Abstract
Mapping kinase-substrate interactions demands robust methods to rapidly and unequivocally identify substrates from complex protein mixtures. Towards this goal we present a method in which a kinase, engineered to utilize synthetic ATPγS analogs, specifically thiophosphorylates its substrates in a complex lysate. The thiophosphate label provides a bio-orthogonal tag that can be used to affinity purify and identify labeled proteins. Following the labeling reaction proteins are digested with trypsin, thiol containing peptides are then covalently captured and non-thiol containing peptides are washed from the resin. Oxidation promoted hydrolysis, at sites of thiophosphorylation, releases phosphopeptides for analysis by tandem mass spectrometry. By incorporating two specificity gates: kinase engineering and peptide affinity purification, this method yields high confidence substrate identifications. This method gives both the identity of the substrates and phosphorylation site localization. With this information investigators can analyze the biological significance of the phosphorylation mark immediately following confirmation of the kinase-substrate relationship. Here we provide an optimized version of this technique to further enable widespread utilization of this technology.
Keywords: phosphorylation, chemical genetics, analog specific kinase, kinase substrate identification, thiophosphate labeling
Introduction
Members of the kinase superfamily are integral components of many signaling pathways. Kinases play pivotal roles in regulating growth and development and are misregulated in many diseases including cancer. Kinases transduce extracellular signals by altering the functions of substrate proteins through phosphorylating specific sites on these proteins. It is estimated that a third of all proteins are phosphorylated, and therefore regulated by kinases, however there are only 518 known human kinases (Cohen, 2001; Manning et al., 2002). Therefore many kinases must phosphorylate multiple substrates. The mapping of these relationships is complicated by the shared enzymology of kinases and therefore it is extremely difficult to assign a phospho-site to a specific kinase.
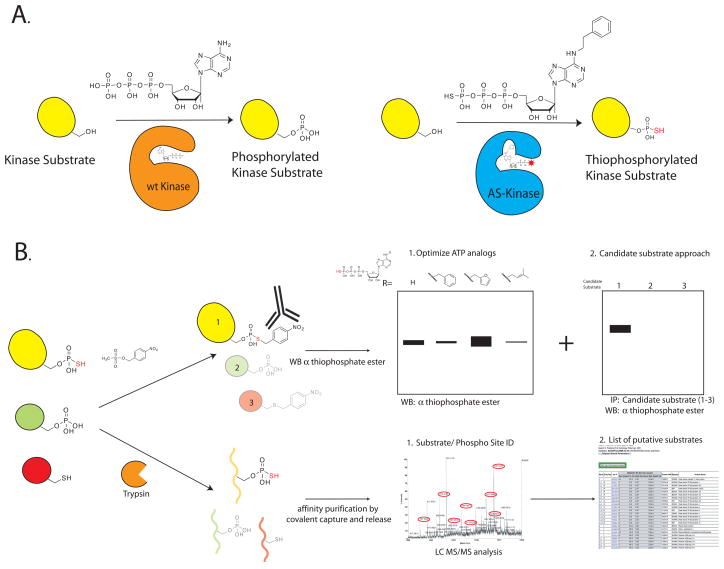
We have developed bio-orthogonal chemical reactions that allow the assignment of a particular phospho-site to a specific kinase (Bishop et al., 2000; Shah et al., 1997). In this approach the active site of the kinase of interest (KOI) is engineered by creating a new active site pocket that allows the kinase to accept a bulky ATP analog (Figure 1A). This engineered “lock and key” provides selectivity for the KOI, as the engineered kinase will accept the N6 substituted ATP analog whereas the vast majority of wild-type kinases cannot use these analogs.
Figure 1.
Thiophosphopeptide purification scheme. A. wt kinase utilizes ATP to phosphorylate its substrates. In contrast, an engineered AS kinase has a new active site pocket that allows it to accept an unnatural bulky ATP analog. This “lock and key” allows the AS kinase to transfer the thiophospho group to its substrates. B. Detail of the two different affinity purification methods: The thiophosphorylated substrates are found in a background of phosphorylated and unlabeled proteins. In the first technique we react the thiophosphorylated proteins with a thiol specific alkylating agent that generates a bio-orthogonal thiophosphate ester. Labeled proteins are detected by using a thiophosphate ester specific antibody. In the second approach the lysate is first digested to generate tryptic peptides. Thiol containing peptides are captured by reaction with iodoacetyl agarose beads. All non-thiol containing peptides are then washed away, and the remaining peptides are treated with Oxone, which releases thiophosphate ester linked peptides by spontaneous hydrolysis.
In order to identify the substrates of a particular enzyme, many of the most widespread proteomic methods rely on affinity based approaches to enrich for proteins of interest. To apply this approach to kinase-substrate mapping, we developed a modified gamma-phosphate analog (thiophosphate) capture and release strategy for thiophosphate purification. The ATP analog used to identify direct kinase substrates is modified in two ways: a bulky group is added in the N6 position providing exclusive recognition by the engineered kinase of interest, and the gamma phosphate is replaced with a thiophosphate moiety (Figure 1). The modified analog-specific (AS) kinase will use these analogs most efficiently, therefore the substrates of the AS kinase will be uniquely labeled with the thiophosphate affinity tag. This tag can then be used to purify and identify the tagged proteins (Figure 1B).
We have recently published two different approaches for the affinity purification of thiophosphorylated substrates (Allen et al., 2007; Blethrow et al., 2008). In this report we provide an updated protocol for the application of these techniques, and demonstrate how they can be used in a complementary manner to rapidly identify the substrates of one kinase from a complex protein mixture.
In the first technique we react the thiophosphorylated proteins with a thiol specific alkylating agent that generates a bio-orthogonal thiophosphate ester. We affinity purify the tagged proteins by using a thiophosphate ester specific antibody 51-8 (Allen et al., 2005; Allen et al., 2007) also see Figure 2B. The antibody is able to distinguish between a thiophosphate ester and a cysteine thioether, and in this way we are able to specifically visualize and identify tagged substrates by immunoblotting (IB) and immunoprecipitation (IP). This approach cannot reliably allow for the identification of the site of phosphorylation because the alkylating agent can prevent normal collision induced dissociation of the modified peptide. Given that it is currently difficult to produce the monoclonal antibody 51-8 in sufficient amounts for IP, we now routinely use this technique for two optimization steps. 1. To determine the optimal N6 substituted ATPγS analog for a KOI and 2. To optimize N6 substituted ATPγS, ATP, and GTP concentrations to achieve the lowest background labeling in cell lysates. Once labeling conditions are optimized, a candidate protein may also be verified as a KOI substrate by immunoprecipitating the candidate substrate and immunoblotting with monoclonal antibody 51-8.
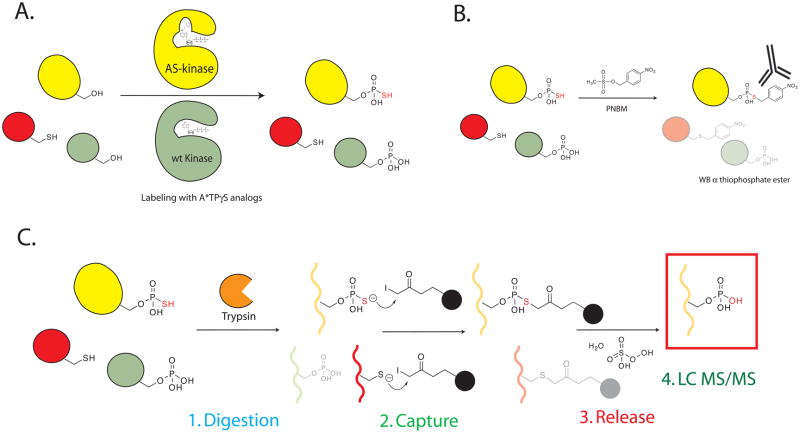
Figure 2.
