundle. There is no evidence of this helical region in ORs. Other kinks (a proline-induced kink in TM 6 of rhodopsin) and bends that are consequences of the rhodopsin sequence which are not conserved in ORs have to be removed as these protein structural features can easily influence static docking as well as dynamic simulations. In addition, OR helices are shorter than those of rhodopsin; ORs have correspondingly longer loops.
TM refinement and minimization
We used the GROMACS 4.5 software (van der Spoel et al. 2010) to perform molecular dynamics simulations on each TM helix individually. The helices were parameterized using the CHARMM36 (Chemistry at HARvard Macromolecular Mechanics) (MacKerell et al. 1998; Hogberg et al. 2008; Brooks et al. 2009; Klauda et al. 2010) force field with CMAP corrections (MacKerell et al. 2004). Each helix was hydrated with TIPS3P water. Short molecular dynamics were used to relax the geometry of each TM. Distance restraints were used to parameterize the canonical hydrogen bond configurations that sustain the helices. After the geometry was minimized, the waters were removed. This step served to remove kinks and bends from the initial homology-modeled structure that are not necessarily structural features of ORs. Our methodology relied on a structural alignment of the transmembrane helices of the OR with those of rhodopsin. Next, the hydrophobic packing of the helical bundle had to be addressed, that is, the hydrophilic side of the amphipathic helix should be pointed towards the binding regions of ORs, while the hydrophobic side has to be pointed towards the surrounding plasma membrane. We have developed a tool, Hydro-Eff (Crasto 2010) that, for a helix, will determine the aggregate hydrophobicities at 20 degree intervals around an idealized helical wheel for that helix. The aggregate hydrophobicity, Θ θ at every angle θ is given by Equation (1).
 |
(1) |
 is the point hydrophobicity of the residue at angle θ summed over the hydrophobicity contributions (given by the cosine value) of other residues around the helical wheel. Hydro-Eff is freely available over the World Wide Web (http://bioinfo.genetics.uab.edu/hydro.pl). Using the sequence of each TM region, the effective hydrophobicities were determined for angles on a helical wheel, at 20° intervals, starting from 0°. The helices were then rotated such that the angle at which the highest hydrophilicity resided was pointed into the center of the OR binding region. We used the visualization software Visual Molecular Dynamics (VMD) (Humphrey et al. 1996) (http://www.ks.uiuc.edu/Research/vmd) to rotate each helix along its longitudinal axis to fit its ideal hydrophobic profile within the TM bundle.
is the point hydrophobicity of the residue at angle θ summed over the hydrophobicity contributions (given by the cosine value) of other residues around the helical wheel. Hydro-Eff is freely available over the World Wide Web (http://bioinfo.genetics.uab.edu/hydro.pl). Using the sequence of each TM region, the effective hydrophobicities were determined for angles on a helical wheel, at 20° intervals, starting from 0°. The helices were then rotated such that the angle at which the highest hydrophilicity resided was pointed into the center of the OR binding region. We used the visualization software Visual Molecular Dynamics (VMD) (Humphrey et al. 1996) (http://www.ks.uiuc.edu/Research/vmd) to rotate each helix along its longitudinal axis to fit its ideal hydrophobic profile within the TM bundle.
After the atoms in helical bundle were parameterized, minimized and rotated to meet hydrophilicity constraints, the loops were added back onto the TM assembly. Homology modeling, using the entire rhodopsin structure as template, cannot effectively model loops, because, as mentioned previously, rhodopsin loops are far shorter than the loop regions identified for ORs using Hidden Markov Models. The loops were modeled independently using Modeller by creating appropriate gaps in the shorter loops of the rhodopsin template. The N- and C- termini of both structures were truncated so that the completed model of S79 only included residues 30 through 303 and the model of S86 only included residues 29 through 305. There is no evidence that the termini are involved in ligand binding.
This computationally derived OR model was then rehydrated, re-parameterized, and minimized using molecular dynamics in GROMACS with CHARMM36 force-field and CMAP corrections, and TIPS3P water model with the TM alpha-carbon atoms position-restrained.
Building the membrane bilayer
We built a lipid bilayer consisting of molecules of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (POPC). We used the “Membrane” plug-in in the VMD software to construct this bilayer. A cubic box, which defines the dimensions of our system, of each side of 12nm was constructed and hydrated to fill the top and bottom of the bilayer. GROMACS was used to equilibrate the bilayer for 120 ns under NPT (constant temperature and pressure) ensemble conditions at 310 K and 1 atm. A GROMACS tool, g_membed, was used to embed the OR into the resulting bilayer consisting of 226 (for S79) to 228 (for S86) POPC molecules, over 21000 TIPS3P water molecules, and appropriate number of neutralizing ions.
Ligand construction
The ligands used in our computational experiments: heptanoic acid, octanoic acid, nonanedioic acid, nonanoic acid, and heptanol were constructed using the ArgusLab (http://www.arguslab.com/) software. Only the molecule construction component of this software was used, which is one of its many features. ArgusLab allows the user to build a molecule by assembling atoms roughly and creating bonds (double bonds, triple bonds, etc.), using the mouse to point-and-click. Once a rough ligand model was created, we used the geometry optimization component of the software to idealize the structural features of the ligand molecule, that is, the bond distances and angles.
Docking
Each of the odor ligands constructed as described above were positioned into the binding region of the OR using the docking program GRAMM (http://vakser.bioinformatics.ku.edu/main/resources_gramm.php). (Vakser 1997; Vakser and Jiang 2002) In order for the docking ligands to access every solvent-accessible portion of the binding regions, loop residues were removed from the protein model prior to docking. The top 100 docked conformations were considered. We rejected conformations that were outside the solvent accessible regions, outside the receptor molecule, on the lipid-facing surfaces of the receptor and on any region on the cytoplasmic side of the receptor. The remaining odor-ligand docked conformations were then reassessed. During docking, the highest resolved grid-steps allowed in GRAMM were used. Repulsive and attractive forces were considered. Each ligand positional conformation underwent a3-axis rotational sampling at 10° intervals. The GRAMM protein-ligand docking software, while docking molecules, does not sample bond rotamer libraries. It merely returns the best 100 rigid ligand configurations. We considered this sufficient as a prelude to our simulations of the OR–odor dynamic interactions in which every atom and structural feature of our entire simulation system is parameterized and all ligand conformations are sampled. Figure 2 shows the results of the docking for the nonanedioic acid ligand in S79 in 2 possible binding regions, indicated by arrows.
Figure 2.
The structure of OR S79 showing the 2 putative binding pockets. The circle bound by TMs III, IV, V and VI is the region that favors OR activation, and the circle bound by TMs I, II, III and VII is the nonpreferred binding region. The radius of the close-packed spheres of the odor reflects how many ligands favorably docked in one region as opposed to the other, based on preliminary docking using the GRAMM docking software.
Molecular dynamics
A random ligand position from one of the preferred binding region conformations was selected as the starting position of the ligand in the molecular dynamics simulation. Hydrogen atoms were added and the ligand was parameterized using the CGenFF 2.6b (CHARMM Generalized Force Field) force field in GROMACS in order to generate compatible intermolecular interactions with the OR. The ligand molecule conformation was then minimized in vacuum and added to the embedded OR–lipid–water complex. Waters in the binding region overlapping with ligand atoms in the binding region were removed. The resultant system was minimized with the TM alpha-carbons position-restrained and then equilibrated with molecular dynamics for 10 ns with position restraints set on both the TM alpha-carbons and ligand heavy atoms. Figure 3 shows such an equilibrated system. This system was then subjected to unrestrained molecular dynamics simulations for another 2 ns.
