Abstract
Background
Identification of MET genetic alteration, mutation, or amplification in oropharyngeal squamous cell carcinoma (OPSCC) could lead to development of MET selective kinase inhibitors. The aim of this study was to assess the frequency and prognostic value of MET gene mutation, amplification, and protein expression in primary OPSCC.
Methods
A retrospective chart review was conducted of patients treated for single primary OPSCC between January 2007 and December 2009. Pre-treatment OPSCC tissue samples were analyzed for MET mutations, gene amplification, and overexpression using Sanger sequencing, FISH analysis, and immunohistochemistry respectively. Univariate and multivariate analyses were used to analyze correlations between molecular abnormalities and patient survival.
Results
143 patients were included in this study. Six cases (4%) were identified that had a genetic variation, but previously described mutations such as p.Tyr1235Asp (Y1235D) or p.Tyr1230Cys (Y1230C) were not detected. There were 15 high polysomy cases, and only 3 cases met the criteria for true MET amplification, with ≥10% amplified cells per case. Immunohistochemistry evaluation showed 43% of cases were c-MET negative and in 57% c-MET was observed at the tumor cell level. Multivariate analysis showed no significant association between MET mutation, amplification, or expression and survival.
Conclusions
Our study shows a low frequency of MET mutations and amplification in this cohort of OPSCC. There was no significant correlation between MET mutations, amplification, or expression and patient survival. These results suggest that patient selection based on these MET genetic abnormalities may not be a reliable strategy for therapeutic intervention in OPSCC.
Introduction
Head and neck cancer (HNC) is the sixth most common cancer in the world, with approximately 635,000 new cases worldwide in 2008.[1] Oropharyngeal cancer (OPC) was responsible for an estimated 36,540 new cases and 7,880 deaths in 2010 in the United States[2], and 91,900 new cases and 41,700 deaths in 2008 in Europe[3]. The incidence is higher in males (3.9% of total cancers) than females (1.6% of total cancers).[3] OPC includes tumors arising within the posterior pharyngeal wall, soft palate, tonsillar region, and the base of the tongue. The majority of these tumors (>95%) are squamous cell carcinomas (SCC) of mucosal origin.[4] A rise in the incidence of OPC has been observed in the developed world, particularly in North America, which may be explained by oncogenic Human Papilloma Virus (HPV) infection. HPV-positive and HPV-negative OPC's represent distinct subgroups. The first has a better prognosis, whereas the second is associated with tobacco and alcohol abuse and has a poorer prognosis.[5]
MET is a receptor tyrosine kinase located on chromosome 7q31 that encodes several functional domains: the semaphoring (SEMA) domain (ligand-binding), juxtamembrane (JM) domain (regulatory), and the tyrosine kinase (TK) domain.[6], [7] The only known ligand for MET is the hepatocyte growth factor/scatter factor (HGF/SF).[8] HGF/MET signalling leads to various cellular responses, such as increased cell growth, cell motility, survival, angiogenesis, wound healing, tissue regeneration, and invasion/metastasis.[6], [7] MET activation can occur via multiple molecular events including: gene mutation, gene amplification, protein overexpression, or ligand dependent autocrine and paracrine loop.[9]–[12]
MET is overexpressed in various solid tumors (eg. small cell lung cancer, non-small cell lung cancer, gastric cancer, renal cell carcinoma, and breast cancer)[6], [13]–[17] and its overexpression is often associated with an aggressive phenotype and poor prognosis. In head and neck squamous cell carcinoma (HNSCC) specifically, several reports indicate 70% to 100% MET overexpression, suggesting that the HGF/MET pathway plays a role in carcinogenesis.[8], [12], [18]–[21] HGF/MET signalling could also interfere with response of oropharyngeal squamous cell carcinoma (OPSCC) to radiotherapy (RT).[22]
Furthermore, MET genetic alterations have been previously reported in HNC. MET Y1235D mutation (also known as Y1253D) was found in 15 out of 138 patients (11%) with OPSCC[23], and in 21 out of 152 patients (14%) with HNSCC[24], suggesting a relatively high prevalence of this mutant in the OPC. This MET Y1235D mutation and another mutation in the MET activation loop (Y1230C) were also detected in neck lymph nodes metastases from a primary HNSCC tumor.[25] Recently, another epidemiological study reported novel mutations in the SEMA, JM, and TK domains of MET in 13.5% of the cases, and an increased MET gene copy number (>10 copies) in 3 out of 23 cases.[8]
The identification of a cancer type in which MET genetic alteration, mutation, or amplification is present in a significant subset such as OPSCC is of high interest. This could lead to development of MET selective kinase inhibitors, since genetic alterations are often responsible for oncogenic addiction. There are various recent clinical trials that have already demonstrated the effect of MET inhibitors in patients with a variety of advanced or metastatic tumors, including non-small-cell lung cancer, and breast, prostate, liver, and renal cancer.[10] The aim of this study is to determine the frequency and prognostic value of MET gene mutation, amplification, and protein expression in OPSCC.
Materials and Methods
Ethics Statement
The Gustave Roussy Cancer Institute (IGR) Institutional Review Board gave approval for this study. Written informed consent was obtained from included patients, and patient confidentiality was protected throughout the study.
Patient selection
A retrospective chart review was conducted for patients treated for a single primary OPSCC between January 2007 and December 2009 at the IGR. Patients with recurrences, multiple, synchronous, or metachronous lesions were excluded from this study.
Pretreatment tissue samples were retrieved from the IGR Biobank. Most tissue samples were obtained by surgical biopsy and fixed using AFA, while tissue samples obtained during surgical resection were fixed with 10% formalin. Based on hematoxylin and eosin (H&E) staining, tissue samples with at least 70% tumor cells were selected. DNA sequencing, fluorescence in situ hybridization (FISH) assay, immunohistochemistry (IHC), and HPV detection were done in different laboratories, in a strictly blind fashion.