A. Labeling of a cell lysate by incubating AS kinase and N6 substituted ATPγS analogs generates thiophosphorylated and phosphorylated proteins. B. Alkylating thiol containing proteins generates the bio-orthogonal thiophosphate ester. Thiophosphate ester labeled proteins are detected by utilizing the thiophosphate ester specific antibody (clone 51–8). C. Thiol containing peptides are captured by reaction with iodoacetyl beads. During washing all other non-thiol containing peptides are washed away. The bound peptides are treated with Oxone to convert the thiol group to a sulfoxide. The presence of an electrophilic phosphoester in the thiophosphate ester linked peptides catalyzes the spontaneous hydrolysis of these peptides, and after concentration these peptides are analyzed by tandem LC MS/MS.
After optimization of labeling conditions using the thiophosphate specific antibody, our second technique provides unambiguously assigned sites of modification on substrates of the engineered kinase. In this technique, the modified peptides are directly affinity purified, and then subjected to tandem mass spectrometry to identify the parent protein and to assign phosphorylation sites (Blethrow et al., 2008) also see Figure 2B. To accomplish the affinity purification, we first digest the labeled protein lysate and then covalently capture the peptides by reacting the digested peptides with iodoacetyl agarose beads, thus capturing all thiol containing peptides. Unbound peptides are washed away, and the beads are then treated with Oxone to oxidize the sulfur in the thiophosphate ester to a sulfoxide. The peptides linked by a thiophosphate ester bond to the agarose beads are then released by spontaneous hydrolysis, while those linked by a cysteine thioether are retained on the resin. In this protocol we provide the most updated and optimized form of these techniques and provide detailed positive and negative controls to ensure their successful implementation by any lab.
Strategic Planning
Engineering the kinase of interest
To apply this method the KOI must be able to catalytically transfer thiophosphate to substrate proteins, and tolerate the mutation of the normally bulky gatekeeper to a smaller residue. We believe most kinases will be able to use ATPγS as a phosphorylation cofactor; in preliminary experiments 13 out of 15 kinases were able to use ATPγS to thiophosphorylate substrate proteins (Allen et al., 2007). To determine whether a KOI can transfer thiophosphate, the thiophosphate ester specific antibody is used to rapidly determine the thiophosphorylation state of a substrate protein after incubation with ATPγS and the KOI. The first step is therefore to set up an in vitro kinase reaction with the KOI, substrate protein and ATPγS followed by IB with the thiophosphate ester specific antibody (Support Protocol 1). The bioinformatic technique for identifying the gatekeeper as well as the generation of an AS kinase have been described previously (Blethrow et al., 2004; Buzko and Shokat, 2002; Gregan et al., 2007) As described in (Buzco and Shokat, 2002), the kinase database is a online tool that can be used to easily find the gatekeeper residue for their kinase (http://sequoia.ucsf.edu/ksd/). After identifying the gatekeeper residue (equivalent to I338 in v-Src) conventional site directed mutagenesis techniques can be used to mutate the gatekeeper to either a glycine (analog sensitive-1, as1) or alanine (analog-sensitive-2, as2). These space creating mutations will allow the kinase to utilize bulky ATP analogs (Buzko and Shokat, 2002; Gregan et al., 2007).
Certain protein kinases lose catalytic activity upon mutation of their gatekeeper residue to Gly or Ala. In such cases, second-site mutations can be used to rescue activity of the kinase [Zhang, Nature Methods 2005]. Identification of second-site susppressor mutations have been successfully executed through bioinformatic analysis or genetic selection. Mutations at a few positions were found to rescue activity in mutltiple kinases and thus can be considered as top candidate sites for rescue mutations [CZ, KS unpublished results]. For example, one position is immediately N-terminal from the conserved DFG motif (DFG-1) and Ala was found beneficial for kinases activity this position. If the natural residue in a kinase is different from Ala at DFG-1, one can change it to Ala attempting to rescue activity of the kinase. Another position is typically 11 residues N-terminal from the DFG motif (DFG-11) and Leu was found beneficial for kinase activity in general here. If different from Leu, the residue at DFG-11 could be mutated to Leu for rescue of activity. It should be noted that successful rescue mutations varies from kinase to kinase and that there seems to be no universal rescue mutation as of now. Once the AS kinase protein has been expressed and purified, the kinase assay is repeated to identify the N6 substituted ATP analog that is preferentially utilized by the AS-KOI (Support Protocol 1a and Figure 1B).
Lysate Optimization
Optimization of N6 substituted ATPγS analog concentration in each specific lysate is also required to ensure the lowest levels of background and therefore the highest confidence in the identified substrates. There are several different parameters required to optimize the labeling conditions. Although the N6 substituted ATPγS analogs are preferentially used by the engineered AS kinase, at high enough concentrations other kinases can also utilize these analogs. Providing sufficient unmodified ATP and GTP ensure that these kinases are occupied and will therefore decrease the non-specific use of the N6 substituted ATPγS analogs. In addition, each lysate will have a different N6 substituted ATPγS analog that will produce the lowest level of background. The conditions that give the highest signal-to-noise should be determined empirically by varying the levels of AS kinase, N6 substituted ATPγS analog, ATP, and GTP in the presence of AS kinase. Utilizing the thiophosphate ester specific antibody to detect background thiophosphorylated proteins is the best method for lysate optimization. The amount of kinase is critical to the efficiency of the labeling. When using a recombinant form of the AS kinase, up to 1% (by total weight) of the AS kinase can be added. For example 10 μg of AS kianse in 1 mg of total lysate (100 μl of 10 mg/mL lysate). If using a transfected cell than the level of kinase can be varied by using different expression vectors, but usually the highest level of expression is necessary to achieving the best signal to noise. Varying the levels of ATP (50–200 μM), GTP (1–3 mM), and N6 substituted ATPγS analog (100–500 μM) should give an optimal set of specific conditions in which the lowest background and the highest levels of AS specific labeling is observed (Support Protocol 2).
Generating positive control peptides and protein
We recommend generating a hyper-thiophosphorylated control protein (myelin basic protein (MBP)) and control peptide (GSK3 substrate peptide CREB) which can be accomplished using commercially available materials (Support Protocol 3). The labeled protein will serve as a control for the retention of the thiophosphate modification during the digestion steps, and both controls will serve to ensure that the covalent capture, release, and mass spectrometric analysis of thiophosphorylated peptides are working properly.
During the labeling procedure two negative controls are essential; minus N6 substituted ATPγS analog and minus AS kinase. These controls will provide information about which background substrates are detected and therefore excluded from the list of identified substrates. Using these controls to troubleshoot and optimize each step of the procedure is critical to the success of the protocol.
Basic Protocol: Digestion and Covalent Capture of Thiophosphorylated Peptides
Materials
Solutions and Reagents:
2X Denaturation Buffer: 200 mM NH4HCO3, 4 mM EDTA store indefinitely at 22°C
99% pure Urea store indefinitely at 22°C
Siliconized microcentrifuge tubes
1 M tris 2-carboxyethyl phosphine (TCEP) in H2O store for 6 month at −80°C
1 M Dithiothreitol (DTT) store for 6 months at −80°C
Iodoacetyl Agarose Beads (Pierce) store for 6 months at 4°C
200 mM HEPES pH 7.0 store for 4 months at 22°C
50% Acetonitrile 50% 20 mM HEPES pH 7.0 store for 4 months at 22°C
Ziptips (10 μL and 100 μL) store for indefinitely at 22°C
Small Disposable Columns (Isolute SPE Accessories Double Fritted Column 120–1021-A and
Single Fritted Res 120-1111-A) Biotage store for indefinitely at 22°C
5 M NaCl store for indefinitely at 22°C
5% Formic acid store for 3 months at 22°C
50% Acetonitrile: 50% H2O store for indefinitely at 22°C
C-18 Sep Pak (Waters) store for indefinitely at 22°C in dessicator
Oasis SPE (Waters)
0.1 % TFA 50 % Acetonitrile H20 store for indefinitely at 22°C
0.1 % TFA H20 store for indefinitely at 22°C
Special Equipment:
QSTAR Elite Mass spectrometer (Applied Biosystems Foster City, Ca.) or another Tandem LC
MS/MS capable mass spectrometer
Step 1. Digestion of labeled protein lysate
In addition to the experimental samples, we recommend the following five control samples:
100 pmol of hyper-thiophosphorylated MBP (see Support Protocol 3)
Unlabeled lysate.