Figure 3.
The system involved in the dynamic simulations. The OR protein is embedded in the lipid bilayer—shown along the exterior of the TM regions. The water molecules are clearly identified, surrounding the protein-odorant-bilayer system. The odorant ligand can be identified on the extracellular side of the OR in the preferred binding region.
Results and discussion
The key finding in this computational study is that “ORs can possess 2 or more binding regions” (Figure 2) and, more importantly, in ORs the region where the odor ligand is more likely to bind is a determinant of whether an odor can activate an OR leading to recognition of that odor.
In this study, following 6 computational ligand-docking experiments, 4 for S79 and 2 for S86, 2 putative binding regions emerge; Figure 2 illustrates the 2 regions: the first is bound by TMs III, IV, V, and VI, and the second is bound by TMs I, II, III, and VII. It appears that ligands that excite an OR, based on the results of experimental analysis, are likely to bind in the first region, and those that either inhibit a receptor or do not excite it are more likely to bind in the second region. After each docking experiment, ligand configurations improbable during a binding event (as identified earlier) were rejected. The remaining ligands in each binding region were counted to determine likelihood of binding in those regions.
The docking results are consistent for the 2 ORs (S79 and S86) tested. Ligands heptanoic acid, octanoic acid, and nonanedioic acid for S79, and, nonanoic acid for S86 dock in the region bounded by TMs III, IV, V, and VI and heptanol for S79 and heptanoic acid for S86 overwhelmingly dock in the region bounded by TMs I, II, III, and VII. The ratio of the docked conformations in each binding region was used to determine the radius of the ligands’ space-filling sphere representations in Figure 3. In S86, following docking with heptanoic acid (an inhibitory ligand), no ligand conformations out of 100 were docked in the region that favors excitation.
A key development in the computational assessment of OR–odorant binding is our seminal use of dynamic simulations (methodological details are described in the Methods section and elsewhere; Lai and Crasto 2012). Because the positions of the atoms have been recorded every 2 ps in a 2 ns simulation, 1000 different conformations (for the protein, the ligand, the atoms of the lipid bilayer as well as the water molecules) are obtained. We can quantify the role of the amino acids involved in interactions with the ligand—by determining the relative likelihoods of an odorant being in close proximity with amino acids in the OR binding regions, over the duration of the simulation. In every resulting frame obtained from the simulation, the amino acids whose atoms are within a sphere of 6 Å radii from the ligand atoms are identified. We define this sphere as the “close-contact.” The number of close-contact occurrences for each amino acid are aggregated (counted) over the duration of the simulation. The amino acid residues that are in close-contact most often and possessing the appropriate electronic character (a possible electrostatic interaction with the odorant) are more likely to influence odor binding and consequent OR activation.
Figures 4–9 show a visual perspective of the results of the simulation studies, a novel representation of OR amino acid contribution to odor–ligand binding. In each figure, the size of the spheres representing the amino acid side chains is reflective of the time spent in close contact with the ligand. These residues that have the highest total aggregate close-contacts with the odorant ligand are evident in the figures. These amino acids are listed in Table 1. In Figure 4, for example, the residue side chains highlighted are Arg169, Tyr256, Ser263, Val212 and Ile264. Column 1 in Table 1 shows that Arg169 is associated with an aggregate close-contact count of 895. Considering that 2000 frames of data from each simulation were collected, a heavy atom of non anedioic acid has been within the 6 Å sphere of a heavy atom of Arg169 in S79 for over 44% of the simulation. The next highest contact is Y256 (66). This means that Y256 is close to the odor for 66 of the 1000 recorded frames. Using the aggregate counts of close-contacts, for example, the displayed radii of the Arg169 side chain atoms are approximately 13 times the displayed size of those of Y256. This explanation is extensible to the other simulations, illustrated in columns 2–6 in Table 1 and in Figures 5–9
Figure 4.
Results of a 2 ns dynamic simulation of mouse OR S79 and nonanedioic acid odor ligand—a very strong activator. The side chains of the amino acids shown are those that are most likely to interact with with 6 Å sphere radius around the odorant ligand over the duration of the simulation. The size of the atoms in the side-chains reflect the aggregated time spent in close contact with the ligand. The top 5 closest contacted amino acid residues in order of time spent in interactions with the odorant ligand are (from highest to lowest): ARG169, TYR256, SER263, VAL212, and ILE264.
Figure 9.
Results of a 2 ns dynamic simulation of mouse OR S86 and heptanoic acid odor ligand—a nonactivator. The close-packed spherical rendering of the side chains of the amino acids are those that are most likely to interact with with 6 Å sphere radius around the odorant ligand over the duration of the simulation. The size of the spheres reflect the aggregated time spent in close contact with the ligand. The top 5 closest contacted amino acid residues in order of time spent in interactions with the odorant ligand are (from highest to lowest): SER82, TYR41, SER83, PHE109, and ILE110.
Table 1.
The table reflect the aggregates (the summation) of the time steps for which the OR amino acid residues have spent in close contact (within 6 Å) of the ligand over the 2 ns duration of the simulation of the dynamic interactions between the OR and the odor ligands.
| S79 | S79 | S 79 | S79 | S86 | S86 |
|---|---|---|---|---|---|
| Nonanedioic acid (strong activator) | Heptanoic acid (strong activator) | Octanoic acid (weak activator) | Heptanol (non activator) | Nonanoic acid (strong activator) | Heptanoic acid (non activator) |
| 895 ARG169 | 437 TYR284 | 334 CYS107 | 274 VAL113 | 542 SER 274 | 376 SER82 |
| 66 TYR256 | 258 THR205 | 256 ILE108 | 231 PHE114 | 167 PHE207 | 303 TYR41 |
| 13 SER263 | 126 LEU111 | 120 TYR266 | 229 LEU287 | 114 LEU 210 | 271 SER83 |
| 11 VAL212 | 82 LEU280 | 87 THR205 | 90 ASN117 | 84 SER273 | 49PHE109 |
| 10 ILE264 | 50 PHE209 | 59 ALA269 | 81 PHE110 | 60 ARG272 | 2 ILE110 |
| 3 ILE118 | 16 PHE283 | 52 PHE209 | 32 PHE283 | 14 LEU206 | GLY78 |
| 1 ARG267 | 15 PHE114 | 25 ILE208 | 28 VAL291 | 8 SER271 | LEU79 |
| 1 PHE209 | 6 ILE208 | 24 GLY103 | 26 PRO288 | 5 ALA270 | ILE81 |
| 1 ASP115 | 6 CYS107 | 12 LEU111 | 8 CYS76 | 5 LEU269 | THR86 |
| PHE110 | 2 LEU279 | 10 THR201 | 1 VAL73 | 2 ILE276 | 111HIS |
| PHE114 | 2 ALA269 | 9 VAL268 | 1LEU43 | ILE213 | PHE113 |
| LEU119 | 1 TYR266 | 4 VAL165 | LEU111 | LEU214 | SER114 |
| VAL164 | PHE110 | 3 PHE114 | ASP115 | ILE276 | ILE284 |
| ALA167 | PHE114 | 2 PHE283 | LEU166 | VAL288 | |
| SER168 | PHE209 | 2 LEU204 | ALA167 | PRO289 | |
| ARG171 | VAL212 | 1 ARG270 | ILE208 | ||
| ILE208 | THR259 | 1 VAL212 | PHE209 | ||
| ARG211 | SER 262 | VAL212 | |||
| LEU213 | SER263 | THR259 | |||
| THR259 | ILE264 | ||||
| SER262 | VAL265 | ||||
| VAL265 | LEU287 | ||||
| TYR266 | |||||
| LEU279 | |||||
| PHE283 | |||||
| LEU287 |
The residues in bold are those that are in close contact with the ligand as a result of static docking, which within the context of this work, is at the start of the simulation.
