Abstract
Through screening by NMR spectroscopy, we discovered a novel scaffold (DPQ: 6,7-dimethoxy-2-(1-piperazinyl)-4-quinazolinamine) that binds specifically to the influenza A virus promoter RNA. The solution structure of the RNA-DPQ complex reported here demonstrates that the internal loop is the binding site of DPQ. The scaffold has antiviral activity against influenza viruses.
Influenza A viruses have a genome comprising eight negative-sense, segmented, single-stranded RNAs encoding 13 proteins. The viral RNAs (vRNA) are transcribed to messenger RNAs (mRNA) and replicated into fully complementary RNAs (cRNA). Both transcription and replication of vRNAs are performed by the same viral ribonucleoprotein (RNP) complex consisting of three RNA-dependent polymerase subunits (PB1, PB2 and PA) and nucleoprotein (NP).1 In the vRNA genome, the 13 nucleotides at the 5’-end and 12 nucleotides at the 3’-end of each RNA segment are highly conserved among most influenza A virus variants and fold together to form partial duplexes (Fig. 1A). The RNA-dependent RNA polymerase (RdRp) recognizes specifically the partial duplex RNA structure (referred to as the promoter) to regulate the initiation of transcription and replication.2 The promoter structure determined by NMR is panhandle-like and include a unique (A-A)-U internal loop and a bent terminal helix.3 The panhandle-like duplex structure is stable at physiological temperature, which is of great advantage to the initiation of RNA synthesis and polymerase recognition.4 In addition, the promoter sequences are highly conserved and are not involved in the gene variations leading to pathogenesis and antiviral resistance. Based on these observations, the influenza promoter can be considered a new attractive target for anti-influenza drug development.
Fig. 1.
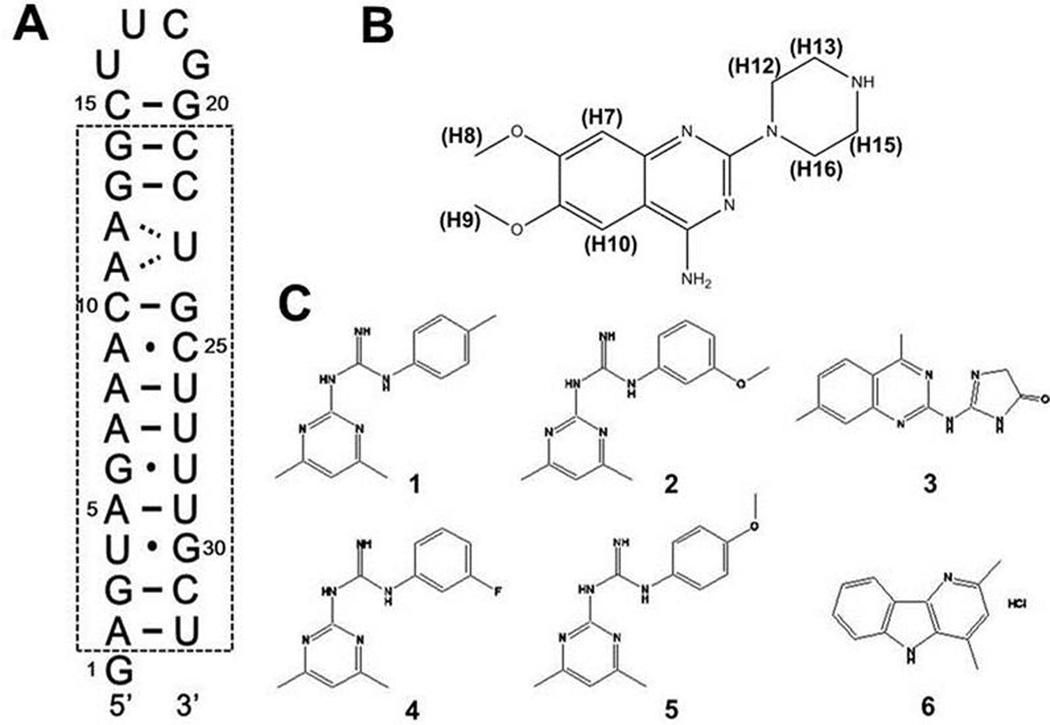
(A) Secondary structure of the RNA promoter of the influenza A virus. The dashed box indicates nucleotides highly conserved among all influenza A virus strains. (B) Structure of the small molecule 6,7-dimethoxy-2-(1-piperazinyl)-4-quinazolinamine (DPQ). (C) Compounds 1–6 had weaker affinity for the RNA compared to DPQ.