Unlabeled lysate plus 100 pmol hyper thiophosphorylated MBP.
Lysate plus ATPγS analog minus kinase.
-
An additional covalent capture reaction control (see step 2-1) and the experimental sample: Lysate plus ATPγS analog plus kinase.
These controls will ensure that all the steps of this protocol are working correctly. The MBP alone and MBP plus lysate will lead to recovery of only one peptide (see Expected Results Section for MW). The lysate controls will provide a list of non-specific substrates.
-
1
Prepare 500 μL complete (1X) denaturation buffer (8M Urea, 10 mM TCEP, 100 mM
NH4HCO3,2 mM EDTA) fresh by mixing the following:
250 μL 2X Denaturation Buffer
5μL 1 M TCEP
68μL H2O
240 mg Urea
Making the denaturation buffer fresh will limit carbamate formation and will ensure the TCEP is fully reduced. Using a non-thiol reducing agent will reduce non-peptide side reactions with the iodoacetyl beads. Performing the digestion in siliconized tubes will ensure that minimal sample is lost to non-specific adsorption to the tube walls. -
2
Add 1X denaturation buffer to the labeled lysate or to the sample of labeled MBP such that the final concentration is 6 M Urea (20 μL of lysate should be dissolved in 60 μL of 1X buffer) : Incubate the sample at 55°C for 1 hour then cool the sample at room temperature for 10 minutes.
Do not incubate the sample with an alkylating agent such as iodoacetamide. This will react with the thiophosphate group thereby destroying the reactivity of the tag. -
3
Dilute the sample by adding 50 mM NH4HCO3 such that the final concentration of Urea is 2 M and add 1 M TCEP to a final concentration of 10 mM. Then add trypsin in a 1:50-1:10 ratio by weight (1:10 would be 10 μg trypsin for 100 μg of lysate protein).
As no iodoacetamide is used in this protocol, high concentrations of urea (2 M) and reducing agent (10 mM TCEP) are used to ensure that proteins in the lysate remain unfolded during the digestion. One easy way to optimize the amount of trypsin and the incubation time is to add BSA to the lysate and then to analyze the digested peptides to determine the sequence coverage. Coverage of at least 60–80% of BSA or around 30–35 peptides should be accomplished. -
4
After incubation at 37°C for 6 hrs to overnight, acidify the sample by adding 2.5% TFA to attain 0.1% TFA
The thiophosphate modification is very labile to acid-promoted hydrolysis so do not acidify to 2% TFA as is common. -
5
Wash a C-18 Sep Pak (or a Oasis HLB) with 10 mL of 0.1% TFA 50% Acetonitrile:H2O, followed by 10 mL of 0.1% TFA:H2O. Load the acidified sample to the column and pass it through the column 5 times. Wash the column with 10 mL 0.1% TFA:H2O. Elute the peptides with 1 mL 0.1% TFA in 50% Acetonitrile:H2O. Concentrate to near dryness using a speed vacuum.
Different Solid Phase Extraction columns can lead to differential retention of peptides. We have seen success with both types of column (the more hydrophobic C-18 as well as the more hydrophilic Oasis HLB type column) however each lysate will require optimization. Do not take the sample to dryness, as the peptides may be difficult to resuspend into solution. The peptides should be in around 50 μL after evaporation so that the correct reaction volume can be attained.
Step 2. Covalent Capture of Thiophosphorylated Peptides
-
6
Prepare the Iodoacetyl agarose beads for each sample by pipeting 100 μL of the 50% resin slurry into a siliconized 0.5 mL tube. Spin on a tabletop centrifuge for 30 seconds at 10,000 x g and remove the supernatant. Wash with 200 μL 200 mM HEPES pH 7.0, spin and remove supernatant. Add 150 μL 50% Acetonitrile 50% 20 mM HEPES pH 7.0 and add 5 μL 5 mg/mL BSA, mix by vortexing briefly and block the beads for ten minutes in the dark.
The beads are supplied in an acidic buffer; washing with 200 mM HEPES pH 7.0 is critical to achieving the correct final pH. The addition of BSA helps by blocking the agarose beads and prevents sample loss by non-specific adsorption. Mixing the slurry for 5 minutes with gentle rocking before pipeting should mix the slurry to homogeneity.
2-1. In this step set up an additional covalent capture reaction by directly adding several pmols of the thiophosphorylated CREB peptide to the beads in 50% Acetonitrile 50% 20 mM HEPES pH 7.0 with BSA. (10–50 pmol should give a very robust signal)
-
7
Adjust the pH of the digested labeled peptide mixture by adding 200 mM HEPES pH 7.0 to a final concentration of 20 mM (for example add 15 μL 200 mM HEPES pH 7.0, 10 μL H2O and 75μL acetonitrile to 50 μL digested peptides for a final volume of 150 μL). Spin the beads down again, remove the supernatant and add each sample to one tube of bead mixture followed by room temperature incubation in the dark with gentle rocking for 12–16 hours.
Adding the additional labeled peptide control at this point will simplify troubleshooting later. To prevent light from generating radicals within the sample we recommend wrapping the tubes in aluminum foil. -
8
Prepare one disposable column for washing and elution of each reaction by washing with 1 mL 50% Acetonitrile: 50% H2O and 1 mL H2O.
At this point two methods can be used, a column method or a spin method. The protocol is the same in each case however the column method is detailed below. For the spin method all the washes should be performed by addition of the same volume followed by a short 30 second spin at 10,000 x g. -
9
Add the entire reaction to the top of the column and let it drain into a 1.5 mL microcentrifuge tube. Wash the reaction tube with 100 μL 50% Acetonitrile 50% 20 mM HEPES pH 7.0 being sure to resuspend all the beads, and add the residual beads to the column.
-
10
Wash the beads by adding 1 mL of each wash liquid to the top of each column. Then push through the wash liquid using a P1000 pipet.
Wash the beads with 1 mL each in the following order:
H2O
5 M NaCl
50% Acetonitrile
-
5% Formic Acid
If the level of wash liquid drops below the top of the beads the recovery of phosphopeptides is severely reduced. In addition, the order of washing is critical to the success of the washing steps.
-
11
Prepare a solution of 10 mM DTT and add 1 mL to the column, allowing it to drain half way, and then incubating for 10 minutes with the solution in the column.
The addition of DTT to the beads will react with any available iodoacetyl groups left on the beads and will ensure that no I2 forms during the oxidation step which can iodinate tyrosine residues (unpublished data). -
12
Prepare a fresh solution of 1 mg/mL Oxone pH ~3.5 in water and add 100 μL of this solution to the column being sure to resuspend the beads. After draining the column, add an additional 100 μL and incubate for 10 minutes.
Be sure to resuspend the beads at this point to ensure the complete oxidation of the thiophosphate ester. -
13
Immediately desalt and concentrate the phosphopeptides with a 10 μL C-18 ziptip. Wash the ziptip 3X with 0.1 % TFA 50 % acetonitrile H20, then 3X with 0.1 % TFA H20. Attach the ziptip to the end of a P200 tip and draw the sample through the tip using the P200 on a setting of 100 μL passing the sample through the ziptip a total of 4 times. Wash 2X with 20 μL 0.1 % TFA H20 and elute by washing 3X with 20 μL of 0.1 % TFA 50 % Acentonitrile H20.