DNA extraction and sequencing
Genomic DNA was extracted from 10 to 20 µm tissue bloc sections from tumors using the DNeasy Blood and tissue kit (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. The full coding sequences of MET exons 2 and 13 through 21 (NM_000245.2), exons already described as known locations for oncogenic mutations, were analyzed. Sanger sequencing was done following polymerase chain reaction (PCR) amplification of target exons. The primer sequences are reported in Table 1. The sequencing reactions were carried out using the BigDye Terminator Cycle Sequencing Kit as indicated by the manufacturer (Applied Biosystems, Forster City, CA). Sequencing reactions were analyzed on a 48-capillary 3730 DNA Analyzer. The sequence alignment and analysis were performed with SeqScape software (Applied Biosystems). All detected mutations were confirmed at least once with independent PCR amplification.
Table 1. Primer sequences for MET gene* (reference sequence NM 001127500.1, gene ID 4233, Chr7q31).
| Primer Couple ID | Exon Targeted | Foward Primer | Foward Primer Lengh | Reverse Primer | Reverse Primer Lenght |
| S327 | 2 | AATGGTATAGGTCTTTCAGTTTTCTCTTC | 29 | TGTAGAATGACATTCTGGATGGGT | 24 |
| S328 | 2 | AGTCCGAGATGAATGTGAATATGAAG | 26 | AGCCATGTTGATGTTATCTTTCCA | 24 |
| S150 | 2 | GATTGTTTCCCATGTCAGGACTG | 23 | TGGTATTGCCTACAAAGAAGTTGATG | 26 |
| S151 | 2 | CTGTGTGGTGAGCGCCC | 17 | AACCTGATTATTCTTGTGTGAAAAGTCT | 28 |
| S152 | 2 | CGGTCCAAAGGGAAACTCTAGAT | 23 | ACATATTTGATAGGGAATGCACACAT | 26 |
| S153 | 2 | TAGGAGCCAGCCTGAATGATG | 21 | CTGCAACTATTTTGGATAAACACCAT | 26 |
| S329 | 13 | CAACCTGTGTAGTACAAATATCTATCATGG | 30 | GACAATCTTAAACTGTAATGACTGTGTTCTTA | 32 |
| S022 | 14 | CACTGGGTCAAAGTCTCCTGG | 21 | TGTCACAACCCACTGAGGTATATGT | 25 |
| S330 | 15 | TTTCAGTCCCCATTAAATGAGGTTT | 25 | GGCCAAAGATAAAATGCTTACTGGA | 25 |
| S023 | 16 | AAAATGAAGCTCATAAAGGGTTTGA | 25 | GGCCCATAATTTCAGTGGTAGC | 22 |
| S154 | 17 | CTCTTCCTATCTAAATTTGACAAAAGTATTCA | 32 | GAAGGGATGGCTGGCTTACA | 20 |
| S155 | 18 | CTTGAGCCATTAAGACCAAACTAATTT | 27 | ACAGTGGGAAACAGATTCCTCC | 22 |
| S024 | 19 | AATTATTCTATTTCAGCCACGGGT | 24 | AAAACTGGAATTGGTGGTGTTGA | 23 |
| S331 | 20 | TTAGTTACCAAGACCTACTGATTTCCTTTC | 30 | TTTGAAGGCAGGCATTTCTGTA | 22 |
| S332 | 21 | TTTACAGAAATGCCTGCCTTCAA | 23 | TCAGGCAGTGAAAAAACCATTG | 22 |
*MET proto-oncogene (hepatocyte growth factor receptor); other aliases: AUTS9, HGFR, RCCP2, c-MET
Detection of HPV DNA viral load using RealTime qPCR
Tests were done to detect the three oncogenic HPV strains (16, 18, and 33) as well as the reference gene gACTB (β-actin) as previously described by Melkane et al.[26] Briefly, quantitative HPV DNA viral load measures were available, and a semi-quantitative viral load categorization was obtained according to individual Cycle Thresholds (CT); CT≥45: −, CT<45: +. All mildly HPV-positive samples (38≤CT<45) were confirmed on independent testing.
MET gene FISH assay
FISH for the MET gene was performed on 4 µm tumor sections, using the ZytoLight SPEC MET/CEN 7 Dual Color Probe (ZytoVision GmbH, Bremerhaven, Germany) according to the manufacturer's instructions. SNU-5 (ATCC® CRL-5973™) cell line xenografts bearing MET amplification were used as positive control samples to set up the FISH staining protocol. Briefly, the probe was co-denatured for five minutes at 84°C on the slide, and then incubated overnight at 44°C. Slides were washed with post-hybridization wash buffer (0.5X SSC/0.1% SDS) for five minutes at 37°C, after which they were air-dried and counterstained with DAPI dissolved in an anti-fade mounting solution. At least 100 tumor cells were analyzed, and the number of fluorescent signals within the nuclear boundary of each evaluable interphase tumor cell was counted using an Axiophot-ZEISS fluorescent microscope at 1000× magnification. Only nuclei with unambiguous centromeric (D7Z1 locus) hybridization red signals (as control) and MET (7q31) probe green signals were scored. Tumors were classified as previously described by Capuzzo et al.[27] Briefly, FISH negative was when there were two or three copies of the MET gene present in major clone of tumor cells, or when four MET gene copies were present in less than 40% of tumor cells. The term ‘high polysomy’ was applied if four to six copies of the MET gene were present in more than 40% of tumor cells. In order to declare ‘gain of gene’, more than one or two supplementary copies of the MET gene (compared to the centromeric probe) had to be present in tumor cells. Amplification was defined as a MET gene to centromere ratio >2.
