Abstract
Eukaryotic cells control their proteome by regulating protein production and protein clearance. Protein production is determined to a large extent by mRNA levels, whereas protein degradation depends mostly upon the proteasome. Dysfunction of the proteasome leads to the accumulation of non-functional proteins that can aggregate, be toxic for the cell, and, in extreme cases, lead to cell death. mRNA levels are controlled by their rates of synthesis and degradation. Recent evidence indicates that these rates have oppositely co-evolved to ensure appropriate mRNA levels. This opposite co-evolution has been correlated with the mutations in the Ccr4-Not complex. Consistently, the deadenylation enzymes responsible for the rate-limiting step in eukaryotic mRNA degradation, Caf1 and Ccr4, are subunits of the Ccr4-Not complex. Another subunit of this complex is a RING E3 ligase, Not4. It is essential for cellular protein solubility and has been proposed to be involved in co-translational quality control. An open question has been whether this role of Not4 resides strictly in the regulation of the deadenylation module of the Ccr4-Not complex. However, Not4 is important for proper assembly of the proteasome, and the Ccr4-Not complex may have multiple functional modules that participate in protein quality control in different ways. In this work we studied how the functions of the Caf1/Ccr4 and Not4 modules are connected. We concluded that Not4 plays a role in protein quality control independently of the Ccr4 deadenylase, and that it is involved in clearance of aberrant proteins at least in part via the proteasome.
Introduction
Messenger RNAs carry the information encoded within DNA to ribosomes where it is translated into the corresponding proteins. Errors regularly occur during protein synthesis. These errors may originate from the mRNA (mutations altering the coding sequence, secondary structure leading to ribosome stalling, etc…), resulting in production of defective proteins. To prevent the accumulation of such aberrant proteins cells have developed RNA quality control mechanisms that recognize defective mRNAs and degrade them efficiently [1], [2]. Errors also may occur at the protein level, when proteins fold inappropriately or interact with aberrant partners. This can be a consequence of aberrant mRNAs or due to unfavorable conditions such as lack of appropriate folding or assembly factors [3], [4].
mRNAs carry polyA tails that protect them from degradation and promotes translation in the cytoplasm. Removal of the polyA tail, deadenylation, is the first and rate-limiting step in mRNA degradation [5], [6]. In eukaryotic cells the Ccr4-Not complex provides the major deadenylation activity [7], [8], [9], [10] and thus it is an important player in RNA quality control. Besides mRNA degradation, this complex has been associated with other cellular activities, such as transcription and protein ubiquitination [11], [12], [13].
In the yeast Saccharomyces cerevisiae the Ccr4-Not complex is composed of nine core subunits, Not1-5, Caf1, Caf40, Caf130 and Ccr4. Not1 is the largest protein of the complex and the other subunits are organized around it. The Not2, 3 and 5 subunits form the Not module [14] and interact with the Not1 C-terminus [15], [16], [17]. Two ribonucleases, Caf1 and Ccr4, compose the deadenylation module [18] and bind a central domain of Not1 [17]. This structural organization is conserved in higher eukaryotes [19]. The Not4 subunit represents an E3 ligase module [20]. In yeast it is a stable subunit of the complex, but this is not the case of higher eukaryotes [21], [22], [23]. Nevertheless, the function of Not4 is certainly conserved, because the human protein complements the absence of the yeast protein [24].
Ccr4 and Caf1 are the subunits of the Ccr4-Not complex that compose the major eukaryotic deadenylase [25], [26], [27], [28]. They belong to 2 different types of deadenylation enzymes, Ccr4 - to the EEP-type family and Caf1 - to the DEDD-type family. In the yeast S. cerevisiae Caf1 contains a substitution in its catalytic site [29], [30]. Thus, only Ccr4 provides deadenylation activity in vivo and it is the primary yeast deadenylase. However in mammals and flies, Caf1 plays an important catalytic role in poly A tail shortening [8], [31], [32]. Caf1 bridges Ccr4 to Not1 [18] and this makes it essential for deadenylation activity in vivo, even in yeast [16].
Not4 is an E3 ligase of the RING family type [33] that catalyzes protein ubiquitination. The RING domain is located at the N-terminus of Not4 [34] and it is important for the ubiquitination activity of Not4, but not for its interaction with the Ccr4-Not complex [20], [35]. Several substrates of Not4 were described [36], amongst which are the ribosomal protein, Rps7A [37] and a ribosome-associated chaperone, NAC (the nascent polypeptide associated complex) [38], [39]. Consistently, Not4 and other subunits of the Ccr4-Not complex were found in translating ribosomes [37]. It was proposed that Not4 may ubiquitinate aborted proteins appearing during translational arrest and that this would lead to the degradation of these peptides by the proteasome [40]. Subsequent studies have indicated instead that the ubiquitination of aberrant products of translation mainly occurs via the E3 ligase Ltn1 [41]. Several other E3 ligases have been described to be involved in ubiquitination and degradation of misfolded proteins, such as Ubr1, Ubr2, San1 and others [42], [43], [44], [45].
A new role for Not4 in protein quality control was suggested by the recent finding that Not4 is involved in proteasome assembly [35], [36]. The proteasome is an important player in protein quality control [46], [47]. It is a large protease that eliminates aberrant proteins in the cell. It is composed of two main subcomplexes, the 20S core particle (CP) and the 19S regulatory particle (RP) [48], [49], [50]. RP is attached to one or both sides of the CP forming single- or double-capped proteasomes, respectively [51]. The RP is responsible for substrate recognition, deubiquitination and their translocation into the CP [52], [53], [54], whilst the CP provides substrate hydrolysis [55], [56], [57]. Appropriate RP and CP interaction and association into 26S proteasomes is important for normal proteasome function. Multiple factors are required for proteasome assembly and normal activity [58]. Not4 was shown to play a role in RP assembly that is important for normal RP-CP association [35].
Accumulation of polyubiquitinated proteins and increased aggregation was observed in the absence of Not4 [37]. To clarify whether that this is due to altered function of the deadenylation module of the Ccr4-Not complex in the absence of Not4, rather than to a problem with the proteasome, we have compared the involvement of the E3 ligase and deadenylase modules in protein quality control. We concluded that Not4 has a specific role in protein quality control that extends beyond regulation of deadenylation. This role consists, at least in part, in Not4’s importance for the functional integrity of the regulatory particle of the proteasome and might additionally include its role as an E3 ligase.
Methods
Cells
The Saccharomyces cerevisiae strains used in this work derive from MY1, BY4741 or SC0000 (Table 1). Single step deletions and gene tagging were performed by PCR. New strains were obtained from crosses. All media were standard.
Table 1. Yeast strains used in this study.
| Strain | Genotype | Source |
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Euroscarf [80] |
| BY4742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Euroscarf [80] |
| SC0000 | MATa ade2 arg4 leu2-3,112 trp1-289 ura3-52 | Euroscarf [81] |
| MY1 | MATa ura3-52 trp1-1 gal2 leu2::PET56 gcn4Δ | [82] |
| MY3417 | Isogenic to BY4742 except not4:: KanMX4 | Euroscarf |
| MY3419 | Isogenic to BY4742 except caf1:: KanMX4 | Euroscarf |
| MY3422 | Isogenic to BY4742 except ccr4:: KanMX4 | Euroscarf |
| MY3593 | Isogenic to MY1 except not4::KanMX4 | [38] |
| MY3621 | Isogenic to MY1 except caf1::TRP1 | [76] |
| MY4747 | Isogenic to MY1 except MATα ccr4::TRP1 | This work |
| MY4857 | Isogenic to BY4741 except not4::NOT4-TapTag-URA3 | [83] |
| MY5559 | Isogenic to SC0000 except rpn11::RPN11-TapTag-URA3 | [35] |
| MY5615 | Isogenic to BY4741 except MATα not4::HIS3 | [35] |
| MY7371 | MATα ade2 arg4 leu2-3,112 trp1-289 ura3-52 rpn11::RPN11-TapTag-URA3 caf1:: HIS3 | [35] |
| MY10004 | MATa ura3-52 leu2-Δ1 his3-Δ200 prc1-1 cim3-1 | From C. Mann [84] |
| MY10005 | MATa ura3 leu2-3,112 his3-11,15 pre1-1 | From D. Wolf [85] |
| MY10107 | MATα rpn11::RPN11-TapTag-URA3 ccr4::TRP1 | From the cross MY5559 x MY4747, this work |
| MY10402 | Isogenic to BY4742 except ubr1:: KanMX4 | Euroscarf |
| MY10404 | Isogenic to BY4742 except ubr2:: KanMX4 | Euroscarf |
| MY10406 | Isogenic to BY4742 except ltn1:: KanMX4 | Euroscarf |
| MY10408 | Isogenic to BY4742 except san1:: KanMX4 | Euroscarf |
Plasmids
A plasmid expressing CPY*-HA under control of the copper dependent promoter, CUP1, was a kind gift of T. Sommer [59]. Plasmids expressing GFP-K0-FLAG-HIS3, GFP-K12(AAA)-FLAG-HIS3, and GFP-R12-FLAG-HIS3, were a kind gift of T. Inada [40]. The plasmids expressing Not4WT, Not4I64A, and Not4ΔRING were described in [35].
Yeast Growth Phenotypes
Yeast strains were grown to exponential phase, diluted to the same OD600 of 0.5. 5 µl of 10-fold serial dilutions were spotted on plates. Standard YPD or –URA media were used. Cycloheximide (CHX), hygromycin B (HygB) or azetidine-2-carboxylic acid (AZC) were added to the final concentrations of 0.05 µg/ml, 0.10 mg/ml or 0.10–0.50 mg/ml, respectively. Plates were incubated for several days at 30°C or, if indicated, at 37°C (heat sensitivity) or 16°C (cold sensitivity).