Passing the entire sample through the ziptip multiple times ensures the retention of low abundance peptides. The 10 μL ziptip should fit on the end of a P200 or P1000 tip to simplify the passage of the entire sample across the C-18 column. -
14
Concentrate the sample to 10 μL on a speed vacuum and analyze 5 μL by tandem mass spectrometry. Typically we use a QSTAR ELITE that includes coupled liquid chromatography on a reverse phase C18 column utilizing a 3–32% acetonitrile gradient in 0.1% formic acid. Analyze the phosphopeptides in positive ion mode. MS spectra are acquired for 1 s. For each MS spectrum the two most intense multiply charged peaks are selected for generation of subsequent collision-induced dissociation MS.
This analysis will provide the identity of both the substrate protein and the phosphorylation site. -
15
Analyze the data by centroiding using Analyst QS software or another appropriate software. Search the MS/MS spectra against the entire UniProt database utilizing Protein Prospector or another appropriate software. Typically we select no constant modifications and allow the following variable modifications: Phospho: Serine, Threonine, Oxidized: Methionine.
Protein Prospector is available free of charge online: http://prospector.ucsf.edu
Support protocols
Materials
N-6 substituted Adenosine 5′-[γ-thio]triphosphate (exclusively from Biolog: www.biolog.de), store 10 mM stock in aliquots at −80C for one year and avoid freeze thaw cycles p-nitrobenzyl mesylate (PNBM) (exclusively from Epitomics: www.epitomics.com) store solid for 1 year at 4°C. A 50 mM stock in DMSO may be stored frozen for one year in one time use aliquots.
Thiophosphate Ester Rabbit Monoclonal Antibody clone 51-8 (1:5,000 in 5% Skim Milk) (exclusively from Epitomics: www.epitomics.com) store for 1 month at 4°C or indefinitely at −20°C
Secondary anti-Rabbit HRP conjugated antibody (Epitomics: www.epitomics.com) store for 1 month at 4°C or indefinitely at −20°C
Materials for Support Protocol 2 (May also require materials from Support Protocol 1)
Plasmid containing KOI suitable for expression of kinase in mammalian cells
Transfection reagents (e.g., Lipofectamine, FuGene)
Phosphate buffered saline (PBS) store indefinitely at 4°C
Cell scrapers
Protease inhibitor cocktail – EDTA (Roche) store for 1 year at 4°C
Phosphatase inhibitor cocktail (Roche) store for 1 year at 4°C
ATP (Sigma) store solid indefinitely at −20°C
GTP (Sigma) store solid indefinitely at −20°C
2X RIPA Buffer (100 mM Tris-HCl pH 8.0, 150 mM NaCl, 1% NP-40, 0.1% SDS) store indefinitely at 4°C
EDTA (Sigma) store solid indefinitely at 22°C
Protein G Magnetic beads (Invitrogen Dynabeads) store for 1 year at 4°C
Magnetic stand which holds 1.5 mL microcentrifuge tubes
Materials for Support Protocol 3 (May also require materials from Support Protocol 1)
Purified active KOI or Plasmid containing KOI suitable for expression of kinase in E.coli
Recombinant GSK3β
CREB peptide (KRREILSRRPS (p) YR) store for 1 year at −80°C
Dephosphorylated Myelin Basic Protein (2.5 mg/mL) (Millipore)
10 mM stock in water of Adenosine 5′-[γ-thio]triphosphate tetralithium salt (Sigma) store for 1 year at −80°C
Myelin Basic Protein, Histone H1, or other general kinase substrate (Millipore)
10X HEPES buffered saline (HBS) (200 mM HEPES pH 7.4, 1.5 M NaCl) store for 6 months at 22°C
100X MgCl2 (1 M MgCl2) store for 6 months at 22°C
Optional: 33X MnCl2 (100 mM MnCl2) store for 6 months at 22°C
5X Laemmli sample buffer store for 1 month at 4°C
OMIX 100 μL Ziptip (Varian)
0.1 % TFA 50 % Acentonitrile store for indefinitely at 22°C
0.1 % TFA H20 store for indefinitely at 22°C
Tris buffered saline 0.5% Triton TX100 (TBST) store for 1 month at 22°C
Speed Vacuum
Equipment necessary for SDS-PAGE and western blotting (gel running apparatus, transfer apparatus, Nitrocellulose or PVDF)
Support Protocol 1: Kinase Reaction with ATPγS followed by Western Blotting Utilizing Thiophosphate Specific Antibody
Detection of thiophosphorylated substrate proteins is a critical analysis tool to determine whether this technique will work for a KOI in a particular lysate. An in-vitro kinase reaction with a KOI, substrate and ATPγS is used here to determine if the kinase will use ATPγS to thiophosphorylate substrate proteins. If the kinase is able to use ATPγS to thiophosphorylate substrate proteins the band corresponding to the substrate should be immunoreactive with the thiophosphate ester specific antibody (51-8).
-
On ice prepare a master mix of 1X HBS (or other kinase reaction buffer suitable for the KOI) 1M MgCl2 (to 10 mM) substrate (or general kinase substrate at 1 mg/mL) and KOI between 50–200 ng per kinase reaction.
Order of addition: H2O, HBS, MgCl2 Substrate, KOI. These general conditions will work for most kinases, but check the literature for kinase specific reaction conditions.
-
Label 4 tubes A-D and aliquot 27 μL of master mix into each tube.
Tubes A-C are controls for non-specific alkylation of cysteine residues by the thiol specific alkylating agent PNBM. -
Add 3 μL 10 mM ATPγS (final concentration of 1 mM) in two of the samples (samples C and D)
Flick the tubes to mix, and then briefly centrifuge to collect the kinase reaction in the bottom of the tube. Let the reaction proceed for 30 minutes at room temperature (or suitable temp for KOI)
-
Add 1.5 μL of 50 mM PNBM to samples B and D to afford a final concentration of 2.5 mM. The alkylating reaction should proceed for one hour at room temperature. If desired the kinase reaction may be quenched by adding EDTA to 20 mM.
The 50 mM PNBM should be prepared immediately before adding to the samples by dissolving PNBM in DMSO (50 mM is equal to 12 mg/mL). Add 5X Laemmli sample buffer to the samples. Run the samples on a SDS-PAGE gel. Transfer to nitrocellulose or PDVF membrane. Block the membrane for 1 hr with 5% Skim milk (in TBST).
-
Incubate the blot overnight at 4°C with 1:5,000 Thiophosphate Ester Rabbit Monoclonal Antibody, clone 51-8 (in 5% skim milk TBST).
Incubating the blot with the antibody for 1–3 hours at room temperature can also lead to detection of modified proteins with lower sensitivity. After washing 4 times with TBST, incubate blot for 1 hr with anti Rabbit HRP secondary, wash 4 times and then add ECL reagent to visualize PNBM alkylated/thiophosphorylated proteins.
Support Protocol 1a. Identifying optimal N6 substituted ATPγS analog
Each analog specific kinase will have a slightly different substrate specificity. To determine the prefered N6 substituted ATPγS each kinase must be tested with several ATP analogs. Following this protocol to perform a kinase assay with several N6 substituted ATPγS analogs will provide the optimal ATP analog for further studies to identify substrates.
-
Follow support protocol 1, utilizing several N6 substituted ATPγS analogs in place of ATPγS to identify which analog is used optimally by the AS KOI.
The optimal analog is the one that results in the most substrate labeling as detected by Thiophosphate Ester Rabbit Monoclonal Antibody, clone 51-8. Wild-type KOI should not be able to utilize this N6 substituted ATPγS analog.