IHC
Immunostaining
We performed MET and p-MET immunostaining using serial tissue sections. Briefly, antigen retrieval was performed with Cell Conditioning 1 (CC1) buffer at 95°C for 8 minutes, and then at 100°C for 28 minutes. After the endogen biotins blocking step, slides were incubated with the primary anti-antibody for one hour at 24°C for the rabbit anti-human c-MET (final dilution 1/50, clone SP44, reference M3444, Spring Bioscience, USA or clone CVD13, reference 18-2257, Invitrogen, USA) and at 37°C for anti-p-MET (Tyr1234/1235) (final dilution 1/50, clone D26, reference 3077, Cell Signaling Technologies, USA). A post-fixation step with glutaraldehyde (0.05% in NaCl 0.9% w/v) for 4 minutes at 24°C was done. For MET detection, the secondary antibody biotin-SP-conjugated Affinipure goat anti-rabbit (reference 111-065-003, batch 84328, Jackson ImmunoResearch Laboratories, Inc, USA) was incubated at 24°C for 32 minutes at 0.5 microg/mL. For p-MET, the secondary antibody biotin free peroxidase multimer anti-rabbit UltraMap™ was incubated at 24°C for 16 minutes. Immunostaining was done with 3,3-diaminobenzidine tetrahydrochloride (DAB) from DABMap™ chromogenic detection kit according to manufacturer's recommendations. A counter-staining step was done with hematoxylin and blueing reagent was applied. Stained slides were dehydrated and coverslipped with cytoseal XYL (8312-4, Richard-Allan Scientific, USA).
Xenograft tumors
Investigation using xenograft tumors was done to determine the impact of fixative type on p-MET detection. SNU-5 (ATCC® CRL-5973™) tumor cell line xenografts were divided into two groups, and placed in neutral buffered formalin (HT50112, Sigma-Aldrich, France) or AFA (acetic acid, formaldehyde, alcohol) fixative.
Image analysis
Immunostained slides were scanned using the ScanScope XT system (Aperio Technologies, Vista, CA). Digitized images were captured using the ImageScope software (version 10.2.2.2319, Aperio Technologies) at 20× magnification.
IHC scoring
Staining evaluation included the histological site of reactivity, main type of reactive cell, staining, intensity, and cell staining frequency. The negative samples were scored as 0+. The positive samples were scored with a scale of intensity from 1+ to 3+. Ranges of intensities were described as weak (0 to 1+), moderate (1+ to 2) and strong (2+ to 3+). Cell frequency was the percentage of immunostained cells, and was estimated by observation by a histologist as the median per sample. The cell frequency was divided into five categories of proportion score: 1 (0–5%), 2 (6–25%), 3 (26–50%), 4 (51–75%) and 5 (76–100%).
Statistical analyses
Descriptive statistics were used to summarize the study cohort. The Kaplan-Meier method was used to estimate overall survival (OS), progression free survival (PFS) and specific survival (SS). The evaluation and selection of covariates to be included in the multivariate model for survival was done as follows. The first step was to assess the correlation between covariates, in order to avoid keeping in the model two covariates when they are highly correlated and bring similar information. A threshold of 0.4 was used for this selection. Then, univariate analyses were conducted using Cox Proportional Hazards Model on the above factors to identify the variables with the highest correlation with OS, PFS, and SS. Lastly, the multivariate Cox Proportional Hazards Model was run using the stepwise procedure with a variable entry of 5% level and a variable removal at each step of 10% level. When two covariates were highly correlated according to the threshold previously defined, only the most significant one in univariate analysis was kept for the multivariate analysis.
Results
Patient and tumor characteristics
Medical files of 150 consecutive patients with OPSCC were reviewed. Of these patients, three presented with synchronous tumor lesions, three had recurrent disease, and one patient had insufficient tissue material available for laboratory studies, and were excluded. There were 143 patients included in this study (Table 2). The majority of patients were male (69%), and the overall mean age of presentation was 59 years (range 27 to 87 years). Most patients had previous tobacco exposure, with 47/143 active smokers and 48/143 former smokers (quit for ≥6 months). Of the patients with previous alcohol use, 42/143 were active consumers and 18/143 were former consumers (quit for ≥6 months). Fifty-nine patients (41%) experienced combined alcohol and tobacco exposure (either active or past), whereas 47 patients (32%) had never been exposed to either carcinogen. Seventy tumors (79%) display p16 staining. Nevertheless, because we have previously shown that HPV DNA viral load status had a higher prognostic value than the p16 expression status, we focus on HPV DNA status for analysis.[26] Eighty-eight tumors (62%) were detected with HPV DNA on qPCR, irrespective of the viral load level, the involved HPV serotype, or the exclusive or combined infection status. There were 55 (38%) cases that were HPV negative.
Table 2. Patient and tumor characteristics.
| Parameter | n (%) | ||
| Total | 143 (100) | ||
| Sex | |||
| Male | 98 (69) | ||
| Female | 45 (31) | ||
| Age | |||
| <50 | 21 (15) | ||
| ≥50 | 122 (85) | ||
| Tobacco exposure | |||
| Yes | 95 (66) | ||
| No | 48 (34) | ||
| Alcohol exposure | |||
| Yes | 60 (42) | ||
| No | 83 (58) | ||
| Tumor location | |||
| Tonsillar fossae and pillars | 90 (63) | ||
| Base of tongue | 21 (14) | ||
| Glosso-tonsillar sulcus | 11 (8) | ||
| Valleculae | 8 (6) | ||
| Soft palate | 7 (5) | ||
| Posterior pharyngeal wall | 6 (4) | ||
| Differentiation | |||
| Well | 70 (49) | ||
| Moderate | 50 (35) | ||
| Poor | 23 (16) | ||
| T stage | |||
| T1/T2 | 59 (41) | ||
| T3/T4 | 84 (59) | ||
| N stage | |||
| N0 | 48 (34) | ||
| N1-N3 | 95 (66) | ||
| AJCC* classification | |||
| Stage I/II | 29 (20) | ||
| Stage III/IV | 114 (80) | ||
*American Joint Committee on Cancer
The majority of tumors were localized in the tonsillar fossae and pillars (63%), or at the base of the tongue (14%). The macroscopic aspect of the tumors was exophytic in 88 cases (62%), and ulcerative or infiltrative in 55 cases (38%). Histopathologically, approximately half of the tumors was well-differentiated, and the other half was moderately or poorly differentiated. The majority of patients presented with T3 or T4 stage tumors, and had N1-N3 lymph node metastases. There were six patients (4%) with distant metastases at the time of evaluation, all with pulmonary metastases. According to the 2010 AJCC classification system, 29/143 patients (20%) presented with early-stage (stage I and II) tumors, while a majority of 114/143 patients (80%) had advanced stage (stage III and IV) disease.