Figure 5.
Results of a 2 ns dynamic simulation of mouse OR S79 and heptanoic acid odor ligand—a moderate activator. The close-packed spherical rendering of the side chains of the amino acids are those that are most likely to interact with with 6 Å sphere radius around the odorant ligand over the duration of the simulation. The size of the spheres reflect the aggregated time spent in close contact with the ligand. The top 5 closest contacted amino acid residues in order of time spent in interactions with the odorant ligand are (from highest to lowest): TYR284, THR205, LEU111, LEU280, and PHE209.
For S86, for the simulation of non anoic acid a strong activator, in Figure 8, the atomic radii in the side chain would be in the order Ser 274 (contact for 542 frames), Phe207 (167 frames), and Leu 210 (114 frames). The close-contact aggregate counts for this can be seen in column 5 of Table 1. Table 1 scores the amino acid residues in order of close-contacts they have with the ligand. The excitatory interactions of S79 and S86 are in columns 1, 2, 3, and 5 and non excitatory interactions are in columns 4 and 6.
Figure 8.
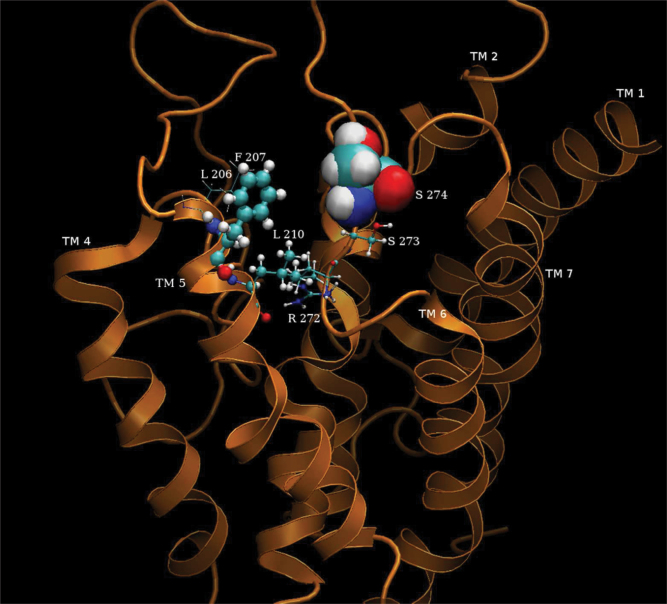
Results of a 2 ns dynamic simulation of mouse OR S86 and nonanoic acid odor ligand—a strong activator. The close-packed spherical rendering of the side chains of the amino acids are those that are most likely to interact with with 6 Å sphere radius around the odorant ligand over the duration of the simulation. The size of the spheres reflect the aggregated time spent in close contact with the ligand. The top 5 closest contacted amino acid residues in order of time spent in interactions with the odorant ligand are (from highest to lowest): SER274, PHE207, LEU210, SER273, and ARG272.
Figures 10–15 are graphs that show traces of the distances between an atom of the odorant (illustrated in the Figures 4–9, respectively) and that of the highest-scored close-contact with an amino acid for each OR-odorant complex. Although the order of amino acids in the columns in Table 1 reflects the time of contact over the duration of the simulation, the amino acid residues in bold in Table 1 are those in closest contact at the initial docked position of the OR. If dynamic simulation studies were not carried out, conjectures as to binding and possible excitation would have to be made based on the proximate amino acids side chains of the OR and the odor ligand following static docking. These results would correspond to time “0” of our simulations. Through static docking, it is not possible to quantify the individual residue-contributions to binding and activation as shown in Figures 4–15.
Figure 10.
The distance changes between the atoms C1 of nonanedioic acid, CZ of Arg169 (black), OH of Tyr256 (light), and C9 of nonanedioic acid and OH of Ser263 (dotted) of S79, over the duration of the simulation. The figure shows the sustained strong electrostatic binding between Arg169 and the carbonyl group of the odor ligand, which renders the interatomic distance close to invariant over the duration of the simulation. The inverse correlation between the Tyr256 and Ser263 testifies to the fluctuating interactions while the ligand stays tethered to the Arginine. This is observed until 700 ps, after that, the stronger electrostatic interaction with the Arginine dominates. Nonanedioic acid is a very strong activator of S79.
Figure 15.
The distance changes between the atoms C1 of heptanoic acid, OH of Ser82 (black), OH of Tyr41 (light), and OH of Ser83 (dotted) of S86, over the duration of the simulation. The figure shows competing electrostatic interactions. These interactions are sustained for most of the simulation. The lack of a single strong electrostatic tether is probably what affects the binding of this ligand to S86. Heptanoic acid is a nonactivator of S86.
Non anedioic acid is a strong activator of S79. Interestingly, for this ligand, the interactions during the simulations mimic those observed from static docking. The first column in Table 1 illustrates this—the bolded (static docking facilitating) names of amino acids are also those with the highest number of total aggregate contacts from the simulation. During the 2 ns simulation, 26 amino acid residues in S79 have been identified as making close contact with the ligand. The primary interactions—the high scoring close-contact amino acids—from static docking, however, are sustained. One can assume that the initial docked position is an energetically and electronically favorable one. The ligand position is optimized and sustained by several electrostatic interactions, most of which occur with Arg169 and Arg267 (Figure 4). Tyr256, which is located deeper on TM VI, also contributes electrostatically while the neutral residues (in column 1 of Table 1) support the atoms in the middle of the odorant straight-carbon atom chain via Van der Waals interactions. From the initial docked position there is a possible electrostatic interaction with Ser263 on the extracellular junction between TM VI and extracellular loop 3 (EC3). As the simulation proceeds, the carboxylic acid oxygen atoms on the ligand strongly interact with Arg169 which is located at the extracellular junction of TM IV and extracellular loop 2 (EC2). Ile264 may play a role as part of a hydrophobic “cap” adjacent to SER263.
Heptanoic acid is a moderately strong activator of S79. Other than a strong interaction with Tyr 284 on TM VII (Figure 5), the majority of close contacts during the simulation appear to be with uncharged residues between TMs V, VII, and VII. Interestingly, if one looks at the top ranking contacts during the dynamic simulation— Tyr284, Thr205, Leu111, Leu280, and Phe209—(Table 1, column 2) and compare them to the closest contacts (bold residues) from the energy minimized docking position, it appears that the closest contacts are between Phe209, Phe283, Leu279, and Tyr266. It appears that a specific electronic environment is required to bind heptanoic acid.
Octanoic acid is 1 carbon atom longer than heptanoic acid. Of the high-ranked close-contacts over the course of the simulation, for both odors, there is only one common amino acid residue—Phe209. An electrostatic interaction with Tyr266 is probably the primary electrostatic binding tether. This residue is at the boundary of TM VI and extracellular loop 3. The strength of binding, and consequently, activation, is possibly dampened by the longest close-contact with Cys107 (Figure 6) which is at the extracellular interface of TM III and the extracellular loop EC2 over the simulation. Cys107 is expected to form a conserved disulfide bridge to Cys193 (not shown in the figure).