Isolation of Aggregates
Isolation of aggregates was done as described in [37]. In order to analyze polyubiquitinated proteins, cell cultures were treated with N-ethylmaleimide (NEM) prior to harvesting as described in [60]. For this 10 mM of NEM was added to 50 ml of culture grown to an OD600 of 1.0. Cells were harvested and pellets were washed in 1 ml of cold water containing 10 mM of NEM and 10 mM of phenylmethylsulfonyl fluoride (PMSF). 10 mM of NEM was also added to all the buffers for aggregate isolation.
In order to analyze newly synthesized proteins in the aggregates the cell cultures were grown to an OD600 of 1.0. 50 ml of the cultures were harvested, washed 2 times with 20 ml of the media without methionine and incubated in 50 ml of this media for 1 h at 30°C under agitation. Cells were collected by centrifugation and resuspended in 5 ml of media without methionine. S35-labeled methionine was added at 20 µCi/ml in the media. Cells were incubated for 5 min at 30°C under agitation. 45 ml of cold water supplemented with 300 µg/ml of CHX was added to the cells. After 5 min incubation on ice cells were collected, washed with 1 ml of cold water with 300 µg/ml of CHX and frozen. After isolation of aggregates the samples were migrated on 4–12% gradient SDS gels and analyzed by laser scanner Typhoon FLA 7000 (GE Healthcare). The quantification was done with ImageQuant TL software (GE Healthcare).
CPY* Stability
Cells transformed with the CPY*-HA plasmid [59] were grown exponentially in the presence of 0.1 mM of CuSO4. Cycloheximide was added to the cultures at OD600 of 0.6–1.0 at a final concentration of 200 µg/ml (+CHX). Control cultures were grown without cycloheximide (−CHX). Cells were collected at the indicated times. Samples were prepared by post alkaline lysis [61] and analyzed by western blot with an antibody against the HA tag, to follow CPY*-HA, and with an antibody against Egd2 for the loading control.
Proteasome Activity
Proteasome activity in total extracts was analyzed as described in [62]. Cells were grown exponentially and 50 ml of culture were collected at OD600 of 1.0. Cell pellets were disrupted in the presence of 150 µl of lysis buffer (50 mM Tris-HCl, pH 8.0, 50 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 10% glycerol, 1 mM ATP, 0.5 mM DTT) with 200 µl of glass beads for 15 min at 4°C. After spinning for 20 min at 16000 g the total protein concentration in the supernatant was adjusted to 15 mg/ml. 100 µg of total proteins were loaded on a 3.5% native gel prepared as in [62] and run for 3 h in a cold room. Gels were incubated with 100 µM of N-succinyl-Leu-Leu-Val-Tyr-7-amino-4-methylcoumarin (Suc-LLVY-AMC) for 20 min in the absence or presence of 0.02% SDS in order to detect latent proteasome activity. The activity was monitored at 365 nm.
Proteasome Purification
Proteasome and proteasome subcomplexes purification was done from the strains expressing the Rpn11-ProteinA tagged subunit of the proteasome as described in [63].
Ribosome Fractionation
Ribosomes were fractionated as in [37]. Briefly, 100 ml of yeast in exponential growth phase were treated with 100 µg/ml of CHX for 10 min on ice. Cells were harvested, washed with 50 ml of cold water with CHX, resuspended in 1 ml of buffer A (20 mM Hepes, pH8.0, 50 mM KCl, 10 mM MgCl2, 1% Triton X-100, 1 mM DTT, 1 mM PMSF, 100 µg/ml CHX, and protease inhibitor cocktail (Roche)) and pelleted. Cells were broken with 0.5 ml of glass beads in 0.5 ml of buffer A for 15 min at 4°C. The lysates were clarified by centrifugation at 14000 g for 10 min. 0.2 ml of lysates containing 3 mg of total protein was applied on a 12 ml 7–47% sucrose gradient in 20 mM Hepes, 50 mM KCl, 10 mM MgCl2, 100 µg/ml CHX and centrifuged for 150 min at 220000 g at 4°C. Fractions were collected using a UA/6 detector (ISCO, Inc.), precipitated with TCA and separated by SDS-PAGE.
Antibodies
Anti-HA (anti-influenza hemagglutinin; Sigma) antibodies were used at the dilution 1∶5000. Anti-Egd2 antibodies (described previously [38]) were used at the dilution 1∶15000. Anti-ubiquitin antibodies (Biomol) were used at the dilution 1∶5000. Anti-Ssa1 and anti-Ssb1 antibodies were kindly provided by E. Craig and were used at the dilution 1∶15000. Anti-Rpt1 antibodies (Biomol) were used at the dilution 1∶10000. Anti- α1,2,3,5,6,7 (α1-7) antibodies (Biomol) were used at the dilution 1∶8000. Antibodies against Rpn8 were kindly provided by D. Finley and were used at the dilution 1∶10000. Anti-Rpl35 antibodies were kindly provided by M.Pool and were used at the dilution 1∶20000. PAP-antibodies (Peroxidise-anti-peroxidase soluble complex, Sigma) were used at the dilution 1∶10000. Anti-GFP antibodies (Roche) were used at the dilution 1∶5000.
Results
The Deletion of the Not4 E3 Ligase or the Ccr4/Caf1 Deadenylase have Different Phenotypes
To determine to which extent the two enzymatic modules of the Ccr4-Not complex are functionally connected, we deleted the Not4 E3 ligase on one hand, and the subunits that play a role in deadenylation, either Caf1 or Ccr4, on the other hand. We compared growth of the wild type and mutants under different conditions: high and low temperature, and in the presence of agents affecting translation (cycloheximide (CHX), hygromycin B (HygB) or azetidine-2-carboxylic acid (AZC)) (Fig. 1). CHX inhibits translation and impairs proteasome function [64], [65]. HygB affects translational fidelity and increases read-through of stop codons [66]. AZC competes with proline during amino acid incorporation and induces misfolding of proteins, their degradation by the proteasome, and ribosome pausing [67], [68]. High temperature also affects translation since it leads to stalled ribosomes with translation arrested products [68], [69]. The Not4 deletion caused slow growth at 30°C and sensitivity to high temperature, CHX, HygB, and AZC. The deletion of Ccr4 lead to slight slow growth at 30°C but otherwise displayed no sensitivity or resistance to the conditions tested, except a slight sensitivity to AZC. Caf1 is necessary for the association of the Ccr4 deadenylase with the rest of the Ccr4-Not complex [18]. Its deletion had more severe phenotypes than the deletion of Ccr4. It reduced growth at 30°C on rich media and led to sensitivity to cold, HygB and CHX. Its impact on cell growth was less severe than the deletion of Not4: it did not lead to sensitivity to high temperature and was less sensitive to AZC. Interestingly, the deletion of Ccr4 or Caf1 led to reduced growth at 16°C, while deletion of Not4, in contrast, improved growth at low temperature (Fig. 1).
Figure 1. Deletions of the E3 ligase Not4, and the deadenylase subunits Ccr4 and Caf1, have different phenotypes.
The indicated strains were grown to exponential phase and diluted to the same OD600 of 0.5. 10-fold serial dilutions were spotted on the YPD plates containing, when indicated, HygB 0.1 mg/ml; CHX 0.05 µg/ml; AZC 0.5 mg/ml, and left to grow for 4 days (A, except 16°C), for 17 days (A, 16°C) or for 6 days (B).
It was described that Not4 is involved in clearance of nascent chains upon translational arrest [40]. Several other E3 ligases were reported to have global roles in protein clearance: Ltn1 ubiquitinates nascent chains on the ribosome [41], [70], Ubr1 and Ubr2 play a role in degradation of misfolded cytosolic proteins [42] and San1 is involved in the proteasome-dependent degradation of aberrant nuclear proteins [43], [44]. The deletion of none of these other ligases had phenotypes similar to not4Δ, except for ltn1Δ that displayed decreased growth in the presence of HygB (Fig. S1).
Not4 Deletion Causes Protein Aggregation in the Cell
We have previously reported that the deletion of the several Not subunits of the Ccr4-Not complex (Not2, Not4 or Not5) caused increased protein aggregation in the cell [37]. Hence, we tested protein aggregation in cells in which the deadenylase module was deleted. We analyzed aggregates by SDS-PAGE and Coomassie staining (Fig. 2A, upper panel). The accumulation of protein aggregates in cells lacking Caf1 or Ccr4 was small and comparable to that in wild-type cells, whereas much stronger aggregation was observed in the not4Δ mutant.
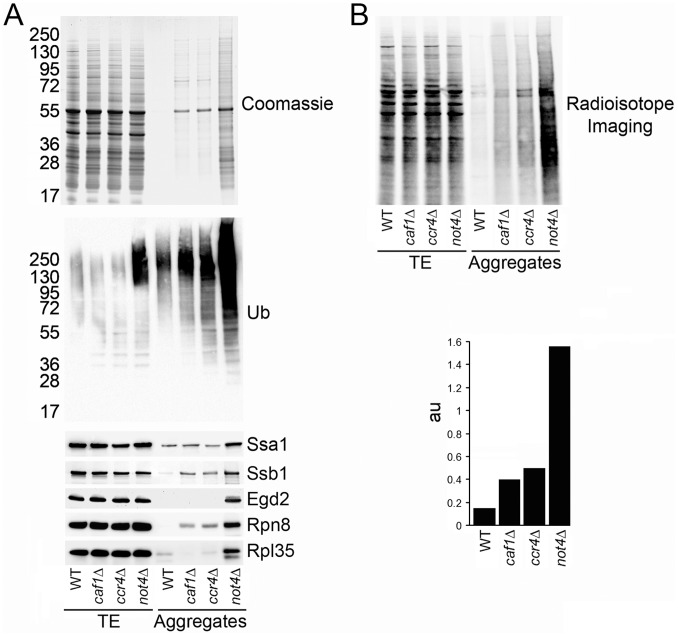
Figure 2. The Not4 deletion caused accumulation of aggregated and polyubiquitinated newly synthesized proteins.