Most of the patients (136/143, 95%) in this study were treated in a curative setting. Patients were treated by RT; either exclusively (31/143 patients, 21%), or by concomitant chemoradiotherapy (CRT) (75/143 patients, 52%), and surgery; either exclusively (15/143 patients, 11%), or followed by adjuvant RT or CRT (16/143 patients, 11%). The remaining six patients (5%) with distant pulmonary metastases were offered treatment with palliative chemotherapy.
MET mutations in OPSCC
Exons 2, 13, 14, 15, 16, 17, 18, 19, 20, and 21 were analyzed in all 143 cases, but due to low tissue sample quality, some cases had several failed PCR's (>5 failures) and were removed from further analyses. Overall, six cases (4%) were identified that had a genetic variation (Table 3).
Table 3. The location and predicted effect of each MET variation found, numbered from the reference sequence NM 000245.2.
| Variation | # of cases | Location | Align-GVGD† [48] | SIFT‡ [49] | PolyPhen-2[50] |
| p.Val136Ile | 1 | Semaphoring domain | Class C0 (benign/no effect) | tolerated | benign/no effect |
| p.Glu312Lys | 1 | Semaphoring domain | Class C0 (benign/no effect) | tolerated | benign/no effect |
| p.Thr1036Ile | 1 | Juxtamembrane domain | Class C15 (damaging) | affects protein function | benign no effect |
| p.Cys1210Arg | 1 | Kinase domain | Class C0 (benign/no effect) | affects protein function | probably damaging |
| p.Ala347Thr | 2 | Semaphoring domain | Class C55 (damaging) | affects protein function | probably damaging |
Mutation Chromatograms include reference sequences and variant description sequence variations described using IUPAC code (http://www.insdc.org/).
Align-Grantham Variation Grantham Deviation.
Sorts Intolerant From Tolerant.
Four of these mutations were located on exon 2, one on exon 15, and one on exon 18. The p.Val136Ile mutation has been previously detected once before in a kidney carcinoma case[6], and the p.Ala347Thr mutation has been described in lung squamous cell carcinoma[28]. The p.Glu312Lys, p.Thr1036Ile, and p.Cys1210Arg variations have not yet been described in literature and are novel somatic variant with unknown pathogenicity.[29] Previously described mutations such as p.Tyr1235Asp (Y1235D) or p.Tyr1230Cys (Y1230C) in exon 19 were not found. Of the six variations, four were expected to affect protein function based on the SIFT classification system, three of which had predicted damaging effects according to the PolyPhen-2 scoring system.
MET amplification in OPSCC
FISH analysis was performed for 128 out of 143 cases, because there was insufficient tissue from the remaining cases for further analysis. Hybridization was successful for 97 out of 128 cases. Of those cases, 39 were considered normal, 17 had monosomy of chromosome 7 or deletion of MET gene, 23 had low polysomy, and 15 had high polysomy of chromosome 7 (Table 4, Figure 1). In the high polysomy category, two cases were detected as gene amplifications, but the amplified clone presented only 4–5% of cells and thus did not meet the criteria to be included in the “amplified” category. Only three cases met the criteria for true MET amplification, with ≥10% amplified cells per case.
Table 4. Fluorescence in situ hybridization results.
| Gene status | # of cases | Classification |
| Gene amplification | 3 | + |
| High polysomy | 15 | + |
| Normal | 39 | − |
| Monosomy/deletion | 17 | − |
| Low polysomy | 23 | − |
| Total | 97 |
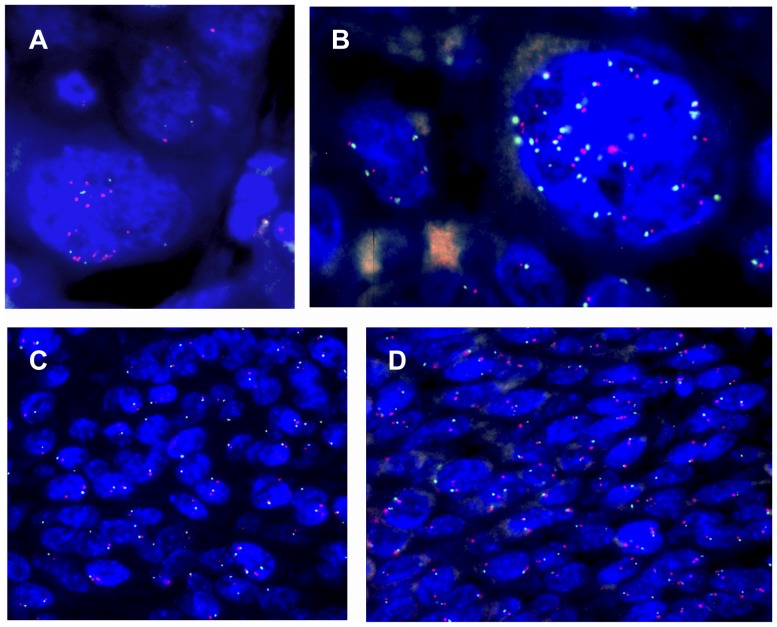
Figure 1. FISH with two color probes: chromosome 7 centromere (green) and MET gene (red).
A) True MET gene amplification in 10% of cells: 4–8 centromere signals and 16–20 MET signals, ratio >2.0. B) High polysomy: the same number of control and MET gene spots were seen in 15% of giant cells, ratio is 1.0. C) Chromosome 7 monosomy: only one control and one MET signal were detected for this case. In some cells there is only one signal (or no signal) due to a nuclei section. D) Normal hybridization pattern: two control spots and two MET gene spots. Again, some cells show only one of two signals due to a nuclei section.