Support Protocol 2: Thiophosphorylation of a candidate kinase substrate in cell lysate
The following method is used to determine if a candidate protein is a substrate of a KOI in a complex protein mixture. First the AS KOI is transiently expressed in cells and activated, then cell lysate is prepared and N6 substituted ATPγS is added to initiate the substrate labeling reaction. Thiophosphorylated proteins are alkylated and the candidate substrate is immunoprecipitated using an antibody against that protein. If a band is detected by IB with Thiophosphate Ester Rabbit Monoclonal Antibody then the candidate is a substrate of the KOI.
Grow appropriate mammalian cells on 10-cm dishes.
Transfect with plasmid encoding WT or AS KOI using transfection reagent of choice, following manufacturer’s protocol.
Allow cells to grow for 24–48 hours.
-
If appropriate, stimulate cells to activate KOI.
For example, mitogen activated protein kinase ERK may be activated by epidermal growth factor. Remove media and rinse once with 5 mL cold PBS.
Using a cell scraper, lyse and scrape cells on ice in 500 μL 1X RIPA buffer + 1X protease inhibitor + 1X phosphatase inhibitor.
Centrifuge at 10,000 x g for 10 min at 4°C to remove cell debris. Keep supernatant.
-
Add 100 μM N6 substituted ATPγS, 100 μM ATP, and 3 mM GTP to each sample (order does not matter).
Recommended concentration ranges for optimizing labeling conditions:- N6 substituted ATPγS: 50–500 μM
- ATP: 50–200μM
- GTP: 1–3 mM
Allow for thiophosphorylation of kinase substrates to occur for 20 minutes at room temperature.
-
Quench reaction with 500 μL 1X RIPA buffer + 40 mM EDTA (final [EDTA] = 20 mM) + 5 mM PNBM (final [PNBM] = 2.5 mM), then alkylate with PNBM for 1 hour at room temperature on a rotator.
Follow Support Protocol 1 for preparation of PNBM stock solution. -
Immunoprecipitate candidate substrate using antibody according to manufacturer’s recommendation. Incubate overnight at 4°C.
Save 20 μL input for western blotting to check immunoprecipitation efficiency. -
Add 40 μL of 50% slurry of appropriate (protein A or G) magnetic beads to each sample. Incubate 3–4 hours at 4°C on a rotator.
Before use, wash beads once with 1 mL 1X RIPA buffer. Then resuspend in original slurry volume and add to each sample. -
Using a magnet to collect the beads, wash 5 times w/1 mL 1X RIPA + protease inhibitor + phosphatase inhibitor.
Note: Before first wash, save 20μl supernatant for western blotting to check immunoprecipitation efficiency. Resuspend beads in 20 μL 1X RIPA + protease inhibitor + phosphatase inhibitor +1X Laemmli sample buffer.
Heat samples at 95°C for 2.5 minutes.
Load the samples onto an SDS-PAGE gel and proceed with IB as described in steps 6–8 of Support Protocol 1, blotting with Thiophosphate Ester Rabbit Monoclonal Antibody.
Support Protocol 3
Making a thiophosphorylated positive control peptide and protein The two control samples, a thiophosphorylated peptide and protein, can be prepared from readily purchasable materials. A kinase reaction is used to make the thiophosphorylated substrates. Mass spectrometric analysis using a MALDI can be used to confirm whether the CREB peptide has been thiophosphorylated. After labeling, the thiophosphorylated myelin basic protein (MBP) can be run on a denaturing polyacrylimide gel followed by an immunoblot using 51-8 (support protocol 1). After this experiment the peptide can be used to evaluate whether the covalent capture reaction is working properly. Labeled MBP can be added to a lysate as a control for the digestion and covalent capture reactions.
-
1
On ice prepare two kinase reactions that each contain:
6 μL 10X HBS
0.6 μL 1M MgCl2 (to 10 mM)
-
6 mL 10 mM ATPgS (to 1 mM final) and either 25 μL 2.5 mg/mL MBP (5 nmol total) or 18μL 0.5 mg/ml CREB peptide (5 nmol) and water to a final volume of 60 μL.
Order of addition: H2O, HBS, MgCl2 Substrate, KOI.
-
2
Add recombinant GSK3β 500 ng and incubate the reaction at room temperature for 4 hours
Incubate for longer reaction times to ensure stoichiometric thiophosphorylation of MBP -
3a
Aliquot the reaction containing MBP into 0.5 mL Eppendorf tubes (5 μL/tube), snap freeze them in liquid N2 and store them at −80°C until needed
-
3b
Use a large capacity ziptip (OMIX 100 μL) to desalt the CREB thiophosphorylation reaction, eluting into 200 μL 0.1% TFA 50% Acetonitrile. Concentrate the sample using a speed vacuum to 60 μL. Aliquot into 0.5 mL Eppendorf tubes (5 μL/tube) and snap freeze them in liquid N2. Store them at −80°C for 3 months or until needed
Utilize the ziptip according to product literature: wash 2X with 0.1% TFA 50% Acetonitrile, 2X with 0.1% TFA H20. Pass the sample through the tip 5X being sure not to introduce any air into the tip. Wash 2X with 0.1% TFA H20 and then elute 2X with 0.1% TFA 50% Acetonitrile.
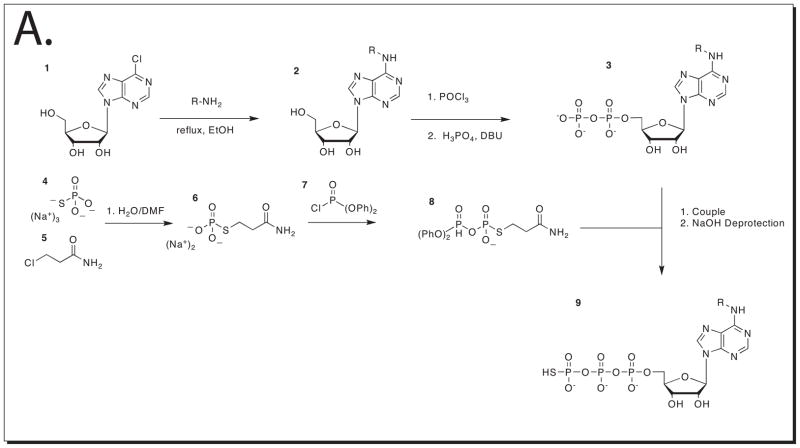
Support Protocol 4: Chemical synthesis of N6 substituted ATPγS
The synthesis of N6 substituted ATPγS is presented to those interested in making additional analogs. The synthesis will require good knowledge of organic chemistry synthesis procedures and a well-equipped laboratory. This protocol will allow the synthesis of larger quantities of N6 substituted ATPγS analogs, and the synthesis of those that are not available from Biolog. This synthesis can be found in the supplementary information in (Allen et al., 2007)
Materials
Ethanol (Sigma)
Alkyl amine containing desired N6 modification
Triethylphosphate (TEP) (Sigma)
POCl3 (Sigma)
H3PO4 (Sigma)
1,8-diazabicyclo[5.4.0]undec-7-en (DBU) (Sigma)
Triethylamine (Sigma)
DOWEX® 501-X8 ion exchange resin
trisodium thiophosphate
3-chloropropionamide
tri-n-octylamine
dimethylformamide DMF
diphenyl phosphorochloridate
tri-n-butylamine
dioxane
petroleum ether
β-mercaptoethanol
NaOH
CO2 Tank
PTFE syringe filter (0.45 μm, Acrodisk)
small plastic column
Buchi Rotovapor Model R-200 or equivalent rotary evaporator
Oil bath
Filter paper
Buchner funnel
Acta FPLC (Amersham Biosciences)
Small molecule capable LCMS instrument (Possibly from Waters)
-
1
Dissolve 1 g (3.5 mmol) of 6-chloropurine ribonucleoside in 21 mL of ethanol while stirring.
-
2
Heat to at 95°C then add seven equivalents of the alkyl-amine desired to be the bulky substituent in the N6 position (in this case phenthylamine, 2.5 g (21 mmol)) and stir under a reflux condenser for 12 hrs.