Figure 6.
Results of a 2 ns dynamic simulation of mouse OR S79 and octanoic acid odor ligand—a weak activator. The close-packed spherical rendering of the side chains of the amino acids are those that are most likely to interact with with 6 Å sphere radius around the odorant ligand over the duration of the simulation. The size of the spheres reflect the aggregated time spent in close contact with the ligand. The top 5 closest contacted amino acid residues in order of time spent in interactions with the odorant ligand are (from highest to lowest): CYS107, ILE108, TYR266, THR205, and ALA269.
For heptanol, a non activator of S79, the docking results favored the region bound by TMs I, II, III, and VII. This binding pocket is overwhelmingly uncharged; the only charged residue is Asn 117 (Figure 7) and there is minimal dynamic contact with Asp115. Because both these amino acids are acidic, any charged interactions in this binding region would be necessarily repulsive. The fact that there is overlap between close contacts from static docking to dynamic interactions for Phe114 and Phe110 indicates that over the course of the simulation, the ligand has displaced significantly from its initial docking position.
Figure 7.
Results of a 2 ns dynamic simulation of mouse OR S79 and heptanol odor ligand—a nonactivator. The close-packed spherical rendering of the side chains of the amino acids are those that are most likely to interact with with 6 Å sphere radius around the odorant ligand over the duration of the simulation. The size of the spheres reflect the relative aggregated time spent in close contact with the ligand. The top 5 closest contacted amino acid residues in order of time spent in interactions with the odorant ligand are (from highest to lowest): VAL113, PHE114, LEU287, ASN117, and PHE110.
Non anoic acid is a strong activator of S86. The primary residues that appear to be involved in ligand binding are located along the extracellular loop EC3. Primary hydrophobic interactions arise from Leu206, Leu210, and Phe207 (Figure 8). All these residues lie on the TM helix V close to the extracellular junction with EC2. Ser274, Arg272, and Ser271 all on EC3 can be considered responsible for polar interactions.
Heptanoic acid is a non activator of S86. The largest cluster of docked conformations is in the region bound by TMs I, II, and III. There are several possible polar contacts: Ser82, Ser83 (TM II), and Tyr41 (TM I) (Figure 9). All other interactions are of a hydrophobic nature.
For the 6 OR–odor model systems, it appears that among the activators, docking is favored in the binding region supported by TMs III, IV, V, and VI. Non activators prefer to dock in the region supported by TMs I, II, III, and VII. Using our methodology that includes dynamic simulations of the OR–odorant interactions, one can identify amino acid residues which form the closest contacts with the ligand over time. It is important to distinguish residues that are in closest contact at an energy minimized state of the OR–odor complex versus contacts that prevail over the duration of the simulation. Here, we challenge the traditionally held notions of drawing conclusions about OR activation following static ligand docking.
Figures 10–15 are of distance calculations between high-ranked amino acid residue contacts (from Table 1) and the odorants. These distance calculations enable us to visualize the stability of the interactions. They also enable us to trace the dynamic pathway of the odorant within the binding pocket. The specific atoms for which distance traces are illustrated are identified in the figure legends. One of the conclusions we draw from this study is that for activation a strong (or sustained) electrostatic interaction is necessary between a charged part of the odorant and a complementarily charged amino acid residue. This appears to occur between non anedioic acid and Arg169 of S79 (Figure 10). After 250 ps, the carbonyl oxygen becomes electrostatically (and permanently) tethered to the arginine. The other carboxylic end “tail” (the distance is associated with the C9 carbon on the ligand) of the odorant appears to alternately swing between Ser263 and Tyr256 which are on adjacent TM helices. This is evidenced by the inverse correlation of their changing distances with the ligand. Figure 11 shows similar characteristics: that the electrostatic interaction between the moderately strong activator heptanoic acid and the Tyr284 of S79 is sustained after 1250 ps. Once the electrostatic tethering is achieved, the carboxylic end of the ligand moves away from Leu111. The weaker force from Thr205 is unable to compete with the one between the ligand and Tyr284. In a testament to the importance of studying OR-odorant interaction behavior through dynamical interactions, the odorant goes through significant shifts from the initial docked position before it stabilizes, facilitated by the electrostatic interaction with Tyr284.
Figure 11.
The distance changes between the atoms C1 of heptanoic acid, OH of Tyr284 (light), OH of Thr205 (dotted), and CD of Leu111 (black) of S79, over the duration of the simulation. The figure shows the establishment and sustenance of the electrostatic interaction between the tyrosine and heptanoic acid after 1 ns of simulation. While the threonine-ligand distance does not show a large shift over the duration of the simulation, the increasing distance between the Leu and the ligand indicate weakening interactions following a sustained electrostatic interaction of heptanoic acid with tyrosine. Heptanoic acid is a good activator of S79.
Figure 12 follows similar behavior for the interactions of S79 and the weak activator, octanoic acid. Significant fluctuations arise among the interactions of the 3 high-ranked contact amino acids with the odor ligand. Cys 107 is the top ranked contact and also is closest in distance to the odorant. It appears that the odorant is stabilized in a pocket created by Cys107, Ile108, and Tyr266. What is also interesting to note is that although the electrostatic interaction with Tyr 266 is established at 1800 ps, the lack of a sustained tether (or one for a longer length during the interactions) can explain the weak activation of S79 by this odorant. The characteristics observed in Figure 11 can be extended to those observed in Figure 13, for the binding of the non activator heptanol with S79. It has already been discussed that this odorant binds in the binding region that is not preferred for activation, and Table 1 shows that there is a lack of a charged residue in the binding region. The distance calculations between heptanol and the uncharged Val113, Phe114, and Leu287 amino acids clearly show the lack of an electrostatic interaction. The distance graphs show that the odorant ligand position fluctuates between the 3 residues all sustained by weaker Van der Waals interactions; there is an inverse correlation among the ligand distances between Val113 and Leu287 as the ligand continually shifts from one TM to the other.
Figure 12.
The distance changes between the atoms C1 of octanoic acid, OH of Tyr266 (light), SG of Cys107 (black), and CD of Ile108 (dotted) of S79, over the duration of the simulation. The figure shows competing binding for the ligand between the 3 residues. Toward 1800 ps, the interactions between the serine tyrosine strengthen and stabilize while the Van der Waals interactions between the ligand, and cysteine and isoleucine weaken. Octanoic acid is a weak activator of S79.
Figure 13.
The distance changes between the atoms O (OH) of heptanol, CB of Val113 (black), ring of Phe114 (dotted), and CG of Leu287 (light) of S79, over the duration of the simulation. These residues are in the nonpreferred (for activation) binding region. The interactions are of a Van der Waals nature. The ligand appears to move between the 3 residues, with greater fluctuations in the aromatic ring of Phe114. The likelihood of nonbinding and consequent nonactivation of S79 is probably because of the lack of an electrostatic interaction.
Figure 14 shows the distance calculations between the strong activator non anoic acid and S86. Here, the electrostatic interaction is sustained, in terms of the closest contact with Ser274. At the initial docked position, it appears that Leu210 is the closest to the ligand, within 200 ps of the beginning of the simulation; the interaction with the serine is strengthened and sustained. It appears for some duration of the simulation, before the eventual electrostatic interaction wins out, that there is competing binding for the ligand between Ser274 and Leu210, shown by the inverse correlation of the interactions. The swinging motion of the long tail of the ligand fluctuates between Phe207 and Leu210, residues that are in close proximity on the same TM.