A. Aggregates were isolated from the indicated cells and analyzed by SDS-PAGE and Coomassie staining (upper panel), or western blot with antibodies against ubiquitin (middle panel), or against Ssa1, Ssb1, Egd2, Rpn8, and Rpl35 (lower panel). B. Aggregates were isolated from the same cells treated with S35-Met for 5 min and analyzed by SDS-PAGE and radioisotope imaging (upper panel). Images were quantified (lower panel). “au” is a ratio of the signal observed in the aggregates to the signal observed in the total protein fraction.
We have also reported that polyubiquitinated proteins accumulate in not4Δ cell extracts [35]. So we compared the level of polyubiquitinated proteins in total extracts and in protein aggregates from wild-type and mutant cells lacking the enzymatic modules of the Ccr4-Not complex (Fig. 2A, middle panel). No increased level of polyubiquitinated proteins was detected in total extracts from caf1Δ or ccr4Δ cells compared to wild type. In contrast, in not4Δ cells polyubiquitinated proteins were observed in total extracts. A slight increase of polyubiquitinated proteins was observed in the aggregates from caf1Δ and ccr4Δ mutants, whereas very high levels of polyubiquitinated proteins were found in the aggregates from not4Δ cells. These aggregates in not4Δ contained the Hsp70 cytoplasmic chaperone, Ssa1; the ribosome associated chaperones Ssb1 and Egd2; the proteasomal protein, Rpn8; and the ribosomal protein, Rpl35 (Fig. 2A, lower panel).
To determine whether de novo synthesized proteins were contributing to the aggregates in the mutants, we did metabolic labeling of the cells with S35-methionine for 5 min. Aggregates were isolated from these cells and analyzed by radioisotope imaging (Fig. 2B, upper panel). Appearance of radioactive signal in the aggregates indicated that, indeed, newly synthesized peptides were aggregating, and this to a much greater extent in cells lacking Not4, than in cells lacking Caf1 or Ccr4 (Fig. 2B, lower panel).
Hence, loss of the ubiquitin ligase module of the Ccr4-Not complex provokes a severe accumulation of de novo synthesized and polyubiquitinated proteins. This cannot be accounted for simply by defective activity of the deadenylation module of the Ccr4-Not complex due to the absence of Not4. Indeed, the deletion of the deadenylation module of the Ccr4-Not complex does not by far have a comparable impact on accumulation of protein aggregates.
Proteasome is Defective in not4Δ
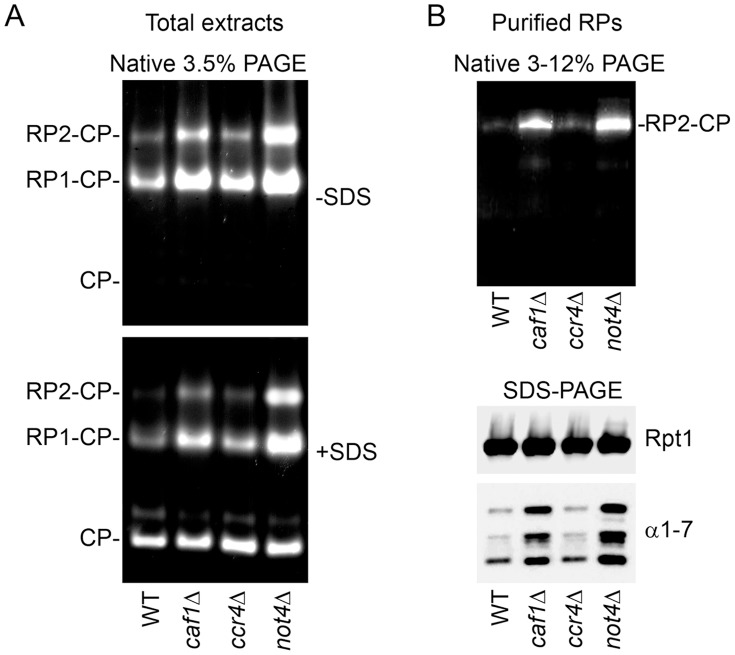
An important role of Not4 in functional assembly of the proteasome has been described [35], suggesting that accumulation of polyubiquitinated aggregated proteins in not4Δ might be partially due to their reduced clearance by the proteasome. Deletion of Not4 results in abnormal salt-resistant interaction between 2 proteasomal subcomplexes, regulatory particle (RP) and core particle (CP). This correlates with a greater level of proteasome activity measured with the substrate Suc-LLVY-AMC in extracts from not4Δ cell compared to wild-type cell extracts [35]. This observation was also true for some other mutants of the Ccr4-Not complex, in particular for caf1Δ (Fig. S6 in [35]). However, the proteasome has not been analyzed in the ccr4Δ mutant. Therefore we compared proteasomes isolated from cells deleted for Not4 or for Ccr4. We followed proteasome activity in the total extracts (Fig. 3A). As expected higher activities of double (RP2-CP) and single (RP1-CP) capped-proteasomes were detected in caf1Δ and not4Δ mutants. In contrast, activity of the proteasome from ccr4Δ was not significantly different than from the wild type, except for a very slight increase of RP-CP proteasome activity.
Figure 3. Proteasome was defective in not4Δ cells but not in ccr4Δ cells.
A. Total cellular extracts were prepared from wild-type, caf1Δ, ccr4Δ, and not4Δ cells and loaded on 3.5% native gels. After electrophoresis gels were incubated with Suc-LLVY-AMC to analyze the proteasome activity in the absence (-SDS) and then in the presence (+SDS) of 0.02% SDS to detect the latent CP activity. The positions of double (RP2-CP) and single (RP1-CP) capped proteasomes and CP alone are indicated on the left. B. RPs were purified from wild-type, caf1Δ, ccr4Δ, and not4Δ cells, loaded on a gradient 3–12% native gel and then analyzed for activity (upper panel). The same purified material was analyzed by SDS-PAGE and western blot with antibodies against the RP subunit (Rpt1) and with antibodies against CP subunits (α1-7) (lower panel).
We also purified RPs from the different strains (Fig. 3B). The same amount of the RP subunit, Rpt1, was isolated from all strains (Fig. 3B, lower panel), indicating that the efficiency of the purification was identical. In wild-type cells the RP-CP interaction is salt-sensitive and incubation with high salt concentrations results in removal of CP subunits from RP. This is why no, or very little, amount of CP subunits (Fig. 3B, lower panel) and activity (Fig. 3B, upper panel) was detected in the purification of RP from wild-type cells under high salt. The same phenotype was observed for RP purified from the ccr4Δ mutant. In contrast, as we previously observed [35], salt-resistant RP-CP active complexes were purified via RP from caf1Δ and not4Δ mutants in high salt and CP subunits were detected (Fig. 3B, upper panel).
Deletion of Not4 Stabilizes the Proteasomal Substrate, CPY*
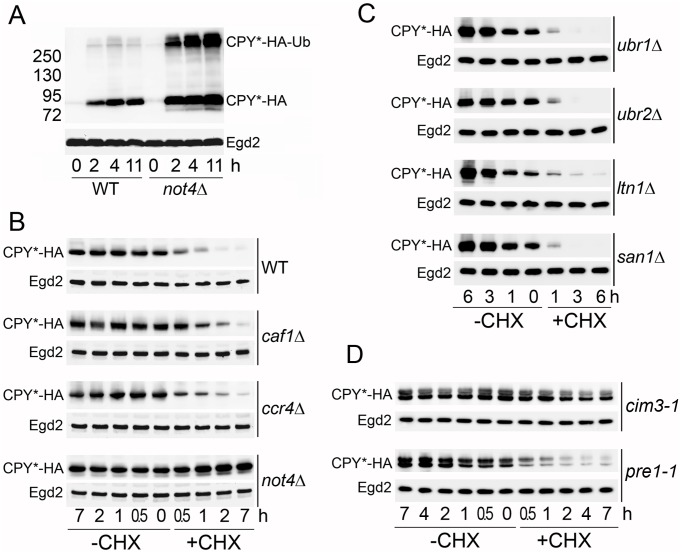
An increase in the proteasome activity with Suc-LLVY-AMC indicates that the interaction between RP and CP is not normal. The Suc-LLVY-AMC is a small artificial substrate that is not specifically targeted for proteasomal degradation like cellular protein substrates. It can be used for estimation of the peptidase activity of the proteasome in vitro. However, increased cleavage of Suc-LLVY-AMC in vitro does not directly reflect in vivo protease activity of the proteasome, which is complex and includes several steps (substrate recognition, deubiquitination, translocation to the CP, and, finally, cleavage). We decided to estimate protease activity of the proteasome in wild-type and mutant cells with an in vivo cellular substrate, CPY*. CPY* is a highly unstable mutated version of carboxypeptidase yscY (CPY), a lysosomal protein that is retarded in the ER lumen and rapidly degraded by the ubiquitin proteasome system [71], [72]. In our experiments we used an HA-tagged version of CPY* expressed under control of the copper dependent promoter, CUP1. In wild-type and not4Δ cells, CPY* was well induced after 2 h of copper treatment, and the level was not dramatically changed after longer induction times (Fig. 4A). So for the rest of the experiments we grew the cells in the constant presence of copper in the media. In both wild-type and not4Δ strains, we detected slower migrating forms of CPY*, that correspond to ubiquitinated CPY*. The level of CPY* was higher in not4Δ compared to wild type, but, more importantly, the level of ubiquitinated CPY* was much greater in not4Δ.