-
3
Remove solvent under reduced pressure in Buchi Rotovapor.
-
4
Add 150 mL of Ethanol and place at 4°C for 5 hours.
-
5
Filter the crystals and wash with cold Ethanol to obtain N6 Phenethyl (or additional alkyl) Adenosine
Synthesize N6 Phenethyl ADP
-
6
Dissolve 2 mmol of the N6 Phenethyl (or additional alkyl) Adenosine (0.76 g) in 5 mL of triethethylphosphate (TEP)
-
7
Cool to 0°C in an ice bath while stirring.
-
8
Add 0.6 g (4 mmol) of POCl3 dropwise and continue to stir at 0°C for two hours.
-
9
In a separate container dissolve first dissolve 8 mmol of 1,8-diazabicyclo[5.4.0]undec-7-en (DBU) in 5 mL TEP then add 8 mmol of H3PO4.
The order of addition is critical here, first dissolve the DBU then add the H3PO4 -
10
Add the entire H3PO4 DBU mixture to the mixture of the N6 substituted Adenosine and POCl3.
A yellow solid should form immediately -
11
Continue to stir for two minutes and then quench the reaction by adding 30 mL of 0.1M triethylammonium bicarbonate (TEAB) and stir for 30 minutes.
Make 2M TEAB by mixing 557.5 mL triethylamine (TEA) with 1,442.5 mL H2O. Bubble CO2 through this mixture until the TEA is dissolved. -
12
Filter the mixture through a PTFE syringe filter (0.45 μm, Acrodisk)
-
13
Load the mixture onto two HiPrep 16/10 QFF anion exchange columns (Amersham Biosciences) in series using a peristaltic pump.
-
14
Run a 80 minute gradient from 100% 0.1M TEAB to 50% 2M TEAB:50% 0.1MTEAB
-
15
Pool the fractions containing the diphosphate
Analyze the fractions by mass spec (LCMS) on a small molecule capable LCMS instrument and pool those fractions containing the diphosphate -
16
Lyophilize fractions containing the diphosphate
N6 phenylethyl ADP calculated mass 530.10 m/z
Synthesize Disodium S-2-Carbamoylethyl Phosphorothioate
-
17
Dissolve 18.6 mmol of trisodium thiophosphate in 28 mL of water
-
18
Dissolve 28 mmol of 3-chloropropionamide in 5.6 mL of DMF and add mixture to trisodium thiophosphate mixture followed by stirring for 48 hours at room temperature.
-
19
Filter the reaction and add 100 mL of ethanol to the filtrate and then cool to 4°C overnight (16 hours)
-
20
Filter the precipitate, wash with ethanol and dry to afford disodium S-2-carbamoylethyl phosphorothioate
Convert disodium S-2-carbamoylethyl phosphorothioate and N6 phenylethyl ADP (TEA salt) to pyridinium salt
-
21
Swell 5 mL of DOWEX® 501-X8 ion exchange resin in 50 mL 5% Pyridine (95% water)
-
22
Add the DOWEX® 501-X8 ion exchange resin to a small plastic column and let the 5% pyridine run through until the pH is greater than 10.
When the pH is above 10 all of the sites on the resin are occupied with pyridine. -
23
Dissolve 2.0 mmol of disodium S-2-carbamoylethyl phosphorothioate or 0.3 mmol N6 phenylethyl ADP (TEA salt) in 10 (or 5) mL 1:1 MeOH: H20
-
24
Add the mixture to the top of the DOWEX® 501-X8 ion exchange resin column (pyridinium form)
-
25
Elute with another 10 (or 5) mL 1:1 MeOH: H20
-
26
Dry in vacuo on a Buchi rotory evaporator to afford the pyridinium salt of S-2-carbamoylethyl phosphorothioate and N6 phenylethyl ADP
Synthesize O-diphenyl phosphoro S-2-carbamoylethyl phosphorothioate
-
27
Add 20 mL methanol to 2.0 mmol of the pyridinium salt of S-2-carbamoylethyl phosphorothioate
-
28
Add 2 mmol of tri-n-octylamine to convert to the mono (tri-n-octylammonium) salt.
-
29
Remove solvent in vacuo on a Buchi rotory evaporator. Repeat evaporation (two times) after addition of 10 mL aliquots of dry DMF.
-
30
Dissolve the residue in 14 mL of dry dioxane, then add 2.9 mmol diphenyl phosphorochloridate followed by 3.8 mmol of tri-n-butylamine and stir for two hours at room temperature
-
31
Remove solvent in vacuo on a Buchi rotory evaporator. Add 20 mL ether and after dissolution add 40 mL warm petroleum ether.
-
32
Place mixture at 4°C for 30 minutes until a higher density oily mixture separates and can be decanted into an additional roundbottom flask
-
33
Dry the oily residue on a high vacuum for 1 hour to afford O-diphenyl phosphoro S-2-carbamoylethyl phosphorothioate
Synthesis of N6 phenylethyl ATPγS
-
34
Add 12 mL methanol to the pyridinium salt of N6 phenylethyl ADP (from step 26)
-
35
Add 0.32 mmol tri-n-octylamine and 0.32 mmol tri-n-butylamine until complete dissolution occurs.
-
36
Remove solvent in vacuo on a Buchi rotory evaporator. Dry the residue by repeated (three times) evaporation of 5 mL aliquots of dry pyridine.
-
37
Dissolve residue in 3.3 mL dry pyridine and add this to the dry residue of O-diphenyl phosphoro S-2-carbamoylethyl phosphorothioate (from step 33).
-
38
Stir at room temperature for two hours,
A precipitate will form during this time -
39
Remove solvent in vacuo on a Buchi rotory evaporator. Add 20 mL 0.2 M NaOH then heat the reaction to 100°C for 10 minutes.
-
40
Quench the reaction by adding 10 ml DOWEX® 501-X8 ion exchange resin (pyridinium form) and 0.4 ml β-mercaptoethanol.
-
41
Filter the reaction and then purify on the two HiPrep 16/10 QFF anion exchange columns (Amersham Biosciences) exactly as in step 13.
-
42
Fractions should be analyzed by LCMS and fractions containing the N6 phenylethyl ATPγS should be pooled and lyophilized.
Redissolving the dried fractions in 0.1M TEAB and another round of lyophilization can be done to concentrate and to produce a white solid. -
43
After lyophilization the compounds should be assumed to be in the TEA form. To obtain accurate concentrations the absorbance at 280 nm should be compared to several standard solutions made up of the N6 modified adenosine.
Commentary
Background Information
Identifying the substrates of a kinase is a critical step towards understanding its biological role. Traditional approaches for kinase substrate mapping depend on knockout experiments followed by phosphopeptide enrichment and mass spectrometry. While useful, this method fails to identify all kinase substrates as there exists redundancy in which a single substrate may be phosphorylated by multiple kinases. Methods for phosphopeptide enrichment include metal affinity chromatography using titanium oxide columns (Trinidad et al., 2008) and identifying new phospho proteins by enriching thiophospho peptides (Kwon et al., 2003). A quantitative mass spectrometry approach can more precisely define the effects of stimulation or knockout of a particular kinase. In an application of this technique, a SILAC labeling approach followed by TiO2 enrichment was used to determine the effects of growth factors on global phosphorylation status (Olsen et al., 2006). All of these approaches fail to specifically assign a phosphorylation site to a particular kinase but are useful for defining the global changes in phosphorylation and for mapping novel phosphorylation sites.
