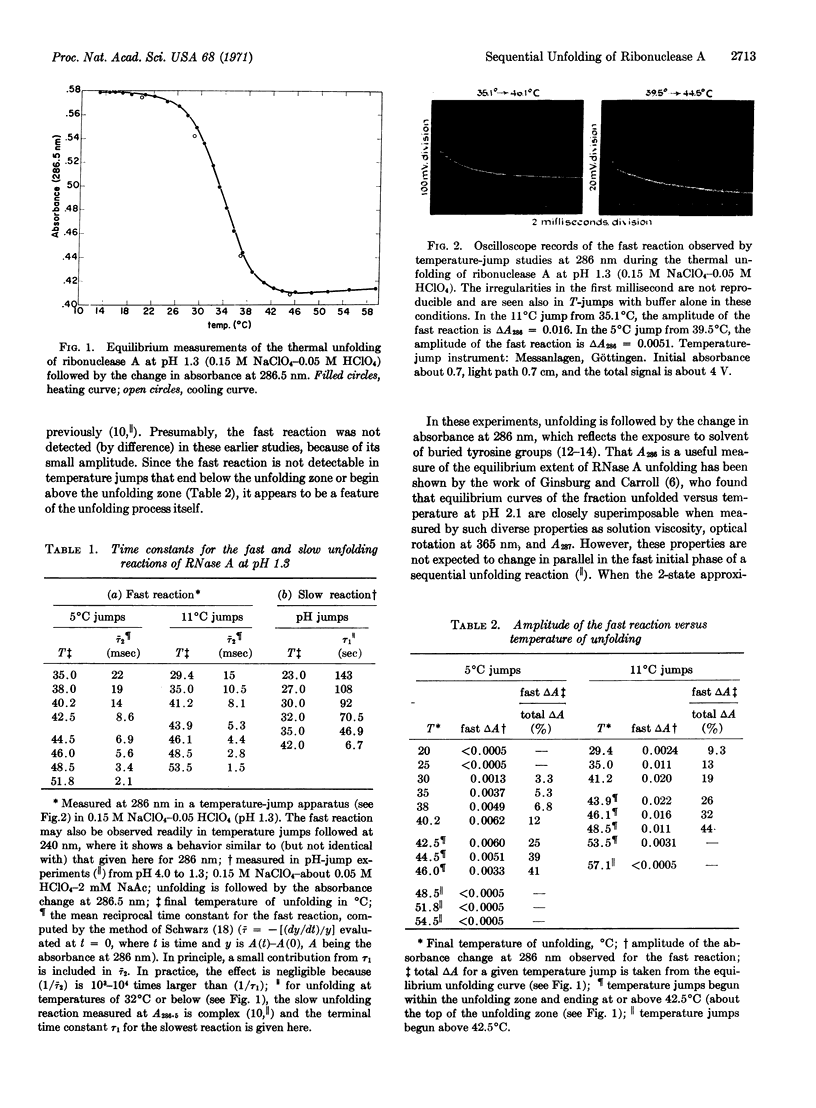
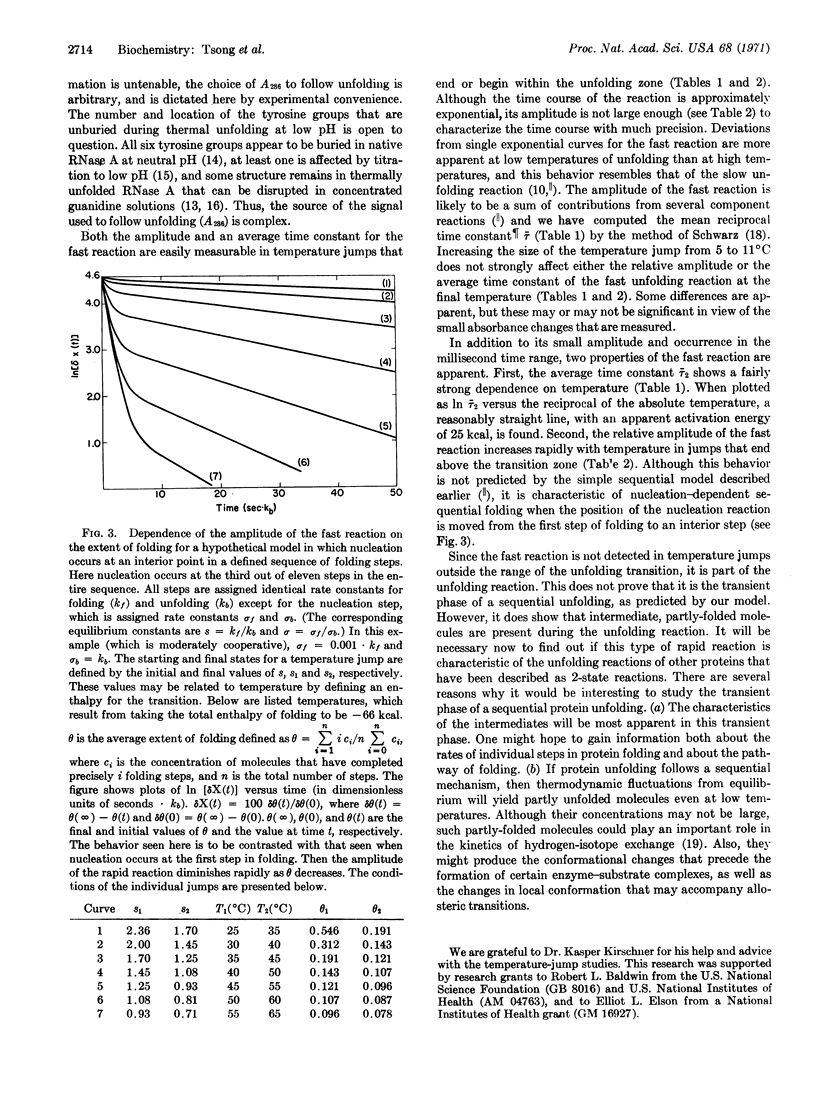
Abstract
Temperature-jump studies have been used to detect a rapid reaction in the thermal unfolding of ribonuclease A (RNase A). The fast reaction occurs over a wide range of pH, and the results of a detailed study at pH 1.3 are reported here. Although its amplitude is small, the reaction is easily measurable over the entire temperature range of thermal unfolding. It occurs in the millisecond time range, and is faster by 3-4 orders of magnitude than the slow unfolding reaction studied previously. Unfolding is measured here by the change in absorbance at 287 nm, which reflects the exposure to solvent of buried tyrosine groups. Since the fast reaction has a detectable amplitude only in the temperature range of unfolding, it apparently detects the presence of intermediate, partly-folded states. Previous equilibrium studies of the unfolding of RNase A in the pH range 1-2 have indicated that it is essentially a 2-state reaction, without detectable intermediates.