We tested binding scaffolds identified by NMR–based fragment screening to interact with the RNA promoter structure and investigated the binding affinity and antiviral activity of the strongest binding compound. We determined the structure of the complex between the RNA promoter and this compound (PDB: 2LWK) and established how the small molecule interacts with the RNA major groove at the (A-A)-U internal loop. For the convenience of structural analysis by NMR, we designed an RNA construct of 32 nucleotides which includes a UUCG hairpin loop to facilitate RNA folding and an additional single G at the 5’ terminus to increase in vitro transcriptional efficiency (Fig. 1A).
In order to screen small molecules binding to influenza A virus promoter, we recorded one-dimensional (1D) spectra in the presence of various small molecular weight compounds derived from a fragment library of 4279 compounds (ESI†). Compound binding induced conformational changes in the RNA, resulting in line broadening or chemical shifts changes for the imino peaks, which can be easily observed even in 1D NMR spectra. We identified seven compounds by comparing the spectra of the free and bound RNAs (Fig. 1B–C). Among them, 6,7-dimethoxy-2-(1-piperazinyl)-4-quinazolinamine (DPQ) induced the most dramatic changes in the 1D NMR imino resonances upon binding to the RNA promoter (Fig. 1B).
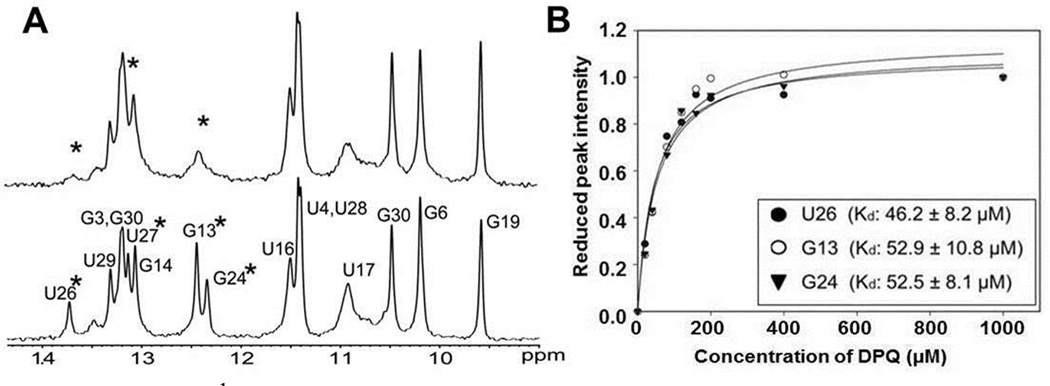
DPQ caused changes in imino peaks of residues near the internal loop (U26, U27, G13, and G24) (Fig. 2A). In particular, the imino peaks of U26 and G24 were dramatically line-broadened due to exchange between the free and bound states and/or to changes in base stability. In contrast, no evidence for DPQ binding to the UUCG tetraloop and terminal stem region was observed. To measure the binding affinity (Kd), 1D NMR spectra were recorded at various concentrations of DPQ. Analysis of the 1D NMR spectra and plotting of the peak intensities produced a Kd of 50.5±9 µM (Fig. 2B).
Fig. 2.
(A) 1D 1H NMR spectra recorded for the free RNA (bottom) and for the 1:1 complex with DPQ (top). Starred residues indicate lines that are significantly broadened. (B) The reduced peak height of the imino proton resonance (U27, G13, and G24) is plotted vs DPQ concentration to extract an apparent Kd for complex formation. The average Kd is 50.5±9 µM. The RNA sample concentration is 200 µM.
