Abstract
Levels of asymmetric dimethylarginine (ADMA), an endogenous inhibitor of nitric oxide synthase, are increased in lung, sputum, exhaled breath condensate and plasma samples from asthma patients. ADMA is metabolized primarily by dimethylarginine dimethylaminohydrolase 1 (DDAH1) and DDAH2. We determined the effect of DDAH1 overexpression on development of allergic inflammation in a mouse model of asthma. The expression of DDAH1 and DDAH2 in mouse lungs was determined by RT-quantitative PCR (qPCR). ADMA levels in bronchoalveolar lavage fluid (BALF) and serum samples were determined by mass spectrometry. Wild type and DDAH1-transgenic mice were intratracheally challenged with PBS or house dust mite (HDM). Airway inflammation was assessed by bronchoalveolar lavage (BAL) total and differential cell counts. The levels of IgE and IgG1 in BALF and serum samples were determined by ELISA. Gene expression in lungs was determined by RNA-Seq and RT-qPCR. Our data showed that the expression of DDAH1 and DDAH2 was decreased in the lungs of mice following HDM exposure, which correlated with increased ADMA levels in BALF and serum. Transgenic overexpression of DDAH1 resulted in decreased BAL total cell and eosinophil numbers following HDM exposure. Total IgE levels in BALF and serum were decreased in HDM-exposed DDAH1-transgenic mice compared to HDM-exposed wild type mice. RNA-Seq results showed downregulation of genes in the inducible nitric oxide synthase (iNOS) signaling pathway in PBS-treated DDAH1-transgenic mice versus PBS-treated wild type mice and downregulation of genes in IL-13/FOXA2 signaling pathway in HDM-treated DDAH1-transgenic mice versus HDM-treated wild type mice. Our findings suggest that decreased expression of DDAH1 and DDAH2 in the lungs may contribute to allergic asthma and overexpression of DDAH1 attenuates allergen-induced airway inflammation through modulation of Th2 responses.
Introduction
Asthma is a chronic inflammatory lung disease characterized by airway hyperresponsiveness (AHR), airway inflammation, excess mucus production and pulmonary remodeling [1]. Arginine metabolism has been found to play a critical role in the pathogenesis of allergic asthma [1], [2] and mounting evidence suggests that nitric oxide (NO) bioavailability plays an important role in the development of allergic inflammation [3], [4]. Arginine is metabolized by arginase yielding urea and L-ornithine that is further metabolized to polyamines and prolines, which can modulate cell proliferation and collagen production [4]. Arginine is also metabolized by nitric oxide synthases (NOS) including neuronal NOS (nNOS), inducible NOS (iNOS) and endothelial NOS (eNOS) [5]. eNOS is expressed in bronchial epithelium and type II alveolar epithelium, nNOS is expressed in airway nervous tissue, and iNOS is expressed in type II alveolar epithelium, lung fibroblasts, airway and vascular smooth muscle cells. The constitutive NO produced by nNOS and eNOS is important for smooth muscle relaxation, bronchodilation and determination of vascular tone and blood pressure while the inducible NO produced by iNOS has pro-inflammatory effects [5].
Asymmetric dimethylarginine (ADMA) is a by-product released from proteolysis of methylated proteins and ADMA competitively inhibits all three NOS by displacing L-arginine from NOS [6]. ADMA also competes with arginine for cellular uptake by cationic amino-acid transporters, affecting the cellular ADMA/arginine ratio [7], [8]. Analysis of methylarginine metabolism in the cardiovascular system showed that the lung is a major source of ADMA [9]. ADMA has been shown to have profound effects on multiple tissues. Microarray studies showed that pathophysiological concentrations of ADMA elicit significant changes in the gene expression in coronary artery endothelial cells [10]. Treatment of primary mouse lung fibroblasts with ADMA induced arginase activity and collagen production [11]. ADMA infusion resulted in increased lung resistance and decreased compliance in response to methacholine in mice, which was associated with significantly increased pulmonary collagen deposition [11]. ADMA potentiates ovalbumin-induced airway inflammation in a mouse model of asthma [12]. In humans, plasma ADMA levels are increased in severe asthma patients compared to nonsevere asthma patients and control subjects [3]. Other studies showed that ADMA levels are increased in sputum and exhaled breath condensate from asthma patients [13]–[15]. A more recent study showed that lower L-arginine/ADMA ratios are associated with reduced lung function and increased respiratory symptom frequency in subjects with late-onset asthma [16].
DDAH activity is a key determinant of intracellular ADMA concentration [17]. Ninety percent of ADMA is metabolized by DDAH and the rest is excreted through the kidneys. DDAH metabolizes ADMA to generate citrulline and dimethylamine. There are two isoforms of DDAH in human, mouse, rat and other species, DDAH1 and DDAH2 [18]–[20]. Immunostaining of human lung tissues showed expression of DDAH1 and DDAH2 in both alveolar and bronchiolar epithelium [21]. One study showed that homozygous DDAH1-deficient mice die before birth while the heterozygous DDAH1-deficient mice have a 20% increased level of ADMA and develop severe endothelial dysfunction [22]. Anther study showed that DDAH1-deficient mice are viable and have significantly increased ADMA levels [23]. ADMA levels are decreased in DDAH1-transgenic mice by 50% compared to wild type mice. Further, DDAH1-transgenic mice display a significant increase in NOS activity and a decreased risk of endothelial dysfunction compared to the wild type mice [24]–[26]. DDAH1 is the major enzyme responsible for metabolizing ADMA whereas DDAH2 has no detectable role in degrading ADMA in vivo [23]. In this paper, we determined the role of DDAH1 in the development of AHR and airway inflammation.
Materials and Methods
Mice
Mice were maintained and handled under Institutional Animal Care and Use Committee-approved procedures (Cincinnati Children's Hospital Medical Center, Protocol Number: 2D10082) and the Guide for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources, National Research Council). C57BL/6 wild type and DDAH1-transgenic mice were purchased from Jackson Laboratory (Bar Harbor, ME).
Immunization
C57BL/6 wild type and DDAH1-transgenic mice were immunized with intratracheal instillation of 100 µg of house dust mite (HDM) (Dermatophagoides pteronyssinus) extract (Greer Laboratories, Lenoir, NC) in 50 µl of PBS or 50 µl of PBS alone 3 times per week for 3 weeks as previously described [27], [28].
Analysis of AHR and airway inflammation
Twenty four hours after the last treatment, AHR in response to methacholine (0, 50, 100 and 200 mg/ml) was measured by flexiVent (SCIREQ, Montreal, Canada) as previously described [29]. After AHR measurement, blood, bronchoalveolar lavage (BAL) samples and lung tissues were harvested as previously described [30]. The BAL total and differential cell counting as well as histological staining of lung sections by hematoxylin and eosin (H&E) or periodic acid Schiff (PAS) (Thermo Fisher Scientific, Waltham, MA) were performed as described by the manufacturer or described previously [30]. ELISAs for total IgE and IgG1 or HDM-specific IgE and IgG1 in BAL fluid (BALF) and serum were done as previously described [31].
RNA-Seq and Bioinformatics Analysis
Total RNA was isolated from mouse lungs using Trizol reagent (Invitrogen, Carlsbad, CA), digested with RNase-free DNase and purified using an RNeasy MinElute kit (QIAGEN, Valencia, CA). Equal amounts of RNA were pooled from each mouse lung in an experimental group (n = 4 per group) and analyzed in duplicate. RNA-Seq was performed by the Genomics Sequencing Core in the University of Cincinnati. The RNA-Seq library was constructed using a PrepX SPIA RNA-Seq kit (IntegenX, Pleasanton, CA) and Apollo 324 NGS Library Prep System (IntegenX). 10 ng of total RNA was converted into cDNA suitable for mRNA sequencing. The cDNA was then sheared by Covaris S2 (Covaris, Woburn, MA) under the conditions recommended by IntegenX, followed by Bioanalyzer assay of the size distribution with Agilent High Sensitivity DNA kit (Agilent, Santa Clara, CA). The properly sheared cDNA fragments were purified by Agencourt AMPure XP magnetic beads (Beckman Coulter, Brea CA). Using the IntegenX PrepX ILM DNA library kit for Illumina and Apollo 324 NGS Library Prep System, 500 ng of purified cDNA fragments were then put through end repair, addition of a single ‘A’ base and ligation of adapters, and indexed individually. The products were purified and enriched by PCR to create the final cDNA library targeting mRNAs. The size of the generated library was validated by Bioanalyzer and the library was quantified using the Kapa Library Quantification kit (Kapa Biosystems, Woburn, MA). Six individually indexed cDNA libraries were equal amount pooled for clustering in cBot system (Illumina, San Diego, CA). Libraries at the concentration of 6.5 pM were clustered onto a flow cell using Illumina's TruSeq SR Cluster Kit v3, and sequenced for 50 cycles using the TruSeq SBS kit on Illumina HiSeq system.
Sequence reads were aligned to the reference genome (mm10) using TopHat aligner [32]. The counts of reads aligning to each gene's coding region were summarized using ShortRead and associated Bioconductor packages for manipulating and analysis of next-generation sequencing data and custom-written R programs [33], [34]. Statistical analysis to identify differentially expressed genes for each comparison was performed using the negative-binomial model of read counts as implemented in the DESeq Biocondoctor package [35]. P-values were adjusted for multiple comparisons based on false discovery rates (FDR) [36]. Differential expressions with adjusted p-values of <0.05 were considered statistically significant.