Figure 14.
The distance changes between the atoms C1 of nonanoic acid, CG of Leu210 (dotted), O (OH) of Ser274 (light), and the ring of Phe207 (black) of S86, over the duration of the simulation. The distance fluctuations indicate an initial competing for binding between the leucine and the serine. After 1200 ps, these fluctuations stabilize, with the closest contact being sustained with the serine. Once the interaction with serine stabilizes, the distance fluctuations of the phenylalanine also stabilize. It is likely that Phe207 supports the ligand binding through Van der Waals interactions. Nonanoic acid is a good activator of S86.
Unlike that of S79, the non preferred binding region of S86 possesses charged amino acid residues. The distance characteristics in the course of the simulations show that the high-ranked contacts are Ser82, Ser83, and Tyr41. As the simulation proceeds, initially (an artifact of static docking) the closest contact is Ser 82, while the Ser 83 is farther away because its position on the helix is initially pointed away from the binding pocket (Figure 14). Tyr41 from the neighboring TM forms an electrostatic interaction with the heptanoic acid which degrades and is then restored. By the end of the simulation, however, all 3 close-contact residues show stable interactions. The behavior of this residue within the non activating binding region is different from that observed in the previous 5 cases. There are 2 possible explanations for this behavior: the interactions of the ligand are weakened because of competing electrostatic interactions that prevent a strong ligand tether to an OR amino acid and, or, irrespective of the nature of the interactions, binding in the region that is not conducive to OR activation precludes the structural change in the OR protein, which is considered a precursor to OR activation and the eventual perception of olfaction.
With the exception of heptanol, all the ligands used in our study are carboxylic acids. A strong electron withdrawing component exists in the carbonyl oxygen of the carboxylic acid functional group. For all activators tested with S79 and S86, the preferred binding pockets contained basic and weakly basic amino acid residues which formed salt bridges with the ligands’ carboxylic groups over the duration of the simulation. The strength of activation is directly correlated with how much time the odor spends in electrostatic contact with a basic amino acid residue. The primary electrostatic interaction for the activators of S79 and S86 appears to be arginines, serines, and tyrosines.
For the strongest activator of S79, Arg169 is the closest contact over the duration of the simulation. Other, relatively weak electrostatic interactions with Ser263 and Tyr256 are also present for significant amounts of time (Table 1). Arg267 is also in close proximity to Ser263. Interestingly, for this complex, it appears that the static docking conformation appears to be the most stable: the closest contacts for the dynamic simulations are the same as those measured after static docking.
One can also see how as the strength of the primary electrostatic tether weakens, the activation strength decreases. For S79’s interaction with both heptanoic acid and octanoic acid, a tyrosine contributes heavily towards the electrostatic interaction. However, in the case of S79 and octanoic acid (its weaker activator), the proximity of Cys107, whose polarity is otherwise reduced due to the presence of a disulfide bridge, dampens the strength of this interaction, resulting in a fewer interactions with Tyr266, though these interactions are established later in the simulation. In the S86–non anoic acid complex, although Ser274 generates the largest contacts, the presence of 2 additional Serines and Arg272 explains strong activation.
For both non activators, there is a significant absence of a primary electrostatic tether in what we have defined as non-preferred binding region, which does not facilitate OR activation. The S79 interaction with heptanol is predominantly hydrophobic, without the presence of even a single charged residue. For heptanoic acid and S86, the competing basicity in the side chains of the binding region precluding the establishment of a sustained electrostatic interaction, probably results in non activation.
We have advanced a notion here that ORs possess 2 viable binding regions, and that the activating ligands are more likely to bind the OR in a preferred binding region. Our protein modeling protocol, conceived and developed to maintain the classic sequence-structural prerogatives of the rhodopsin-like GPCR, plays an important role in how the TMs are positioned, which is critical to the characterization of the 2 binding regions.
A novel aspect of this current study is that using dynamic simulations, we can quantify the likelihood of binding contributions from amino acid residues and potential OR activation. Because we determined the residues that are in closest contact at every time step of the simulation and then summed up (aggregated) the number of times such residues were in closest contact, we could surmise which residues are primarily responsible for the majority of receptor–ligand interactions during binding and activation. This we believe is a major advancement over conventional methodologies that involved static docking. In our protocol (Lai and Crasto 2012), the results of static docking are the first step, a prelude to our dynamic simulations. We make a case here that dynamic simulations studies offer significant advantages over static docking. From static docking, the picture of the OR–odorant interaction is a single snapshot in time. It is likely possible that incorrect conjectures can be made as to which amino acid residues facilitate binding, because of the limited perspective of static docking. A protein–ligand interaction is a dynamic phenomenon. It is likely that different amino acids bind the ligand over time, and one or more of these interactions likely facilitate OR activation. We have shown that our simulations can trace the residues that facilitate the odorant binding as well as identify the conformational changes that the odorant ligand can undergo. The graphs in Figures10–15 illustrate how ligand positions can change as the simulation of the interaction progresses.
OR activation that results in the cascade of steps leading to the perception of olfaction begins with structural changes in the OR (Krautwurst et al. 1998). This activation can occur at a specific point in time during the ligand’s interaction with the OR. Key OR amino acids that are in contact with the odor ligand are likely to facilitate these changes. Our distance determinations largely support this notion.
Table 2 reflects our efforts at viewing our work from the perspective of the efforts of others, in identifying amino acids responsible for odor binding and odor activation. In column 1 in this table are listed the top 3 close-contact amino acid residues for each of our simulations. These amino acids are ordered in this column as in Table 1. Man et al. (2004) by a comparative assessment of orthologs and paralogs of ORs from the repertoires of human and mouse identified 22 amino acid positions, mostly in the TM helices and 2 in the second extracellular interhelical loop, as putatively responsible for binding. The second column of Table 2 shows that most of the top 3 interacting amino acid residues for S79 and S86, irrespective of whether they were excitatory or non-excitatory coincide with a positions determined by Man et al. The positions are represented using the same nomenclature used by Man et al. the TM number, with the TM position as subscript. In some cases the positions are off by one. That is reasonable because, during the creation of the OR model and assigning TM helical regions to match with the structure of rhodopsin, there is often uncertainty in identifying the exact residues which constitute the helix loop boundaries. Of the 18 top interacting residues in our study, 14 matched the positions identified by Man et al.
Table 2.
Results of the the current studies in perspective of other theoretical and experimental OR-activation results.
| This study | Man (2004) | Abaffy (2007) | Schmiedeberg et al. (2007) | Katada (2005) |
|---|---|---|---|---|
| S79 | ||||
| Y256 | ~F252 | |||
| L111 | 37, 38 | |||
| F209 | ~52 | I205V ++ | F206 | |
| C107 | ~34 | |||
| I108 | 34 | |||
| Y266 | 416 | |||
| V113 | ~38 | V113S | N109K 0; K109N ++ | ~Val109 |
| F114 | ~311 | T110A − | ||
| L287 | 75 | |||
| N117 | ~312 | N117A | S112 | ~ S113 |
| F110 | ~37 | |||
| S86 | ||||
| F207 | 59 | |||
| L210 | 510 | ~F206 | ||
| F109 | 34 |
The first column contains results of this study in terms of the residues in both S79 and S86 that are shown to have the closest interactions, both for excitation and non excitation. The other 4 columns show if there is a match in terms of possible interaction sites for residues between S79, S86, and ORs from 4 other studies. The primary binding amino acid residues are included in the table only if there is a match with 1 out of 4 studies. Column 2 shows the position in the format used by Man et al.: of TM number and the subscript as position of the residue on the TM. In Column 3, in each position the specific mutation is illustrated. In Column 4, the result of the mutations are also embedded. A mutation resulting in a large increase in OR excitation is denoted by “++”, a moderate excitation is denoted by “+”, no change in excitation is denoted by “0”, and, decrease in excitation or inhibition is denoted by “−.” In all the columns the “~” denotes that the residue position is possibly one away from an exact positional match.