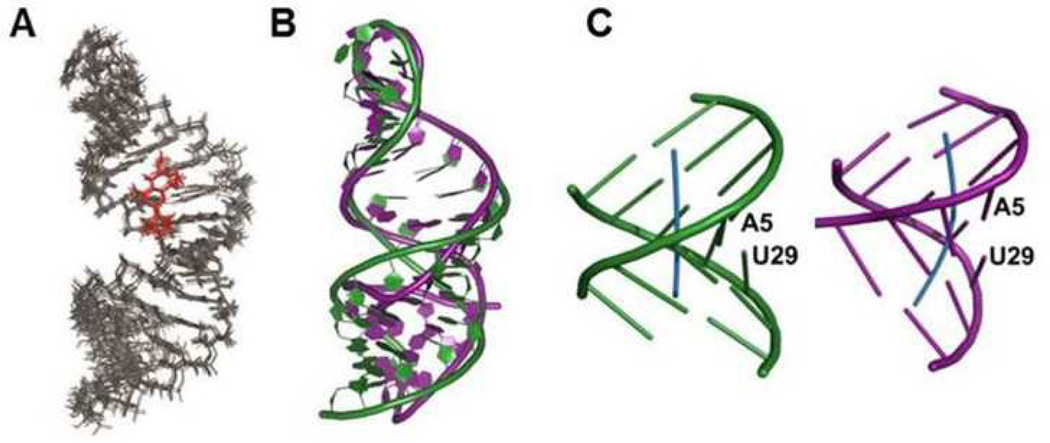
A total of 741 experimental NMR restraints, including RDC data, were used for the RNA-DPQ structure calculations with X-PLOR (Table S1, ESI†). The free RNA promoter showed an internal (A-A)-U internal loop interrupting two A-form helical stems.3 In the DPQ-bound promoter, A-form geometry is maintained and most of the base pairs are well stacked along the helical axes (Fig. 3A). Interestingly, the helical axis of the bound RNA is straight while a bent structure is observed for the free RNA (Fig. 3B); the bending angles of the terminal stems of both RNAs (around the A5-U29 pair) are very different as well (Fig. 3C); the bound RNA was bent at the A5-U29 pair to an angle of 6.7±3.4°, whereas the free RNA has a larger bending angle of 46±10°.3
Fig. 3.
(A) Superposition of the 10 lowest energy structures. (B) Overlay of the backbone structure of the free (purple) and bound (green) promoters. (C) Schematic representation of the helical geometry of the terminal stems from the free (purple) and bound (green) promoters. The bending angle was calculated with the CURVES program.5
The complex structure revealed that DPQ fits snugly into the major groove of the internal loop at the (A-A)-U loop (Fig. 3A). DPQ binding widens the major groove near the proximal base pairs (G13-C22 and G14-C21) and the end of the helical stem compared to the free RNA (Fig. S1). This wider major groove makes the RNA structure better accessible to binding of small molecules such as DPQ. The imino resonance region of the 1D and 2D NMR spectra confirmed that the DPQ-RNA complex has 8 base pairs, whereas the free RNA has 10 base pairs. The missing base pairs from the bound RNA, C10-G24 and A8-U26, are located near the internal loop (A11-A12, U23) and open up when the complex forms. Altogether, the interaction with DPQ requires a conformational change in internal loop opens up, compared to its free form, to accommodate the ligand.