MET protein expression in OPSCC
IHC evaluation was done for 113 cases, and a total of 107 cases passed the quality controls and were included (Table 5). Forty six cases (43%) were c-MET negative. c-MET protein was observed in 61 cases (57%) at the tumor cell level. Of the 61 c-MET positive cases, immunostaining was localized at the membrane ± cytoplasm level in 50 cases (82%), with moderate expression (median intensity 1+ to 2+, and median cell frequency 50% to 75%). In the other 11 cases (18%), immunostaining was observed only at the cytoplasm level, with moderate expression (median intensity 1+, and median cell frequency 75% to 100%; Figure 2). Detection of pYY1234-1235 MET was observed in two cases (3%), but in non-tumor cells (at margin of tissue sample). AFA fixation resulted in complete disappearance of p-MET immunostaining (Figure 2). p-MET analysis was therefore only done for formol-fixed tissue.
Table 5. Immunohistochemistry results[26].
| Parameter | n (%) |
| Analyzable cases | 107 (100) |
| c-MET negative cases | 46 (43) |
| c-MET positive cases | 61 (57) |
| Membrane with or without cytoplasm expression | 50 (82) |
| Cytoplasm expression | 11 (18) |
| p-MET positive cases | 2 (3) |
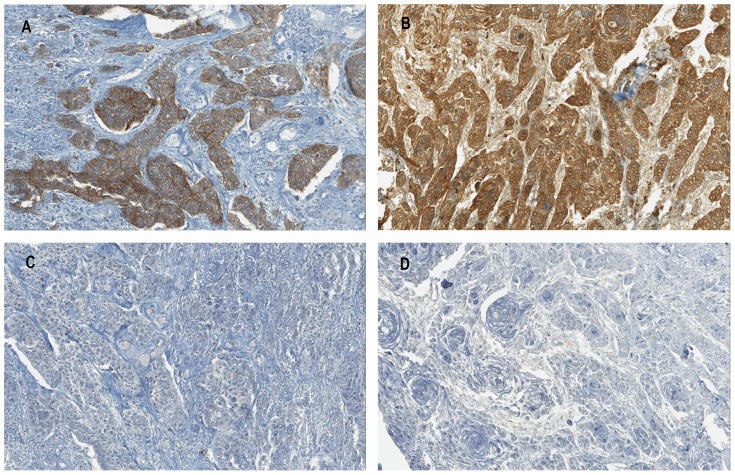
Figure 2. IHC staining for c-MET and pYY1234-1235 MET in OPSCC specimens.
Moderate (A) and strong (B) membranous and cytoplasm c-MET immunostaining in tumor cells. No p-MET immunostaining was observed in serial section (C and D). (original magnification ×20).
Survival analysis
The median follow-up time was 2.6 years (min. 11 days and max. 13.9 years), and there were 57/143 (40%) deaths observed. The median OS was 5.3 years. The probabilities of surviving (95% confidence interval) at one, two, and three years were 82.4% (CI: 76.1–88.7), 67.3% (CI: 59.5–75.1) and 64.5% (56.5–72.6) respectively. There were 71 oncological events (tumor progression, loco-regional or distant recurrence, and death) observed in the 143 patients. The median PFS was 4.4 years. The probabilities of having an oncological event (95% confidence interval) at one, two, and three years were 68.3% (CI: 60.7–76.0), 61.8% (CI: 53.8–69.9), and 57.5% (CI: 49.2–65.8) respectively. There were 30/143 (21%) cancer-related deaths. The positive HPV status, alcohol exposure, high tumor differentiation, T3/T4 staging, N1-3 staging, presence of metastases and advanced disease (AJCC classification III/IV) were significant at 5% for OS in the univariate Cox Proportional Hazards Model. Except N1-3 staging, these variables were also significant for PFS (at 5%). Univariate analysis did not show any significant association between OS, PFS, or SS and MET mutations, MET amplification, or MET overexpression. It is notable that MET positive cases were not specifically distributed in either the HPV positive or HPV negative groups. The multivariate analysis did not show any significant association between OS, PFS, or SS and MET mutations, MET amplification, or MET overexpression.
Discussion
The aim of this study was to determine the frequency and prognostic value of MET abnormalities including DNA mutations, gene amplification, and protein expression in primary OPSCC. Our study shows a low frequency of MET mutations and MET amplification in OPSCC. There was no significant correlation between MET mutations, amplification, or expression and patient survival.
In our study six cases were identified with MET genetic variations, four of which have not been previously described, though the clinical relevance of these variations remains unclear. Unfortunately, non-tumor tissue was not available to confirm if those mutations were somatic. Overall, our MET mutation incidence (4%) was low compared to a number of other studies. For example, Seiwert et al. found a 13.5% mutation incidence in the semaphoring, juxtamembrane, and tyrosine kinase domains of the MET gene in 66 HNSCC tumor tissues, as well as MET over expression in 84% of samples.
Previously described HNSCC mutations such as p.Tyr1235Asp (Y1235D)[23]–[25] and p.Tyr1230Cys (Y1230C)[25] were not detected in our study. In a study investigating the prevalence and clinical impact of the Y1253D mutation in patients with OPSCC treated by radical RT, the mutation was detected in 15/138 tumors (10.9%). Also, survival analysis showed a significant correlation between Y1253D mutation and impaired local tumor control.[23] Another study by Ghadjar et al. detected Y1253D mutation in 21 tumors in 152 patients (14%) with HNSCC and observed an association with decreased distant metastasis-free survival.[24] On the other hand, a set of 12 oral SCC's were examined for mutation and while overexpression of the MET receptor was present in all cases, point mutations were not detected.[20]
Studies investigating MET mutations use different DNA detection techniques. Our Sanger sequencing approach is known to detect mutation from 20–30% of mutated DNA. Our cohort included only samples with more than 70% of tumor cells. Nevertheless, our Sanger approach does not detect minor clones representing less than 20% of the tumor cell, but can detect unexpected mutations. More sensitive techniques such as the technique used by Aebersold et al.[23] can detect minor clones, but can only detect mutations that are targeted specifically. We could hypothesize that the technique used in our study may have yielded a relatively lower DNA mutation frequency than a more sensitive technique, if MET mutations are mainly present in minor clones. One the other hand, we could also stipulate that the present French cohort had a lower rate of MET mutation that other published cohorts.