Figure 4. The deletion of Not4, but not the deletion of the deadenylase, stabilizes the proteasomal substrate CPY*.
A. CPY*-HA was expressed from an episome under control of a copper dependent promoter in wild-type (WT) and not4Δ cells. Cells were exponentially grown without induction to OD600 of 0.6 (time 0). 0.1 mM CuSO4 was added to the media and cells were collected at indicated time points (2, 4 and 11 h) and analyzed by SDS-PAGE and western blot with antibodies against HA, to see CPY*-HA levels, and against Egd2 as a loading control. The positions of CPY*-HA and ubiquitinated CPY*-HA (CPY*-HA-Ub) are indicated on the right. The molecular weight markers are indicated on the left. B. Stability of CPY*-HA was analyzed in wild-type, caf1Δ, ccr4Δ, and not4Δ cells. Cells were grown exponentially and treated (+CHX) or not (−CHX) with CHX. Samples were collected at indicated time points and analyzed as in A. Since CPY*-HA expression was different in mutant strains (see Fig. S2A) 2 times less material was loaded on the gel in the case of not4Δ samples compared to wild type, and 4 times less material was loaded on the gel in the case of the ccr4Δ and caf1Δ samples compared to wild type. C. Stability of CPY*-HA was analyzed in ubr1Δ, ubr2Δ, ltn1Δ, and san1Δ cells as in B. D. Stability of CPY*-HA was analyzed in cim3-1 and pre1-1 cells as in B.
We then compared CPY* stability in wild-type, caf1Δ, ccr4Δ, and not4Δ cells (Fig. 4B). In wild-type cells the level of CPY* was noticeably reduced after 30 min of incubation with CHX, consistent with a described half-life of CPY* of about 24 min [71]. CPY* was similarly unstable in caf1Δ and ccr4Δ mutants (Fig. 4B). Surprisingly, CPY* was expressed at very high levels in caf1Δ and ccr4Δ mutants (Fig. S2A). This increase did not correlate with any comparable increase of the mRNA levels (Fig. S2B) suggesting that, instead, translation of CPY* may be higher in these mutants, since Caf1 and Ccr4 have been associated not only with mRNA deadenylation but also translational repression (reviewed in [13]). CPY* was strongly stabilized when Not4 was deleted. Even after 24 h of protein synthesis arrest, the level of CPY* in not4Δ was not reduced (data not shown). We also tested CPY* stability in cells mutated for the Ubr1, Ubr2, Ltn1, and San1 E3 ligases (Fig. 4C). In all of these mutants CPY* was as unstable as in wild-type cells, indicating that, if these quality control E3 ligases participate in CPY* degradation, they are redundant, while Not4 might have a global role in clearance of CPY*, probably acting via the proteasome.
Because of the role of Not4 in the functional assembly of the proteasome, we considered that the stability of CPY* in not4Δ could be due to altered proteasome function. Hence, we tested the stability of CPY* in two proteasome mutants, cim3-1 and pre1-1, mutant alleles of genes encoding the Rpt6 RP subunit and the β4 CP subunit of the proteasome, respectively. In both mutants proteasome was not active for cleavage of Suc-LLVY-AMC (data not shown). CPY* was stable in the cim3-1 mutant, while in the pre1-1 mutant it was unstable (Fig. 4D). These results indicate that different proteasome mutants differently affect stability of CPY*, as previously observed [73], [74], and, in particular, that functional integrity of RP is required for degradation of CPY*. In this context it is important to underline that cells lacking Not4 displayed 2 distinguishable defects in proteasome integrity: besides carrying salt-resistant RP-CP proteasomes, a phenotype shared by caf1Δ as shown above (Fig. 3), it also led to unstable free RP [35]. Hence, our current findings that CPY* is stabilized in cim3-1 as in not4Δ lead us to conclude that cells lacking Not4 fail to degrade CPY* because of defective RP.
Use of Not4 Mutants to Characterize the Role of Not4 in Protein Quality Control
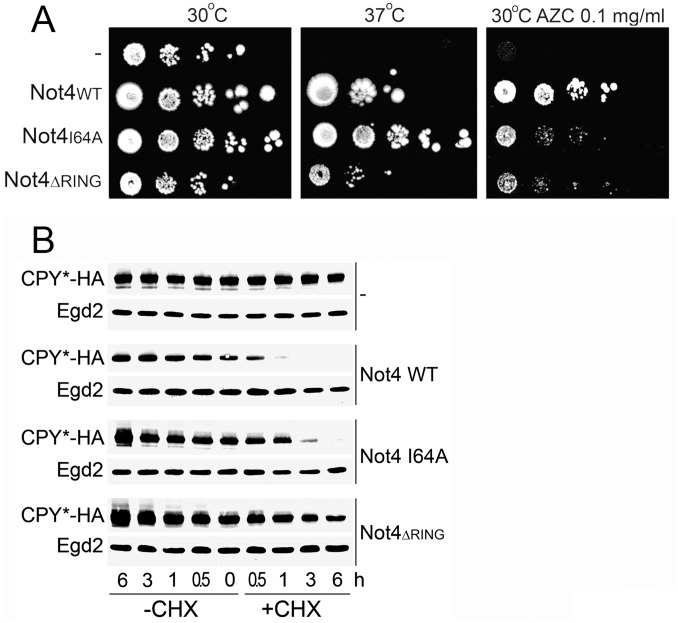
Not4 has been suggested to play a role in co-translational protein quality control by acting directly as an E3 ligase for ubiquitination of translationally-arrested proteins [40]. Not4 could also be important for protein quality control because of its importance for functional integrity of the proteasome [35] that clears aberrant proteins. We studied Not4 mutants to define the relevance of these 2 phenotypes for growth on media affecting translation, namely high temperature and AZC. The RING domain of Not4 is important for substrate ubiquitination and proteasome integrity [35], [37]. In contrast, a point mutation within the RING domain of Not4 (I64A) affects the interaction of Not4 with E2 partners [75] and reduces the ubiquitination of its Egd2 substrate (Fig. S3A in [35]), but it does not influence proteasome integrity [35]. We analyzed the growth phenotypes of either the Not4 I64A point mutant or a mutant bearing an entire deletion of the RING domain (Not4ΔRING) (Fig. 5A). The Not4ΔRING was very sick but distinguishable from the null mutant: it grew better than not4Δ at 37°C and in the presence of AZC. However, CPY* was similarly stabilized in both Not4ΔRING and in the null mutant (Fig. 5B). The I64A mutant grew slightly slower at 37°C and in the presence of AZC compared to the wild type, and CPY* was slightly stabilized (Fig. 5A and B). The I64A mutant was also sensitive to CHX [35] and HygB [75].
Figure 5. Use of Not4 mutants to characterize the role of Not4 in protein quality control.
A. not4Δ cells expressing only vector (−) or vector containing the Not4 derivatives (Not4WT, Not4I64A or Not4ΔRING) were analyzed as in Fig. 1 and spotted on – URA plates and left to grow for 13 days at 30°C or 37°C. When indicated, plates contained 0.1 mg/ml of AZC. B. Stability of CPY*-HA was analyzed in not4 mutants as in Fig. 4B.
These results indicate that Not4 is likely to play a role in protein quality control that extends beyond its impact on the proteasome and involves its function as an E3 ligase.
Not4 Accumulates in Polysomes in Response to AZC and High Temperature
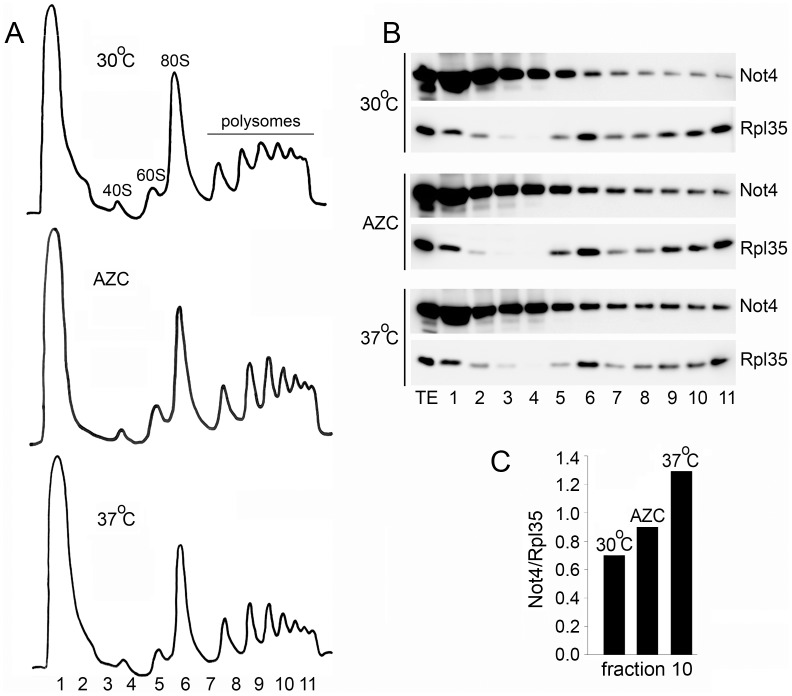
Not4 is important for temperature and AZC resistance (see above, Fig. 1), it is present in polysomes [37], [40] and it is thought to ubiquitinate translationally-arrested protein products [40]. To clarify better the role of Not4 in protein quality control, we analyzed what happens with Not4 under conditions that adversely affect protein synthesis. We analyzed ribosome profiles from cells grown in the presence of AZC and high temperature. Not4 accumulated in polysome fractions upon incubation of cells with AZC and, particularly, upon incubation of cells at high temperature (Fig. 6). These results are consistent with the idea that the presence of Not4 on actively translating ribosomes is important under these conditions.