Functional enrichment analysis was performed using the logistic regression based LRpath methodology [37]. The gene lists used in the functional enrichment analysis were from genes associated with Gene Ontology terms and KEGG pathways. The statistical significance of gene list enrichment was determined by the False Discovery Rate cut-off of 0.1. Genes that were both members of at least one statistically significant gene list and had differential expression p-values of <0.01 were considered to be differentially expressed for the purpose of network analysis. Ingenuity Pathways Analysis (IPA) (Ingenuity Systems, Mountain View, CA) was used to identify integrated and interconnected biological networks and upstream targets for the differentially expressed genes between two groups (with adjusted p-values of <0.05). RNA-Seq data have been deposited with the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE49047.
RT-PCR
Total RNA was isolated from lung tissues using Trizol reagent (Invitrogen), digested with RNase-free DNase and purified using an RNeasy MinElute kit (QIAGEN). Reverse transcription was done using Oligo-dT First-Strand cDNA Synthesis Kit (GE Healthcare, Piscataway, NJ). Quantitative PCR (qPCR) was done using the SYBR Green Master Kit and LightCycler® 480 instrument (Roche Diagnostics, Indianapolis, IN). All primers used are listed in Table S1.
Mass Spectrometry (MS)
MS was performed by the Analytical & Mass Spectrometry of Small Molecules Core at the University of Cincinnati. The procedure to quantify ADMA was adapted from Schwedhelm et al [38]. In brief, BALF and serum samples were filtrated through 0.45 µm filters to remove particles and 200 µl aliquots were then ultra-filtrated through 3 kDa MWCO filters to remove proteins. The samples were derivatized by adding 200 µl of 2 M HCl in 1-butanol for 20 min at 65°C. After evaporation, samples were reconstituted in 200 µl of distilled deionized water and analyzed by nano liquid chromatography chip electrospray ionization ion trap MS (nanoLC-Chip ESI-IT-MS). For nanoLC-Chip ESI-MS/MS analysis, the derivatized ADMA was separated in a microfluidic reverse phase chip column and detected by electrospray ionization with ion trap MS/MS detection. The tandem system consisted of an Agilent 1200 HPLC (Agilent Technologies, Santa Clara, CA), equipped with a capillary and nano pump, used for loading and flushing the on-chip nano column, a chip cube interface that contains the nano-chip column Zorbax SB C-18, 150×0.75 mm (Agilent Technologies), and an Agilent 6300 ion trap XCT system (Agilent Technologies). The mobile phase A consisted of 0.1% formic acid in water while B consisted of 0.1% formic acid in a 7:1 acetonitrile:water solution. A linear gradient from 2% to 25% B was carried out in 10 min and the column was then cleaned with 100% B for 5 min and then regenerated at original conditions for 10 min before the next injection. The flow rate was 0.3 µl per min and the outlet of the column communicates directly with the nano electrospray needle. The analysis was carried out in the MRM mode by following the transition m/z 259.3→214. ADMA standards (Sigma-Aldrich, St. Louis, MO) were derivatized in the same way and quantification was carried out by the external calibration method.
Statistical analysis
All values are expressed as mean ± SD. The data were analyzed with a 2-tailed unpaired student's t-test with Welch's correction or 1-way ANOVA with Newman-Keuls' post test using Prism 5.0c for Mac OS X from GraphPad Software (San Diego, CA). A p-value of <0.05 was considered statistically significant.
Results
Expression of DDAH1 and DDAH2 is decreased in the lungs and ADMA levels are increased in BALF and serum from HDM-treated mice
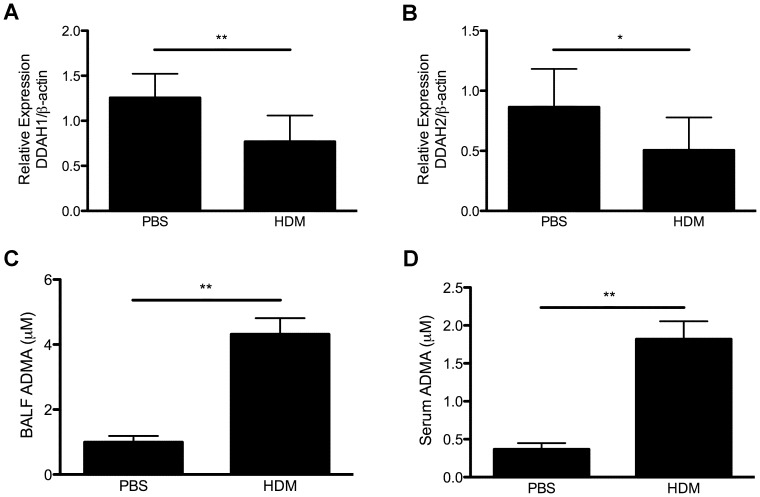
Since ADMA levels are increased in allergic asthma and DDAH is responsible for the majority of ADMA metabolism, we hypothesized that the increased levels of ADMA are due to decreased expression of DDAH following allergen exposure. We determined the expression of DDAH1 and DDAH2 in the lungs in a mouse model of asthma. C57BL/6 mice were challenged by intratracheal instillation of 100 µg HDM, 3 times per week for 3 weeks. This is a more clinically relevant protocol of chronic allergen exposure with an increased dosage of HDM for the C57BL/6 mouse strain. As shown in Fig. 1A and 1B, HDM exposure resulted in decreased expression of DDAH1 and DDAH2 in mouse lungs, which correlated with increased levels of ADMA in BALF and serum (Fig. 1C and 1D).
Figure 1. Expression of DDAH1 and DDAH2 in lungs and ADMA levels in BALF and serum samples from PBS or HDM-treated C57BL/6 mice.
The mice were treated with PBS or HDM by 3 intratracheal challenges per week for 3 weeks. (A–B) DDAH1 and DDAH2 expression in lungs (n = 7–9). (C–D) ADMA levels in BALF and serum samples (n = 3). Data are shown as mean±SD. *, p<0.05; **, p<0.01.
Overexpression of DDAH1 attenuates HDM-induced airway inflammation
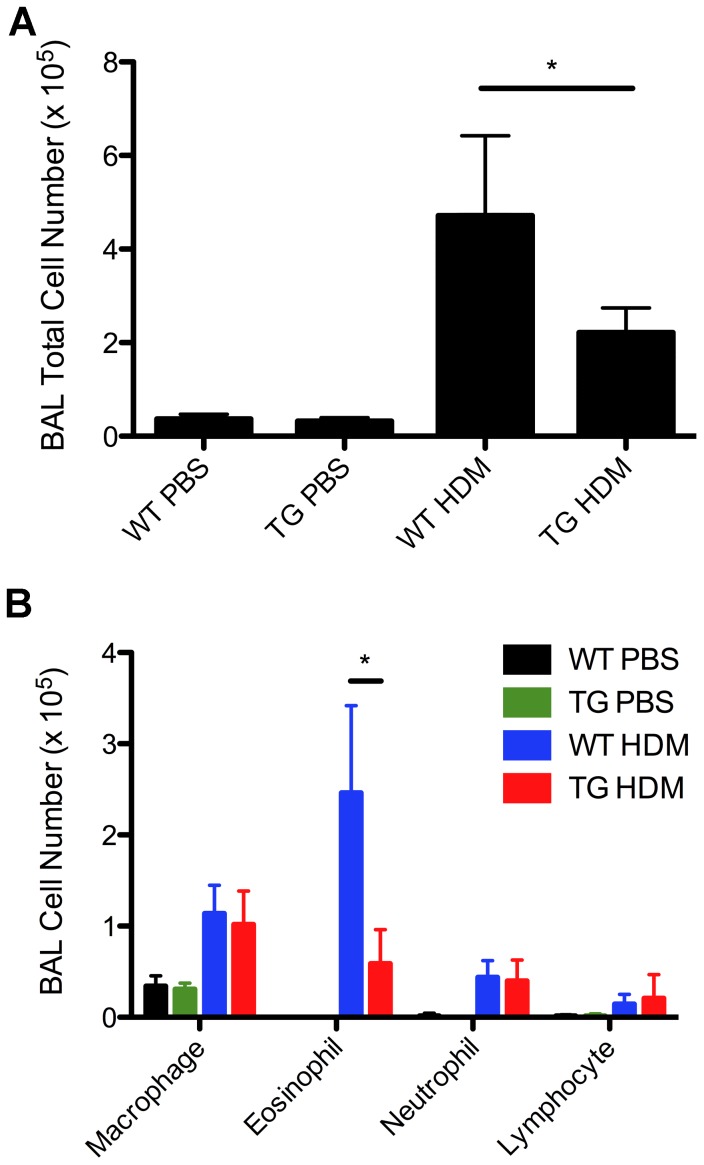
We next determined the effect of DDAH overexpression on airway inflammation and AHR in a HDM-induced asthma model using DDAH1-transgenic mice since DDAH1 is the major enzyme responsible for ADMA metabolism [23]. The mice were challenged by intratracheal instillation of 100 µg HDM 3 times per week for 3 weeks. Twenty-four hours after the last treatment AHR was measured by flexiVent and airway inflammation was assessed by BAL cell counts. BAL total cell and eosinophil numbers were decreased in HDM-treated DDAH1-transgenic mice compared to HDM-treated wild type mice (Fig. 2A-2B). No significant differences in inflammatory cell infiltration in airways determined by H&E staining or in mucus production determined by PAS staining of lung sections were observed between HDM-treated DDAH1-transgenic mice and HDM-treated wild type mice (data not shown). No significant difference in AHR was observed in HDM-treated DDAH1-transgenic mice compared to HDM-treated wild type mice (data not shown).
Figure 2. Airway inflammation in PBS or HDM-treated C57BL/6 wild type and DDAH1-transgenic mice.
BAL total and differential cell counts in wild type and DDAH1-transgenic mice treated with PBS or HDM (3 intratracheal challenges per week for 3 weeks). WT: wide type; TG: DDAH1-transgenic. Data are shown as mean±SD (n = 5–9). *, p<0.05.