We also found a low frequency of amplification, with no correlation with patient survival. Out of 97 cases, there were only three cases that met the criteria for true MET amplification and 15 cases with high polysomy. A previous study found >10 MET gene copies in 3 of 23 (13%) HNSCC tumor tissues.[8] In other studies, MET gene amplification has been associated with poor prognosis in gastric and lung cancer. Lee et al. recently found 61 MET high polysomy and 13 MET gene amplification cases in a cohort of 438 gastric carcinomas. In their study, gene amplification correlated with a poor prognosis.[14] In another study concerning gastric cancer patients, c-MET amplification again correlated and poor survival.[15] In a cohort of 380 non-small cell lung carcinoma cases, Park et al. found that an increase of MET gene copy number, present in 11.1% of patients (high polysomy 8.7% and gene amplification 2.4%), was a negative prognostic factor for survival.[13] The relatively low frequency of MET mutations and amplification in this OPSCC study cohort could be explained, because MET may play a larger role in disease progression in other types of cancer, such as gastric or lung cancer.
Another possible explanation for the low frequency of MET mutations and amplification in our study cohort could be that tissue samples were from primary OPSCC's, and not from metastases. There is evidence from previous studies that MET mutations are primarily involved in tumor progression to the metastatic phase.[9], [30]–[36] A study by Di Renzo et al. showed that in 4/15 HNSCC cases, activating MET mutations undergo clonal expansion during metastatic spread, as their frequency increased from 2% in the primary tumors to 50% in the metastases.[25] MET overexpression was also found to be significantly higher in tumor stages associated with enlarged or multiple (N2-N3) lymph node metastases.[37] These findings indicate that the frequency of MET molecular abnormalities could be higher in metastatic tissue than in primary tumors, such as the tissue samples studied in our cohort.
Protein expression analysis showed that out of 107 analyzable cases, 46 cases (43%) were c-MET negative, and 61 cases (57%) were c-MET positive at the tumor cell level. We found no significant relationship between MET expression and survival. This is in contrast with previous studies showing that MET expression is an early event in HNC carcinogenesis[38], and has an association with a poorer overall survival rate[19], [39]. Later, reports describe that compartment localization of c-Met is linked to differentiation and stage of OPSCC tumors.[19] In one study, HNSCC's and their metastases were analyzed, showing that MET expression was increased from 2- to 50-fold in about 70% of tumors.[37] Kim et al. previously reported that elevated HGF/c-MET expression in HNSCC correlated with tumor progression[40], and later showed that survival was significantly affected in patients with c-MET expression in SCC of the oral tongue[21], [41].
In previous studies analyzing MET expression by IHC, various guidelines were used to classify data. For example, different cut-off values have been used to determine a positive result, such as MET expression in >10% of tumor cells[42] or MET expression in >30% of tumor cells[43]. These variations in data interpretation can cause the same raw data to yield different (possibly misleading) conclusions.
Unfortunately in our study, analysis of p-MET expression was only possible for select cases, since the AFA fixative caused complete disappearance of p-MET immunostaining. In the future, AFA fixative must be avoided when p-MET IHC is done on archival biopsies. Another factor that may have affected the incidence of MET molecular abnormalities was a relatively large number (62%) of HPV positive patients in our sample set. HPV dependant carcinogenesis may partially explain the relatively low incidence of MET abnormalities. It is even possible that HPV is a stronger predictive biological marker than MET. [4], [44]–[46] Our data does not suggest a relationship between HPV status and MET abnormalities.
Our study indicates that MET may not be a reliable prognostic biological marker in primary OPSCC, and therefore that anti-MET therapy may not be the ideal therapeutic option for these types of tumors. Except for HPV status, we lack reliable diagnostic and prognostic molecular markers for HNSCC, including OSCC. Screening for MET molecular abnormalities in primary OPSCC may not be efficient, partly because previous studies have indicated that MET abnormalities are predominantly detected in case of metastases. Detection of MET abnormalities might be more appropriate as a marker of response to treatment. In a pilot study, Druzgal et al. compared pre-treatment and post-treatment cytokine levels in HNSCC, and found evidence for a strong relationship between HGF serum levels and both therapeutic response and survival.[47] Uchida et al. studied HGF/MET in oral SCC and found significantly higher HGF concentrations in metastatic cancer tissues, than non-metastatic or normal tissue. A decline in serum HGF was seen in tumor-free survivors.[30] Dysregulation of the MET pathway plays a role in various human cancers, though ‘MET addiction’ only occurs in a small percentage of these cancers. There are tumors that only partially depend on MET signaling for growth and metastasis, which may be more difficult to detect using biomarkers.[11] Identification of tumors carrying the relevant genetic abnormality and methods for patient stratification according to HGF/MET expression requires further investigation to guarantee clinical benefit.[10]
Conclusions
There was a low frequency of MET mutations and amplification in this OPSCC cohort. There was no association between MET molecular abnormalities and patient survival. Our results indicate that these MET genetic abnormalities may not be reliable prognostic biological markers in OPSCC for patient selection for anti-MET therapy.
Acknowledgments
We would like to thank Nelly Motte for technical support during the experimental work done for this study. Many thanks also go to the Pathology Department at the Gustave Roussy Cancer Institute (Villejuif, France).
Funding Statement
Funding was received from Sanofi-Aventis for laboratory work done at the Gustave Roussy Cancer Institute (labor, assay development, reagents). Sanofi performed the immunohistochemistry, and provided xenograft tumor tissue with different levels of MET amplification for the characterization of the MET FISH probe. Sanofi also provided assistance for statistical analysis and was involved in data analysis.