We also compared our results to those of Katada et al. (2005), Abaffy et al. (2006), and Schmiedeberg et al. (2007). These studies that have used mutagenesis experiments followed by OR activation studies, often choosing amino acids to mutate at positions proposed by the Man et al. study. Unfortunately, exact comparisons are tendentious at best, because these studies used ORs and odorants different from the ones tested here. Although, Abaffy et al. (2006) did use S79 (mOR42-3), the ligands were not the same as used in this study. They used longer 11- and 12-C-atom chain dicarboxylic acids; our largest ligand, nonanedioic acid, consisted of a 9 C-atom chain.
Regardless, to assess the positions of amino acids responsible for binding odorants, a multiple sequence analysis was carried out for S79, S86, MOR-EG (Katada et al. 2005), OR1A1 and OR1A2 (Schmiedeberg et al. 2007), and MOR42-3 and MOR42-1, using the PRALINE web-based software described in the Methods section of this article. In each of the sequences, the amino acid residue positions mutagenized for OR activation studies were identified. These were then matched to corresponding positions in S79 and S86 from our study. Columns 3, 4, and 5 in Table 2 summarize these results. Two positions from the Abaffy (undecane- and dodecanedioic acid ligands), 3 from the Schmiedeberg study ((S)-(-)-citronellol ligand) and 5 from the Katada (eugenol ligand) study match the top binding residues in S79 and S86. As has been mentioned above, because of the different ORs and different odorants use, exact comparisons are not possible. In all cases, as illustrated in Table 2, we observe that the matched residues are on TMs III, IV, V, and VI, which coincides with the conclusions of this study.
Our studies begin with the creation of the model of the OR, which, in turn begins with the identification of a starting GPCR structure that will allow us to correctly establish the transmembrane helical scaffold. Figure 1 illustrates that there are no discernible differences between the structures of the proteins compared, regardless of whether they are active or inactive. Given the poor sequence similarities between rhodopsin and β-2-adrenergic receptor and of these proteins with the ORs studied here, we are confident in our contention that the template structure merely provides us with starting structures, which are subsequently refined, according to our rigorously established protocol. Besides, because every one of the several thousand atoms in our system, consisting of the protein, the ligand, the simulated lipid bilayer and the water molecules are allowed to parameterize in an unconstrained way, our studies sample myriad protein structures, side chain rotamers and ligand conformations.
From the results of our studies, it is possible to identify amino acids for mutagenesis experiments, to validate our methodologies and ascertain the role of the amino acids in odor binding. For both S79 and S86, one might start by mutating the Arg and Tyr residues and replace them with hydrophobic or acidic amino acid residues, creating an electronic environment in the binding region that is not conducive to binding and OR excitation, or mimic the non-preferred binding region.
Functional studies of ORs have shown us that hydrophobic interactions are likely to lessen the strength of excitation but not necessarily remove activation. We believe that hydrophobic residues stabilize the non charged portions of the odor ligand during interactions leading to activation. For this, we recommend that the hydrophobic amino acids with strong close-contact aggregate counts should be mutated; the choice of replacement residues would depend in this case not on charge, but on spatial constraints. A smaller residue that shows numerous contacts could be replaced with a bulky one, a residue with a side chain that is long could be replaced by a shorter one, a ring-based side chain might be replaced by a straight chain side chain, and so forth.
In order to continually improve our methodology, one or more of the steps outlined in the Methods section could be more finely tuned. We encourage experimental functional analysts to test some of the conclusions drawn from the observations of our computational analysis—much like current attempts at computational assessments of OR-odorant binding have been exclusively conducted on OR-ligand systems which have already been previously functionally characterized.
Conclusions
We have presented in this article a novel notion that allows a mechanistic glimpse into OR binding by odorants correlated with OR activation. We have used functional characterization results from the first combinatorial study of ORs, and an established and rigorous computational methodology. We observe for every OR–odorant system studied, that there is higher likelihood of OR activation if the odor binds in the preferred binding region—of 2 potential binding regions.
Our methodology relies on an innovative computational approach: performing long-term simulations for an unrestrained system. This system consists of the OR with odor docked in an energetically feasible conformation, a lipid bilayer which simulates the plasma membrane, and several thousand water molecules. By simulating the dynamic interactions of the OR and the odorant, we determine structural and energetic parameters at every time step of the simulation. This is a significant advance over previous work where conjectures to binding and potential activation were made solely after static docking of the odor in the OR binding region.
By performing dynamic simulation studies we have observed ligand and consequent OR behavior at 2 ps intervals over 2 ns. We quantified the OR contribution to binding by determining, over the duration of the simulation, which residues were in closest contact. This is with a view to providing a basis of experimental validation whereby, specific amino acid residues can be mutated prior to conduction experimental functional analysis.
Our results here show that in order for activation to occur, a primary electrostatic tether in the form of an amino acid with a strong basic side chain should be in the preferred binding region. All the odorants used in our studies were di- or mono-carboxylic acids and one alcohol.
Our results and conclusions are based on experimental results. Additional work assessing the ORs from the same study as well as more recent combinatorial experimental results would be necessary to ascertain and establish the our conclusions.
Funding
The authors gratefully recognize the support for this work from grant 1R21DC011068 (National Institute on Deafness and Other Communicative Disorders, National Institutes of Health). Other support for this work has been from the University of Alabama at Birmingham-Center for Clinical and Translational Studies (UAB-CCTS; 5UL1RR025777).