IgE levels in BALF and serum are decreased in DDAH1-transgenic mice compared to wild type mice after HDM treatment
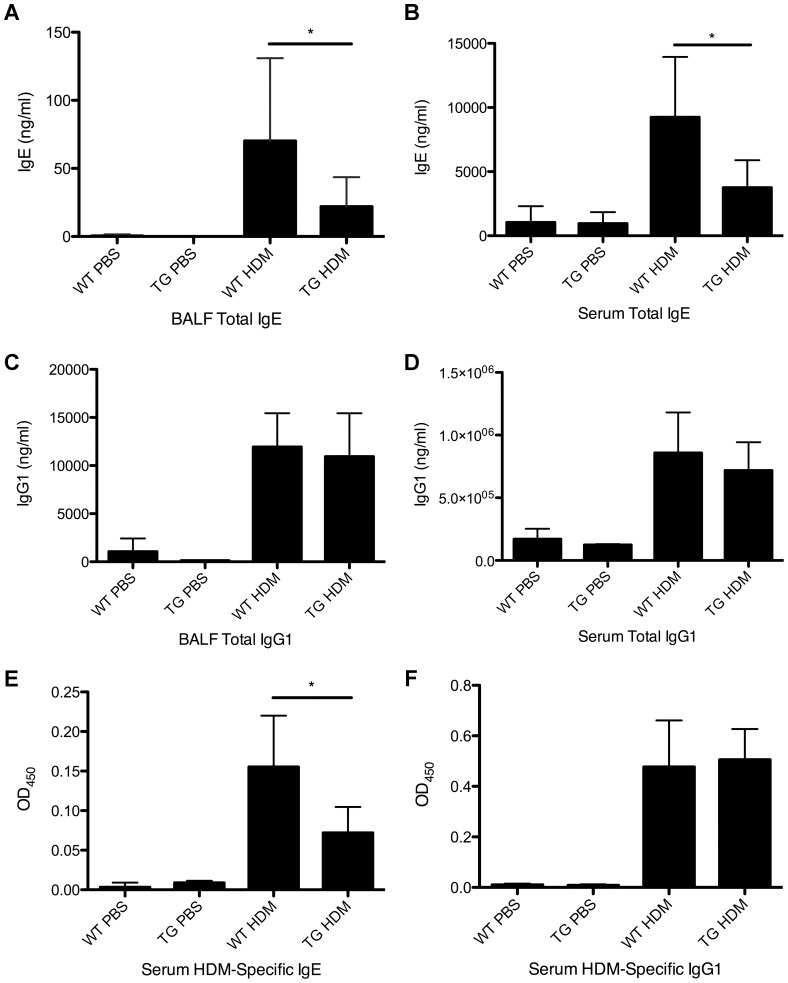
We next tested whether allergen sensitization was affected by the overexpression of DDAH1. As shown in Fig. 3A and 3B, the total IgE levels were decreased in BALF and serum in HDM-treated DDAH1-transgenic mice compared to HDM-treated wild type mice but there was no difference in the levels of total IgG1 in BALF or serum (Fig. 3C and 3D). The level of HDM-specific IgE was decreased in serum from HDM-treated DDAH1-transgenic mice compared to HDM-treated wild type mice but the level of HDM-specific IgG1 in serum was unaffected (Fig. 3E and 3F).
Figure 3. Levels of IgE and IgG1 in BALF and serum in PBS or HDM-treated C57BL/6 wild type and DDAH1-transgenic mice.
(A) BALF total IgE. (B) Serum total IgE. (C) BALF total IgG1. (D) Serum total IgG1. (E) Serum HDM-specific IgE. (F) Serum HDM-specific IgG1. WT: wide type; TG: DDAH1-transgenic. Data are shown as mean±SD (n = 5–9). *, p<0.05.
Gene expression profiles are significantly different in the lungs of wild type and DDAH1-transgenic mice
To elucidate the potential mechanisms underlying attenuated HDM-induced airway inflammation in DDAH1-transgenic mice, we determined gene expression profiles in the lungs from C57BL/6 wild type and DDAH1-transgenic mice treated with PBS or HDM (intratracheal instillation of 100 µg of HDM in 50 µl of PBS or 50 µl of PBS alone 3 times per week for 3 weeks) by RNA-Seq.
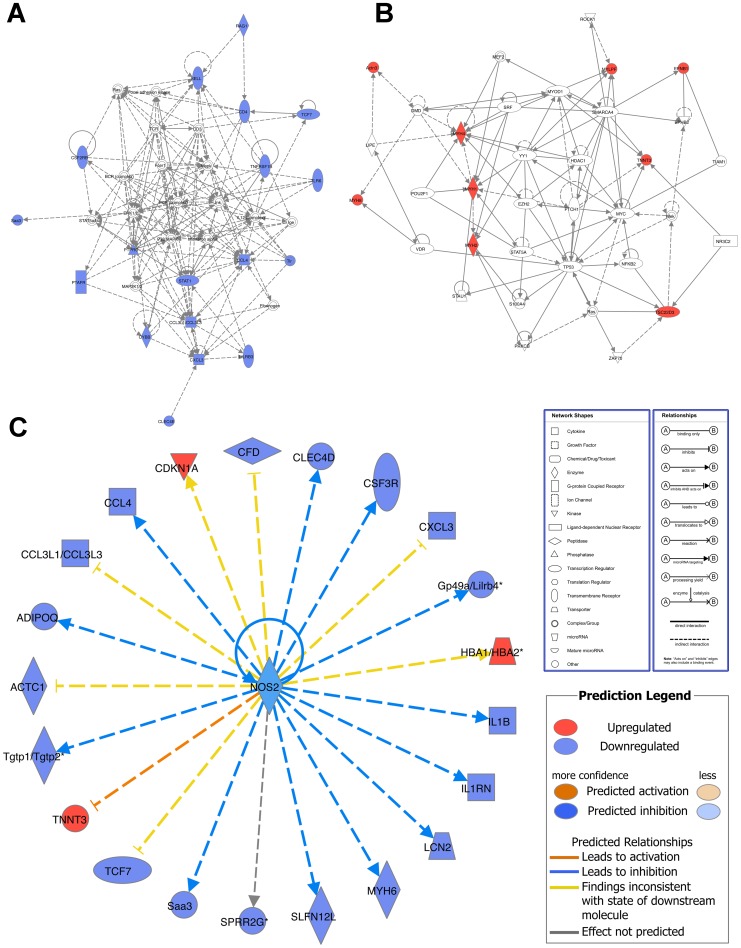
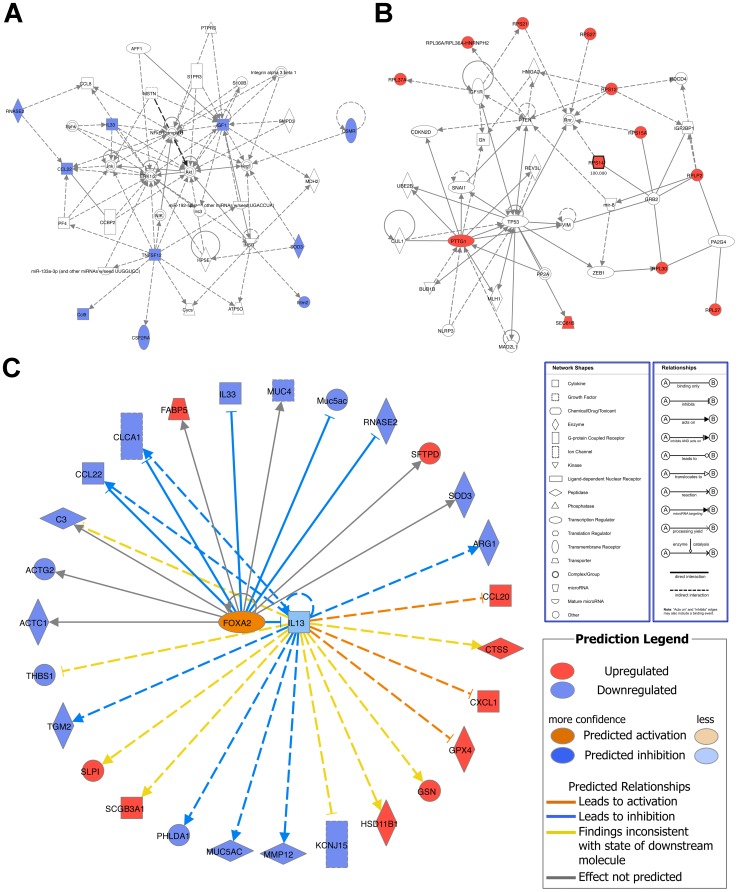
There were 354 genes differentially expressed in the lungs of PBS-treated wild type and PBS-treated DDAH1-transgenic mice (Table S2). The top downregulated genes (with an adjusted p-value of <0.05) are shown in Table 1 (grouped and sorted by fold change), including immune/defense response genes (Rag1, Irg1, Tlr6, etc.), cell structure/adhesion/migration genes (Sprr2a2, Sprr2a1, Adipoq, etc.), cytokine/chemokine genes (Ccl4, Ccl3, Cxcl2 and Il1b), and transcriptional factors (Tcf7 and Stat1). The top upregulated genes (with an adjusted p-value of <0.05) are shown in Table 2 (grouped and sorted by fold change), including muscle/cell structure genes (Myh2, Myh8, Myh4, etc.), ion homeostasis/metabolism genes (Atp2a1, Ckm, Sod1, etc.) and hemopoiesis genes (Hba-a2, Hba-a1, Beta-s, etc.). For the downregulated immune/defense response genes, cytokine/chemokine genes and transcriptional factor genes, IPA analysis showed that the top network was associated with inflammatory response, immunological disease, and respiratory disease (Fig. 4A). The top network for the upregulated muscle/cell structure genes was associated with organ morphology, skeletal and muscular system development and function, and cancer (Fig. 4B). The top upstream target for both downregulated and upregulated genes was iNOS (NOS2) (Fig. 4C).