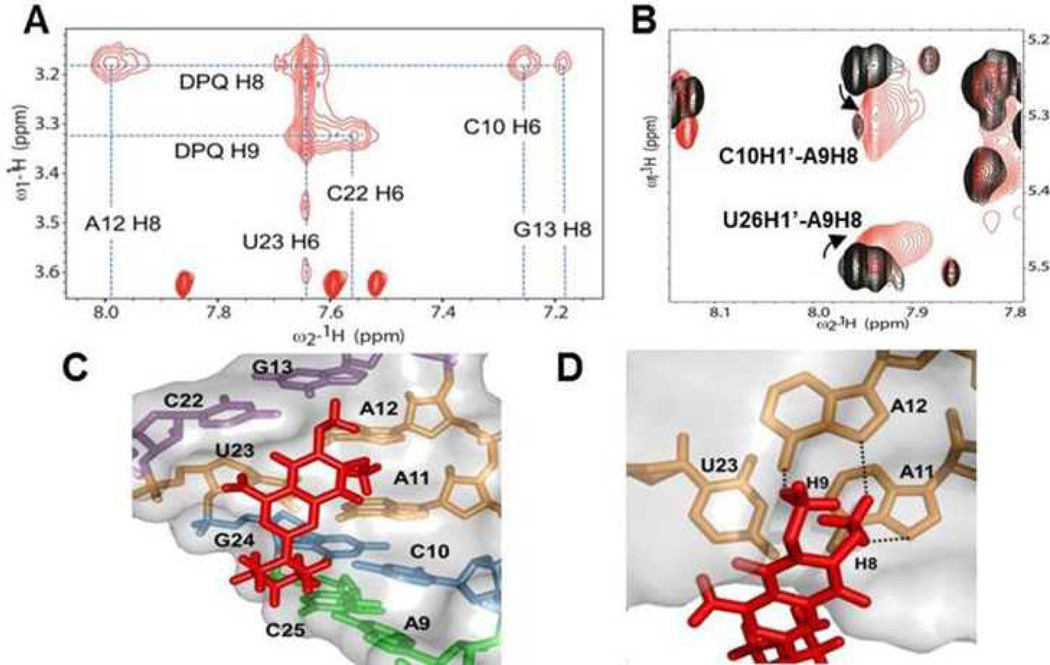
In the 1H-1H NOESY spectrum obtained from a partially deuterated RNA sample, most of the intermolecular NOEs originate from cross peaks between the dimethoxy group of DPQ and residues within the internal loop of the RNA (C10, A12, G13, C22, and U23) (Fig. 4A). The strong A12H8-DPQH8 and U23H6-DPQH8/H9 cross peaks demonstrate direct DPQ binding at the (A11-A12)-U23 internal loop. In contrast, the piperazinyl protons (H12s, H13s, H15s, and H16s) provide weaker NOE interactions with the RNA. In addition to intermolecular NOE cross-peaks, the changes in sequential NOEs for the internal loop and proximal base pairs provide evidence of the specific binding of DPQ to the RNA. Peaks of the A9, C10, G24, C25, and U26 nucleotides especially C10H1’-A9H8 and U26H1’-A9H8, exhibited considerable changes in chemical shifts and line-broadening (Fig. 4B). In the free RNA structure, the H5-H6 resonance of C10 showed significant field-dependent broadness implying that C10 or its neighbour are involved in dynamic motion on a millisecond time scale, and A9 forms a base-pair with C25 (Fig. S1).3 In the NMR structure of the complex, the C10-G24 and A9-C25 pairs are positioned near the DPQ molecule and are thus able to engage in direct interactions with it (Fig. 4C). DPQ likely exploits the dynamic nature of A9 and C10 to induce an optimal conformation for its interaction with the RNA promoter. Another proximal base pair (G13-C22) and the (A11-A12)-U23 internal loop are involved in DPQ binding as well (Fig. 4C–D). The dimethoxy protons of DPQ are within hydrogen bonding distance to G13, C22, A11, and A12 in the internal loop.
Fig. 4.
(A) Intermolecular NOEs between RNA and DPQ in the NOESY spectrum. (B) Overlay of 2D NOESYs of the free RNA (black) and complex (red). (C) DPQ interacts with the internal loop and its proximal base pairs. (D) The interaction of (A11-A12)-U23 and DPQ. Dotted lines indicate hydrogen bonds based on close contacts observed in the structure.