Figure 6. Not4 accumulates in polysomes in response to AZC and high temperature.
A. Polysome profiles from the cells expressing Not4-ProteinA. Cells were exponentially grown on YPD media at 30°C or 37°C, as indicated, and collected at OD600 of 1.0. When indicated, cells were treated with 0.4 mg/ml of AZC. AZC was added at OD600 of 0.15 and cells were grown till OD600 of 1.0 and collected. Extracts, containing 3 mg of total proteins, were subjected to 7–47% sucrose gradient centrifugation and analyzed by UV reading at 254 nm. Fraction numbers and the positions of 40S, 60S, 80S, and polysomes are indicated. B. Fractions were collected and analyzed by western blot with PAP and Rpl35 antibodies. C. Not4 content in polysomes was quantified. For this the Not4 signal in polysomes (fraction 10) was quantified with ImageQuant TL software (GE Healthcare) and normalized on the Rpl35 signal.
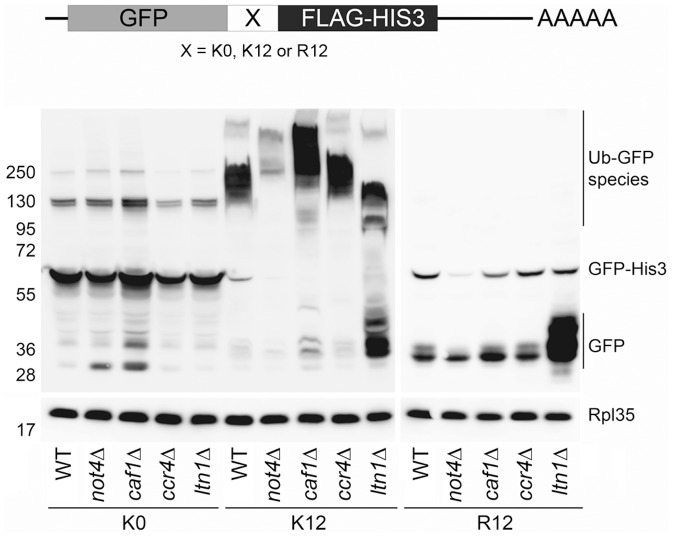
We then investigated how Not4 and the deadenylase subunits Ccr4 and Caf1 might be important for dealing with translationally-arrested products. We transformed wild-type, not4Δ, ltn1Δ, caf1Δ, and ccr4Δ cells with constructs described in [40]. In these constructs GFP fused to FLAG-HIS3 is expressed without (K0), or with a positively charged stretch of 12 lysines (K12) or 12 arginines (R12) inserted between GFP and the FLAG-HIS3 moieties. With these constructs it was shown that translationally-arrested products accumulated in not4Δ cells, when a positively charged stretch of amino acids was present in the middle of the open reading frame (ORF) [40]. We observed that relatively equal levels of full length GFP-K0-HIS3 product were detected in all strains (Fig. 7). Full length GFP-R12-HIS3 was also detectable for all strains, but it was less abundant compared to the levels of GFP-K0-HIS3, and it was particularly lower in not4Δ than in the other cells. In all R12 transformants translationally-arrested GFP (described in [40]) was additionally detected. The levels of this arrested product were relatively equal for all strains except for ltn1Δ, where they were dramatically increased. This observation is compatible with the previous reports indicating that Ltn1 is responsible for ubiquitination and degradation of translationally-arrested proteins [41], though it is surprising that no ubiquitinated forms of GFP from the R12 transformants were detected in any of the strains.
Figure 7. Role of the Not4 and Caf1/Ccr4 modules in co-translational degradation.
Indicated strains were transformed with GFP-X-FLAG-HIS3 plasmids, that either contains no positively charged stretch between GFP and FLAG-HIS3 moieties (K0), or contains 12 lysine residues (K12), or 12 arginine residues (R12) [40]. Cells were grown to exponential phase, collected and analyzed by post alkaline lysis. The same amount of the samples were loaded on 4–12% SDS gels and after electrophoresis analyzed by western blot with antibodies against GFP and against Rpl35 as a loading control. Positions of GFP arrested products, full length GFP-X-FLAG-HIS3 products and GFP containing ubiquitinated species are indicated on the right.
The levels of full length GFP-K12-HIS3 were significantly reduced compared to GFP-K0-HIS3 or GFP-R12-HIS3. A very low level of translationally-arrested GFP was detected in the wild type and even less in not4Δ. Slightly more of arrested GFP was detected in caf1Δ. Significantly greater amounts of this product were detected in ltn1Δ cells, an observation consistent with the role of Ltn1 in degradation of translationally-arrested proteins [41]. Interestingly, in the case of these K12 constructs, many high molecular weight GFP species were detected in all strains. They probably correspond to ubiquitinated derivatives (Fig. 7, Ub-GFP). The migration of the Ub-GFP smear was similar in wild-type and ccr4Δ cells, but it migrated slower in not4Δ and caf1Δ. This correlates with altered proteasome integrity in these 2 mutants, and may indicate that the mostly highly polyubiquitinated forms of GFP do not get deubiquitinated and degraded by the proteasome in these mutants. In contrast, the smear was smaller and faster migrating in ltn1Δ cells, in good correlation with the role of Ltn1 in ubiquitination of translationally-arrested proteins. Clearly, however, Ltn1 cannot be the sole E3 enzyme involved, since much residual ubiquitination is observed in the absence of Ltn1. It is noticeable that the amount of these Ub-GFP forms was significantly reduced in not4Δ. In fact, the detectable total amount of protein produced from the K12 construct in not4Δ was reduced compared to the wild type. Total levels of protein detected from the R12 construct were also reduced in not4Δ compared to the wild type, whereas the amount of protein produced from K0 was similar in wild type and not4Δ. This indicates a specific role of Not4 for preserving translation, and stability of mRNA or protein from constructs that lead to translational arrest in the middle of the ORF. No such role could be observed for Ccr4, and in this context it is important to note that while polysomes were reduced in not4Δ [14], no such reduction was observed in ccr4Δ (Fig. S3).
Discussion
In this report we compared the function of the 2 enzymes of the Ccr4-Not complex, the Ccr4 deadenylase, and the Not4 E3 ligase. We also analyzed the Caf1 subunit of the complex, which does not have deadenylation activity in baker’s yeast, but is important for the connection of the Ccr4 deadenylase with the rest of the Ccr4-Not complex and, therefore, is important for the deadenylation activity of Ccr4. We came to the conclusion, that the function of Not4 in protein quality control is separable from the role of the deadenylase, and it is specific: 1) the Ccr4 deletion led to different growth phenotypes than the Not4 deletion and, in particular, was not sensitive to media affecting translation like the deletion of Not4. 2) Protein aggregates accumulated in cells when Not4 was deleted, but much less so when Ccr4 or Caf1 were deleted. 3) Polyubiquitinated proteins accumulated in not4Δ, but not in ccr4Δ or caf1Δ cells. 4) The decay of the proteasomal substrate CPY* in ccr4Δ or caf1Δ cells was similar to wild-type cells. In contrast, CPY* was stabilized when Not4 was deleted. 5) Finally, proteasome integrity was altered in not4Δ, but not in ccr4Δ cells.
Ccr4-Not Complex has Two Different Modules Acting in Quality Control
Many studies have indicated that the Ccr4-Not complex is composed of distinct functional modules. If we consider the 2 enzymes of the complex, microarray analyses revealed that genes deregulated in the ligase not4Δ mutant only marginally overlapped with genes deregulated in the deadenylase ccr4Δ mutant [76]. In humans Not4 was found outside of the Ccr4-Not complex [21], indicating that some functions of Not4 may not even require its association with the rest of the complex. Nevertheless it has to be considered that, within the Ccr4-Not complex, the E3 ligase might play a role in regulating the deadenylation activity provided by Ccr4. No evidence of this regulatory function is available. In fact there are several indications that the Not4 E3 ligase module does not regulate the deadenylase module: the deletion of Not4 does not dramatically change the composition of the Ccr4-Not complex; Caf1 and Ccr4 still interact with Not1 [76]; mostly normal deadenylase activity occurs in vivo in the not4Δ mutant [16], and deadenylation of reporter mRNAs was only slightly decreased [30]. One exception is a report, which describes that the accumulation of aberrant translation products in cells lacking Not4 correlates with a slight increase of the related mRNAs [40]. This raised the possibility that Not4 might be necessary to activate the deadenylase module in the context of co-translational quality control, and that this may explain fully that aggregated proteins accumulate in cells lacking Not4.
To address this possibility, we compared the phenotypes of cells lacking one enzyme or the other enzyme. In particular, we wanted to determine whether the phenotypes, which are associated with the loss of Not4 and connected to protein quality control, could be recapitulated by the loss of the deadenylase. This is what one would expect, if the phenotypes in not4Δ were due to the lack of activation of the deadenylase. To investigate consequences that are strictly due to loss of deadenylation in yeast, ccr4Δ is a better mutant to study than caf1Δ. Indeed, in yeast, although Caf1 like Ccr4 can deadenylate substrates in vitro [9], [77], [78], only Ccr4 plays a catalytic role in vivo, as mentioned above. Caf1 is nevertheless indispensable for deadenylation activity in vivo [16], because it functions to bridge Ccr4 to Not1 [18]. However, it seems to play additional roles in the structure of the Ccr4-Not complex [76] and cells lacking Caf1 have more severe phenotypes than cells lacking Ccr4 only.