References
- 1. Hunter KD, Parkinson EK, Harrison PR (2005) Profiling early head and neck cancer. Nat Rev Cancer 5: 127–135. [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Siegel R, Xu J, Ward E (2010) Cancer statistics, 2010. CA Cancer J Clin 60: 277–300. [DOI] [PubMed] [Google Scholar]
- 3. Ferlay J, Parkin DM, Steliarova-Foucher E (2010) Estimates of cancer incidence and mortality in europe in 2008. Eur J Cancer 46: 765–781. [DOI] [PubMed] [Google Scholar]
- 4. van Monsjou HS, Balm AJ, van den Brekel MM, Wreesmann VB (2010) Oropharyngeal squamous cell carcinoma: A unique disease on the rise? Oral Oncol 46: 780–785. [DOI] [PubMed] [Google Scholar]
- 5. Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, et al. (2010) Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med 363: 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ma PC, Tretiakova MS, MacKinnon AC, Ramnath N, Johnson C, et al. (2008) Expression and mutational analysis of MET in human solid cancers. Genes Chromosomes Cancer 47: 1025–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Peruzzi B, Bottaro DP (2006) Targeting the c-met signaling pathway in cancer. Clin Cancer Res 12: 3657–3660. [DOI] [PubMed] [Google Scholar]
- 8. Seiwert TY, Jagadeeswaran R, Faoro L, Janamanchi V, Nallasura V, et al. (2009) The MET receptor tyrosine kinase is a potential novel therapeutic target for head and neck squamous cell carcinoma. Cancer Res 69: 3021–3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF (2003) Met, metastasis, motility and more. Nat Rev Mol Cell Biol 4: 915–925. [DOI] [PubMed] [Google Scholar]
- 10. Gherardi E, Birchmeier W, Birchmeier C, Vande Woude G (2012) Targeting MET in cancer: Rationale and progress. Nat Rev Cancer 12: 89–103. [DOI] [PubMed] [Google Scholar]
- 11.Peters S, Adjei AA (2012) MET: A promising anticancer therapeutic target. Nat Rev Clin Oncol. [DOI] [PubMed]
- 12. Knowles LM, Stabile LP, Egloff AM, Rothstein ME, Thomas SM, et al. (2009) HGF and c-met participate in paracrine tumorigenic pathways in head and neck squamous cell cancer. Clin Cancer Res 15: 3740–3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Park S, Choi YL, Sung CO, An J, Seo J, et al. (2012) High MET copy number and MET overexpression: Poor outcome in non-small cell lung cancer patients. Histol Histopathol 27: 197–207. [DOI] [PubMed] [Google Scholar]
- 14. Lee HE, Kim MA, Lee HS, Jung EJ, Yang HK, et al. (2012) MET in gastric carcinomas: Comparison between protein expression and gene copy number and impact on clinical outcome. Br J Cancer 107: 325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shi J, Yao D, Liu W, Wang N, Lv H, et al. (2012) Frequent gene amplification predicts poor prognosis in gastric cancer. Int J Mol Sci 13: 4714–4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miyata Y, Kanetake H, Kanda S (2006) Presence of phosphorylated hepatocyte growth factor receptor/c-met is associated with tumor progression and survival in patients with conventional renal cell carcinoma. Clin Cancer Res 12: 4876–4881. [DOI] [PubMed] [Google Scholar]
- 17. Garcia S, Dales JP, Jacquemier J, Charafe-Jauffret E, Birnbaum D, et al. (2007) c-met overexpression in inflammatory breast carcinomas: Automated quantification on tissue microarrays. Br J Cancer 96: 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cortesina G, Martone T, Galeazzi E, Olivero M, De Stefani A, et al. (2000) Staging of head and neck squamous cell carcinoma using the MET oncogene product as marker of tumor cells in lymph node metastases. Int J Cancer 89: 286–292. [PubMed] [Google Scholar]
- 19. Lo Muzio L, Farina A, Rubini C, Coccia E, Capogreco M, et al. (2006) Effect of c-met expression on survival in head and neck squamous cell carcinoma. Tumour Biol 27: 115–121. [DOI] [PubMed] [Google Scholar]
- 20. Morello S, Olivero M, Aimetti M, Bernardi M, Berrone S, et al. (2001) MET receptor is overexpressed but not mutated in oral squamous cell carcinomas. J Cell Physiol 189: 285–290. [DOI] [PubMed] [Google Scholar]
- 21. Kim CH, Moon SK, Bae JH, Lee JH, Han JH, et al. (2006) Expression of hepatocyte growth factor and c-met in hypopharyngeal squamous cell carcinoma. Acta Otolaryngol 126: 88–94. [DOI] [PubMed] [Google Scholar]
- 22. Aebersold DM, Kollar A, Beer KT, Laissue J, Greiner RH, et al. (2001) Involvement of the hepatocyte growth factor/scatter factor receptor c-met and of bcl-xL in the resistance of oropharyngeal cancer to ionizing radiation. Int J Cancer 96: 41–54. [DOI] [PubMed] [Google Scholar]
- 23. Aebersold DM, Landt O, Berthou S, Gruber G, Beer KT, et al. (2003) Prevalence and clinical impact of met Y1253D-activating point mutation in radiotherapy-treated squamous cell cancer of the oropharynx. Oncogene 22: 8519–8523. [DOI] [PubMed] [Google Scholar]
- 24. Ghadjar P, Blank-Liss W, Simcock M, Hegyi I, Beer KT, et al. (2009) MET Y1253D-activating point mutation and development of distant metastasis in advanced head and neck cancers. Clin Exp Metastasis 26: 809–815. [DOI] [PubMed] [Google Scholar]
- 25. Di Renzo MF, Olivero M, Martone T, Maffe A, Maggiora P, et al. (2000) Somatic mutations of the MET oncogene are selected during metastatic spread of human HNSC carcinomas. Oncogene 19: 1547–1555. [DOI] [PubMed] [Google Scholar]