Table 1. Genes Downregulated in the Lungs of PBS-treated DDAH1-transgenic Mice versus PBS-treated Wild Type Mice.
| Symbol | Name | Fold Change |
| Immune/Defense Response | ||
| Rag1 | recombination activating gene 1 | 0.0020 |
| Irg1 | immunoresponsive gene 1 | 0.0042 |
| Tlr6 | toll-like receptor 6 | 0.0224 |
| Saa3 | serum amyloid A 3 | 0.0265 |
| Clec4e | C-type lectin domain family 4, member e | 0.0360 |
| Ptafr | platelet-activating factor receptor | 0.0656 |
| Pram1 | PML-RAR alpha-regulated adaptor molecule 1 | 0.1024 |
| Clec4d | C-type lectin domain family 4, member d | 0.1306 |
| Cfd | complement factor D (adipsin) | 0.1491 |
| Cd4 | CD4 antigen | 0.2667 |
| Tnfrsf1b | tumor necrosis factor receptor superfamily, member 1b | 0.3327 |
| Il1rn | interleukin 1 receptor antagonist | 0.3365 |
| Lilrb3 | leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 | 0.3596 |
| Sell | selectin, lymphocyte | 0.3826 |
| Acsl1 | acyl-CoA synthetase long-chain family member 1 | 0.4620 |
| Cybb | cytochrome b-245, beta polypeptide | 0.4694 |
| Lcn2 | lipocalin 2 | 0.4804 |
| Tgtp1 | T-cell specific GTPase 1 | 0.4907 |
| Lilrb4 | leukocyte immunoglobulin-like receptor, subfamily B, member 4 | 0.4931 |
| Tgtp2 | T-cell specific GTPase 2 | 0.4986 |
| Csf2rb | colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) | 0.5378 |
| Ptprc | protein tyrosine phosphatase, receptor type, C | 0.6052 |
| Cell Structure/Adhesion/Migration | ||
| Sprr2a2 | small proline-rich protein 2A2 | 0.0179 |
| Sprr2a1 | small proline-rich protein 2A1 | 0.0179 |
| Adipoq | adiponectin, C1Q and collagen domain containing | 0.0702 |
| S100a9 | S100 calcium binding protein A9 (calgranulin B) | 0.0736 |
| Pla2g7 | phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) | 0.1651 |
| S100a8 | S100 calcium binding protein A8 (calgranulin A) | 0.2070 |
| H2-M2 | histocompatibility 2, M region locus 2 | 0.2210 |
| Sema4d | sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D | 0.2948 |
| Itgb2 | integrin beta 2 | 0.4746 |
| Myh6 | myosin, heavy polypeptide 6, cardiac muscle, alpha | 0.4752 |
| Itgal | integrin alpha L | 0.4913 |
| Actc1 | actin, alpha, cardiac muscle 1 | 0.5662 |
| Csf3r | colony stimulating factor 3 receptor (granulocyte) | 0.5942 |
| Cytokine/Chemokine | ||
| Ccl4 | chemokine (C-C motif) ligand 4 | 0.0000 |
| Ccl3 | chemokine (C-C motif) ligand 3 | 0.0114 |
| Cxcl2 | chemokine (C-X-C motif) ligand 2 | 0.0236 |
| Il1b | interleukin 1 beta | 0.1342 |
| Transcriptional Factor | ||
| Tcf7 | transcription factor 7, T-cell specific | 0.4134 |
| Stat1 | signal transducer and activator of transcription 1 | 0.5833 |
| Other | ||
| Arpp21 | cyclic AMP-regulated phosphoprotein, 21 | 0.0036 |
| Gxylt2 | glucoside xylosyltransferase 2 | 0.0112 |
| 1700071M16Rik | RIKEN cDNA 1700071M16 gene | 0.0427 |
| Trim30b | tripartite motif-containing 30B | 0.0516 |
| Cd177 | CD177 antigen | 0.0746 |
| Niacr1 | niacin receptor 1 | 0.0993 |
| Mmp8 | matrix metallopeptidase 8 | 0.1307 |
| Slfn4 | schlafen 4 | 0.1469 |
| Acpp | acid phosphatase, prostate | 0.2390 |
| Steap4 | STEAP family member 4 | 0.2412 |
| F13a1 | coagulation factor XIII, A1 subunit | 0.2631 |
| Gpnmb | glycoprotein (transmembrane) nmb | 0.3398 |
| Slpi | secretory leukocyte peptidase inhibitor | 0.3547 |
| Bpifa1 | BPI fold containing family A, member 1 | 0.3796 |
| Gp49a | glycoprotein 49 A | 0.4032 |
| Car3 | carbonic anhydrase 3 | 0.4062 |
| Gm1966 | predicted gene 1966 | 0.5319 |
| Ctss | cathepsin S | 0.6001 |
| Fth1 | ferritin heavy chain 1 | 0.6449 |
NCBI Gene Expression Omnibus accession numbers GSE49047 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE49047).
Table 2. Genes Upregulated in the Lungs of PBS-treated DDAH1-transgenic Mice versus PBS-treated Wild Type Mice.
| Symbol | Name | Fold Change |
| Muscle/Cell Structure | ||
| Myh2 | myosin, heavy polypeptide 2, skeletal muscle, adult | 181.3505 |
| Myh8 | myosin, heavy polypeptide 8, skeletal muscle, perinatal | 109.1566 |
| Myh4 | myosin, heavy polypeptide 4, skeletal muscle | 82.8001 |
| Myh1 | myosin, heavy polypeptide 1, skeletal muscle, adult | 65.6757 |
| Tnnt3 | troponin T3, skeletal, fast | 44.0881 |
| Actn3 | actinin alpha 3 | 26.0940 |
| Mylpf | myosin light chain, phosphorylatable, fast skeletal muscle | 14.9051 |
| Neb | nebulin | 10.4391 |
| Ion Homeostasis/Metabolism | ||
| Atp2a1 | ATPase, Ca++ transporting, cardiac muscle, fast twitch 1 | 45.0004 |
| Ckm | creatine kinase, muscle | 9.6517 |
| Sod1 | superoxide dismutase 1, soluble | 1.8226 |
| Errfi1 | ERBB receptor feedback inhibitor 1 | 1.7924 |
| Dpep1 | dipeptidase 1 (renal) | 1.6845 |
| Sgms1 | sphingomyelin synthase 1 | 1.6440 |
| Glul | glutamate-ammonia ligase (glutamine synthetase) | 1.4929 |
| Hemopoiesis | ||
| Hba-a2 | hemoglobin alpha, adult chain 2 | 1.6691 |
| Hba-a1 | hemoglobin alpha, adult chain 1 | 1.6691 |
| Beta-s | hemoglobin subunit beta-1-like | 1.5436 |
| Hbb-b1 | hemoglobin, beta adult major chain | 1.5242 |
| Hbb-b2 | hemoglobin, beta adult minor chain | 1.5242 |
| MicroRNA | ||
| Mir5109 | microRNA 5109 | 2.7839 |
| Other | ||
| Flrt2 | fibronectin leucine rich transmembrane protein 2 | 11.5849 |
| Gm13375 | predicted gene 13375 | 8.1901 |
| Gm3893 | predicted gene 3893 | 5.6492 |
| Rmrp | RNA component of mitochondrial RNAase P | 4.8670 |
| Angptl4 | angiopoietin-like 4 | 2.7019 |
| Efnb1 | ephrin B1 | 2.3349 |
| Zbtb16 | zinc finger and BTB domain containing 16 | 2.2658 |
| Cdkn1a | cyclin-dependent kinase inhibitor 1A (P21) | 2.2030 |
| Tsc22d3 | TSC22 domain family, member 3 | 2.1869 |
| 6430548M08Rik | RIKEN cDNA 6430548M08 gene | 2.1316 |
| Krt7 | keratin 7 | 2.0891 |
| Bmp6 | bone morphogenetic protein 6 | 1.8952 |
| Gm10393 | predicted gene 10393 | 1.8802 |
| Lars2 | leucyl-tRNA synthetase, mitochondrial | 1.8365 |
| Plxna2 | plexin A2 | 1.8323 |
| Eif3e | eukaryotic translation initiation factor 3, subunit E | 1.7913 |
| Cpm | carboxypeptidase M | 1.7450 |
| Rn45S | 45S pre-ribosomal RNA | 1.7382 |
| Vps54 | vacuolar protein sorting 54 (yeast) | 1.5716 |
| Dnaja1 | DnaJ (Hsp40) homolog, subfamily A, member 1 | 1.5133 |
| Tmbim6 | transmembrane BAX inhibitor motif containing 6 | 1.4524 |
| Sepp1 | selenoprotein P, plasma, 1 | 1.3714 |
| Sftpa1 | surfactant associated protein A1 | 1.3527 |
NCBI Gene Expression Omnibus accession numbers GSE49047 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE49047).
Figure 4. Network analysis of differentially expressed genes in the lungs of PBS-treated C57BL/6 wild type and PBS-treated DDAH1-transgenic mice.
(A) Top network of the downregulated immune/defense response genes, cytokine/chemokine genes and transcriptional factor genes in PBS-treated DDAH1-transgenic mice. (B) Top network of the upregulated muscle/cell structure genes in PBS-treated DDAH1-transgenic mice. (C) Upstream target analysis of genes differentially expressed in PBS-treated wild type and DDAH1-transgenic mice. The genes with an adjusted p-value of <0.05 were included for analysis.