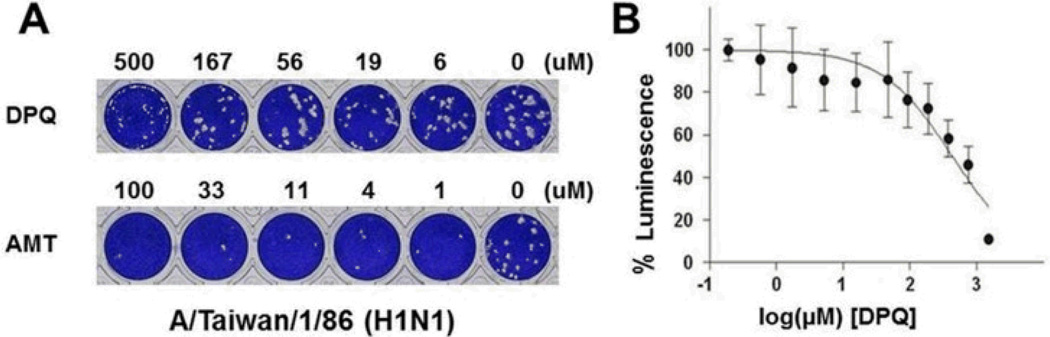
To investigate whether DPQ possess anti-influenza virus activity, we performed antiviral activity (Cytopathic effect inhibition) and cellular toxicity assays and compared the results with those of approved anti-influenza (Table 1).6 MDCK cells were infected with different strains of influenza A (H1N1: TW; H3N2: HK) and influenza B (PNM) virus followed by treatment with either DPQ or the antivirals. Cell viability was measured by the FDA-based CPE assay to estimate the CC50 and EC50 (ESI†). The results showed that DPQ was non-toxic to cells at concentrations up to 500 µM, and, although much less efficient than the standard antiviral compounds, it inhibited replication of both influenza A and B viruses with EC50 values of 71.7±29, 275.5±98, and 113.7±9 µM against TW, HK, and PNM, respectively. This result suggests that DPQ inhibits influenza A and B virus infection in cell-based assays. A plaque reduction assay further supports the inhibitory activity of DPQ based on reduced viral plaque formation (Fig. 5A, Fig. S2, ESI†). Plaque formation by wild-type influenza A virus strains was inhibited by DPQ at concentration ranging from 56 to 500µM. The plaque size was remarkably decreased with treatment of DPQ, although reduction in plaque numbers was much less compared to amantadine. To further corroborate these findings, we employed an orthogonal viral replication assay. We adopted a WSN-Ren luciferase assay, which uses a modified H1N1 influenza virus, producing Renilla luciferase in the same fashion of other viral proteins (ESI†). Therefore, by measuring the luminescence, we can assess the level of influenza replication in the presence and absence of our compounds. We performed this assay in an engineered MDCK cell line expressing influenza hemagglutinin (MDCK-HA) to allow multiple cycles of viral replication. In the non-cytotoxic concentration range of MDCK-HA cells, we observe an IC50 value for DPQ of 435 µM (Fig. 5B), in agreement with the activity detected in the plaque assay. The IC50 values of compounds 3 and 4 were 581 µM and 535 µM, respectively. Compounds 1, 2, 5 and 6 exerted cytotoxic effects in MDCK-HA cells at the concentrations tested (Fig. 1C).
Table 1.
Antiviral activity of DPQ and standard antiviral agents against influenza A and B viruses in cell-based assays
| Compound | CC50 (µM) |
TW | EC50 (µM) HK |
PNM |
|---|---|---|---|---|
| DPQ | >500 | 71.6±28.8 | 275.5±97.6 | 113.7±8.9 |
| Amantadine | >100 | 0.6±0.04 | 0.09±0.01 | >100 |
| Oseltamivir | >100 | <0.01 | <0.01 | 0.02±0.00 |
| Ribavirin | >100 | 3.5±2.7 | 4.4±1.8 | 1.7±0.8 |
Fig. 5.
(A) Plaque reduction assays. The effect of DPQ and amantadine (AMT) on viral plaque formation of A/Taiwan/1/86 (H1N1) strain was measured. (B) The IC50 value of anti-influenza activity of DPQ measured in WSN-Ren luciferase assay was determined to be 435 µM.
To initiate influenza viral RNA synthesis, three subunits of RdRp and NP must bind to the RNA promoter.7 The RdRp binding site was proposed to be the A9-A11, C21-G24, and U29-U32.8 Mutations in these nucleotides inhibit transcription and replication, and naturally occurring sequence variation within the terminal helix caused down-regulated transcription as well as significant structural changes.9 The structure of the free RNA promoter suggested that the (A-A)-U internal loop and the bent terminal helix have unique structural and dynamic properties that could be recognized specifically by the RdRp.3 Thus, identification of inhibitory molecules that interact with the internal loop and/or terminal helix of the promoter would be an attractive approach to developing new anti-influenza drugs.