Our approach allows for the labeling of the substrates of a single kinase in a chemical genetic approach, and can therefore unambiguously assign kinase substrate relationships. Originally the N6 modified ATP analogs were radioactively labeled in the gamma phosphate position. Putative substrates were then identified by coupling this approach with affinity tagged libraries. This approach was successfully used to identify Cdk1 and Pho 85 substrates in S. cerevisiae (Dephoure et al., 2005; Ubersax et al., 2003). Although this engineered “lock and key” provides selectivity for a single kinase, the transfer of a radioactively tagged phosphate by the N6 modified ATP substrate only assists in visualization and traditional biochemical purification of direct kinase substrates from tagged libraries that are not available in more complex eukaryotes.
Introduction of a bio-orthogonal tag onto the substrate proteins of a particular kinase simplifies the affinity purification and identification of the substrates of any protein kinase in a complex lysate. This approach relies on two separate specificity gates: the kinase is engineered to accept bulky ATP analogs, and the substrates are tagged with a bio-orthogonal phosphate analog. The affinity purification of the tagged substrates also depends on two specificity gates: only thiol containing peptides are pulled down, and of those peptides that are covalently bound to the affinity matrix, only those linked by thiophosphate ester bonds are cleaved off the resin. These specificity elements allow for the purification of very low abundance substrates, while also increasing the confidence in the identification of the identified substrates.
Critical Parameters
Utilization of the N6 modified ATP analogs by other kinases can lead to false identification of kinase substrate relationships. It is therefore crucial to fully optimize the labeling reaction. Using a wide range of ATP, GTP, and N6 modified ATP analogs during the lysate optimization steps will provide the best conditions for these reactions. Nonspecific binding of peptides to the agarose beads can also lead to the false identification of substrate proteins. Generating several positive and negative control samples will help control for the non-specific purification of peptides.
Several controls are essential for distinguishing background from AS kinase specific labeling. First, it is essential to demonstrate the AS kinase dependence of purification of any phosphopeptides. When labeling is observed, or phosphopeptides are affinity purified from samples that do not contain AS kinase but do contain the N6 modified ATP analog, these proteins must be subtracted from any list of putative substrates. When non-phosphorylated peptides are identified in the absence of AS kinase it may indicate that the washing steps are not effective. Increasing the stringency of the wash steps by increasing the volume or incubating the washes for 10 minutes should lower background. Secondly, the positive control thiophosphorylated peptide and protein are very useful for diagnosing the digestion and affinity purification steps. In typical tryptic digests, complex lysates are reduced and alkylated before digestion to ensure the complete digestion of all proteins. The alkylation step with iodoacetamide is incompatible with our technique, thereby complicating the digestion of thiophosphorylated protein lysates. Also the thiophosphate mark is susceptible to acid promoted hydrolysis. It is therefore important to include the hyper-thiophosphorylated MBP as a digestion control to ensure the mark is retained on substrate proteins. A rapid way to determine whether the covalent capture and release reaction is working is by including a separate covalent capture reaction with the CREB peptide followed by MALDI TOF analysis.
Troubleshooting
A trouble-shooting guide is presented in Table 1.
Table 1.
| Problem | Cause | Solution |
|---|---|---|
| No thiophosphorylation of substrate seen during in-vitro ATPγS kinase reaction | Inactive kinase or kinase doesn’t utilize ATPγS | Generate sufficient quantities of pure kinase–check activity with ATPγS35 kinase assay |
| Inactive kinase | Generate a constitutively active kinase | |
| Singly charged polymer or detergent ions seen during mass spectrometric analysis | Concentration of detergents is too high in the lysis buffer | Typically the lysis buffer should only contain low amounts of a non-ionic non denaturing detergent like 0.1% NP-40 |
| No phospho-CREB detected after test covalent capture reaction | pH is too low or high in covalent capture buffer | Check the pH of the flow through after the covalent capture reaction. Ensure the pH of the agarose beads is correctly adjusted by washing the agarose beads carefully with 200 mM HEPES pH 7.0 |
| Insufficient reaction time for the covalent capture reaction | Incubate the thiophosphopeptides overnight (11–16 hrs, especially if using a complex lysate). | |
| Insufficient incubation with 10 mM DTT | Incubate with 10 mM DTT for at least 120 minutes following the the5% Formic acid wash step | |
| Non-specific adsorption of peptides to Eppendorf tubes | Using siliconized Eppendorf tubes and BSA will reduce non-specific adsorption | |
| Thiophospho CREB levels are too low | Increase thiophospho CREB to see a signal, and then adjust other parameters to diagnose where the sample loss is occurring | |
| No thiophospho peptide is detected after digesting the labeled MBP | Levels of TCEP are too low during digestion | Make a fresh solution of 1 M TCEP before the digestion and add to 10 mM concentration in both the reduction and the digestion steps. |
| Levels of urea are too low during digestion | Do not dilute to 1M urea, only dilute to 2M as the additional denaturant will assist in unfolding proteins during the digestion | |
| Digestion not allowed to proceed long enough | Incubate digestion for a longer time (11–16 hrs) | |
| Insufficient trypsin added | Add trypsin to 1:10 w/w | |
| Phosphopeptides detected in no AS kinase control | Background kinases are utilizing the ATP analogs | Follow all optimization steps to determine the correct amount of ATP, GTP and N6 modified ATP to use during the labeling |
| Background kinases are utilizing the ATP analogs | Identify which background kinases are utilizing the ATP analogs and include inhibitors for these kinases during the labeling reaction | |
| No phosphopeptides detected in the plus AS kinase reaction | Kinase activity is low | Increase the amount of AS kinase in the labeling reaction |
| Stoichiometry of labeling is low | Increase the effective concentration of substrates by fractionating the lysate before the labeling reaction | |
| Stoichiometry of labeling is low | Increase the effective concentration of substrates by isolating specific sub cellular compartments prior to labeling | |
| N6 ATP analog concentration is too low | Increase the amount of ATP analog in both the negative control and the sample with AS kinase |
Anticipated results
The application of this technique to appropriately labeled lysates should result in the identification of phosphopeptides. After subtraction of any phosphopeptides found in the control lysates, these peptides should predominantly be the substrates of the AS kinase that was used in the labeling reaction. The location of the phosphorylation mark on the substrate proteins will also allow the investigator to produce non-phosphorylatable substrates and thus to analyze the biological significance of the phosphorylation mark immediately following confirmation of the kinase-substrate relationship.
Thiophosphorylation of CREB (KRREILSRRPS (p) YR) (1795.84 MH+) by GSK3B should lead to a predominant peak at 1891.986 (MH+). After covalent capture with iodoacetyl agarose beads followed by specific hydrolysis the only product ion present should be a double phosphorylated CREB peak at 1875.986 (MH+).
The hyper-thiophosphorylation of Myelin Basic Protein followed by digestion with trypsin and analysis by LC MS/MS will lead to the detection of a thiophosphorylated peptide 1800.8939 (MH+) NIVT (Thiophospho) PRTPPPSQGKGRor 1587.7951 NIVT (Thiophospho) PRTPPPSQGK or 796.4044 (MH+) NIVT (Thiophospho) PR. After covalent capture with iodoacetyl agarose beads followed by specific hydrolysis the only multiply charged ions present will be the same peptide a phospho group instead of a thiophospho modification, a loss of 16 Daltons. 1784.8939 (MH+) NIVT (phospho) PRTPPPSQGKGR, 1587.8001 (MH+), NIVT (phospho) PRTPPPSQGK, or 780.403 (MH+) NIVT (phospho) PR. In addition, incubating the labeled MBP with the lysate to be used for substrate ID and then performing the covalent capture reaction should yield these same peptides, but no background should be seen as there should be no source of thiophosphate in the lysate for background labeling to occur.
Time considerations
A significant investment of time and resources is required for the design and production of an AS kinase. Once a kinase has been engineered, optimization of labeling conditions and identification of putative substrate proteins should be rapidly achieved in 1–2 weeks. Production of the labeled positive controls should take 1–2 days depending on how long the digestion of labeled MBP is permitted. Once comfortable with labeling, digestion and preparation of covalent capture reactions an experienced investigator should be able to prepare and process several samples in 2–3 days.