The deletion of Ccr4 did not have the complete set of phenotypes connected to protein quality control observed in the absence of Not4. It did not impact on proteasome integrity, and it did not result in stabilization of proteasomal substrates, nor did it lead to accumulation of aggregated proteins. Thus, it is highly unlikely that defective deadenylation explains the accumulation of aberrant and aggregated proteins in not4Δ.
In contrast to ccr4Δ, the Caf1 deletion revealed many phenotypes similar to the deletion of Not4 and not shared by the deletion of Ccr4. The caf1Δ mutant was sensitive to HygB and to CHX, and displayed proteasome defects. This might be due to the fact that Caf1 plays structural roles within the Ccr4-Not complex unlike Ccr4, and its absence might impinge on Not4 function. However, caf1Δ was not as sensitive to AZC as not4Δ, nor was it temperature sensitive, demonstrating that not all functions of Not4 are compromised in caf1Δ.
Not4 Contributes to Protein Quality Control
What then is the role of Not4 in protein quality control and why do aggregated proteins accumulate in the absence of Not4? We show that a proteasomal substrate, CPY*, fails to be degraded in not4Δ, compatible with the defective functional integrity of the proteasome in not4Δ that we have previously reported. However, in addition, we show that when cells are exposed to proteotoxic shock with a mistranslating agent, AZC, or when temperature is increased and translation stalls, the presence of Not4 in polysomes increases, indicating that Not4 is needed where co-translational responses take place. And indeed, the deletion of Not4 leads to sensitivity of the cells to growth under conditions in which translation is compromised (this article and [35], [75]): HygB affects translational fidelity and increases read-through of stop codons [66]. AZC induces protein misfolding and proteotoxic stress [67], [68]. CHX is a translation inhibitor. All these agents lead to appearance of protein quality control substrates. Sensitive growth phenotypes in the presence of these agents support an important physiological role for Not4 during translation. Consistently, we show that expression from no-go mRNAs is altered in not4Δ, indicating that Not4 is important for the co-translational regulation of no-go mRNAs. Aggregates found in not4Δ cells contain newly synthesized and polyubiquitinated proteins, supporting the idea that Not4 functions in quality control of de novo synthesized proteins. Finally, in good agreement with the idea that both Ltn1 and Not4 are required for co-translation quality control, is the observation that double mutant not4Δ ltn1Δ displays a synthetic slow growth phenotype (Fig. S4).
An important question is the co-translational role played by Not4. It was proposed that no-go mRNA translation arrest was accompanied by Not4-dependent ubiquitination and proteasomal degradation of aberrant products [40], but later studies revealed that ubiquitination of arrested proteins mainly occurred by the E3 ligase Ltn1 [41], [79]. Our own previous work showed that Not4 is important for proteasome assembly [35]. In this study we have comparatively analyzed the levels of translationally-arrested proteins and read-through full-length proteins in wild-type, not4Δ, ccr4Δ, caf1Δ, and ltn1Δ cells from no-go mRNAs. We observed that, while in the absence of Ltn1 translationally-arrested proteins accumulated and were less ubiquitinated, as described previously [41], in not4Δ they were ubiquitinated to a greater extent. This phenotype was shared by caf1Δ, and, hence, may be indicative of defective proteasome activity detected in both caf1Δ and not4Δ. It is also possible that Not4 and Caf1 limit Ltn1 activity. We also observed that the total level of protein produced from constructs with a stalling amino-acid basic stretch, but not without, was reduced specifically in not4Δ. This indicates that Not4 plays a specific role in preserving translation efficiency or mRNA levels from the constructs that lead to translational arrest. An alternative possibility could be that the proteins produced are less stable in the absence of Not4, especially since for one construct, K12, the arrested products were more ubiquitinated. However, this seems unlikely because clearance of proteins by the proteasome in not4Δ is less efficient, as indicated by stabilization of CPY*, and, moreover, in caf1Δ the K12 arrest products are also more ubiquitinated and yet they accumulate as in wild-type cells.
Bengtson and Joazeiro reported, like us, a reduction of full-length protein product from their no-go K12 constructs in not4Δ compared to wild type [41]. However, they discarded this observation as not significant, because they saw a similar decrease from the construct without a stalling sequence [41]. We have carefully looked at this point in our study and did not see the decrease in K0 (Fig. 7). In fact, Dimitrova et al. also observed a reduction of full-length product from their no-go constructs [40]. They did not discuss this at all, because in contrast to us, they saw an increased accumulation of arrested protein from the K12, and even more R12 constructs, in not4Δ, and they focused their discussion on this accumulation. This is where our results differ from those of Dimitrova et al., despite the fact that we used the same constructs but in a different strain background (they used W303 and we used BY4741). Bengtson and Joazeiro, who used BY4741 strain background, like us, did not observe increased levels of K12-induced translationally-arrested products in the absence of Not4 only, but they did see such an increase if Ltn1 was deleted.
To understand the inconsistencies and similarities between the results of the 2 previous studies and our current work, it is important to mention that W303 background has sequence differences in genes compared to S288C, from which BY4741 was derived, and these are, in particular, in many stress resistance factors (as explained in the [36]). The deletion of Ltn1 can also be sensed as a stressful situation for the cell. Hence, it could be that translationally-arrested proteins increase in the absence of Not4 upon stress (that could be in W303 background), but decrease in the absence of stress. In such a model, Not4 acts as a switch important to preserve the proteome: in the absence of stress and presence of Ltn1, if translation of an mRNA momentarily stalls, Not4 acts initially to preserve production of full-length protein: it increases translation and/or represses deadenylation. Both functions could be though regulation of the Ccr4/Caf1 module of the Ccr4-Not complex, but could also involve other proteins such as the Dhh1 DEAD box RNA helicase (discussed in [36]). Not4 might also moderate Ltn1 function or have a positive impact on the deubiquitination activity of the proteasome RP, to give a chance for the stalled protein not to be degraded and to be translated into full-length protein. In contrast, upon stress, or if Ltn1 is deleted, Not4 is important to mobilize the deadenylase module of the Ccr4-Not complex to repress translation and/or induce mRNA degradation. Hence, if Not4 is deleted, stalled protein accumulates, and the level of polyubiquitination of this protein will depend upon the presence or not of Ltn1.
In conclusion, in this work we have shown that Not4 is important for cellular protein quality control first, because it is globally important for appropriate clearance of aberrant proteins, because it is important for functional integrity of the proteasome, but also through its function as an E3 ligase, that does not affect proteasome function. Additionally, Not4 is important during translation where it acts as a switch to promote or inhibit production of proteins from stalled mRNAs depending upon the cellular conditions. Determining how the Not4 switch is regulated and exactly operates are obviously now exciting questions to tackle.
Supporting Information
Growth phenotypes of the E3 ligases mutants. The indicated strains were grown to exponential phase and diluted to the same OD600 of 0.5. 10-fold serial dilutions were spotted on the YPD plates containing, when indicated, HygB 0.1 mg/ml or CHX 0.05 µg/ml; and left to grow for 6 days (except 16°C) or for 21 days (16°C).
(TIF)
CPY*-HA mRNA and protein levels in wild-type, not4Δ, ccr4Δ , and caf1Δ cells. A. CPY*-HA protein levels in wild-type, not4Δ, ccr4Δ, and caf1Δ cells. CPY*-HA was expressed from an episome under control of copper dependent promoter in wild-type (WT), not4Δ, ccr4Δ, and caf1Δ cells. Cells were exponentially grown in the constant presence of 0.1 mM of CuSO4 and collected at OD600 of 1.0. Different amount of the cells (0.5 OD units (lane 1), 0.125 OD units (lane 2) and 0.05 OD units (lane 3)) were analyzed by SDS-PAGE and western blot with antibodies against HA, to see CPY*-HA levels, and against Egd2 as a loading control. B. CPY*-HA mRNA levels in wild-type, not4Δ, ccr4Δ, and caf1Δ cells. Cells were grown as described in A. 50 OD units of the cultures were collected. Pellets were resuspend in 400 µl of acid phenol and 400 µl of TES buffer (10 mM Tris-HCl pH 7.5, 10 mM EDTA, 0.5% SDS) and incubated at 65°C for 10 min. Samples were chilled on ice for 5 min and spun at 4°C for 10 min. Aqueous phase was extracted with 400 µl of acid phenol and then with chloroform. Finally, RNA was collected by ethanol/sodium acetate precipitation. 4 µg of the RNA were treated with DNAse (Promega) and than reverse transcribed with M-MLV RT (Promega) according to the manufacturer’s instructions and using oligo d(T) primers (Qiagen). SYBR green based quantitative RT-PCR was performed using BioRad cycler. ACT1 was used as a housekeeping gene and CPY*-HA signals were normalized on ACT1 level. The ratio CPY*-HA/ACT1 in wild type was normalized to 1. Primers used for analysis: The forward primer: 5′-TCCCCGGGTTAATTAACATC-3′ and reverse primer: 5′-TCGCTTATTTAGAAGTGGCG-3′ amplify 149 bp fragment of HA tag of CPY*-HA gene. The forward primer: 5′-TTGTCCGTGACATCAAGGAA-3′ and reverse primer: 5′-ACCCAAAACAGAAGGATGGA-3′ amplify 182 bp fragment of ACT1 gene.