References
- Abaffy T, Matsunami H, Luetje CW. 2006. Functional analysis of a mammalian odorant receptor subfamily. J Neurochem. 97:1506–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abaffy T, Malhotra A, Luetje CW. 2007. Molecular basis for ligand specificity in a mouse olfactory receptor—a network of functionally important residues. J Biol Chem. 282:1216–1224 [DOI] [PubMed] [Google Scholar]
- Araneda RC, Kini AD, Firestein S. 2000. The molecular receptive range of an odorant receptor. Nat Neurosci. 3:1248–1255 [DOI] [PubMed] [Google Scholar]
- Arata-Kawai H, Singer MS, Bistrup A, Zante A, Wang YQ, Ito Y, Bao X, Hemmerich S, Fukuda M, Rosen SD. 2011. Functional contributions of N- and O-glycans to L-selectin ligands in murine and human lymphoid organs. Am J Pathol. 178:423–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barresi M, Ciurleo R, Giacoppo S, Foti Cuzzola V, Celi D, Bramanti P, Marino S. 2012. Evaluation of olfactory dysfunction in neurodegenerative diseases. J Neurol Sci. 323:16–24 [DOI] [PubMed] [Google Scholar]
- Bertsch RA, Vaidehi N, Chan SI, Goddard WA., 3RD. 1998. Kinetic steps for alpha-helix formation. Proteins 33:343–357 [DOI] [PubMed] [Google Scholar]
- Boekhoff I, Touhara K, Danner S, Inglese J, Lohse MJ, Breer H, Lefkowitz RJ. 1997. Phosducin, potential role in modulation of olfactory signaling. J Biol Chem. 272:4606–4612 [DOI] [PubMed] [Google Scholar]
- Bozza T, Feinstein P, Zheng C, Mombaerts P. 2002. Odorant receptor expression defines functional units in the mouse olfactory system. J Neurosci. 22:3033–3043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks BR, Brooks CL, 3RD, Mackerell AD, JR., Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, et al. 2009. CHARMM: the biomolecular simulation program. J Comput Chem. 30:1545–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasto CJ. 2009. Computational biology of olfactory receptors. Curr Bioinform. 4: 8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasto CJ. 2010. Hydrophobicity profiles in G protein-coupled receptor transmembrane helical domains. J Receptor Ligand Channel Res. 2010:123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasto C, Singer MS, Shepherd GM. 2001. The olfactory receptor family album. Genome Biol. 2:REVIEWS1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies A, Schertler GF, Gowen BE, Saibil HR. 1996. Projection structure of an invertebrate rhodopsin. J Struct Biol. 117:36–44 [DOI] [PubMed] [Google Scholar]
- Doty RL, Perl DP, Steele JC, Chen KM, Pierce JD, JR., Reyes P, Kurland LT. 1991. Olfactory dysfunction in three neurodegenerative diseases. Geriatrics 46:Suppl 1, 47–51 [PubMed] [Google Scholar]
- Eswar N, Eramian D, Webb B, Shen MY, Sali A. 2008. Protein structure modeling with MODELLER. Methods Mol Biol. 426:145–159 [DOI] [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. 2006. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. Chapter 5, Unit 5, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. 2007. Comparative protein structure modeling using MODELLER. Curr Protoc Protein Sci, Chapter 2, Unit 2 9. [DOI] [PubMed] [Google Scholar]
- Ewing TJA, Makino S, Skillman AG, Kuntz ID. 2001. DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J. Comput-Aided Molec. Design. 15:411–428 [DOI] [PubMed] [Google Scholar]
- Fiser A, Sali A. 2003. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 374:461–491 [DOI] [PubMed] [Google Scholar]
- Floriano WB, Vaidehi N, Goddard WA., 3rd 2004. Making sense of olfaction through predictions of the 3-D structure and function of olfactory receptors. Chem Senses 29:269–290 [DOI] [PubMed] [Google Scholar]
- Floriano WB, Vaidehi N, Goddard WA, 3RD, Singer MS, Shepherd GM. 2000. Molecular mechanisms underlying differential odor responses of a mouse olfactory receptor. Proc Natl Acad Sci USA 97:10712–10716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y, Bustamante CD, Lancet D, Paabo S. 2003. Natural selection on the olfactory receptor gene family in humans and chimpanzees. Am J Hum Genet. 73:489–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glusman G, Bahar A, Sharon D, Pilpel Y, White J, Lancet D. 2000a. The olfactory receptor gene superfamily: data mining, classification, and nomenclature. Mamm Genome 11:1016–1023 [DOI] [PubMed] [Google Scholar]
- Glusman G, Sosinsky A, Ben-Asher E, Avidan N, Sonkin D, Bahar A, Rosenthal A, Clifton S, Roe B, Ferraz C, et al. 2000b. Sequence, structure, and evolution of a complete human olfactory receptor gene cluster. Genomics 63:227–245 [DOI] [PubMed] [Google Scholar]
- Grosmaitre X, Fuss SH, Lee AC, Adipietro KA, Matsunami H, Mombaerts P, Ma M. 2009. SR1, a mouse odorant receptor with an unusually broad response profile. J Neurosci. 29:14545–14552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall SE, Floriano WB, Vaidehi N, Goddard WA., 3RD 2004. Predicted 3-D structures for mouse I7 and rat I7 olfactory receptors and comparison of predicted odor recognition profiles with experiment. Chem Senses 29:595–616 [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. 1996. VMD – Visual Molecular Dynamics. J Molec Graphics 14.1:33–38 [DOI] [PubMed] [Google Scholar]
- Hawkes C. 2006. Olfaction in neurodegenerative disorder. Adv Otorhinolaryngol. 63:133–151 [DOI] [PubMed] [Google Scholar]
- Hogberg CJ, Nikitin AM, Lyubartsev AP. 2008. Modification of the CHARMM force field for DMPC lipid bilayer. J Comput Chem.29:2359–2369 [DOI] [PubMed] [Google Scholar]
- Kajiya K, Inaki K, Tanaka M, Haga T, Kataoka H, Touhara K. 2001. Molecular bases of odor discrimination: reconstitution of olfactory receptors that recognize overlapping sets of odorants. J Neurosci. 21:6018–6025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katada S, Hirokawa T, Oka Y, Suwa M, Touhara K. 2005. Structural basis for a broad but selective ligand spectrum of a mouse olfactory receptor: mapping the odorant-binding site. J Neurosci. 25:1806–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katada S, Nakagawa T, Kataoka H, Touhara K. 2003. Odorant response assays for a heterologously expressed olfactory receptor. Biochem Biophys Res Commun. 305:964–969 [DOI] [PubMed] [Google Scholar]
- Kato A, Touhara K. 2009. Mammalian olfactory receptors: pharmacology, G protein coupling and desensitization. Cell Mol Life Sci. 66:3743–3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauda JB, Venable RM, Alfredo Freites J, O’connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, Mackerell AD, JR, Pastor RW. 2010. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. J Phys Chem B. 114:7830–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs T. 2004. Mechanisms of olfactory dysfunction in aging and neurodegenerative disorders. Ageing Res Rev. 3:215–232 [DOI] [PubMed] [Google Scholar]
- Krautwurst D, Yau K-W, Reed R. 1998. Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell 91:917–923 [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, Von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- Kufareva I, Rueda M, Katritch V, Stevens RC, Abagyan R, Participants GD. 2011. Status of GPCR modeling and docking as reflected by community-wide GPCR Dock 2010 assessment. Structure 19:1108–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai PC, Crasto CJ. 2012. Beyond modeling: all-atom olfactory receptor model simulations. Front Genet. 3:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai PC, Singer MS, Crasto CJ. 2005. Structural activation pathways from dynamic olfactory receptor-odorant interactions. Chem Senses 30:781–792 [DOI] [PubMed] [Google Scholar]
- Li J, Haddad R, Chen S, Santos V, Luetje CW. 2012. A broadly tuned mouse odorant receptor that detects nitrotoluenes. J Neurochem. 121:881–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackerell AD, JR., Bashford D, Bellott M, Dunbrack RL, JR., Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, et al. 1998. All-atom empirical potential for molecular modeling and dynamics Studies of proteins. J Phys Chem B. 102:3586–3616 [DOI] [PubMed] [Google Scholar]