The existence of a rapid transient phase in the unfolding of RNase A had been predicted previously from a model for this unfolding reaction, based on nucleation-dependent sequential folding. The model served to reconcile kinetic and equilibrium studies of the thermal unfolding reaction of RNase A at neutral pH. Kinetic studies had shown that the slow unfolding reaction, measured at 287 nm, could be represented as a single exponential process, as expected for a 2-state reaction. However, earlier equilibrium measurements, especially the calorimetric studies of Sturtevant and coworkers, had revealed significant deviations from the 2-state behavior at neutral pH. These conflicting observations are explained by the model, which satisfies closely many criteria for a 2-state unfolding, even when appreciable concentrations of partly folded molecules are present. In particular, it predicts that the final, and major, portion of the kinetic reaction will occur as a single process characterized by an exponential time course.
Keywords: temperature jump, 2-state reaction, thermal unfolding
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aune K. C., Salahuddin A., Zarlengo M. H., Tanford C. Evidence for residual structure in acid- and heat-denatured proteins. J Biol Chem. 1967 Oct 10;242(19):4486–4489. [PubMed] [Google Scholar]
- BIGELOW C. C. THE DENATURED STATES OF RIBONUCLEASE. J Mol Biol. 1964 May;8:696–701. doi: 10.1016/s0022-2836(64)80118-2. [DOI] [PubMed] [Google Scholar]
- Brandts J. F., Hunt L. The thermodynamics of protein denaturation. 3. The denaturation of ribonuclease in water and in aqueous urea and aqueous ethanol mixtures. J Am Chem Soc. 1967 Sep 13;89(19):4826–4838. doi: 10.1021/ja00995a002. [DOI] [PubMed] [Google Scholar]
- HARRINGTON W. F., SCHELLMAN J. A. Evidence for the instability of hydrogen-bonded peptide structures in water, based on studies of ribonuclease and oxidized ribonuclease. C R Trav Lab Carlsberg Chim. 1956;30(3):21–43. [PubMed] [Google Scholar]
- Ikai A., Tanford C. Kinetic evidence for incorrectly folded intermediate states in the refolding of denatured proteins. Nature. 1971 Mar 12;230(5289):100–102. doi: 10.1038/230100a0. [DOI] [PubMed] [Google Scholar]
- Lumry R., Biltonen R. Validity of the "two-state" hypothesis for conformational transitions of proteins. Biopolymers. 1966 Sep;4(8):917–944. doi: 10.1002/bip.1966.360040808. [DOI] [PubMed] [Google Scholar]
- Pohl F. M. Einfache Temperatursprung-Methode im Sekunden-bis Stundenbereich und die reversible denaturierung von Chymotrypsin. Eur J Biochem. 1968 Apr;4(3):373–377. doi: 10.1111/j.1432-1033.1968.tb00221.x. [DOI] [PubMed] [Google Scholar]
- SCHWARZ G. ON THE KINETICS OF THE HELIX-COIL TRANSITION OF POLYPEPTIDES IN SOLUTION. J Mol Biol. 1965 Jan;11:64–77. doi: 10.1016/s0022-2836(65)80171-1. [DOI] [PubMed] [Google Scholar]
- Schechter A. N., Chen R. F., Anfinsen C. B. Kinetics of folding of staphylococcal nuclease. Science. 1970 Feb 6;167(3919):886–887. doi: 10.1126/science.167.3919.886. [DOI] [PubMed] [Google Scholar]
- Simons E. R., Schneider E. G., Blout E. R. Thermal effects on the circular dichroism spectra of ribonuclease A and of ribonuclease S-protein. J Biol Chem. 1969 Aug 10;244(15):4023–4026. [PubMed] [Google Scholar]
- Tanford C. Protein denaturation. Adv Protein Chem. 1968;23:121–282. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- Tsong T. Y., Hearn R. P., Wrathall D. P., Sturtevant J. M. A calorimetric study of thermally induced conformational transitions of ribonuclease A and certain of its derivatives. Biochemistry. 1970 Jun 23;9(13):2666–2677. doi: 10.1021/bi00815a015. [DOI] [PubMed] [Google Scholar]