A new inhibitor we identified, DPQ, interacts with the major groove in the internal loop of the promoter, a prominent binding feature.10 We observed that the internal loop structure of the promoter widens upon DPQ binding to form an enlarged binding pocket. This structural change and the competitive DPQ binding to the internal loop might inhibit the association of the RdRp subunits. Furthermore, the orientation of the terminal helix is altered as well when DPQ binds to the promoter sequence. Since the bent structure of the terminal stem has been proposed to be an important feature for RdRp recognition,3 preventing the natural bending of the terminal helix and rigidifying the structure could also inhibit RdRp assembly and therefore influenza virus transcription and/or replication.
In summary, our structural results and cellular activity tests of the RNA-DPQ complex identify a novel scaffold for developing anti-influenza drugs with novel molecular target and mechanism of action. Our studies clearly suggest that further characterizations of DPQ, and its chemical elaboration to improve its affinity for the promoter and consequently anti-viral activity, are warranted with the long term goal of identifying and developing a novel class of anti-influenza agents.
Supplementary Material
Acknowledgments
This work was supported by National Research Foundation of Korea [B.-S.C]. Research in Seattle was funded from the National Institute of Allergy and Infectious Diseases, the National Institutes of Health (NIH), and Department of Health and Human Services. M.P. acknowledges the NIH for generous financial support.
Footnotes
Electronic Supplementary Information (ESI) available: additional data and materials and methods are provided. See DOI: 10.1039/b000000x/
Notes and references
- 1.(a) Compans RW, Content J, Duesberg PH. J Virol. 1972;10:795. doi: 10.1128/jvi.10.4.795-800.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Coloma R, Valpuesta JM, Arranz R, Carrascosa JL, Ortin J, Martin-Benito J. PLoS Pathog. 2009;5:e1000491. doi: 10.1371/journal.ppat.1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Desselberger U, Racaniello VR, Zazra JJ, Palese P. Gene. 1980;8:315. doi: 10.1016/0378-1119(80)90007-4. [DOI] [PubMed] [Google Scholar]; (b) Fodor, Pritlove DC, Brownlee GG. J Virol. 1994;68:4092. doi: 10.1128/jvi.68.6.4092-4096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Brownlee GG, Sharps JL. J Virol. 2002;76:7103. doi: 10.1128/JVI.76.14.7103-7113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae SH, Cheong HK, Lee JH, Cheong C, Kainosho M, Choi BS. Proc Natl Acad Sci U S A. 2001;98:10602. doi: 10.1073/pnas.191268798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Noble E, Mathews DH, Chen JL, Turner DH, Takimoto T, Kim B. J Biol Chem. 2011;286:22965. doi: 10.1074/jbc.M111.239509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanchet C, Pasi M, Zakrzewska K, Lavery R. Nucleic Acids Res. 2011;39:W68. doi: 10.1093/nar/gkr316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Schols D, Pauwels R, Vanlangendonck F, Balzarini J, De Clercq E. J Immunol Methods. 1988;114:27. doi: 10.1016/0022-1759(88)90149-4. [DOI] [PubMed] [Google Scholar]; (b) Kim M, Yim JH, Kim SY, Kim HS, Lee WG, Kim SJ, Kang PS, Lee CK. Antiviral Res. 2012;93:253. doi: 10.1016/j.antiviral.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 7.Noda T, Sugita Y, Aoyama K, Hirase A, Kawakami E, Miyazawa A, Sagara H, Kawaoka Y. Nat Commun. 2012;3:639. doi: 10.1038/ncomms1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Fodor E, Seong BL, Brownlee GG. J Gen Virol. 1993;74(Pt 7):1327. doi: 10.1099/0022-1317-74-7-1327. [DOI] [PubMed] [Google Scholar]; (b) Tiley LS, Hagen M, Matthews JT, Krystal M. J Virol. 1994;68:5108. doi: 10.1128/jvi.68.8.5108-5116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Lee MK, Bae SH, Park CJ, Cheong HK, Cheong C, Choi BS. Nucleic Acids Res. 2003;31:1216. doi: 10.1093/nar/gkg214. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Jiang H, Zhang S, Wang Q, Wang J, Geng L, Toyoda T. Virology. 2010;408:190. doi: 10.1016/j.virol.2010.09.022. [DOI] [PubMed] [Google Scholar]
- 10.Davidson A, Patora-Komisarska K, Robinson JA, Varani G. Nucleic Acids Res. 2011;39:248. doi: 10.1093/nar/gkq713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.