(TIF)
Polysome profiles from wild-type, not4Δ , and ccr4Δ cells. Extracts from wild-type (black), not4Δ (red), and ccr4Δ (blue) cells, containing 3 mg of total proteins, were subjected to 7–47% sucrose gradient centrifugation and analyzed by UV reading at 254 nm (left). Profiles were superposed (right). The positions of 40S, 60S, 80S, and polysomes are indicated.
(TIF)
Double mutant ltn1Δ not4Δ grows slowly compare to single ltn1Δ or not4Δ mutants.
(TIF)
Acknowledgments
We thank T. Sommer, D. Finley, M. Pool, T. Inada, C. Mann, D. Wolf, and E. Craig for the kindly provided materials. We thank Dr Irene Dunn-Siegrist for the help with some experiments.
Funding Statement
This work was supported by grants from the Ernst and Lucie Schmidheiny (http://www.fondation-schmidheiny.ch/lafondation.html) and Pierre Mercier (http://www.fondationmercier.ch/wb/) Foundations awarded to OOP and a grant 31003A_135794 from the Swiss National Science Foundation (http://www.snf.ch/E/Pages/default.aspx) awarded to MAC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Shoemaker CJ, Green R (2012) Translation drives mRNA quality control. Nat Struct Mol Biol 19: 594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Doma MK, Parker R (2007) RNA quality control in eukaryotes. Cell 131: 660–668. [DOI] [PubMed] [Google Scholar]
- 3. Hartl FU, Hayer-Hartl M (2009) Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol 16: 574–581. [DOI] [PubMed] [Google Scholar]
- 4. Chen B, Retzlaff M, Roos T, Frydman J (2011) Cellular strategies of protein quality control. Cold Spring Harb Perspect Biol 3: a004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Caponigro G, Parker R (1996) Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol Rev 60: 233–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Decker CJ, Parker R (1993) A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev 7: 1632–1643. [DOI] [PubMed] [Google Scholar]
- 7. Nousch M, Techritz N, Hampel D, Millonigg S, Eckmann CR (2013) The Ccr4-Not deadenylase complex constitutes the major poly(A) removal activity in C. elegans. J Cell Sci 126: 4274–4285. [DOI] [PubMed] [Google Scholar]
- 8. Temme C, Zaessinger S, Meyer S, Simonelig M, Wahle E (2004) A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J 23: 2862–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, et al. (2001) The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104: 377–386. [DOI] [PubMed] [Google Scholar]
- 10. Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, et al. (2005) Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol 12: 1054–1063. [DOI] [PubMed] [Google Scholar]
- 11. Miller JE, Reese JC (2012) Ccr4-Not complex: the control freak of eukaryotic cells. Crit Rev Biochem Mol Biol 47: 315–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Collart MA (2003) Global control of gene expression in yeast by the Ccr4-Not complex. Gene 313: 1–16. [DOI] [PubMed] [Google Scholar]
- 13. Collart MA, Panasenko OO (2012) The Ccr4–not complex. Gene 492: 42–53. [DOI] [PubMed] [Google Scholar]
- 14. Collart MA, Panasenko OO, Nikolaev SI (2013) The Not3/5 subunit of the Ccr4-Not complex: A central regulator of gene expression that integrates signals between the cytoplasm and the nucleus in eukaryotic cells. Cell Signal 25: 743–751. [DOI] [PubMed] [Google Scholar]
- 15. Maillet L, Tu C, Hong YK, Shuster EO, Collart MA (2000) The essential function of Not1 lies within the Ccr4-Not complex. J Mol Biol 303: 131–143. [DOI] [PubMed] [Google Scholar]
- 16. Basquin J, Roudko VV, Rode M, Basquin C, Seraphin B, et al. (2012) Architecture of the nuclease module of the yeast ccr4-not complex: the not1-caf1-ccr4 interaction. Mol Cell 48: 207–218. [DOI] [PubMed] [Google Scholar]
- 17. Bhaskar V, Roudko V, Basquin J, Sharma K, Urlaub H, et al. (2013) Structure and RNA-binding properties of the Not1-Not2-Not5 module of the yeast Ccr4-Not complex. Nat Struct Mol Biol 20: 1281–1288. [DOI] [PubMed] [Google Scholar]
- 18. Bai Y, Salvadore C, Chiang YC, Collart MA, Liu HY, et al. (1999) The CCR4 and CAF1 proteins of the CCR4-NOT complex are physically and functionally separated from NOT2, NOT4, and NOT5. Mol Cell Biol 19: 6642–6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Boland A, Chen Y, Raisch T, Jonas S, Kuzuoglu-Ozturk D, et al. (2013) Structure and assembly of the NOT module of the human CCR4-NOT complex. Nat Struct Mol Biol 20: 1289–1297. [DOI] [PubMed] [Google Scholar]
- 20. Albert TK, Hanzawa H, Legtenberg YI, de Ruwe MJ, van den Heuvel FA, et al. (2002) Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. Embo J 21: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lau NC, Kolkman A, van Schaik FM, Mulder KW, Pijnappel WW, et al. (2009) Human Ccr4-Not complexes contain variable deadenylase subunits. Biochem J 422: 443–453. [DOI] [PubMed] [Google Scholar]
- 22. Jeske M, Meyer S, Temme C, Freudenreich D, Wahle E (2006) Rapid ATP-dependent deadenylation of nanos mRNA in a cell-free system from Drosophila embryos. J Biol Chem 281: 25124–25133. [DOI] [PubMed] [Google Scholar]
- 23. Temme C, Zhang L, Kremmer E, Ihling C, Chartier A, et al. (2010) Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. RNA 16: 1356–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Albert TK, Lemaire M, van Berkum NL, Gentz R, Collart MA, et al. (2000) Isolation and characterization of human orthologs of yeast CCR4-NOT complex subunits. Nucleic Acids Res 28: 809–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wiederhold K, Passmore LA (2010) Cytoplasmic deadenylation: regulation of mRNA fate. Biochem Soc Trans 38: 1531–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bartlam M, Yamamoto T (2011) The structural basis for deadenylation by the CCR4-NOT complex. Protein Cell 1: 443–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wahle E, Winkler GS (2013) RNA decay machines: deadenylation by the Ccr4-not and Pan2-Pan3 complexes. Biochim Biophys Acta 1829: 561–570. [DOI] [PubMed] [Google Scholar]
- 28. Parker R, Song H (2004) The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol 11: 121–127. [DOI] [PubMed] [Google Scholar]
- 29. Chen J, Chiang YC, Denis CL (2002) CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J 21: 1414–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R (2002) Ccr4p is the catalytic subunit of a Ccr4p/Pop2p/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J 21: 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mauxion F, Faux C, Seraphin B (2008) The BTG2 protein is a general activator of mRNA deadenylation. EMBO J 27: 1039–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sandler H, Kreth J, Timmers HT, Stoecklin G (2011) Not1 mediates recruitment of the deadenylase Caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic Acids Res 39: 4373–4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Deshaies RJ, Joazeiro CA (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78: 399–434. [DOI] [PubMed] [Google Scholar]
- 34. Hanzawa H, de Ruwe MJ, Albert TK, van Der Vliet PC, Timmers HT, et al. (2001) The structure of the C4C4 ring finger of human NOT4 reveals features distinct from those of C3HC4 RING fingers. J Biol Chem 276: 10185–10190. [DOI] [PubMed] [Google Scholar]
- 35. Panasenko OO, Collart MA (2011) Not4 E3 Ligase Contributes to Proteasome Assembly and Functional Integrity in Part through Ecm29. Mol Cell Biol 31: 1610–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collart MA (2013) The NOT4 RING E3 ligase: a relevant player in co-translational quality control. Molecular Biology ID 548359. [DOI] [PMC free article] [PubMed]
- 37. Panasenko OO, Collart MA (2012) Presence of Not5 and ubiquitinated Rps7A in polysome fractions depends upon the Not4 E3 ligase. Mol Microbiol 83: 640–653. [DOI] [PubMed] [Google Scholar]
- 38. Panasenko O, Landrieux E, Feuermann M, Finka A, Paquet N, et al. (2006) The yeast Ccr4-Not complex controls ubiquitination of the nascent-associated polypeptide (NAC-EGD) complex. J Biol Chem 281: 31389–31398. [DOI] [PubMed] [Google Scholar]
- 39. Panasenko OO, David FP, Collart MA (2009) Ribosome association and stability of the nascent polypeptide-associated complex is dependent upon its own ubiquitination. Genetics 181: 447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dimitrova LN, Kuroha K, Tatematsu T, Inada T (2009) Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J Biol Chem 284: 10343–10352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bengtson MH, Joazeiro CA (2010) Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 467: 470–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nillegoda NB, Theodoraki MA, Mandal AK, Mayo KJ, Ren HY, et al. (2010) Ubr1 and Ubr2 function in a quality control pathway for degradation of unfolded cytosolic proteins. Mol Biol Cell 21: 2102–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sommer T, Hirsch C (2005) San1p, checking up on nuclear proteins. Cell 120: 734–736. [DOI] [PubMed] [Google Scholar]
- 44. Gardner RG, Nelson ZW, Gottschling DE (2005) Degradation-mediated protein quality control in the nucleus. Cell 120: 803–815. [DOI] [PubMed] [Google Scholar]
- 45. Chhangani D, Joshi AP, Mishra A (2012) E3 ubiquitin ligases in protein quality control mechanism. Mol Neurobiol 45: 571–585. [DOI] [PubMed] [Google Scholar]
- 46. Ciechanover A (1998) The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J 17: 7151–7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Goldberg AL (2003) Protein degradation and protection against misfolded or damaged proteins. Nature 426: 895–899. [DOI] [PubMed] [Google Scholar]