There were 707 genes differentially expressed in the lungs of HDM-treated wild type and HDM-treated DDAH1-transgenic mice (Table S3). The top downregulated genes (with an adjusted p-value of <0.05) are shown in Table 3 (grouped and sorted by fold change), including metabolism/transport genes (Fbp1, Dhrs9, Arg1, etc.), immune/defense response genes (Ear11, Chia, Sod3, etc.), cytokine/chemokine genes (Ccl22, Tnfsf12, Ccl9 and Il33), extracellular matrix genes (Col6a2, Muc5ac, Muc4 and Col1a1) and microRNAs (Mir5109 and Mir5107). The top upregulated genes (with an adjusted p-value of <0.05) are shown in Table 4 (grouped and sorted by fold change), including ribosomal complex genes (Rps21, Rps12, Rpl30, etc.) and ribosomal pseudogenes (Gm11968, Gm12191, Gm13253, etc.). The top network for the downregulated immune/defense genes and cytokine/chemokine genes was associated with inflammatory response, cellular movement, and hematological system development and function (Fig. 5A). The top network for the upregulated ribosomal complex genes and ribosomal pseudogenes is associated with cellular development, cancer, and cellular assembly and organization (Fig. 5B). The top upstream targets for both downregulated and upregulated genes were IL-13 and FOXA2 (Fig. 5C).
Table 3. Genes Downregulated in the Lungs of HDM-treated DDAH1-transgenic Mice versus HDM-treated Wild Type Mice.
| Symbol | Name | Fold Change |
| Metabolism/Transport | ||
| Fbp1 | fructose bisphosphatase 1 | 0.1302 |
| Dhrs9 | dehydrogenase/reductase (SDR family) member 9 | 0.1595 |
| Arg1 * | arginase, liver | 0.1808 |
| Slc26a4 | solute carrier family 26, member 4 | 0.2789 |
| Cox15 | COX15 homolog, cytochrome c oxidase assembly protein (yeast) | 0.2889 |
| Chi3l4 * | chitinase 3-like 4 | 0.3373 |
| Atp1a3 | ATPase, Na+/K+ transporting, alpha 3 polypeptide | 0.3547 |
| Slc5a1 | solute carrier family 5 (sodium/glucose cotransporter), member 1 | 0.3701 |
| Clca3 | chloride channel calcium activated 3 | 0.3773 |
| Kcnj15 | potassium inwardly-rectifying channel, subfamily J, member 15 | 0.4207 |
| Chi3l3 * | chitinase 3-like 3 | 0.4246 |
| Slco4c1 | solute carrier organic anion transporter family, member 4C1 | 0.4487 |
| Man2b2 | mannosidase 2, alpha B2 | 0.4520 |
| Immune/Defense Response | ||
| Ear11 | eosinophil-associated, ribonuclease A family, member 11 | 0.2201 |
| Chia * | chitinase, acidic | 0.3222 |
| Sod3 | superoxide dismutase 3, extracellular | 0.3353 |
| Csf2ra | colony stimulating factor 2 receptor, alpha, low-affinity (granulocyte-macrophage) | 0.3959 |
| Ifitm2 | interferon induced transmembrane protein 2 | 0.4031 |
| Igf1 | insulin-like growth factor 1 | 0.4424 |
| Osmr | oncostatin M receptor | 0.4612 |
| Cytokine/Chemokine | ||
| Ccl22 | chemokine (C-C motif) ligand 22 | 0.2776 |
| Tnfsf12 | tumor necrosis factor (ligand) superfamily, member 12 | 0.3065 |
| Ccl9 | chemokine (C-C motif) ligand 9 | 0.4578 |
| Il33 * | interleukin 33 | 0.4640 |
| Extracellular Matrix | ||
| Col6a2 | collagen, type VI, alpha 2 | 0.3976 |
| Muc5ac | mucin 5, subtypes A and C, tracheobronchial/gastric | 0.4245 |
| Muc4 | mucin 4 | 0.4415 |
| Col1a1 | collagen, type I, alpha 1 | 0.4641 |
| Transcriptional Factor | ||
| Srebf2 | sterol regulatory element binding factor 2 | 0.3294 |
| Foxp4 | forkhead box P4 | 0.3743 |
| Nfic | nuclear factor I/C | 0.4391 |
| MicroRNA | ||
| Mir5109 | microRNA 5109 | 0.2007 |
| Mir5107 | microRNA 5107 | 0.4523 |
| Other | ||
| Syn2 | synapsin II | 0.0688 |
| 4833422F24Rik | RIKEN cDNA 4833422F24 gene | 0.0775 |
| LOC100048885 | major urinary protein LOC100048885 | 0.1392 |
| Corin | corin | 0.1992 |
| Zfp366 | zinc finger protein 366 | 0.2274 |
| Zfp385b | zinc finger protein 385B | 0.2356 |
| Epha7 | Eph receptor A7 | 0.2597 |
| Bahcc1 | BAH domain and coiled-coil containing 1 | 0.2673 |
| Actg2 | actin, gamma 2, smooth muscle, enteric | 0.2694 |
| Phlda1 | pleckstrin homology-like domain, family A, member 1 | 0.2729 |
| Actc1 | actin, alpha, cardiac muscle 1 | 0.2852 |
| BC048546 | cDNA sequence BC048546 | 0.2998 |
| Trim65 | tripartite motif-containing 65 | 0.3015 |
| Bin2 | bridging integrator 2 | 0.3016 |
| Fcgbp | Fc fragment of IgG binding protein | 0.3248 |
| Prrc2a | proline-rich coiled-coil 2A | 0.3288 |
| Dusp8 | dual specificity phosphatase 8 | 0.3328 |
| Rhob | ras homolog gene family, member B | 0.3346 |
| Atn1 | atrophin 1 | 0.3370 |
| Rpph1 | ribonuclease P RNA component H1 | 0.3560 |
| Midn | midnolin | 0.3735 |
| Notch3 | Notch gene homolog 3 (Drosophila) | 0.3899 |
| Sema5a | sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A | 0.3949 |
| Gm15401 | predicted gene 15401 | 0.3967 |
| Tgfb1i1 | transforming growth factor beta 1 induced transcript 1 | 0.4005 |
| Taok2 | TAO kinase 2 | 0.4159 |
| Samd4b | sterile alpha motif domain containing 4B | 0.4693 |
Verified by RT-qPCR.
NCBI Gene Expression Omnibus accession numbers GSE49047 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE49047).
Table 4. Genes Upregulated in the Lungs of HDM-treated DDAH1-transgenic Mice versus HDM-treated Wild Type Mice.
| Symbol | Name | Fold Change |
| Ribosomal Complex | ||
| Rps21 | ribosomal protein S21 | 5.4796 |
| Rps12 | ribosomal protein S12 | 5.4397 |
| Rpl30 | ribosomal protein L30 | 4.2587 |
| Mrps16 | mitochondrial ribosomal protein S16 | 4.1518 |
| Rpl34 | ribosomal protein L34 | 3.8427 |
| Sec61b | Sec61 beta subunit | 3.7764 |
| Rps14 | ribosomal protein S14 | 3.6956 |
| Rpl36a | ribosomal protein L36A | 3.6885 |
| Rps15a | ribosomal protein S15A | 3.6498 |
| Rpl37a | ribosomal protein L37a | 3.6295 |
| Pttg1 | pituitary tumor-transforming gene 1 | 3.5963 |
| Rplp2 | ribosomal protein, large P2 | 3.4879 |
| Rps27 | ribosomal protein S27 | 3.4532 |
| Rpl27 | ribosomal protein L27 | 3.4067 |
| Rps28 | ribosomal protein S28 | 3.3511 |
| Mrpl20 | mitochondrial ribosomal protein L20 | 3.2634 |
| Ribosomal Pseudogene | ||
| Gm11968 | Rps15a pseudogene | 4.4686 |
| Gm12191 | ribosomal protein L30 pseudogene | 4.2620 |
| Gm13253 | ribosomal protein S15a pseudogene | 4.0395 |
| Rpl34-ps1 | ribosomal protein L34, pseudogene 1 | 3.8391 |
| Rplp2-ps1 | ribosomal protein, large P2, pseudogene 1 | 3.3696 |
| Metabolism/Oxidative Process | ||
| Qdpr | quinoid dihydropteridine reductase | 5.7248 |
| Glrx5 | glutaredoxin 5 homolog (S. cerevisiae) | 5.0037 |
| Ndufa2 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 | 4.9044 |
| Ndufs5 | NADH dehydrogenase (ubiquinone) Fe-S protein 5 | 4.7132 |
| Sumf1 | sulfatase modifying factor 1 | 4.4089 |
| Cox7a2l | cytochrome c oxidase subunit VIIa polypeptide 2-like | 4.2862 |
| Cox17 | cytochrome c oxidase, subunit XVII assembly protein homolog (yeast) | 4.1528 |
| Cytokine/Chemokine | ||
| Ccl20 | chemokine (C-C motif) ligand 20 | 6.3854 |
| Cxcl3 | chemokine (C-X-C motif) ligand 3 | 3.9814 |
| Other | ||
| Dtwd1 | DTW domain containing 1 | 14.2305 |
| Flrt2 | fibronectin leucine rich transmembrane protein 2 | 10.6123 |
| 1110038B12Rik | RIKEN cDNA 1110038B12 gene | 8.9809 |
| Scgb3a2 | secretoglobin, family 3A, member 2 | 7.4584 |
| Fpr2 | formyl peptide receptor 2 | 6.5479 |
| Fkbp14 | FK506 binding protein 14 | 6.3078 |
| Hbxip | hepatitis B virus x interacting protein | 4.8871 |
| BC002163 | NADH dehydrogenase Fe-S protein 5 pseudogene | 4.7683 |
| S100a13 | S100 calcium binding protein A13 | 4.6935 |
| H3f3a | H3 histone, family 3A | 4.6727 |
| Commd1 | COMM domain containing 1 | 4.6707 |
| Osgep | O-sialoglycoprotein endopeptidase | 4.2386 |
| 2010001M09Rik | RIKEN cDNA 2010001M09 gene | 4.1526 |
| Mettl7a2 | methyltransferase like 7A2 | 4.1202 |
| 0610007C21Rik | RIKEN cDNA 0610007C21 gene | 3.9575 |
| Tmbim4 | transmembrane BAX inhibitor motif containing 4 | 3.9312 |
| Snrpe | small nuclear ribonucleoprotein E | 3.9247 |
| Plac9 | placenta specific 9 | 3.7440 |
| Gngt2 | guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 | 3.5724 |
| Tomm7 | translocase of outer mitochondrial membrane 7 homolog (yeast) | 3.5352 |
| Gm9846 | predicted gene 9846 | 3.4544 |
| Ccdc84 | coiled-coil domain containing 84 | 3.4251 |
| S100a6 | S100 calcium binding protein A6 (calcyclin) | 3.3956 |
| Vamp8 | vesicle-associated membrane protein 8 | 3.3746 |
| Shfm1 | split hand/foot malformation (ectrodactyly) type 1 | 3.3414 |
| Cd52 | CD52 antigen | 3.3025 |
| Mettl7a1 | methyltransferase like 7A1 | 3.2925 |
| Pcolce2 | procollagen C-endopeptidase enhancer 2 | 3.2669 |
| Fam166a | family with sequence similarity 166, member A | 3.2667 |
| Neat1 | nuclear paraspeckle assembly transcript 1 (non-protein coding) | 3.2657 |
NCBI Gene Expression Omnibus accession numbers GSE49047 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE49047).