- 26.Melkane AE, Auperin A, Saulnier P, Lacroix L, Vielh P, et al.. (2013) Human papillomavirus prevalence and prognostic implication in oropharyngeal squamous cell carcinomas. Head Neck. [DOI] [PubMed]
- 27. Cappuzzo F, Marchetti A, Skokan M, Rossi E, Gajapathy S, et al. (2009) Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J Clin Oncol 27: 1667–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krishnaswamy S, Kanteti R, Duke-Cohan JS, Loganathan S, Liu W, et al. (2009) Ethnic differences and functional analysis of MET mutations in lung cancer. Clin Cancer Res 15: 5714–5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, et al.. (2008) The catalogue of somatic mutations in cancer (COSMIC). Curr Protoc Hum Genet Chapter 10: Unit 10.11. [DOI] [PMC free article] [PubMed]
- 30. Uchida D, Kawamata H, Omotehara F, Nakashiro K, Kimura-Yanagawa T, et al. (2001) Role of HGF/c-met system in invasion and metastasis of oral squamous cell carcinoma cells in vitro and its clinical significance. Int J Cancer 93: 489–496. [DOI] [PubMed] [Google Scholar]
- 31. Zhang YW, Vande Woude GF (2003) HGF/SF-met signaling in the control of branching morphogenesis and invasion. J Cell Biochem 88: 408–417. [DOI] [PubMed] [Google Scholar]
- 32. Benvenuti S, Comoglio PM (2007) The MET receptor tyrosine kinase in invasion and metastasis. J Cell Physiol 213: 316–325. [DOI] [PubMed] [Google Scholar]
- 33. Lai AZ, Abella JV, Park M (2009) Crosstalk in met receptor oncogenesis. Trends Cell Biol 19: 542–551. [DOI] [PubMed] [Google Scholar]
- 34. Ma PC, Maulik G, Christensen J, Salgia R (2003) c-met: Structure, functions and potential for therapeutic inhibition. Cancer Metastasis Rev 22: 309–325. [DOI] [PubMed] [Google Scholar]
- 35. Organ SL, Tsao MS (2011) An overview of the c-MET signaling pathway. Ther Adv Med Oncol 3: S7–S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. De Herdt MJ, Baatenburg de Jong RJ (2008) HGF and c-MET as potential orchestrators of invasive growth in head and neck squamous cell carcinoma. Front Biosci 13: 2516–2526. [DOI] [PubMed] [Google Scholar]
- 37. Galeazzi E, Olivero M, Gervasio FC, De Stefani A, Valente G, et al. (1997) Detection of MET oncogene/hepatocyte growth factor receptor in lymph node metastases from head and neck squamous cell carcinomas. Eur Arch Otorhinolaryngol 254 Suppl 1S138–43. [DOI] [PubMed] [Google Scholar]
- 38. Chen YS, Wang JT, Chang YF, Liu BY, Wang YP, et al. (2004) Expression of hepatocyte growth factor and c-met protein is significantly associated with the progression of oral squamous cell carcinoma in taiwan. J Oral Pathol Med 33: 209–217. [DOI] [PubMed] [Google Scholar]
- 39.Lim YC, Han JH, Kang HJ, Kim YS, Lee BH, et al.. (2012) Overexpression of c-met promotes invasion and metastasis of small oral tongue carcinoma. Oral Oncol. [DOI] [PubMed]
- 40. Kim CH, Lee JS, Kang SO, Bae JH, Hong SP, et al. (2007) Serum hepatocyte growth factor as a marker of tumor activity in head and neck squamous cell carcinoma. Oral Oncol 43: 1021–1025. [DOI] [PubMed] [Google Scholar]
- 41. Kim CH, Koh YW, Han JH, Kim JW, Lee JS, et al. (2010) c-met expression as an indicator of survival outcome in patients with oral tongue carcinoma. Head Neck 32: 1655–1664. [DOI] [PubMed] [Google Scholar]
- 42. Nakajima M, Sawada H, Yamada Y, Watanabe A, Tatsumi M, et al. (1999) The prognostic significance of amplification and overexpression of c-met and c-erb B-2 in human gastric carcinomas. Cancer 85: 1894–1902. [DOI] [PubMed] [Google Scholar]
- 43. Taniguchi K, Yonemura Y, Nojima N, Hirono Y, Fushida S, et al. (1998) The relation between the growth patterns of gastric carcinoma and the expression of hepatocyte growth factor receptor (c-met), autocrine motility factor receptor, and urokinase-type plasminogen activator receptor. Cancer 82: 2112–2122. [PubMed] [Google Scholar]
- 44. D'Souza G, Kreimer AR, Viscidi R, Pawlita M, Fakhry C, et al. (2007) Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med 356: 1944–1956. [DOI] [PubMed] [Google Scholar]
- 45. Mehanna H, Jones TM, Gregoire V, Ang KK (2010) Oropharyngeal carcinoma related to human papillomavirus. BMJ 340: c1439. [DOI] [PubMed] [Google Scholar]
- 46. van Monsjou HS, van Velthuysen ML, van den Brekel MW, Jordanova ES, Melief CJ, et al. (2012) Human papillomavirus status in young patients with head and neck squamous cell carcinoma. Int J Cancer 130: 1806–1812. [DOI] [PubMed] [Google Scholar]
- 47. Druzgal CH, Chen Z, Yeh NT, Thomas GR, Ondrey FG, et al. (2005) A pilot study of longitudinal serum cytokine and angiogenesis factor levels as markers of therapeutic response and survival in patients with head and neck squamous cell carcinoma. Head Neck 27: 771–784. [DOI] [PubMed] [Google Scholar]
- 48. Mathe E, Olivier M, Kato S, Ishioka C, Hainaut P, et al. (2006) Computational approaches for predicting the biological effect of p53 missense mutations: A comparison of three sequence analysis based methods. Nucleic Acids Res 34: 1317–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kumar P, Henikoff S, Ng PC (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4: 1073–1081. [DOI] [PubMed] [Google Scholar]
- 50. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, et al. (2010) A method and server for predicting damaging missense mutations. Nat Methods 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]