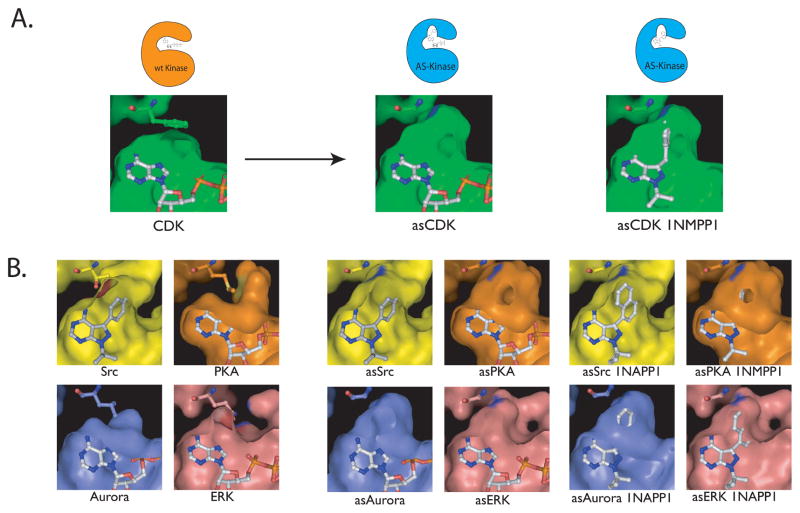
Figure 3.
A. The gatekeeper residue is shown in CDK1 as a phenylalanine above the bound ATP in the CDK1 active site. After mutation to the much smaller glycine, a pocket is opened that allows for a bulky substituent to bind (pictured as asCDK1). A “bumped” (N6 substituted) ATP analog or inhibitor (in this case 1NMPP1) can now bind to the engineered kinase by utilizing the extra space above the N7 position. B. The active sites of SRC, PKA, aurora and ERK are shown along with their gatekeeper residues and the successful mutation of these bulky residues to smaller ones. The mutation of the gatekeeper to a smaller residue creates a similar pocket in each of these four kinases. These kinases are shown with ATP bound or with bumped inhibitors that target these engineered kinases, but not the wild type. The similarity in the engineered pocket among disparate kinases demonstrates the wide applicability of this method.
Figure 4.
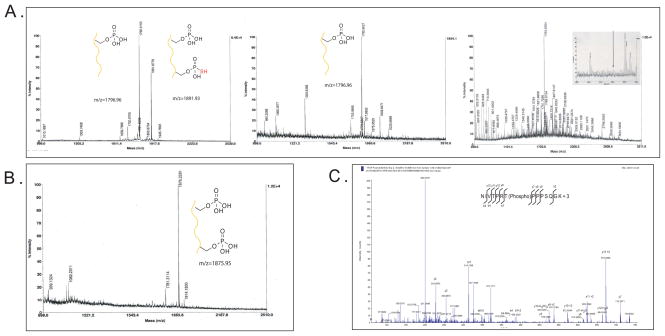
A. Raw data showing an expected MALDi analysis of thiophosphorylated CREB peptide. Both the singly phosphorylated (KRREILSRRPS(p)YR) (1795.84 MH+) and the thiophosphorylated (KRREILS(thiophosphate)RRPS(p)YR) 1891.986 (MH+) peptide can be seen in the first pane. Analysis of the flow-through after covalent capture with iodoacetyl agarose beads in the second pane shows the complete reaction of the thiophosphorylated (1891.986) peptide, but no reaction with the parent peptide (1795.84 MH+). Analysis of the flow-through with additional HeLa lysate after covalent capture with iodoacetyl agarose beads in the third pane shows many peptides, but the CREB peptide is not seen. B. Following hydrolysis the only peptide present is the double phosphorylated CREB peak at 1875.986 (MH+) demonstrating the specificity of the technique. C. After the covalent capture reaction with digested thiophosphorylated myelin basic protein only one peptide (NIVT(phospho)PRTPPPSQGK) is observed. MSMS analysis of the 1587.8001 (MH+), NIVT(phospho)PRTPPPSQGK (seen as the triply phosphorylated peptide at 524.6 m/z) shows good sequence coverage of this peptide with loss of phosphate from the y10 ion.
Figure 5.
A. Synthetic scheme for the production of N6 substituted ATPγS analogs
Literature Cited
- Allen JJ, Lazerwith SE, Shokat KM. Bio-orthogonal affinity purification of direct kinase substrates. J Am Chem Soc. 2005;127:5288–5289. doi: 10.1021/ja050727t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JJ, Li M, Brinkworth CS, Paulson JL, Wang D, Hubner A, Chou WH, Davis RJ, Burlingame AL, Messing RO, et al. A semisynthetic epitope for kinase substrates. Nat Methods. 2007;4:511–516. doi: 10.1038/nmeth1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop A, Buzko O, Heyeck-Dumas S, Jung I, Kraybill B, Liu Y, Shah K, Ulrich S, Witucki L, Yang F, et al. Unnatural ligands for engineered proteins: new tools for chemical genetics. Annu Rev Biophys Biomol Struct. 2000;29:577–606. doi: 10.1146/annurev.biophys.29.1.577. [DOI] [PubMed] [Google Scholar]
- Blethrow J, Zhang C, Shokat KM, Weiss EL. Design and use of analog-sensitive protein kinases. Curr Protoc Mol Biol. 2004;Chapter 18(Unit 18):11. doi: 10.1002/0471142727.mb1811s66. [DOI] [PubMed] [Google Scholar]
- Blethrow JD, Glavy JS, Morgan DO, Shokat KM. Covalent capture of kinase-specific phosphopeptides reveals Cdk1-cyclin B substrates. Proc Natl Acad Sci U S A. 2008;105:1442–1447. doi: 10.1073/pnas.0708966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzko O, Shokat KM. A kinase sequence database: sequence alignments and family assignment. Bioinformatics. 2002;18:1274–1275. doi: 10.1093/bioinformatics/18.9.1274. [DOI] [PubMed] [Google Scholar]
- Cohen P. The role of protein phosphorylation in human health and disease. The Sir Hans Krebs Medal Lecture. Eur J Biochem. 2001;268:5001–5010. doi: 10.1046/j.0014-2956.2001.02473.x. [DOI] [PubMed] [Google Scholar]
- Dephoure N, Howson RW, Blethrow JD, Shokat KM, O’Shea EK. Combining chemical genetics and proteomics to identify protein kinase substrates. Proc Natl Acad Sci U S A. 2005;102:17940–17945. doi: 10.1073/pnas.0509080102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregan J, Zhang C, Rumpf C, Cipak L, Li Z, Uluocak P, Nasmyth K, Shokat KM. Construction of conditional analog-sensitive kinase alleles in the fission yeast Schizosaccharomyces pombe. Nat Protoc. 2007;2:2996–3000. doi: 10.1038/nprot.2007.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SW, Kim SC, Jaunbergs J, Falck JR, Zhao Y. Selective enrichment of thiophosphorylated polypeptides as a tool for the analysis of protein phosphorylation. Mol Cell Proteomics. 2003;2:242–247. doi: 10.1074/mcp.M300039-MCP200. [DOI] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Shah K, Liu Y, Deirmengian C, Shokat KM. Engineering unnatural nucleotide specificity for Rous sarcoma virus tyrosine kinase to uniquely label its direct substrates. Proc Natl Acad Sci U S A. 1997;94:3565–3570. doi: 10.1073/pnas.94.8.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinidad JC, Thalhammer A, Specht CG, Lynn AJ, Baker PR, Schoepfer R, Burlingame AL. Quantitative analysis of synaptic phosphorylation and protein expression. Mol Cell Proteomics. 2008;7:684–696. doi: 10.1074/mcp.M700170-MCP200. [DOI] [PubMed] [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]