Figure 5. Network analysis of differentially expressed genes in the lungs of HDM-treated C57BL/6 wild type and HDM-treated DDAH1-transgenic mice.
(A) Top network of the downregulated immune/defense response genes and cytokine/chemokine genes in HDM-treated DDAH1-transgenic mice. (B) Top network of the upregulated ribosomal complex genes and ribosomal pseudogenes in HDM-treated DDAH1-transgenic mice. (C) Upstream target analysis of genes differentially expressed in HDM-treated wild type and DDAH1-transgenic mice. The genes with an adjusted p-value of <0.05 were included for analysis.
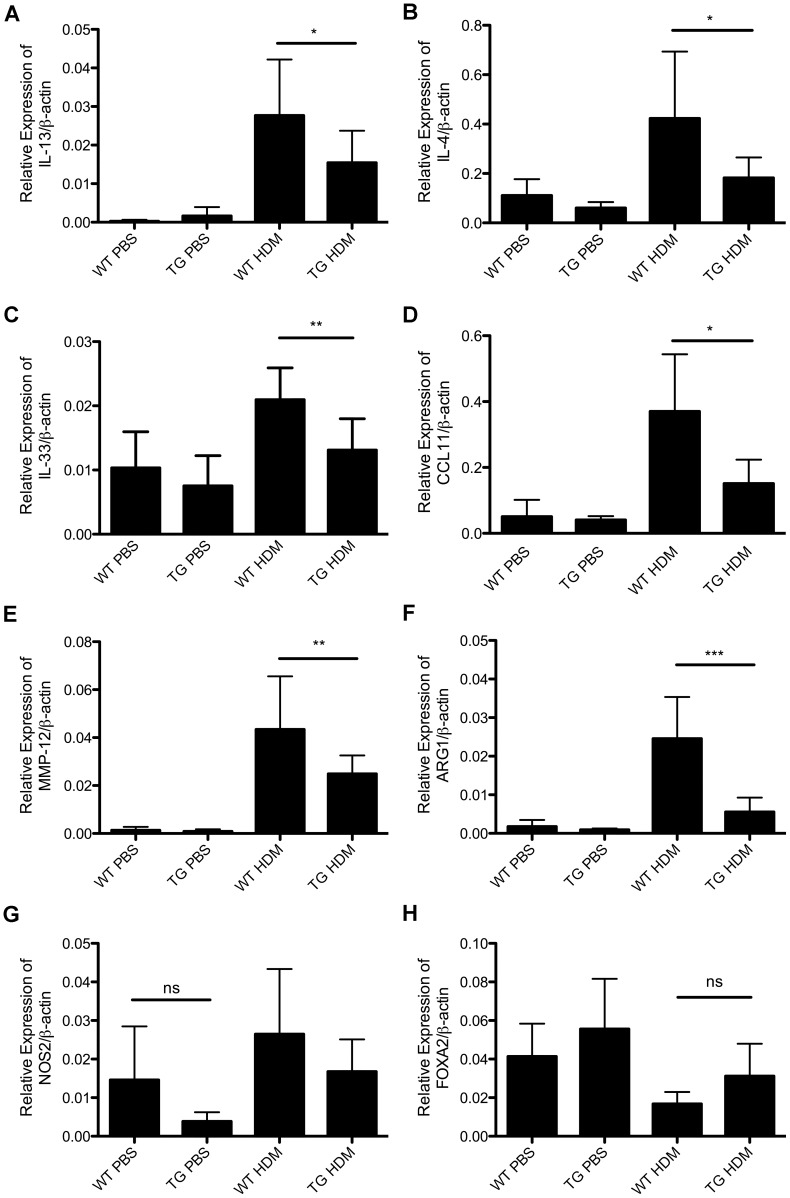
The expression of IL-13, IL-4, IL-33, CCL11, ARG1, MMP-12, CHIA, CHI3L3 and CHI3L4 is decreased in the lungs from DDAH1-transgenic mice compared to wild type mice following allergen challenge
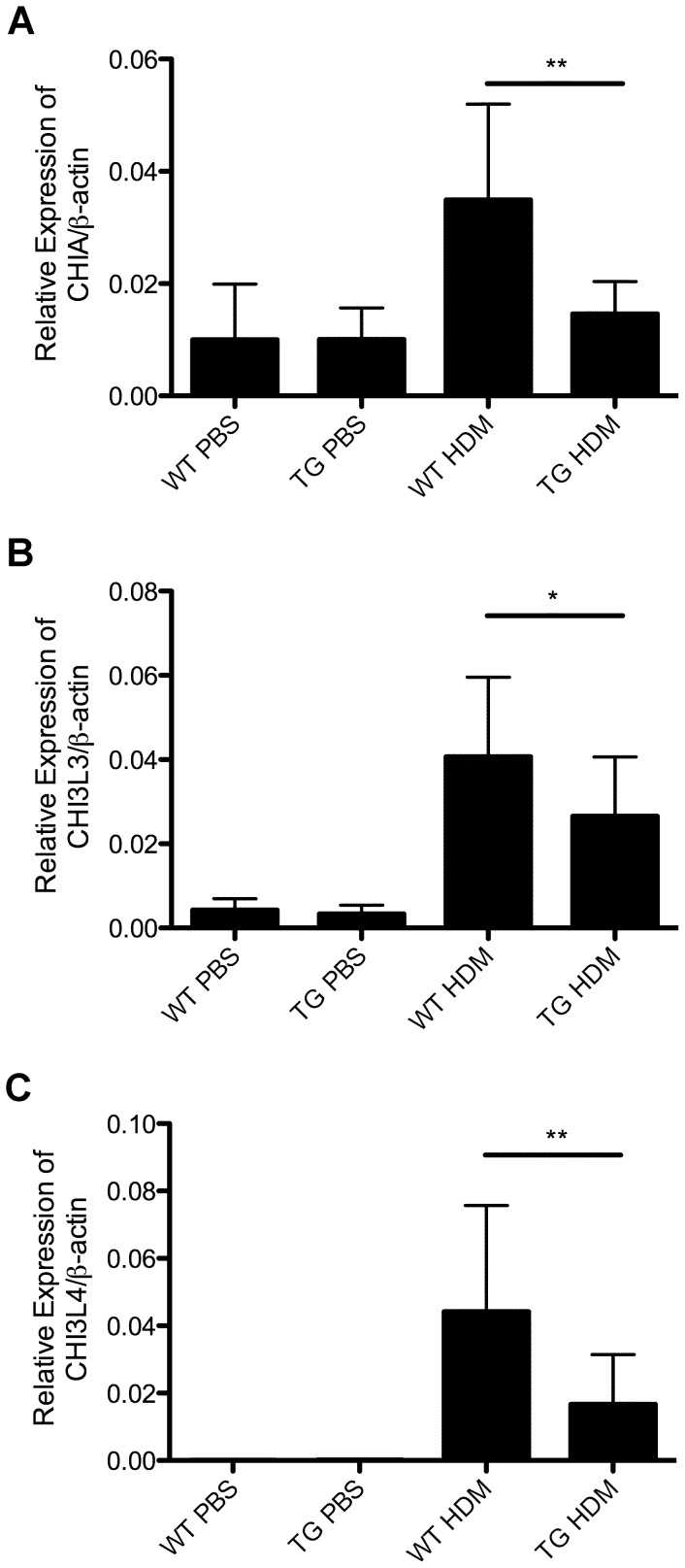
Based on RNA-Seq results, we verified the expression of some key cytokines, chemokines, matrix metallopeptidases and chitinases including IL-13, IL-4, IL-33, CCL11, MMP-12 and CHIA. As shown in Fig. 6A and 6B, induction of IL-13 and IL-4 expression in the lungs was attenuated in HDM-treated DDAH1-transgenic mice compared to wild type mice. Expression of IL-33 was decreased in lungs from DDAH1-transgenic mice after HDM treatment compared to wild type mice (Fig. 6C). We found that the expression of CCL11 and MMP-12 was decreased in HDM-treated DDAH1-transgenic mice compared to HDM-treated wild type mice (Fig. 6D, 6E). The expression of ARG1, a key enzyme in arginine/NO metabolism, was significantly decreased in HDM-treated DDAH1-transgenic mice compared to HDM-treated wild type mice (Fig. 6F). The difference in expression of iNOS (NOS2) or FOXA2 was not significant between wild type and DDAH1-transgenic mice (Fig. 6G, 6H). The expression of acidic chitinase (CHIA) and chitinase like proteins (CHI3L3 and CHI3L4) was decreased in the lungs of HDM-treated DDAH1-transgenic mice compared to HDM-treated wild type mice (Fig. 7A–7C).