- 48. Finley D (2009) Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem 78: 477–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Finley D, Ulrich HD, Sommer T, Kaiser P (2012) The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics 192: 319–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kish-Trier E, Hill CP (2013) Structural biology of the proteasome. Annu Rev Biophys 42: 29–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Beck F, Unverdorben P, Bohn S, Schweitzer A, Pfeifer G, et al. (2012) Near-atomic resolution structural model of the yeast 26S proteasome. Proc Natl Acad Sci U S A 109: 14870–14875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lam YA, Lawson TG, Velayutham M, Zweier JL, Pickart CM (2002) A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature 416: 763–767. [DOI] [PubMed] [Google Scholar]
- 53. Glickman MH, Rubin DM, Fried VA, Finley D (1998) The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol Cell Biol 18: 3149–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Glickman MH, Rubin DM, Fu H, Larsen CN, Coux O, et al. (1999) Functional analysis of the proteasome regulatory particle. Mol Biol Rep 26: 21–28. [DOI] [PubMed] [Google Scholar]
- 55. Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, et al. (2000) A gated channel into the proteasome core particle. Nat Struct Biol 7: 1062–1067. [DOI] [PubMed] [Google Scholar]
- 56. Kohler A, Bajorek M, Groll M, Moroder L, Rubin DM, et al. (2001) The substrate translocation channel of the proteasome. Biochimie 83: 325–332. [DOI] [PubMed] [Google Scholar]
- 57. Heinemeyer W, Fischer M, Krimmer T, Stachon U, Wolf DH (1997) The active sites of the eukaryotic 20 S proteasome and their involvement in subunit precursor processing. J Biol Chem 272: 25200–25209. [DOI] [PubMed] [Google Scholar]
- 58. Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, et al. (2002) Multiple associated proteins regulate proteasome structure and function. Mol Cell 10: 495–507. [DOI] [PubMed] [Google Scholar]
- 59. Friedlander R, Jarosch E, Urban J, Volkwein C, Sommer T (2000) A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat Cell Biol 2: 379–384. [DOI] [PubMed] [Google Scholar]
- 60. Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, et al. (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15: 963–972. [DOI] [PubMed] [Google Scholar]
- 61. Kushnirov VV (2000) Rapid and reliable protein extraction from yeast. Yeast 16: 857–860. [DOI] [PubMed] [Google Scholar]
- 62. Elsasser S, Schmidt M, Finley D (2005) Characterization of the proteasome using native gel electrophoresis. Methods Enzymol 398: 353–363. [DOI] [PubMed] [Google Scholar]
- 63. Leggett DS, Glickman MH, Finley D (2005) Purification of proteasomes, proteasome subcomplexes, and proteasome-associated proteins from budding yeast. Methods Mol Biol 301: 57–70. [DOI] [PubMed] [Google Scholar]
- 64. Finley D, Ozkaynak E, Varshavsky A (1987) The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell 48: 1035–1046. [DOI] [PubMed] [Google Scholar]
- 65. Hanna J, Leggett DS, Finley D (2003) Ubiquitin depletion as a key mediator of toxicity by translational inhibitors. Mol Cell Biol 23: 9251–9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Brodersen DE, Clemons WM Jr, Carter AP, Morgan-Warren RJ, Wimberly BT, et al. (2000) The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell 103: 1143–1154. [DOI] [PubMed] [Google Scholar]
- 67. Goldberg AL, Dice JF (1974) Intracellular protein degradation in mammalian and bacterial cells. Annu Rev Biochem 43: 835–869. [DOI] [PubMed] [Google Scholar]
- 68. Liu B, Han Y, Qian SB (2013) Cotranslational response to proteotoxic stress by elongation pausing of ribosomes. Mol Cell 49: 453–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Shalgi R, Hurt JA, Krykbaeva I, Taipale M, Lindquist S, et al. (2013) Widespread regulation of translation by elongation pausing in heat shock. Mol Cell 49: 439–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, et al. (2012) A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151: 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Knop M, Finger A, Braun T, Hellmuth K, Wolf DH (1996) Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J 15: 753–763. [PMC free article] [PubMed] [Google Scholar]
- 72. Hiller MM, Finger A, Schweiger M, Wolf DH (1996) ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science 273: 1725–1728. [DOI] [PubMed] [Google Scholar]
- 73. Jarosch E, Taxis C, Volkwein C, Bordallo J, Finley D, et al. (2002) Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat Cell Biol 4: 134–139. [DOI] [PubMed] [Google Scholar]
- 74. Park SH, Bolender N, Eisele F, Kostova Z, Takeuchi J, et al. (2007) The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol Biol Cell 18: 153–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mulder KW, Inagaki A, Cameroni E, Mousson F, Winkler GS, et al. (2007) Modulation of Ubc4p/Ubc5p-mediated stress responses by the RING-finger-dependent ubiquitin-protein ligase Not4p in Saccharomyces cerevisiae. Genetics 176: 181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Azzouz N, Panasenko OO, Deluen C, Hsieh J, Theiler G, et al. (2009) Specific roles for the Ccr4-Not complex subunits in expression of the genome. RNA 15: 377–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Daugeron MC, Mauxion F, Seraphin B (2001) The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res 29: 2448–2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Thore S, Mauxion F, Seraphin B, Suck D (2003) X-ray structure and activity of the yeast Pop2 protein: a nuclease subunit of the mRNA deadenylase complex. EMBO Rep 4: 1150–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lyumkis D, Doamekpor SK, Bengtson MH, Lee JW, Toro TB, et al. (2013) Single-particle EM reveals extensive conformational variability of the Ltn1 E3 ligase. Proc Natl Acad Sci U S A 110: 1702–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, et al. (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132. [DOI] [PubMed] [Google Scholar]
- 81. Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, et al. (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440: 631–636. [DOI] [PubMed] [Google Scholar]
- 82. Collart MA, Struhl K (1994) NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev 8: 525–537. [DOI] [PubMed] [Google Scholar]
- 83. Lau NC, Mulder KW, Brenkman AB, Mohammed S, van den Broek NJ, et al. (2010) Phosphorylation of Not4p functions parallel to BUR2 to regulate resistance to cellular stresses in Saccharomyces cerevisiae. PLoS One 5: e9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ghislain M, Udvardy A, Mann C (1993) S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature 366: 358–362. [DOI] [PubMed] [Google Scholar]
- 85. Gerlinger UM, Guckel R, Hoffmann M, Wolf DH, Hilt W (1997) Yeast cycloheximide-resistant crl mutants are proteasome mutants defective in protein degradation. Mol Biol Cell 8: 2487–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth phenotypes of the E3 ligases mutants. The indicated strains were grown to exponential phase and diluted to the same OD600 of 0.5. 10-fold serial dilutions were spotted on the YPD plates containing, when indicated, HygB 0.1 mg/ml or CHX 0.05 µg/ml; and left to grow for 6 days (except 16°C) or for 21 days (16°C).
(TIF)
CPY*-HA mRNA and protein levels in wild-type, not4Δ, ccr4Δ , and caf1Δ cells. A. CPY*-HA protein levels in wild-type, not4Δ, ccr4Δ, and caf1Δ cells. CPY*-HA was expressed from an episome under control of copper dependent promoter in wild-type (WT), not4Δ, ccr4Δ, and caf1Δ cells. Cells were exponentially grown in the constant presence of 0.1 mM of CuSO4 and collected at OD600 of 1.0. Different amount of the cells (0.5 OD units (lane 1), 0.125 OD units (lane 2) and 0.05 OD units (lane 3)) were analyzed by SDS-PAGE and western blot with antibodies against HA, to see CPY*-HA levels, and against Egd2 as a loading control. B. CPY*-HA mRNA levels in wild-type, not4Δ, ccr4Δ, and caf1Δ cells. Cells were grown as described in A. 50 OD units of the cultures were collected. Pellets were resuspend in 400 µl of acid phenol and 400 µl of TES buffer (10 mM Tris-HCl pH 7.5, 10 mM EDTA, 0.5% SDS) and incubated at 65°C for 10 min. Samples were chilled on ice for 5 min and spun at 4°C for 10 min. Aqueous phase was extracted with 400 µl of acid phenol and then with chloroform. Finally, RNA was collected by ethanol/sodium acetate precipitation. 4 µg of the RNA were treated with DNAse (Promega) and than reverse transcribed with M-MLV RT (Promega) according to the manufacturer’s instructions and using oligo d(T) primers (Qiagen). SYBR green based quantitative RT-PCR was performed using BioRad cycler. ACT1 was used as a housekeeping gene and CPY*-HA signals were normalized on ACT1 level. The ratio CPY*-HA/ACT1 in wild type was normalized to 1. Primers used for analysis: The forward primer: 5′-TCCCCGGGTTAATTAACATC-3′ and reverse primer: 5′-TCGCTTATTTAGAAGTGGCG-3′ amplify 149 bp fragment of HA tag of CPY*-HA gene. The forward primer: 5′-TTGTCCGTGACATCAAGGAA-3′ and reverse primer: 5′-ACCCAAAACAGAAGGATGGA-3′ amplify 182 bp fragment of ACT1 gene.
(TIF)
Polysome profiles from wild-type, not4Δ , and ccr4Δ cells. Extracts from wild-type (black), not4Δ (red), and ccr4Δ (blue) cells, containing 3 mg of total proteins, were subjected to 7–47% sucrose gradient centrifugation and analyzed by UV reading at 254 nm (left). Profiles were superposed (right). The positions of 40S, 60S, 80S, and polysomes are indicated.
(TIF)
Double mutant ltn1Δ not4Δ grows slowly compare to single ltn1Δ or not4Δ mutants.
(TIF)