- Mackerell AD, JR, Feig M, Brooks CL., III 2004. Detailed presentation of the new CMAP procedure and its use to treat the conformational properties of the protein backbone: extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comp Chem. 25:1400–1415 [DOI] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB. 1999. Combinatorial receptor codes for odors. Cell 96:713–723 [DOI] [PubMed] [Google Scholar]
- Man O, Gilad Y, Lancet D. 2004. Prediction of the odorant binding site of olfactory receptor proteins by human-mouse comparisons. Prot Sci. 13:240–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matarazzo V, Clot-Faybesse O, Marcet B, Guiraudie-Capraz G, Atanasova B, Devauchelle G, Cerutti M, Etievant P, Ronin C. 2005. Functional characterization of two human olfactory receptors expressed in the baculovirus Sf9 insect cell system. Chem Senses 30:195–207 [DOI] [PubMed] [Google Scholar]
- Mombaerts P. 2001. The human repertoire of odorant receptor genes and pseudogenes. Annu Rev Genom Hum Genet. 2:493–510 [DOI] [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. 2009. Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity. J Comp Chem. 16:2785–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller G. 2000. Towards 3D structures of G protein-coupled receptors: a multidisciplinary approach. Curr Med Chem. 7:861–888 [DOI] [PubMed] [Google Scholar]
- Oka Y, Katada S, Omura M, Suwa M, Yoshihara Y, Touhara K. 2006. Odorant receptor map in the mouse olfactory bulb: in vivo sensitivity and specificity of receptor-defined glomeruli. Neuron 52:857–869 [DOI] [PubMed] [Google Scholar]
- Oka Y, Omura M, Kataoka H, Touhara K. 2004. Olfactory receptor antagonism between odorants. EMBO J. 23:120–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. 2004. The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J Mol Biol. 342:571–583 [DOI] [PubMed] [Google Scholar]
- Olender T, Fuchs T, Linhart C, Shamir R, Adams M, Kalush F, Khen M, Lancet D. 2004. The canine olfactory subgenome. Genomics 83:361–372 [DOI] [PubMed] [Google Scholar]
- Ondachi P, Castro A, Luetje CW, Damaj MI, Mascarella SW, Navarro HA, Carroll FI. 2012. Synthesis and nicotinic acetylcholine receptor in vitro and in vivo pharmacological properties of 2’-fluoro-3’-(substituted phenyl)deschloroepibatidine analogues of 2’-fluoro-3’-(4-nitrophenyl)deschloroepibatidine. J Med Chem. 55:6512–6522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, et al. 2000. Crystal structure of rhodopsin: A G protein-coupled receptor. Science 289:739–745 [DOI] [PubMed] [Google Scholar]
- Rasmussen SG, Devree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, et al. 2011. Crystal structure of the β(2) adrenergic receptor-Gs protein complex. Nature 477:549–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repicky SE, Luetje CW. 2009. Molecular receptive range variation among mouse odorant receptors for aliphatic carboxylic acids. J Neurochem. 109:193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum DM, Zhang C, Lyons JA, Holl R, Aragao D, Arlow DH, Rasmussen SG, Choi HJ, Devree BT, Sunahara RK, et al. 2011. Structure and function of an irreversible agonist-β(2) adrenoceptor complex. Nature 469:236–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouquier S, Blancher A, Giorgi D. 2000. The olfactory receptor gene repertoire in primates and mouse: Evidence for reduction of the functional fraction in primates. Proc Natl Acad Sci USA 97:2870–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Chi Q, Zhuang H, Matsunami H, Mainland JD. 2009. Odor coding by a Mammalian receptor repertoire. Sci Signal 2:ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R, Sali A. 2000. Comparative protein structure modeling. Introduction and practical examples with modeller. Methods Mol Biol. 143:97–129 [DOI] [PubMed] [Google Scholar]
- Schertler GF, Hargrave PA. 1995. Projection structure of frog rhodopsin in two crystal forms. Proc Natl Acad Sci USA 92:11578–11582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertler GF, Villa C, Henderson R. 1993. Projection structure of rhodopsin. Nature 362:770–772 [DOI] [PubMed] [Google Scholar]
- Schmiedeberg K, Shirokova E, Weber HP, Schilling B, Meyerhof W, Krautwurst D. 2007. Structural determinants of odorant recognition by the human olfactory receptors OR1A1 and OR1A2. J Struct Biol. 159:400–412 [DOI] [PubMed] [Google Scholar]
- Singer MS. 2000. Analysis of the molecular basis for octanal interactions in the expressed rat 17 olfactory receptor. Chem Senses 25:155–165 [DOI] [PubMed] [Google Scholar]
- Singer MS, Hughes TE, Shepherd GM, Greer CA. 1998. Identification of olfactory receptor mRNA sequences from the rat olfactory bulb glomerular layer. Neuroreport 9:3745–3748 [DOI] [PubMed] [Google Scholar]
- Singer MS, Oliveira L, Vriend G, Shepherd GM. 1995. Potential ligand-binding residues in rat olfactory receptors identified by correlated mutation analysis. Receptors Channels 3:89–95 [PubMed] [Google Scholar]
- Touhara K. 2001. Functional cloning and reconstitution of vertebrate odorant receptors. Life Sci. 68:2199–2206 [DOI] [PubMed] [Google Scholar]
- Touhara K. 2002. Odor discrimination by G protein-coupled olfactory receptors. Microsc Res Tech. 58:135–141 [DOI] [PubMed] [Google Scholar]
- Touhara K, Sengoku S, Inaki K, Tsuboi A, Hirono J, Sato T, Sakano H, Haga T. 1999. Functional identification and reconstitution of an odorant receptor in single olfactory neurons. Proc Natl Acad Sci USA 96:4040–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovchigrechko A, Vakser IA. 2006. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 34:W310–W314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabanino RJ, Hall SE, Vaidehi N, Floriano WB, Kam VW, Goddard WA., 3rd 2004. First principles predictions of the structure and function of g-protein-coupled receptors: validation for bovine rhodopsin. Biophys J. 86:1904–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnady GE, Simon I. 2001. The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849–850 [DOI] [PubMed] [Google Scholar]
- Vaidehi N, Floriano WB, Trabanino R, Hall SE, Freddolino P, Choi EJ, Zamanakos G, Goddard WA., 3rd 2002. Prediction of structure and function of G protein-coupled receptors. Proc Natl Acad Sci USA 99:12622–12627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakser IA. 1997. Evaluation of GRAMM low-resolution docking methodology on the hemagglutinin-antibody complex. Proteins Suppl 1: 226–230 [PubMed] [Google Scholar]
- Vakser IA, Jiang S. 2002. Strategies for modeling the interactions of transmembrane helices of G protein-coupled receptors by geometric complementarity using the GRAMM computer algorithm. Methods Enzymol. 343:313–328 [DOI] [PubMed] [Google Scholar]
- Van Der Spoel D, Lindahl E, Hess B, Van Buuren AR, Apol E, Meulenhoff PJ, Tieleman DP, Sijbers ALTM, Feenstra KA, Van Drunen R, et al. 2010. Gromacs User Manual version 4.5.4. Available from: http://manual.gromacs.org/ [Google Scholar]
- Wu H, Wacker D, Mileni M, Katritch V, Han GW, Vardy E, Liu W, Thompson AA, Huang XP, Carroll FI, et al. 2012. Structure of the human κ-opioid receptor in complex with JDTic. Nature 485:327–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Wu H, Katritch V, Han GW, Jacobson KA, Gao ZG, Cherezov V, Stevens RC. 2011. Structure of an agonist-bound human A2A adenosine receptor. Science 332:322–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, Trask BJ. 2002. The sense of smell: genomics of vertebrate odorant receptors. Hum Mol Genet. 11:1153–1160 [DOI] [PubMed] [Google Scholar]
- Zhao H, Ivic L, Otaki JM, Hashimoto M, Mikoshiba K, Firestein S. 1998. Functional expression of a mammalian odorant receptor. Science 279:237–242 [DOI] [PubMed] [Google Scholar]
- Zozulya S, Echeverri F, Nguyen T. 2001. The human olfactory receptor repertoire. Genome Biol. 2:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]