Figure 6. Expression of IL-13, IL-4, IL-33, CCL11, MMP-12, ARG1, NOS2 and FOXA2 in lungs from PBS or HDM-treated C57BL/6 wild type and DDAH1-transgenic mice.
(A) IL-13. (B) IL-4. (C) IL-33. (D) CCL11 (E) MMP-12. (F) ARG1. (G) NOS2. (H) FOXA2. WT: wide type; TG: DDAH1-transgenic. Data are shown as mean±SD (n = 5-9). *, p<0.05; **, p<0.01; ***, p<0.001; ns, not significant.
Figure 7. Expression of acidic chitinase (CHIA) and chitinase like proteins (CHI3L3 and CHI3L4) in the lungs of PBS or HDM-treated C57BL/6 wild type and DDAH1-transgenic mice.
(A) CHIA. (B) CHI3L3. (C) CHI3L4. WT: wide type; TG: DDAH1-transgenic. Data are shown as mean±SD (n = 5–9). *, p<0.05; **, p<0.01.
Discussion
Our data reveal that expression of DDAH1 and DDAH2 is decreased in the lungs in a mouse model of asthma, and overexpression of DDAH1 attenuates allergen-induced airway inflammation. Asthma is a condition of decreased NO bioavailability. ADMA is an endogenous inhibitor of NOS, which is a major source of NO. DDAH1 and DDAH2 are responsible for metabolism of over 90% of ADMA in vivo, and our data support a role for DDAH downregulation in asthma pathogenesis.
The mechanism of the observed downregulation of DDAH may be epigenetic modulation. Studies using mouse trophoblast stem cells and trophoblastic tissues of postimplantation mouse embryos showed DNA methylation-dependent epigenetic regulation of DDAH2 gene expression. The CpG island in the DDAH2 promoter was hypermethylated in trophoblast stem cells but hypomethylated in differentiated cells [39]. We found that the mouse DDAH1 promoter also contains a CpG island [40], but their methylation status and relationship to gene expression are unknown. Previous studies have shown that allergen exposure results in altered methylation status of IL-4 and IFNγ promoter CpG islands [41]. It is likely that allergen exposure results in hypermethylation of mouse DDAH1 and DDAH2 promoter CpG islands, which results in decreased expression of DDAH1 and DDAH2 in mouse lungs.
Our data showed that overexpression of DDAH1 attenuated allergen-induced airway inflammation although it had no significant effect on AHR. It is not surprising as studies showed uncoupled airway inflammation and AHR in allergen challenged C57BL/6 mice due to strain-dependent genomic factors [42]. The infiltration of eosinophils into the lungs was decreased in HDM-exposed DDAH1-transgenic mice, which is consistent with the result that expression of CCL11 was decreased in lungs from HDM-treated DDAH1-transgenic mice. The total IgE and HDM-specific IgE levels in BALF or serum were decreased in DDAH1-transgenic mice after HDM exposure, suggesting that overexpression of DDAH1 affected Ig class switch.
The RNA-Seq data showed that overexpression of DDAH1 results in decreased lung expression of multiple immune/defense response genes that are associated with a network of inflammatory responses. The top upstream target is iNOS. A previous study showed that increased ADMA levels increase the expression of iNOS in mouse lungs [12], suggesting the effect of DDAH1 overexpression on iNOS expression/activity may be mediated by altered ADMA levels. Following HDM exposure, the expression of ARG1 is significantly decreased in the lungs from DDAH1 transgenic mice, suggesting arginine/NO pathways play important roles in the effect of DDAH1 overexpression on allergic airway inflammation. We found the expression of genes involved in mucus production (Clca3, Muc5ac and Muc4) and collagen synthesis (Col6a2 and Col1a1) is also decreased. Another previous study showed that increased levels of ADMA resulted in increased pulmonary collagen deposition [11], suggesting DDAH1 may regulate collagen synthesis through modulation of ADMA levels. Network analysis suggests that DDAH1 regulates mucus production gene expression through IL-13/FOXA2. RT-qPCR data further showed that the expression of IL-13, IL-4 and CCL11 is decreased in HDM-treated DDAH1-transgenic mice, which is consistent with attenuated eosinophil infiltration in airways and decreased serum and BALF IgE levels. Although the difference in expression of iNOS and FOXA2 is not significant between the wild type and DDAH1-transgenic mice, overexpression of DDAH1 may directly or indirectly affect the activity of iNOS and FOXA2. Interestingly, the expression of acidic chitinase (CHIA) and chitinase like proteins (CHI3L3 and CHI3L4) was decreased in the lungs of HDM-treated DDAH1-transgenic mice. Acidic chitinase has been shown to paly important roles in asthma [43]. CHI3L3 and CHI3L4 are rodent specific chitinase like proteins that are induced by Th2 cytokines or allergen challenge and have chemotactic activity [44]–[46]. Whether acidic chitinase and chitinase like proteins can modulate the expression of DDAH1 requires further studies. Overexpression of DDAH1 may have non-specific effects on gene expression. Generation of cell type specific transgenic mice with different expression levels of the DDAH1-transgene would help minimize the non-specific effects.
In summary, our data suggest that decreased expression of DDAH1 and DDAH2 in lungs may contribute to allergic asthma and overexpression of DDAH1 attenuates allergen-induced airway inflammation through modulation of Th2 responses.
Supporting Information
Primers for PCR.
(DOC)
Genes differentially expressed in the lungs of PBS-treated wild type and PBS-treated DDAH1-transgenic mice. For RNA-Seq, equal amounts of RNA were pooled from each mouse lung in an experimental group (n = 4 per group) and analyzed in duplicate. The bioinformatics analyses were described in Materials and Methods. The differentially expressed genes with a p-value of <0.01 were shown. WT: wild type; TG: DDAH1-transgenic; Inf: infinity.
(XLS)
Genes differentially expressed in the lungs of HDM-treated wild type and HDM-treated DDAH1-transgenic mice. For RNA-Seq, equal amounts of RNA were pooled from each mouse lung in an experimental group (n = 4 per group) and analyzed in duplicate. The bioinformatics analyses were described in Materials and Methods. The differentially expressed genes with a p-value of <0.01 were shown. WT: wild type; TG: DDAH1-transgenic; Inf: infinity.
(XLS)
Acknowledgments
The authors thank Dr. Xiang Zhang for help with RNA-Seq.
Funding Statement
This work was supported by National Institutes of Health grants NIAID R01-AI058157 (GKKH), NIAMS R01-AR054490 (GKKH), NHLBI P01-HL076383 (GKKH), NIAID R56-AI084414 (GKKH) and Center for Environmental Genetics NIEHS P30-ES006096 (WC). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Warrier MR, Hershey GK (2008) Asthma genetics: personalizing medicine. J Asthma 45: 257–264. [DOI] [PubMed] [Google Scholar]
- 2. Malerba G, Pignatti PF (2005) A review of asthma genetics: gene expression studies and recent candidates. J Appl Genet 46: 93–104. [PubMed] [Google Scholar]
- 3. Lara A, Khatri SB, Wang Z, Comhair SA, Xu W, et al. (2008) Alterations of the arginine metabolome in asthma. Am J Respir Crit Care Med 178: 673–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zimmermann N, Rothenberg ME (2006) The arginine-arginase balance in asthma and lung inflammation. Eur J Pharmacol 533: 253–262. [DOI] [PubMed] [Google Scholar]
- 5. Ricciardolo FL, Sterk PJ, Gaston B, Folkerts G (2004) Nitric oxide in health and disease of the respiratory system. Physiol Rev 84: 731–765. [DOI] [PubMed] [Google Scholar]
- 6. Vallance P, Leone A, Calver A, Collier J, Moncada S (1992) Accumulation of an endogenous inhibitor of nitric oxide synthesis in chronic renal failure. Lancet 339: 572–575. [DOI] [PubMed] [Google Scholar]
- 7. Schmidt K, Klatt P, Mayer B (1993) Characterization of endothelial cell amino acid transport systems involved in the actions of nitric oxide synthase inhibitors. Mol Pharmacol 44: 615–621. [PubMed] [Google Scholar]
- 8. Bogle RG, Moncada S, Pearson JD, Mann GE (1992) Identification of inhibitors of nitric oxide synthase that do not interact with the endothelial cell L-arginine transporter. Br J Pharmacol 105: 768–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bulau P, Zakrzewicz D, Kitowska K, Leiper J, Gunther A, et al. (2007) Analysis of methylarginine metabolism in the cardiovascular system identifies the lung as a major source of ADMA. Am J Physiol Lung Cell Mol Physiol 292: L18–24. [DOI] [PubMed] [Google Scholar]
- 10. Smith CL, Anthony S, Hubank M, Leiper JM, Vallance P (2005) Effects of ADMA upon gene expression: an insight into the pathophysiological significance of raised plasma ADMA. PLoS Med 2: e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wells SM, Buford MC, Migliaccio CT, Holian A (2009) Elevated asymmetric dimethylarginine alters lung function and induces collagen deposition in mice. Am J Respir Cell Mol Biol 40: 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein E, Weigel J, Buford MC, Holian A, Wells SM (2010) Asymmetric Dimethylarginine Potentiates Lung Inflammation in a Mouse Model of Allergic Asthma. Am J Physiol Lung Cell Mol Physiol. [DOI] [PMC free article] [PubMed]
- 13. Sott JA, North ML, Rafii M, Huang H, Pencharz P, et al. (2011) Asymmetric dimethylarginine is increased in asthma. Am J Respir Crit Care Med 184: 779–785. [DOI] [PubMed] [Google Scholar]
- 14. Di Gangi IM, Pirillo P, Carraro S, Gucciardi A, Naturale M, et al. (2012) Online trapping and enrichment ultra performance liquid chromatography-tandem mass spectrometry method for sensitive measurement of “arginine-asymmetric dimethylarginine cycle” biomarkers in human exhaled breath condensate. Anal Chim Acta 754: 67–74. [DOI] [PubMed] [Google Scholar]
- 15.Carraro S, Giordano G, Piacentini G, Kantar A, Moser S, et al.. (2013) Asymmetric Dimethylarginine (Adma) in Exhaled Breath Condensate and Serum of Asthmatic Children. Chest. [DOI] [PubMed]
- 16. Holguin F, Comhair SA, Hazen SL, Powers RW, Khatri SS, et al. (2013) An association between L-arginine/asymmetric dimethyl arginine balance, obesity, and the age of asthma onset phenotype. Am J Respir Crit Care Med 187: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Palm F, Onozato ML, Luo Z, Wilcox CS (2007) Dimethylarginine dimethylaminohydrolase (DDAH): expression, regulation, and function in the cardiovascular and renal systems. Am J Physiol Heart Circ Physiol 293: H3227–3245. [DOI] [PubMed] [Google Scholar]
- 18. Kimoto M, Miyatake S, Sasagawa T, Yamashita H, Okita M, et al. (1998) Purification, cDNA cloning and expression of human NG,NG-dimethylarginine dimethylaminohydrolase. Eur J Biochem 258: 863–868. [DOI] [PubMed] [Google Scholar]
- 19. Leiper JM, Santa Maria J, Chubb A, MacAllister RJ, Charles IG, et al. (1999) Identification of two human dimethylarginine dimethylaminohydrolases with distinct tissue distributions and homology with microbial arginine deiminases. Biochem J 343 Pt 1: 209–214. [PMC free article] [PubMed] [Google Scholar]
- 20. Tran CT, Fox MF, Vallance P, Leiper JM (2000) Chromosomal localization, gene structure, and expression pattern of DDAH1: comparison with DDAH2 and implications for evolutionary origins. Genomics 68: 101–105. [DOI] [PubMed] [Google Scholar]
- 21. Pullamsetti S, Kiss L, Ghofrani HA, Voswinckel R, Haredza P, et al. (2005) Increased levels and reduced catabolism of asymmetric and symmetric dimethylarginines in pulmonary hypertension. FASEB J 19: 1175–1177. [DOI] [PubMed] [Google Scholar]
- 22. Leiper J, Nandi M, Torondel B, Murray-Rust J, Malaki M, et al. (2007) Disruption of methylarginine metabolism impairs vascular homeostasis. Nat Med 13: 198–203. [DOI] [PubMed] [Google Scholar]
- 23. Hu X, Atzler D, Xu X, Zhang P, Guo H, et al. (2011) Dimethylarginine dimethylaminohydrolase-1 is the critical enzyme for degrading the cardiovascular risk factor asymmetrical dimethylarginine. Arterioscler Thromb Vasc Biol 31: 1540–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dayoub H, Achan V, Adimoolam S, Jacobi J, Stuehlinger MC, et al. (2003) Dimethylarginine dimethylaminohydrolase regulates nitric oxide synthesis: genetic and physiological evidence. Circulation 108: 3042–3047. [DOI] [PubMed] [Google Scholar]
- 25. Jacobi J, Sydow K, von Degenfeld G, Zhang Y, Dayoub H, et al. (2005) Overexpression of dimethylarginine dimethylaminohydrolase reduces tissue asymmetric dimethylarginine levels and enhances angiogenesis. Circulation 111: 1431–1438. [DOI] [PubMed] [Google Scholar]
- 26. Dayoub H, Rodionov RN, Lynch C, Cooke JP, Arning E, et al. (2008) Overexpression of dimethylarginine dimethylaminohydrolase inhibits asymmetric dimethylarginine-induced endothelial dysfunction in the cerebral circulation. Stroke 39: 180–184. [DOI] [PubMed] [Google Scholar]
- 27. Lee GB, Brandt EB, Xiao C, Gibson AM, Le Cras TD, et al. (2013) Diesel exhaust particles induce cysteine oxidation and s-glutathionylation in house dust mite induced murine asthma. PLoS One 8: e60632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brandt EB, Kovacic MB, Lee GB, Gibson AM, Acciani TH, et al.. (2013) Diesel exhaust particle induction of IL-17A contributes to severe asthma. J Allergy Clin Immunol. 2013 Sep 20. doi:pii: S0091-6749(13)01208-6. 10.1016/j.jaci.2013.06.048. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 29. Kramer EL, Mushaben EM, Pastura PA, Acciani TH, Deutsch GH, et al. (2009) Early growth response-1 suppresses epidermal growth factor receptor-mediated airway hyperresponsiveness and lung remodeling in mice. Am J Respir Cell Mol Biol 41: 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen W, Ericksen MB, Levin LS, Khurana Hershey GK (2004) Functional effect of the R110Q IL13 genetic variant alone and in combination with IL4RA genetic variants. J Allergy Clin Immunol 114: 553–560. [DOI] [PubMed] [Google Scholar]
- 31. Sivaprasad U, Warrier MR, Gibson AM, Chen W, Tabata Y, et al. (2010) IL-13Ralpha2 has a protective role in a mouse model of cutaneous inflammation. J Immunol 185: 6802–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Morgan M, Anders S, Lawrence M, Aboyoun P, Pages H, et al. (2009) ShortRead: a bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics 25: 2607–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ihaka R, Gentleman R (1996) R: A Language for Data Analysis and Graphics. J Comput Graph Stat 5: 299–314. [Google Scholar]
- 35. Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Storey JD, Tibshirani R (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sartor MA, Leikauf GD, Medvedovic M (2009) LRpath: a logistic regression approach for identifying enriched biological groups in gene expression data. Bioinformatics 25: 211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schwedhelm E, Tan-Andresen J, Maas R, Riederer U, Schulze F, et al. (2005) Liquid chromatography-tandem mass spectrometry method for the analysis of asymmetric dimethylarginine in human plasma. Clin Chem 51: 1268–1271. [DOI] [PubMed] [Google Scholar]
- 39. Tomikawa J, Fukatsu K, Tanaka S, Shiota K (2006) DNA methylation-dependent epigenetic regulation of dimethylarginine dimethylaminohydrolase 2 gene in trophoblast cell lineage. J Biol Chem 281: 12163–12169. [DOI] [PubMed] [Google Scholar]
- 40. Sujuan Y, Asaithambi A, Liu Y (2008) CpGIF: an algorithm for the identification of CpG islands. Bioinformation 2: 335–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kwon NH, Kim JS, Lee JY, Oh MJ, Choi DC (2008) DNA methylation and the expression of IL-4 and IFN-gamma promoter genes in patients with bronchial asthma. J Clin Immunol 28: 139–146. [DOI] [PubMed] [Google Scholar]
- 42. Kelada SN, Wilson MS, Tavarez U, Kubalanza K, Borate B, et al. (2011) Strain-dependent genomic factors affect allergen-induced airway hyperresponsiveness in mice. Am J Respir Cell Mol Biol 45: 817–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhu Z, Zheng T, Homer RJ, Kim YK, Chen NY, et al. (2004) Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science 304: 1678–1682. [DOI] [PubMed] [Google Scholar]
- 44. Chang NC, Hung SI, Hwa KY, Kato I, Chen JE, et al. (2001) A macrophage protein, Ym1, transiently expressed during inflammation is a novel mammalian lectin. J Biol Chem 276: 17497–17506. [DOI] [PubMed] [Google Scholar]
- 45. Webb DC, McKenzie AN, Foster PS (2001) Expression of the Ym2 lectin-binding protein is dependent on interleukin (IL)-4 and IL-13 signal transduction: identification of a novel allergy-associated protein. J Biol Chem 276: 41969–41976. [DOI] [PubMed] [Google Scholar]
- 46. Welch JS, Escoubet-Lozach L, Sykes DB, Liddiard K, Greaves DR, et al. (2002) TH2 cytokines and allergic challenge induce Ym1 expression in macrophages by a STAT6-dependent mechanism. J Biol Chem 277: 42821–42829. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers for PCR.
(DOC)
Genes differentially expressed in the lungs of PBS-treated wild type and PBS-treated DDAH1-transgenic mice. For RNA-Seq, equal amounts of RNA were pooled from each mouse lung in an experimental group (n = 4 per group) and analyzed in duplicate. The bioinformatics analyses were described in Materials and Methods. The differentially expressed genes with a p-value of <0.01 were shown. WT: wild type; TG: DDAH1-transgenic; Inf: infinity.
(XLS)
Genes differentially expressed in the lungs of HDM-treated wild type and HDM-treated DDAH1-transgenic mice. For RNA-Seq, equal amounts of RNA were pooled from each mouse lung in an experimental group (n = 4 per group) and analyzed in duplicate. The bioinformatics analyses were described in Materials and Methods. The differentially expressed genes with a p-value of <0.01 were shown. WT: wild type; TG: DDAH1-transgenic; Inf: infinity.
(XLS)